Abstract
Free full text

The structure of the β-barrel assembly machinery complex
Abstract
β-barrel outer membrane proteins (OMPs) are found within the outer membranes (OM) of Gram-negative bacteria and are essential for nutrient import, signaling, and adhesion. While the exact mechanism is unknown, a 200 kDa five component complex called the β-barrel assembly machinery (BAM) complex has been implicated in the biogenesis of OMPs. Here, we report the structure of the BAM complex from E. coli, revealing that binding of the accessory proteins BamCDE modulates the conformation of BamA, the central component of the complex, which may regulate the function of the BAM complex. The periplasmic domain of BamA was found in a closed state that prevents access to the barrel lumen from the periplasm, indicating substrate OMPs likely do not enter the barrel during biogenesis. Further, the first eight strands of the β-barrel domain undergo an unprecedented conformational shift leading to opening of the exit pore and rearrangement at the lateral gate.
Gram-negative bacteria contain both an inner membrane (IM) and an outer membrane (OM) which serve important roles in nutrient import, cell signaling, waste export, and protection. The IM exclusively contains integral membrane proteins that have an α-helical fold consisting of one or more α-helices which anchor the protein into the membrane. The OM, however, contains almost exclusively integral membrane proteins that have a β-barrel fold consisting of 8–26 antiparallel β-strands which anchor the protein in the OM. Like α-helical membrane proteins, β-barrel outer membrane proteins (OMPs) serve many functions and therefore have diverse structural and biochemical properties. In pathogenic strains of bacteria, some OMPs can also serve as virulence factors that mediate infection. In nature, OMPs are only found in the OMs of Gram-negative bacteria, mitochondria, and chloroplasts (1, 2).
Despite their essential roles, the exact mechanism explaining how OMPs are folded and inserted into the OM remains unknown. In the last decade, studies have identified a general pathway and conserved machinery that is responsible for the biogenesis of OMPs (3–5). A majority of these advances have been made working with Gram-negative bacteria (3, 6, 7). Here, the nascent OMPs are first synthesized in the cytoplasm with an N-terminal leader sequence which directs them to the Sec translocon for transport across the IM, into the periplasm (Figure 1A). Chaperones such as SurA and Skp then further escort the nascent OMPs to a multicomponent complex called the β-barrel assembly machinery (BAM) complex, which has been shown to be responsible for folding and inserting OMPs into the OM (4, 8). In E. coli, the BAM complex consists of five components called BamA, BamB, BamC, BamD, and BamE. BamA, a 16-stranded OMP itself, is the central and essential component of the complex and is conserved both in mitochondria and chloroplasts. BamB, BamC, BamD, and BamE are all lipoproteins which are anchored to the OM via lipidation of the N-terminal cysteine residue. BamA and BamD are essential for viability; however, all components are required for efficient OMP folding/insertion (9–11). Studies have shown that both BamB and BamD interact directly with BamA via non-overlapping binding sites while BamC and BamE interact directly with BamD to stabilize the complex (9, 12).
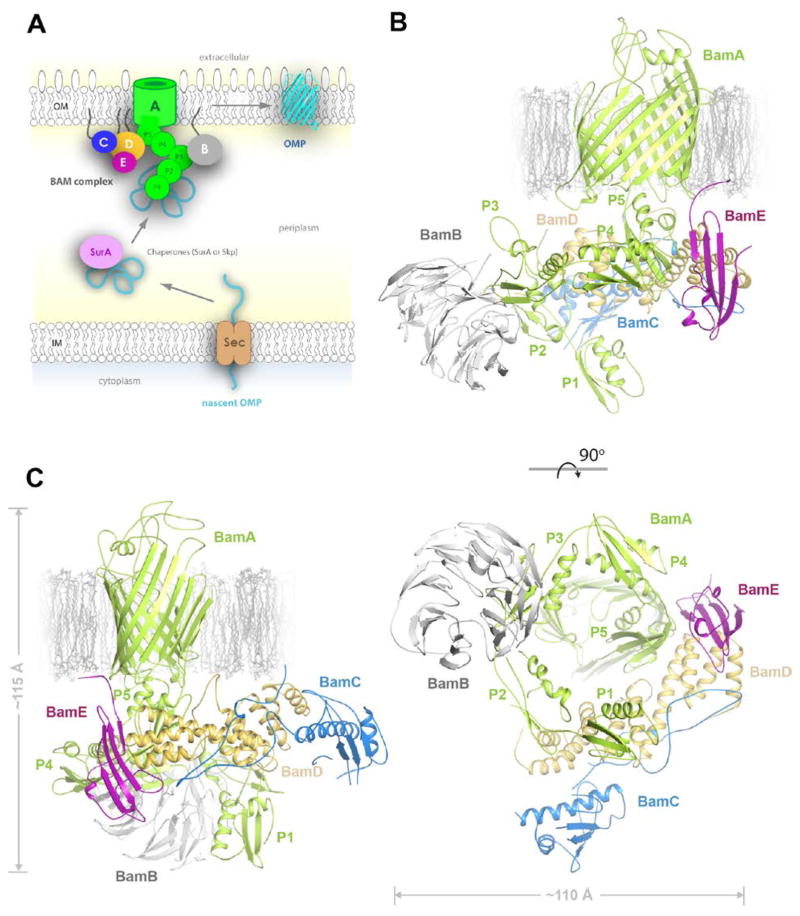
A. The pathway for the biogenesis of β-barrel outer membrane proteins in Gram-negative bacteria. B. A membrane view of the structure of the full BAM complex, formed from merging the crystal structures of BamACDE and BamAB (PDB ID 4PK1). The bottom panel shows the periplasmic view, rotated 90º along the x-axis relative to the top panel. BamA is shown in green, BamB in gray, BamC in blue, BamD in gold, and BamE in purple. C. The β-barrel domain of BamA undgoes a dramatic conformational change along strand β1-β8, which can be seen here. This panel is rotated ~90º along the y-axis relative to the top view of panel B. The overall measurements of the BAM complex are ~115 × 115 × 115 Å.
Structures of all the Bam components have now been reported including partial complexes of BamAB and BamCD (7, 13–23). The full-length structure of BamA from Neisseria gonorrhoeae revealed a large periplasmic domain consisting of five polypeptide transport associated (POTRA) domains and a C-terminal 16-stranded β-barrel domain. Subsequent studies showed that lateral opening of the barrel domain was required for function in BamA, strengthening an existing hypothesis that the barrel domain must open laterally in the membrane to allow insertion of the substrate OMPs into the OM (16, 18, 24). It has been proposed that BamB might serve as a scaffold, assisting in the handoff of nascent OMPs by SurA/Skp to BamA, while BamC, BamD, and BamE may serve support roles in regulating the function of BamA (14, 17, 25). The structures have offered clues to how each component may function within the complex; however, the lack of structural information regarding the fully assembled complex has hindered progress towards exploring the mechanism further. To address this, we report here the structure of the BAM complex from E. coli. We used X-ray crystallography to solve the crystal structure of the BamACDE subcomplex to 3.4 Å resolution and utilized the previously reported partial structure of BamAB to form a model for the fully assembled BAM complex. Our structure shows the periplasmic domain of BamA in a closed state that prevents access to the barrel lumen from the periplasm. Furthermore, binding of BamCDE to BamA causes an unprecedented conformational twisting of strands β1-β8, leading to opening of the top of the barrel domain along the exit pore and structural rearrangement of the lateral opening site. These structural changes suggest that the role of BamCDE may be to modulate the conformational states of BamA, thereby serving as a regulatory step in the function of the BAM complex.
The BAM complex from E. coli was expressed from a single plasmid and purified as previously described with some modifications (11). SDS-PAGE analysis verified the presence of the full complex which produced a monodisperse peak by size exclusion chromatography (Figure S1). We were able to crystallize the complex by detergent screening using C8E4 and collected data at the SER-CAT ID22 beamline at the Advanced Photon Source. We solved the crystal structure of the complex to 3.4 Å resolution by molecular replacement (Figure S2 and Table S1). However, it was clear that BamB was not present, possibly due to proteolytic degradation during incubation. This was verified by SDS-PAGE analysis of crystals and of the original protein sample, both of which lacked BamB after extended incubation/storage (Figure S1). Our crystal structure contains full length BamA, BamD, and BamE, but only the N-terminal flexible domain and the first globular helix-grip domain of BamC with the second helix-grip domain presumably being disordered. To model of the fully assembled BAM complex, we utilized the previously reported structure of the partial BamAB complex (PDB ID 4PK1) (14) to dock BamB into our crystal structure by aligning along POTRA3 of BamA (Figures 1B, C, Movie S1, and Model S1). The overall dimensions of the BAM complex are ~115 Å in width and height. BamD interacts with POTRA5 and POTRA2 of BamA while BamE interacts with both BamD and with POTRA5. BamC was found to interact with BamD in the same manner as has been previously reported (13). For all accessory lipoproteins, the N-terminal residues are positioned in close proximity to where the OM would sit; however, no lipid anchors were observable in our crystal structure.
While the structure of the BamCD complex had been reported previously (13), no structure of the BamCDE complex has been described. In our structure, BamC interacts with BamD primarily along its elongated intrinsically disordered, N-terminal domain with a buried surface area of ~2,300 Å2, while BamE interacts along the C-terminal end of BamD with a buried surface area of ~800 Å2 (Figure 2A and Tables S2, S3). BamCD in our structure aligned very well with the previously reported complex (PDB ID 3TGO), having an RMSD of 1.25 Å across both chains (Figure 2B). While BamC and BamE interact with BamD via non-overlapping binding sites, they do make contact with one another through a buried surface area of ~140 Å2 along residues 67–69 of BamE and residues 55–59 of BamC. Residues 215–344 of BamC consisting of a linker and the second helix-grip domain were not observed in our crystal structure and presumed to be disordered. This region of the structure sits adjacent to a very large open cavity in the crystal lattice which does not provide sufficient contacts for stabilizing this flexible region of the structure. Previous studies have shown that the two helix-grip domains of BamC are surface exposed (26), suggesting that one or both may interact directly with the surface of BamA. In our crystal structure, no direct interaction was observed indicating that if the two helix-grip domains of BamC do indeed interact with BamA, a membrane bilayer and/or substrate may be required to release BamC from BamD so it can be presented on the surface.
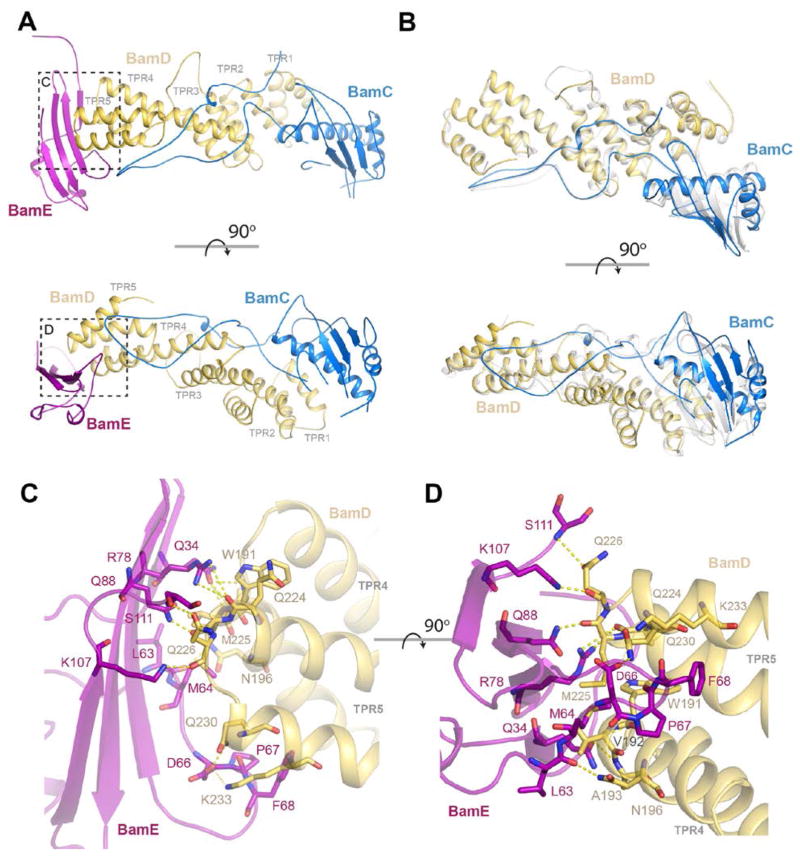
A. The complex structure of BamCDE is shown here with BamD in gold, BamC in blue, and BamE in purple. The TPR domains of BamD are indicated. The bottom panel is rotated 90º along the x-axis relative to the top. B. The interactions between BamC and BamD are nearly the same as that observed in the previously reported crystal structure (PDB ID 3TGO) with an RMSD of 1.25 Å across both chains. The bottom panel is rotated 90º along the x-axis relative to the top. C. View showing the interactions of BamE with BamD TPR4 and 5 through an extensive interface with a buried surface area of ~800 Å2, containing a mix of hydrogen bonds, salt bridges, and hydrophobic interactions. D. A view from the top which is rotated 90º along the x-axis to further illustrate the extensive binding interface.
The structure of BamD consists of five tetratricopeptide-repeat (TPR) motifs (15, 21, 27), with TPR4 and 5 forming the binding site for BamE, which was found oriented perpendicular to the membrane (Figure 2A). The binding interface agrees well with previously published work, where BamD was found to bind BamE along the interface containing residues R29, I32, R78, F68, N71, T72, and T92 (23). In our structure, the extensive binding interface has numerous hydrogen bonds using both side chain and backbone atoms, and a salt bridge between D66 of BamE with K233 of BamD (Figures 2C, D). The binding is further strengthened by interactions of M64 and F68 of BamE into hydrophobic pockets in BamD. The minimal interaction between BamC and BamE is mediated primarily through hydrophobic interactions; the loop of BamC helps to form the large hydrophobic pocket where F68 of BamE binds BamD (Table S4).
In agreement with previous studies (28), BamD was found to bind BamA along POTRA5 almost exclusively via TPR3 and 4, sitting parallel to the membrane with a buried surface area of ~1,100 Å2 (Figures 3A, B and Table S5). Interestingly, TPR1 and 2 of BamD also interact with POTRA2 of BamA through a buried surface area of ~115 Å2. Whether this minimal interaction is real or an artifact of crystallization remains to be determined. The extensive binding interface between BamD and POTRA5 of BamA is mediated by many hydrogen bonds using both side chain and backbone atoms and contains three salt bridges between residues H139, R197, and R188 of BamD and residues D358, E373, and D481 of BamA, respectively (Figures 3B, C). The salt bridge formed by R197 of BamD and E373 of BamA is central to this interaction and these residues have previously been determined to play an essential role in binding via a putative salt bridge interaction, as our structure confirms (29, 30). The extended loop of BamD TPR3 (residues 123–130) also interacts with periplasmic loop 1 of BamA along residues 449–452 using primarily hydrophobic interactions, with a buried surface area of ~120 Å2 (Figure S3). BamD also interacts with POTRA2 along TPR1 and 2 (Figures 3B, D). While this region is less ordered in our structure, there is clearly the potential for additional electrostatic interactions here in which BamD could also participate in modulating the conformation of the POTRA domains of BamA during OMP biogenesis.
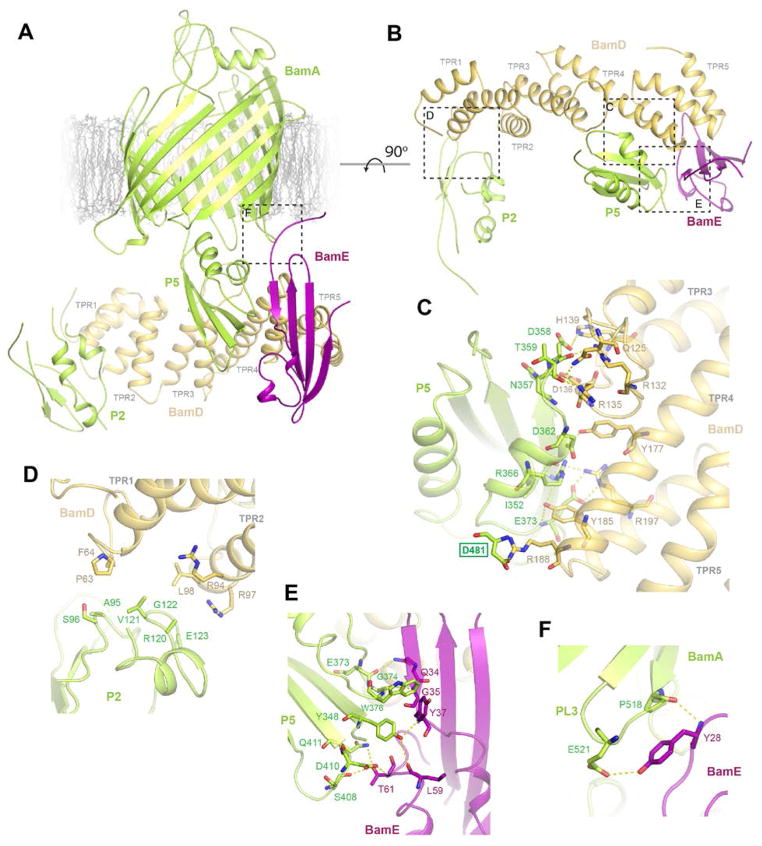
A. BamD interacts primarily along POTRA5 of BamA via TPR4 and 5, but also along POTRA2 via TPR1 and 2, while BamE interacts along POTRA5. B. A rotated view without the β-barrel domain of BamA highlighting the interacting regions. C. View of the interaction of BamD with POTRA5 of BamA, showing an extensive binding interface with a buried surface area of ~1,100 Å2, containing a mix of hydrogen bonds, salt bridges, and hydrophobic interactions. Residue R197 of BamD clearly be forms a salt bridge with E373 of BamA. Residue D481 (green box) is from periplasmic loop 2 of BamA. D. View of the interactions between TPR1 and 2 of BamD with POTRA2 of BamA. While minimal here, these interactions could assist in modulating the conformation of BamA. E. View of the interactions of BamE with POTRA5 of BamA, an extensive interaction with a buried surface area of ~750 Å2, containing a mix of hydrogen bonds, salt bridges, and hydrophobic interactions. F. The N-terminal region of BamE was found anchored via hydrogen bonding interactions of residue Y28 to residues E521 and P518 of BamA periplasmic loop 3.
While it has been well documented that BamD interacts with BamA, it has not been shown that BamE also interacts with BamA, although the association of BamE with BamD has been shown to enhance interaction with BamA (9, 12). In our crystal structure, we observe not only the interaction of BamE with BamD, but also an extensive interaction of BamE with POTRA5 of BamA through a buried surface area of ~750 Å2 (Figures 3B, E and Table S6). This binding interface is composed of numerous hydrogen bonds via side chain and backbone atoms, as well as hydrophobic interactions primarily from W376 of BamA which constitutes ~130 Å2 of buried surface area alone. Residue Y28 of BamE also interacts with periplasmic loop 3 of BamA forming hydrogen bonds with E521 and P518. These interactions anchor the N-terminus of BamE in close proximity to the barrel domain of BamA, which may serve to orient BamCDE optimally for interacting with BamA (Figures 3A, F). The observation that BamE bridges an additional contact between BamD and BamA agrees well with previous work indicating that the role of BamE may be to enhance the interaction of BamD with BamA (12). Previous studies using NMR identified BamE residues that were affected by BamD binding (23); however, not all these residues mapped to the interface observed in our crystal structure. Instead, residues Y37, L38, L63, and T61 lie along the interface which interacts with POTRA5 of BamA, suggesting that binding of BamD may lead to conformational changes in BamE that promote association with BamA. BamE has also been shown to bind phosphatidylglycerol (PG), suggesting that this may contribute to its role within the BAM complex (23). The BamE residues that were identified to interact most extensively with PG (G60, T70, N71, V76, and F95) were found sitting within the periplasm as far as 35 Å from the OM. Therefore, it is unlikely that the PG-binding role of BamE contributes directly to the role of the BAM complex; however, it is still possible that the recruitment of PG into proximity of the BAM complex could increase the efficiency of OMP biogenesis.
No structure of full-length BamA from E. coli has been reported previously, although a structure containing the β-barrel domain and POTRA5 is available (16). In this structure, POTRA5 was oriented away from the barrel domain in an open conformation allowing direct access to the barrel lumen from the periplasm (Figure S4). However, in our structure, which contains full-length BamA, POTRA5 was found in a closed conformation relative to the β-barrel domain, where it fully occludes access to the barrel lumen from the periplasm. Compared to the full-length BamA structure from N. gonorrhoeae (18), the POTRA domains undergo ~90º rotation along the plane of the membrane upon interaction with BamCDE. POTRA5 makes numerous contacts with periplasmic loops 1, 2, 3, 4, and 6, with the most extensive interactions through periplasmic loops 1, 2 and 4 (Figure S5). This conformation constitutes ~45º hinge-like conformational change of POTRA5 to the closed state (Figure 4A). POTRA5 interacts most extensively with periplasmic loop 4, which sits tucked inside the barrel domain (Figure S5 and Movie S2). Here, two important interactions stabilize the conformation observed in our crystal structure. These include a salt bridge between E396 (POTRA5) and R583 (periplasmic loop 4) and pi stacking between R421 (POTRA5) and Y585 (periplasmic loop 4). Conformational flexibility of the POTRA domains has been observed for many years (28, 31, 32), so it remains to be determined whether the association of BamCDE is solely responsible for the closed conformation observed in our crystal structure.
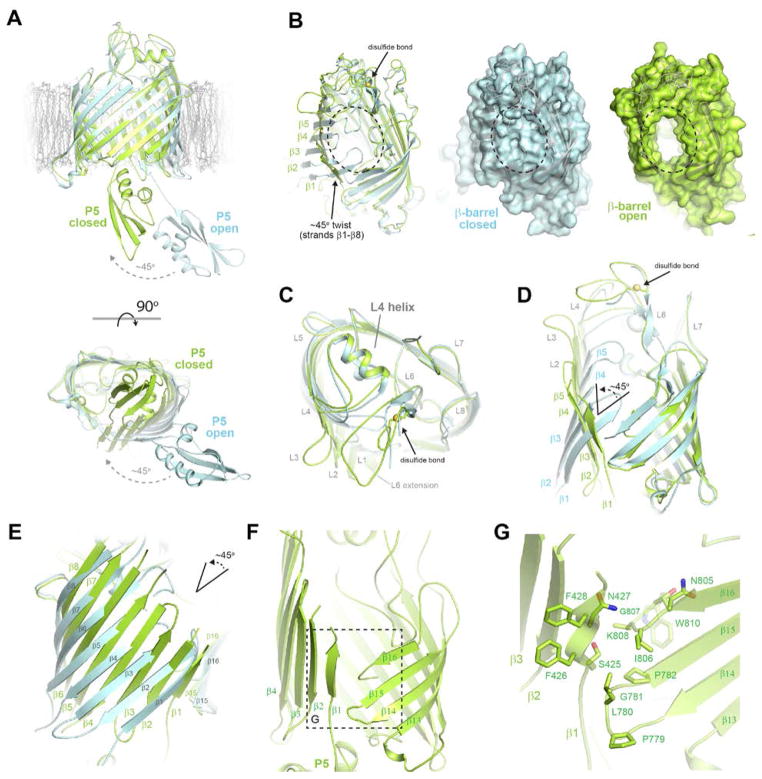
A. Superposition of BamA from our structure (green) with PDB ID 4C4V (cyan) reveals ~45º hinge-like conformational change of the POTRA domains to a closed, periplasm occluded state. B. Binding of BamCDE to BamA leads to an unprecedented twist of strands β1-β8 with the most dramatic change emanating from strand β1 (~45º) and gradually diminishing until strand β9 (Movie S2). This leads to opening of the exit pore and lateral gate. C. In response to the shift of the barrel strands, an opening of surface loops 1, 2, and 3 was observed; however, surface loops 4, 5, 6, 7, and 8 were mostly unchanged. D. Rotated view highlighting the strand shift along strands β1-β8 of the barrel domain of BamA. E. View of the strand shift along strandsβ1-β8 rotated ~90º relative to panel D illustrating a twist of the strands rather than just a simple rotation. F. View of the lateral gate in BamA where strand β1 and β16 no longer form a β-sheet interaction to close the barrel, rather, strands β15 and β16 are situated at ~45º angle relative to strand β1. G. View showing residues along the lateral gate with β16 sitting tucked inside the barrel and periplasmic loop 7 interacting with strand β1.
A surprising finding of this work is that binding of BamCDE leads to an unprecedented shift of the β-barrel domain of BamA, where strands β1 – β8 undergo a conformational twist with the most dramatic change emanating from strand β1 (~45º) and gradually diminishing until strand β9. This conformation is likely due to the strong interactions of POTRA5 with BamCDE and with periplasmic loop 4, which impose mechanical strain on the first half of the barrel strands (Figures 4B, D, E and Movie S2). This in effect, leads to a dramatic change in the angle of the strands in the membrane such that strands β1 and β16 no longer associate as a β-sheet, thereby leading to opening of the exit pore along extracellular loops 1, 2 and 3. Here, the exit pore is fully open with dimensions of ~15 Å × ~27 Å. This agrees with recent studies that have shown that disulfide crosslinking the exit pore in a closed state renders the BAM complex nonfunctional (24). Aligning BamA from our complex structure with the previously reported BamA structure containing POTRA5 only (PDB ID 4C4V), we calculated an RMSD of 1.07 Å for the entire β-barrel domain. However, the calculated RMSD of strands β1-β8 alone was 2.74 Å, while the RMSD for strands β9-β16 was 0.67 Å. These differences highlight the drastic shift observed in our structure.
While the conformational change of the β-barrel domain led to opening of extracellular loops 1, 2, and 3 along the exit pore, the remaining loops 4, 5, 6, 7, and 8 remained largely unchanged. The exception is that loop 4 undergoes a slight shift to stabilize loop 6, such that loop 6 is now entirely visible in our crystal structure, including the disulfide bond formed by residues C690 and C700 (Figures 4C, D). The rest of loop 6, including the conserved VRGF motif, was unchanged in conformation (Figure S6). Another consequence of the conformational twist of strands β1-β8 is structural rearrangement at the lateral gate such that strand β1 no longer interacts with strand β16 (last strand) in barrel closure, in contrast to what has been observed in all other OMPs with known structure (Figures 4F, G). Rather, most of strand β16 sits tucked inside the barrel domain, with the C-terminus stabilized by R632, while periplasmic loop 7 and strand β15 contact strand β1 at an offset angle of ~45º (Figures 4D, E, F, G). This is in agreement with recent studies that rendered the BAM complex non-functional by using disulfide crosslinking to prevent such conformational changes at the lateral gate (24).
Based on our crystal structure of BamACDE and the existing crystal structure of BamAB (14), here we report the structure of the fully assembled BAM complex from E. coli (Movies S1, S2, and Model S1). As further validation, our structure is in agreement with a model for the BAM complex that we recently reported which was based on all structural, functional, genetics, and biochemical studies to date (33). The exceptions are that the model contained an open conformation of the periplasmic domain of BamA and no conformational changes were predicted in the β-barrel domain. Our structure reveals that upon binding BamCDE, the β-barrel domain of BamA undergoes an unprecedented conformational twist that dramatically changes the angle of the strands (shear) in the membrane, leading to opening of the exit pore and rearrangement at the lateral gate. This suggests that binding of BamCDE modulates the conformation of BamA, possibly serving as a regulatory step to increase the efficiency of OMP biogenesis by the BAM complex. The conformational changes in the β-barrel domain of BamA may serve a number of functions. For example, a longstanding question is how the BAM complex is able to fold and insert OMPs with diverse properties such as differing strand and shear numbers. It is possible that association of BamCDE may serve to ‘tune’ the β-barrel domain of BamA to fold OMPs with differing shear numbers by adjusting the angle of the strands in the membrane to match that of the substrate OMPs. This would be especially important if newly forming OMPs indeed utilize the exposed strands of BamA for strand templating by β-augmentation which has been suggested previously. Another possible function of the conformational changes observed in the β-barrel domain of BamA may be to further destabilize or even part the local membrane, thereby reducing the kinetic barrier for OMP insertion (34).
It has been suggested that substrate OMPs may be threaded through the barrel domain of BamA during biogenesis. However, in our crystal structure of the BamACDE complex, access to the interior of the barrel is completely occluded from the periplasm. Therefore, OMPs are likely inserted into the membrane at the lateral gate of BamA rather than first being threaded through the barrel domain. The lateral gate is positioned central to the BAM complex and would be directly accessible for substrate handoff by the accessory lipoproteins and/or by the POTRA domains of BamA.
Materials and Methods
Expression of recombinant BamABCDE complex
A single plasmid (pJH114) containing all five Bam proteins (BamA, B, C, D, and E) was obtained from Harris Bernstein and used for expression and purification (11). The plasmid was transformed into BL21(DE3) cells (NEB), plated onto LB-carbenicillin agar plates (Teknova) and incubated overnight at 37°C. A single colony was used to inoculate a 5-mL LB-ampicillin culture and incubated overnight at 37°C. The overnight culture was then used to inoculate a 50 mL starter culture of LB-ampicillin which was allowed to grow to saturation. The cells were then centrifuged, washed three times with 1x PBS and then re-suspended in 12 mL 1x PBS. The resuspended cells (1 mL) were then added to twelve 2 L baffled flasks containing 1 L of 2xYT medium supplemented with ampicillin (50 μg/ml). These cultures were incubated at 37°C with shaking at 180 rpm, grown to an OD600 between 0.8–1.0, and then induced with 0.5 mM IPTG. Cultures were grown an additional 4 hours at 37°C before harvesting. Cell were either used immediately or flash frozen and stored at −20°C.
Purification and crystallization
Cells were resuspended in lysis buffer (1x PBS supplemented with DNase I (10 μg/ml) and PMSF (500 μM)) and lysed with three passes through an Emusiflex C-3 high pressure homogenizer (Avestin) at 18,000 psi. The cell lysate was then centrifuged at 6000 x g for 10 min at 4°C to remove cell debris, and the resulting supernatant was centrifuged at 200,000 x g for 90 min at 4°C to isolate cell membranes. The membranes were then re-suspended in solubilization buffer (1x PBS, 1% DDM, and 37 mM imidazole) using a dounce homogenizer and stirred at medium speed overnight at 4°C. The solubilized sample was then centrifuged again at 200,000 x g for 60 min at 4°C and the supernatant collected.
Solubilized BamABCDE complex was purified by affinity chromatography using a 5 mL HiTrap Nickel column (Qiagen) and an ÄKTA Pure system (GE Healthcare). The column was equilibrated with Buffer A (1x PBS, 0.03% DDM, and 37 mM imidazole) and the sample automatically loaded using the sample pump with an in-line air sensor. Protein was eluted with a linear gradient of 37–500 mM imidazole using Buffer A and Buffer B (1x PBS, 0.03% DDM, and 1 M imidazole). Fractions containing BamABCDE were pooled, concentrated to ~2 mg/mL, and passed through a 16/60 Sephacryl S-300 HR column (GE Healthcare) at a flow rate of 0.5 mL/min using 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.6% C8E4. All five Bam proteins (BamA, B, C, D, and E) eluted from the gel filtration column as a single monodisperse peak as verified by SDS-PAGE analysis. Fractions containing BamABCDE were pooled and concentrated to ~12 mg/mL.
Broad crystallization screening was performed using hanging drop method on a Mosquito LCP crystallization robot (TTP Labtech) with commercially available crystallization screens. An initial hit was improved by additive screening using the AdditiveHT screen (Hampton Research) with final crystals grown at 22°C in 100 mM Tris-HCl, pH 8.5, 200 mM MgCl2, 10 mM MnCl2, and 8% PEG 4000.
Data collection, structure determination, and modeling
Crystals were harvested by quick transfer directly into a cryoprotectant solution containing 20% glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected to 3.4 Å resolution at the SER-CAT beamline (ID22) at the Advanced Photon Source at Argonne National Laboratory and processed using HKL2000 (35). The structure was solved by molecular replacement using Phaser (36) within PHENIX (37) using previously reported crystal structures of the Bam components. Search order was key here for success, first starting with the BamCD complex (PDB ID 3TGO) followed by the barrel domain of BamA (PDB ID 4C4V), POTRA5 and then POTRA4 of BamA (PDB ID 3Q6B). BamE (PDB ID 2KM7) and POTRA1 (PDB ID 3EFC) were then placed based on density within a difference (Fo-Fc) map. After several rounds of building and refinement, POTRA2 and 3 (PDB ID 3EFC) were then manually placed in weak density followed by rigid body refinement for all components for final placement. The structure was refined to R/Rfree values of 0.22/0.27. All model building and refinement were performed using COOT (38) and PHENIX (37), respectively. Final placement of side chains was based on evaluation of 2Fo-Fc, Fo-Fc, and feature-enhanced (FEM) density maps (39). RMSD analysis was performed within PyMOL (Schrödinger) for C-α atoms using default settings. Surprisingly, BamB was not found within our crystal structure despite it being present in our purification. The absence of BamB was confirmed by analyzing crystals and our initial sample of the complex by SDS-PAGE analysis, which also were lacking BamB presumably due to proteolysis during storage/incubation.
To see how BamB interacts with the BamACDE complex, we modeled BamB into our complex using the previous reported BamAB crystal structure (PDB ID 4PK1) to produce the modeled structure of the fully assembled BamABCDE complex. Here, we were able to place BamB in our structure by performing a superposition of the two structures along POTRA3 of BamA. Analysis of interacting interfaces was performed using the PDBePISA (40). All figures were made with PyMOL (Schrödinger) and annotated and finalized with Adobe Illustrator.
Acknowledgments
We thank Harris Bernstein for providing the pJH114 plasmid. JB and NN are supported by the Department of Biological Sciences at Purdue University and by the National Institute of Allergy and Infectious Diseases (1K22AI113078-01). SKB is supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases. We thank the staff at the Southeast Regional Collaborative Access Team (SER-CAT) beamline at the Advanced Photon Source, Argonne National Laboratory for their assistance during data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38 (SER-CAT).
Footnotes
Materials and Methods
References
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aad3460
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/351/6269/180.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Breaking Barriers: Exploiting Envelope Biogenesis and Stress Responses to Develop Novel Antimicrobial Strategies in Gram-Negative Bacteria.
Pathogens, 13(10):889, 11 Oct 2024
Cited by: 0 articles | PMID: 39452760 | PMCID: PMC11510100
Review Free full text in Europe PMC
The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15(1):8718, 08 Oct 2024
Cited by: 0 articles | PMID: 39379361 | PMCID: PMC11461620
Dual client binding sites in the ATP-independent chaperone SurA.
Nat Commun, 15(1):8071, 14 Sep 2024
Cited by: 0 articles | PMID: 39277579 | PMCID: PMC11401910
Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15(1):7612, 01 Sep 2024
Cited by: 1 article | PMID: 39218969 | PMCID: PMC11366764
A conserved C-terminal domain of TamB interacts with multiple BamA POTRA domains in Borreliella burgdorferi.
PLoS One, 19(8):e0304839, 29 Aug 2024
Cited by: 0 articles | PMID: 39208212 | PMCID: PMC11361582
Go to all (151) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 6 of 6)
-
(3 citations)
PDBe - 3TGOView structure
-
(3 citations)
PDBe - 4C4VView structure
-
(3 citations)
PDBe - 4PK1View structure
-
(2 citations)
PDBe - 3EFCView structure
-
(1 citation)
PDBe - 3Q6BView structure
-
(1 citation)
PDBe - 2KM7View structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Conformational Changes That Coordinate the Activity of BamA and BamD Allowing β-Barrel Assembly.
J Bacteriol, 199(20):e00373-17, 19 Sep 2017
Cited by: 18 articles | PMID: 28760846 | PMCID: PMC5637172
Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine.
J Biol Chem, 290(4):2126-2136, 02 Dec 2014
Cited by: 32 articles | PMID: 25468906 | PMCID: PMC4303665
Structural insight into the formation of lipoprotein-β-barrel complexes.
Nat Chem Biol, 16(9):1019-1025, 22 Jun 2020
Cited by: 27 articles | PMID: 32572278 | PMCID: PMC7610366
The β-Barrel Assembly Machinery Complex.
Methods Mol Biol, 1329:1-16, 01 Jan 2015
Cited by: 7 articles | PMID: 26427672
Review
Funding
Funders who supported this work.
Department of Biological Sciences at Purdue University
Intramural NIH HHS (1)
Grant ID: ZIA DK036139-09
Intramural Research Program of the NIH
NIAID NIH HHS (2)
Grant ID: 1K22AI113078-01
Grant ID: K22 AI113078
National Institute of Allergy and Infectious Diseases (1)
Grant ID: 1K22AI113078-01





