Abstract
Free full text

Lck Mediates Th2 Differentiation through Effects on T-bet and GATA-3
Abstract
The Src family kinase Lck has been shown to be crucial in T cell signaling and development. However, its role in Th effector functions is not well understood. Lck has previously been shown to play a role in the cytokine expression of Th2 cells, but the mechanism by which Lck influences Th2 effector functions is unknown. Using a mouse model, we report that Lck is important in regulating the expression of IL-4 in Th2 skewed cells but is not as necessary for the expression of Th2 cytokines IL-5, IL-10, and IL-13. Furthermore, in the absence of Lck, T-bet and GATA-3 expression is aberrant. Moreover, this atypical expression pattern of T-bet and GATA-3 correlates with increased histone 3 acetylation at the Ifng locus and production of the Th1 cytokine IFN-γ. We find overexpression of GATA-3 restores IL-4 expression in lck−/− Th2 cells; this indicates that the decreased IL-4 expression is due in part to reduced amounts of GATA-3. Taken together, these data imply that Lck mediates Th2 differentiation through effects on T-bet and GATA-3.
CD4+ T cells have been traditionally divided into the Th1 and Th2 lineages, although several other effector lineages, such as Th17, T regulatory, and T follicular, have been described in recent years. Th cells play a crucial role in regulating immune responses by activating other immune cells. Th1 cells protect against intracellular pathogens, whereas Th2 cells protect against extracellular pathogens. Th1 cells are typically characterized by production of cytokines IFN-γ and TNF-α; in contrast, Th2 cells secrete cytokines IL-4, IL-5, IL-13, and IL-10 (1–3). A careful balance of Th1 and Th2 responses must be maintained, because CD4+ T cells can also mediate inappropriate immune responses leading to diseases, such as Th2-mediated allergy and asthma.
Several factors can affect Th differentiation. The presence of IL-4 leads to upregulation of the transcription factor GATA-3, whereas the presence of IL-12 leads to increased expression of the transcription factor T-bet (1). T-bet and GATA-3 levels are maintained via cytokine expression in an autocrine loop, and once upregulated, T-bet and GATA-3 then function to establish Th1 and Th2 differentiation by directly and/or indirectly downregulating each other and mediating activation of epigenetic modifications at the appropriate cytokine loci, such as Ifng and Il4, respectively (4–9).
Engagement of the CD4 coreceptor following TCR/MHC class II interactions leads to the recruitment of the Src family kinase Lck. This results in the activation of a variety of proteins and signaling cascades required for Th1 and Th2 cytokine expression, such as NF-κB, the Ras-Erk MAPK pathway, the p38 MAPK pathway, Itk, and NFATc1 (10–16). Taken together, engagement of these pathways may amplify the initial signal from Lck to drive effector cell differentiation. Furthermore, there is evidence of a direct role for Lck in Th differentiation. Expression of a dominant-negative form of Lck in cells skewed under Th2 conditions results in the production of IFN-γ, a hallmark of Th1 differentiation, and drastically reduced IL-4 (17). This study also found that Abs associated with a Th2 response, IgG1 and IgE, are decreased in mice expressing dominant-negative Lck. Paradoxically, IL-4 expression is maintained in established Th2 cells when Lck is reduced by antisense RNA (18). These data suggest that Lck may be important for the induction of the Th2 lineage but that Lck is not necessary for maintenance of Th2 effector function in established Th2 cells. Although these studies indicate a role for Lck in inducing Th2 function, they do not explain the mechanism by which Lck mediates Th2 responses.
In this study, we explore the mechanism(s) by which Lck mediates Th2 differentiation. We use a mouse system in which Lck is present in the thymus but not in the periphery (19).We find Lck-deficient CD4+ T cells skewed under Th2 conditions have a reduction in IL-4 protein levels, whereas other Th2 cytokines are produced at near wild-type (wt) levels. We also find that Lck-deficient Th2 skewed cells express T-bet, a transcription factor needed for Th1 differentiation, and have reduced levels of GATA-3, a transcription factor required for Th2 differentiation. Despite the reduction in GATA-3, the cells have a normal histone 3 (H3) acetylation pattern at the Th2 cytokine locus (Il4, Il5, and Il13). However, unlike wt Th2 cells, there is increased H3 acetylation at the Ifng promoter. These results suggest that Lck may promote Th2 differentiation by suppressing T-bet expression.
Materials and Methods
Mice
C57BL/6, lck−/− LGF (19), lck−/− LGF OTII, and OTII mice were maintained and bred under pathogen-free conditions in the Northwestern University animal facilities according to Institutional Animal Care and Use Committee regulations. All mice are maintained on the C57BL/6 background.
CD4+ T cell purification and in vitro differentiation of Th cells
CD4+ T cells were positively selected using biotinylated anti-CD4 (RM4-5), followed by streptavidin microbeads on a MACS column (Miltenyi Biotec, Auburn, CA). Purity, as assessed by flow cytometry, was typically ≥92%. CD4+CD62L+ cells were isolated by negative selection for CD4 and then by positive selection using anti-CD62L MACS beads (Miltenyi Biotec). Purified T cells were plated in 24-well dishes (1 × 106/well) that were coated with 5 µg/ml anti-CD28 (clone 2.43 rat IgG) and 0.5 µg/ml anti-TCR (H57-H59). The cells were cultured under Th1 (10 U/ml IL-2, 5 ng/ml IL-12, and 3.3 µg/ml anti–IL-4) or Th2 (10 U/ml IL-2, 10 ng/ml IL-4, 0.12 µg/ml anti–IL-12, and 5 µg/ml anti–IFN-γ) skewing conditions in 2 ml RPMI 1640 complete T cell media (RPMI 1640 + l-glutamine, 10% FBS, 50 µM2-ME, 10 mM HEPES, 1 mM sodium pyruvate, and 0.05 mg/ml gentamycin). In some experiments, cells were differentiated using HL-1 complete T cell media (HL-1, ± 1% FBS, 2 mM l-glutamine, 50 µM 2-ME, 10 mM HEPES, 1× MEM nonessential amino acids, and 0.05 mg/ml gentamycin). After 4 d, the cells were further expanded for 3 d under Th1 or Th2 skewing conditions without anti-CD28 and anti-TCR. Recombinant mouse IL-4 and IL-12 were purchased from PeproTech (Rocky Hill, NJ). Low endotoxin-grade anti–IL-12 (C17.8), anti–IFN-γ (XMG1.2), anti-TCR, and anti–IL-4 (11B11) Abs were purchased from eBioscience (San Diego, CA). In some experiments, cells were differentiated 3 or 4 d with anti-TCR and anti-CD28 stimulation in complete media. Secondary stimulations were performed by incubating day 7 cells with PMA (5 ng/ml) and ionomycin (500 ng/ml) for 4 h.
For experiments using OTII mice, the cells (1 × 106) were differentiated in the presence of 10 µM OVA peptide 323–339 (ISQAVHAAHAEINEAGR), a gift from Dr. W. J. Karpus (Northwestern University, Chicago, IL), and T cell-depleted irradiated APCs (2 × 106) for 5 d in 2 ml complete media in a 24-well dish. The cells were then expanded for 2 d in complete media under skewing conditions in the absence of OVA peptide and APCs.
Intracellular cytokine staining and cytokine capture assays
Cells skewed for 7 d were incubated with monensin (eBioscience) or GolgiStop (BD Biosciences, San Jose, CA), 5 ng/ml PMA (Roche, Basel, Switzerland), and 500 ng/ml ionomycin (Roche) for 4 h at 37°C in complete media. Cells skewed for 3 d were incubated with monensin in the presence of plate-bound anti-TCR (0.5 µg/ml) and anti-CD28 (5 µg/ml) for 5 h. The cells were then fixed and permeabilized with the BD Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences) following the manufacturer’s guidelines. The cells were stained with fluorochrome-conjugated CD4 (RM4-5), IFN-γ (XMG1.2), IL-4 (11B1), IL-10 (Jes5-16E3), and IL-13 (ebio13A) Abs at 4°C for 30 min, washed, resuspended in FACS buffer, and analyzed on a FACSCalibur or FACSCanto (BD Biosciences). With regards to the GATA-3 staining, cells were fixed and permeabilized using the eBioscience FoxP3 Staining buffer kit (eBioscience), and then the cells were incubated with GATA-3 (clone TWAJ) Ab or an isotype control for 30 min, washed, resuspended in FACS buffer, and analyzed on a FACSCanto (BD Biosciences). All Abs for intracellular cytokine staining (ICCS) were obtained from eBioscience or BD Pharmingen (San Diego, CA). IL-5 cytokine capture assays (Miltenyi Biotec) were performed according to the manufacturer’s guidelines.
For experiments using OTII mice, the cells were restimulated in the presence of OVA-pulsed APCs and monensin for 5 h. The cells were then analyzed for IL-4 and IFN-γ as above.
RNA isolation and quantitative RT-PCR
RNA was isolated from day 4 and day 7 skewed cells using TRIzol and reverse transcribed with Reverse Transcriptase III, according to the manufacturer’s protocol (Invitrogen Life Technologies, Carlsbad, CA). Day 7 skewed cells were restimulated prior to RNA isolation. Quantitative RT-PCR (qRT-PCR) was performed using Sybr Green incorporation on an ABI prism 7000 detection system (Applied Biosystems). Target levels were normalized to hypoxanthine phosphoribosyltransferase (HPRT). The relative expression for the qRT-PCR samples is calculated as 2−(ΔCTsample), where CT is the cycle number it took for the sample to reach the analysis threshold. ΔCT is the CT of the sample for the gene of interest minus the CT of the normalizing gene, HPRT. The primers used for qRT-PCR can be found in Table I.
Table I
Primer sequences used for qRT-PCR
| Primer Set | Sequence |
|---|---|
| GATA-3 | 5′-GAACCGCCCCTTATCAAG |
| 3′-CAGGATGTCCCTGCTCTCCTT | |
| T-bet | 5′-AATCGACAACAACCCCTTTG |
| 3′-AACTGTGTTCCCGAGGTGTC | |
| IL-4 | 5′-AACTCCATGCTTGAAGAAGAACTC |
| 3′-CCAGGAAGTCTTTCAGTGATGTG | |
| IL-5 | 5′-GTTGACAAGCAATGAGACGATGAG |
| 3′-CCCACGGACAGTTTGATTCTTC | |
| IFN-γ | 5′-ACAATGAACGCTACACACTGC |
| 3′-CTTCCACATCTATGCCACTTGAG | |
| IL-13 | 5′-GCAACATCACACAAGACCAGAC |
| 3′-GAATCCAGGGCTACACAGAACC | |
| Runx3 | 5′-AGGGAAGAGTTTCACGCTCA |
| 3′-CTTCTATCTTCTGCCGGTGC | |
| HPRT | 5′-TGGGCTTACCTCACTGCTTTC |
| 3′-CCTGGTTCATCATCGCTAATCAC | |
| c-Maf | 5′-GGTCAGCAAGGAGGAGGTG |
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the manufacturer’s protocol (Upstate Biotechnology, Lake Placid, NY). Briefly, 4 × 106 cells were fixed with formaldehyde, and the nuclear lysates were sonicated and subjected to immunoprecipitation overnight at 4°C with anti-acetyl H3 (Millipore, Bedford, MA) or rabbit IgG isotype (The Jackson Laboratory, Bar Harbor, ME). qRT-PCR was performed using Sybr Green incorporation on equal volumes of immunoprecipitated DNA. The relative expression for the ChIP samples is calculated as 2(CTinput − CTIP), where CT is the cycle number it took for the samples to reach the analysis threshold. The primers used for ChIP are found in Table II. qRT-PCR was performed using primers for the Rad50 promoter as a positive control for the ChIP samples.
Table II
Primer sequences used for ChIP analysis
| Primer Set | Sequence |
|---|---|
| IL-4 promoter | F: CCTGGGGAAAGACAGAGTAATATC |
| R: CCCAAGATATCAGAGTTTCCAAGG | |
| CNS1 | F: TTCGAGTGGCTTCGTCATGG |
| R: TCCACCCAAAAGCGACAGAG | |
| IFN-γ promoter | F: CGAGGAGCCTTCGATCAGGT |
| R: GGTCAGCCGATGGCAGCTA | |
| IL-5 promoter | F: ACCCTGAGTTTCAGGACTCG |
| R: TCCCCAAGCAATTTATTCTCTC | |
| Rad50 promoter | F: CAGAGCTAGACCCGATCTCA |
| R: CGAGCCAGCAACCGTAAG | |
| IL-13 promoter | F: AGGGTCTGGGACAGGGTTTC |
| R: CGGTCAGCGGGTGGAATTAC |
CNS, conserved noncoding sequence.
Protein isolation and Western blots
The cells were lysed in radioimmunoprecipitation assay buffer, and debris was pelleted by centrifugation at 100,000 rpm for 10 min at 4°C. Day 7 skewed cells were restimulated for 4 h prior to harvesting. The lysates were run on a 10% SDS gel, transferred to polyvinylidene difluoride, and immunoblotted with the following Abs: anti-mouse GATA-3 (clone HG3-31; Santa Cruz Biotechnology, Santa Cruz, CA), anti-mouse β-actin (clone C4; Santa Cruz Biotechnology), and anti-mouse T-bet (clone ebio4b10; eBioscience).
Retrovirus infections
CD4+ splenocytes were isolated and stimulated under Th1 and Th2 conditions as above and infected with MigR1 or GATA-3 MigR1 (20) retroviruses at 24 h as described previously (21).
Statistical analysis
The data were analyzed using either a two-tailed Student paired or unpaired t test. Use of the different tests were based on whether the samples to be analyzed were dependent or independently related. In the case of unpaired test, sample sets were analyzed to make sure they had the same variance by the F test or Brown-Forsythe test. Samples are considered to differ significantly if the result of the test is a value of p ≤ 0.05.
Results
Lck-deficient Th2 skewed cells produce IFN-γ and have reduced IL-4
It was previously shown that Lck is important in Th2 responses; however, the mechanism by which Lck mediates Th2 differentiation was not explored (17). To address this, we used a mouse model in which Lck is expressed in the thymus but not the periphery (19). This allows for the isolation of mature Lck-deficient cells. These mice are referred to as lck−/− in this study. Bulk CD4+ cells from B6 and lck−/− mice were skewed under Th1 and Th2 conditions for 7 d. We measured IFN-γ and IL-4 expression, hallmark cytokines of Th1 and Th2 differentiation, respectively. We found that Lck-mutant cells had a normal Th1 profile with regards to IFN-γ and IL-4, but when skewed under Th2 conditions, these cells expressed less IL-4 than wt Th2 cells and also produced IFN-γ (Fig. 1A, 1B, and data not shown). Furthermore, we noted that many of the cells were dual IL-4 and IFN-γ expressors. Similar results were obtained when naive (CD4+CD62L+) cells were used (Fig. 2A). In addition, we observed that the Th2 defect became significantly more pronounced with age (Supplemental Fig. 1) in that a greater proportion of the Th2 skewed cells produce IFN-γ.
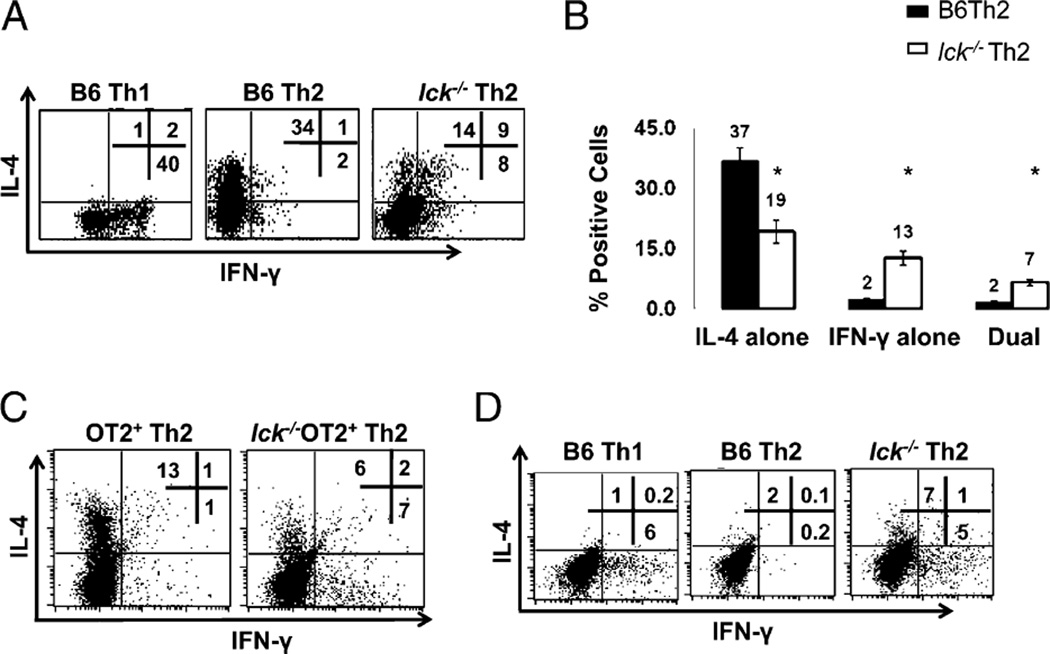
Lck-deficient Th2 skewed cells produce IFN-γ and have reduced IL-4. A, lck−/− Th2 skewed cells have abnormal IL-4 and IFN-γ expression. CD4+ splenocytes were stimulated under Th1 and Th2 conditions for 7 d and analyzed for IL-4 and IFN-γ by ICCS. B, Percentage of cells producing IL-4 and IFN-γ in B6 and lck−/− Th2 cells skewed for 7 d. The graph illustrates the average of >20 ICCS experiments performed on CD4+ cells. The asterisk indicates that the data are statistically significant (Student unpaired t test; p < 0.05). C, Ag-specific skewing. CD4+ splenocytes from OTII and lck−/− OTII mice were skewed in the presence of APCs and OVA peptide. The cells were then restimulated with APCs and OVA peptide for 5 h and analyzed for IL-4 and IFN-γ as in A. The data are representative of three experiments. D, IFN-γ protein expression is elevated in day 3 lck−/− Th2 cultures. Bulk CD4+ cells were skewed under Th1 and Th2 conditions and then analyzed for IL-4 and IFN-γ by ICCS. The cells were initially gated on CD4 expression. The results are representative of three experiments.
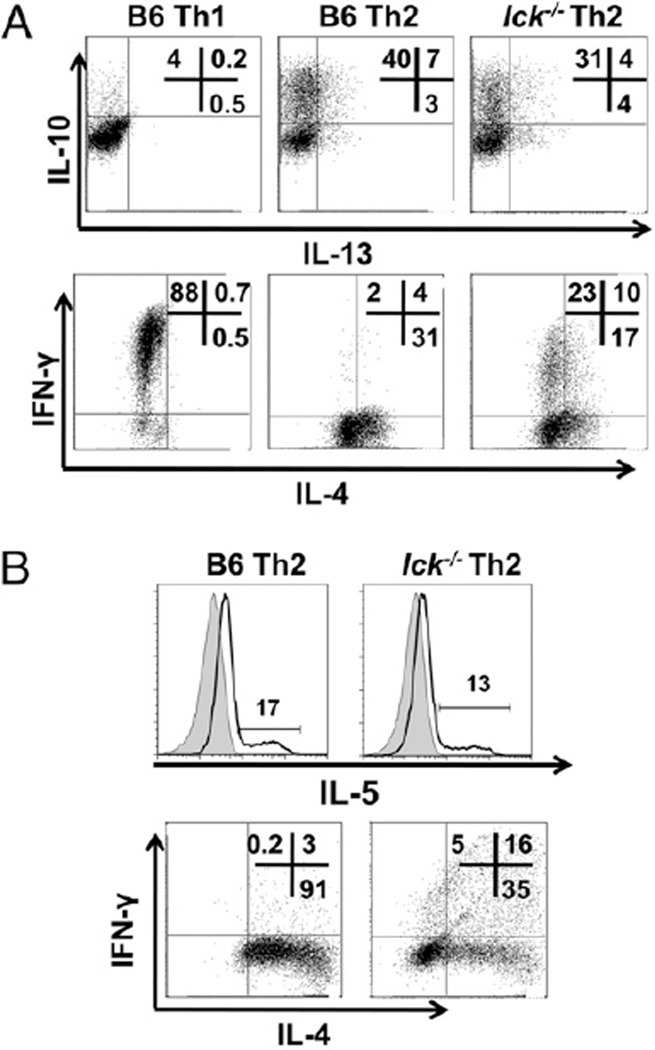
Lck-deficient Th2 skewed cells produce Th2 cytokines IL-10, IL-13, and IL-5. A, lck−/− Th2 cells express IL-10 and IL-13. CD4+CD62L+ T cells were stimulated under Th1 and Th2 conditions for 7 d and IL-13 and IL-10 (upper panels), and IL-4 and IFN-γ (lower panels) were analyzed by ICCS. Cells were initially gated on CD4 expression. B, lck−/− Th2 cells express IL-5. CD4+ T cells were stimulated under Th2 conditions for 7 d, and IL-5 expression was detected by a mouse IL-5 secretion assay (upper panels), and IL-4 and IFN-γ were measured by ICCS (lower panels). The shaded peaks in the IL-5 plots represent the no stimulation control, and the line represents the stimulated sample. The cells were initially gated on CD4 expression. The results are representative of two experiments.
The in vitro skewing protocol performed in Fig. 1A and 1B takes advantage of strong agonist Abs that do not completely mimic interactions between T cells and APCs. The MHC complex engages both the TCR plus the CD4 coreceptor, and the APC binds several costimulatory molecules, in addition to CD28; all of these interactions can affect Th differentiation (reviewed in Ref. 22). Therefore, we wanted to analyze IL-4 and IFN-γ expression under conditions that more closely mimic a physiological response. Lck-mutant mice were bred to OTII mice. These mice express a transgene specific for chicken OVA residues (323–339) in the context of the MHC class II I-Ab molecule (23). We find that lck−/− OTII Th2 skewed cells have reduced IL-4 and express IFN-γ following differentiation with OVA peptide and APCs (Fig. 1C). This indicates that Lck facilitates Ag-induced skewing.
We postulated that the failure of Lck-deficient Th2 skewed cells to appropriately express IL-4 and IFN-γ could be due to defective differentiation into the Th2 lineage or to an inability to respond to a secondary stimulus, as observed in mice deficient in Itk—a Tec family kinase activated by Lck (10, 24). Itk-deficient Th2 skewed cells differentiate into the Th2 lineage but do not express IL-4 upon secondary TCR stimulation because of impaired Ca+2 mobilization; restimulating these cells with ionomycin bypasses this block and restores IL-4 expression (10). In contrast, ionomycin stimulation does not induce normal IL-4 expression in lck−/− Th2 skewed cells upon secondary stimulation (Fig. 1A, 1B), suggesting that the defects are not due to impaired Itk activation. Therefore, we addressed the possibility that the lck−/− Th2 skewed cells may exhibit defects in the early Th2 differentiation program. To test this, lck−/− bulk CD4+ cells were skewed under Th2 conditions for 3 d, and then IL-4 and IFN-γ expression was measured by flow cytometry (Fig. 1D). Although lck−/− Th2 skewed cells express IL-4 early in the differentiation process, they are unable to suppress IFN-γ expression (Fig. 1D). This indicates that the failure of Lck-deficient cells to produce the appropriate cytokines under Th2 skewing conditions may be due to an inability to properly differentiate into the Th2 lineage.
CD4+ T cells can also differentiate into two other lineages important for immune responses: Th17 and T regulatory (Treg) cells (reviewed in Ref. 25). We found lck−/− cells upregulate lineage-specific markers IL-17 and FoxP3 appropriately when differentiated under Treg- and Th17-inducing conditions (data not shown). Thus, it appears that Lck is necessary for proper Th2 differentiation, but it is not required for the establishment of other known CD4+ effector lineages.
Lck-deficient Th2 skewed cells produce Th2 cytokines IL-10, IL-13, and IL-5
The cytokine IL-4 is genetically linked to the cytokines IL-5 and IL-13 at what is termed the Th2 cytokine gene locus (26), and these cytokines have been shown to be important for Th2-mediated responses. In addition, IL-10, which is located elsewhere in the genome, is also expressed by Th2 cells (2). Therefore, we analyzed IL-5, IL-10, and IL-13 expression by qRT-PCR and flow cytometry in both bulk CD4+ cells and CD4+CD62L+ cells (Fig. 2 and data not shown). We found that the lck−/− Th2 skewed cells have modestly decreased levels of IL-10 and normal IL-13 expression compared with wt Th2 cells (Fig. 2A). Furthermore, we found that IL-5 expression is relatively normal in lck−/− Th2 skewed cells compared with wt Th2 cells (Fig. 2B). Overall, these data indicate that the defect in lck−/− Th2 skewed cells predominately affects IL-4 with minimal impact on the other Th2 cytokines.
The expression of T-bet and GATA-3 is aberrant in lck−/− Th2 skewed cells
Th1 and Th2 differentiation is dependent on transcription factors T-bet and GATA-3, respectively (reviewed in Refs. 1, 3, 22). Therefore, we analyzed T-bet and GATA-3 mRNA and protein expression in wt and mutant Th2 and Th1 skewed cells at days 3 and 4, a time when the cells are committing to their respective lineages, and at day 7, upon secondary stimulation of fully differentiated cells (Fig. 3A, 3B). T-bet protein expression is present in lck−/− Th2 skewed cells in the early stages of differentiation, and this correlates with elevated levels of T-bet mRNA (Fig. 3A, 3B). GATA-3 protein is reduced in lck−/− Th2 skewed cells early in the differentiation process, but mRNA levels are similar to wt; this suggest that the decrease in GATA-3 is occurring posttranscriptionally (Fig. 3A, 3B). These aberrant protein expression patterns are more pronounced in day 7 lck−/− Th2 skewed cells compared with B6 Th2 cells (Fig. 3B). These data again emphasize that the loss of Lck results in a failure of cells to properly commit to the Th2 lineage.
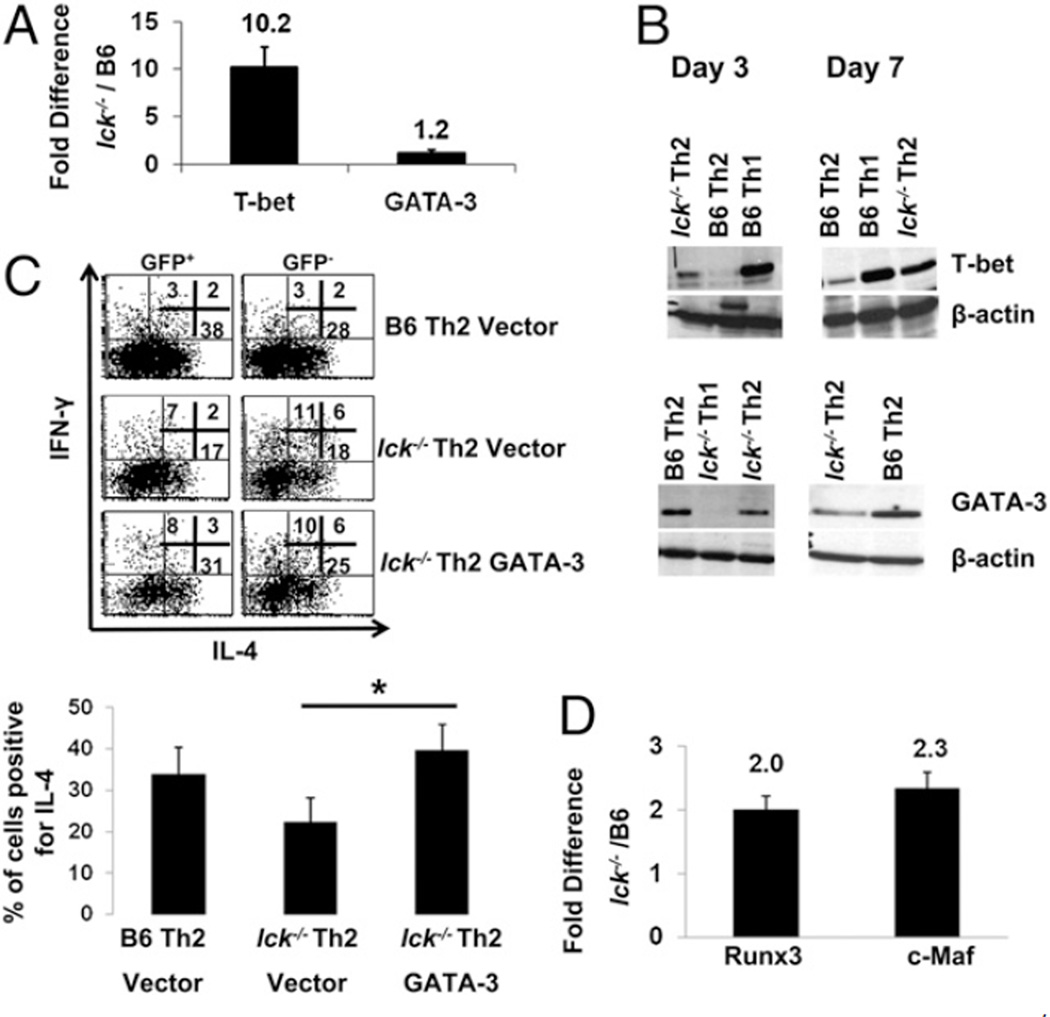
The expression of T-bet and GATA-3 is aberrant in lck−/− Th2 skewed cells. The sequences used for qRT-PCR are listed in Table I. A, GATA-3 and T-bet mRNA expression from wt and lck−/− Th2 cultures. CD4+CD62L+ T cells were stimulated under Th2 conditions for 4 d, and qRT-PCR was performed for T-bet and GATA-3. The data are graphed as the ratio of the expression of the target gene in lck−/− Th2 cells relative to B6 Th2 skewed cells. Note that T-bet is overexpressed in these cultures. The graphs illustrate the average of a minimum of three experiments. B, GATA-3 and T-bet protein expression is altered in lck−/− Th2 cultures. CD4+ cells were skewed under Th1 and Th2 conditions for 3 and 7 d, and protein expression was evaluated by immunoblotting. The data are representative of two (day 3) and three (day 7) experiments. C, GATA-3 overexpression normalizes IL-4 cytokine expression in lck−/− Th2 cells. lck−/− and B6 cells were infected with GATA-3 or empty vector 24 h after initiation of Th2 skewing and then skewed for an additional 6 d. Infected cells were identified by GFP expression and then analyzed for IL-4 and IFN-γ expression by ICCS. The flow plots show a representative experiment. The graph illustrates the average for the percentage of cells expressing IL-4 in GFP+ cells (four experiments). The expression of IL-4 in Lck Th2 vector versus Lck Th2 GATA-3–infected cells is statistically different. The asterisk indicates that p < 0.05 (Student paired t test). Note: IL-4 expression is similar in the wt Th2 and lck−/− GATA-3–infected cells. D, c-Maf and Runx3 are elevated in lck−/− Th2 cultures. lck−/− and B6 CD4+CD62L+ cells were skewed under Th2 conditions for 7 d, and qRT-PCR was performed for c-Maf and Runx3. The data are graphed as the ratio of the expression of the target gene in lck−/− Th2 cells relative to B6 Th2 skewed cells. The graph illustrates the average of three experiments.
T-bet can antagonize GATA-3 expression and function (6, 9). Therefore, we hypothesized that T-bet may be inhibiting GATA-3 expression and/or function in lck−/− Th2 skewed cells and that overexpression of GATA-3 would normalize the Th2 phenotype in lck−/− cells. CD4+ T cells from wt and lck−/− mice were skewed under Th2 or Th1 conditions and infected at 24 h after initial stimulation with a retrovirus encoding GATA-3 or an empty vector control. We observed that GATA-3 overexpression, unlike the empty vector control, resulted in similar levels of IL-4 expression in wt and lck−/− Th2 skewed cells (Fig. 3C). In addition, the increased IL-4 expression following infection with GATA-3 retrovirus is not due to gross overexpression of GATA-3. Although GATA-3 expression in mutant cells transduced with the GATA-3 retrovirus (GFP+) is slightly higher than wt B6 Th2 cells, the overall expression of GATA-3 in these cells is less than that of wt cells infected with the empty vector (Supplemental Fig. 2). These data suggest that the reduction of GATA-3 expression in lck−/− Th2 skewed cells may result in decreased IL-4 cytokine levels.
Previous studies have shown that the loss of GATA-3 results in reduced Th2 cytokines (2). Lck−/− Th2 skewed cells have decreased IL-4 expression but relatively normal levels of other known Th2 cytokines (Figs. 1, ,2);2); this implies that other transcription factors involved in IL-4 gene regulation are affected. Knockout studies have shown that c-Maf is important for IL-4 expression but not for the production of other Th2 cytokines (27). We found c-Maf mRNA expression is elevated in lck−/− Th2 skewed cells at days 4 and 7 compared with wt Th2 cells, and this suggests that the IL-4 defect we observe is not due to c-Maf (Fig. 3D and data not shown). We also analyzed the expression of Runx3, which has been shown to interact with T-bet to silence IL-4 expression in Th1 cells by binding to the Il4 silencer or HSIV (28). We found Runx3 mRNA expression to be elevated 2-fold in lck−/− Th2 skewed cells (Fig. 3D). Significantly, the level of Runx3 expression in lck−/− Th2 skewed cells was equivalent to that of wt Th1 cells (data not shown). These data suggest that the decrease in IL-4 expression may be due to a combination of diminished GATA-3 as well as Runx3/T-bet complexes binding to the Il4 silencer.
lck−/− Th2 cells have increased H3 acetylation at the IFN-γ locus but normal levels of acetylation at the Th2 cytokine gene locus
Epigenetic changes in chromatin structure, such as H3 acetylation, help activate gene expression by allowing loci to become accessible for transcription. The Th2 cytokine gene locus (Il4-Il13-Il5) undergoes demethylation and acetylation at many sites during Th differentiation (reviewed in Ref. 26). GATA-3 is required for permissive modifications, such as H3 acetylation, at the Th2 cytokine gene locus, whereas T-bet is required for permissive modifications at the Ifng locus (reviewed in Ref. 29). Because lck−/− Th2 skewed cells have aberrant expression of T-bet and GATA-3, we entertained the possibility that these cells have an abnormal H3 acetylation state at the Th2 cytokine gene locus and/ or the Ifng locus and that defects in epigenetic modifications at these loci result in aberrant cytokine production.
We focused our attention on the promoter regions of Il4, Il13, Ifng, and Il5, as well as CNS1, a region known to be important for IL-4 and IL-13 production (Fig. 4A) (reviewed in Ref. 26). Regions associated with the Th2 cytokine gene locus should be hyperacetylated in Th2 cells, whereas the Ifng locus should show reduced acetylation in Th2 cells (5, 30–32). The acetylation status of the Th2 cytokine gene locus (CNS1 region and the promoter regions of Il4, Il13, and Il5) was similar in both wt and mutant Th2 cells at day 7 (Fig. 4B). This indicates that the decreased IL-4 expression present in lck−/− Th2 skewed cells is not due to the loss of permissive chromatin remodeling events, such as H3 acetylation. However, we did observe a significant increase in H3 acetylation at the Ifng promoter in lck−/− Th2 skewed cells compared with wt (Fig. 4B). Furthermore, we did not find a significant difference in H3 acetylation at the Th2 cytokine gene and Ifng loci in freshly isolated wt and mutant CD4+ splenocytes (data not shown). This signifies that the abnormal cytokine expression in lck−/− Th2 skewed cells is not due to an inherent defect in the acetylation status of the Ifng locus. These data indicate that Lck is important in negatively regulating hyperacetylation at the Ifng locus in Th2 cells, possibly by suppressing T-bet expression.
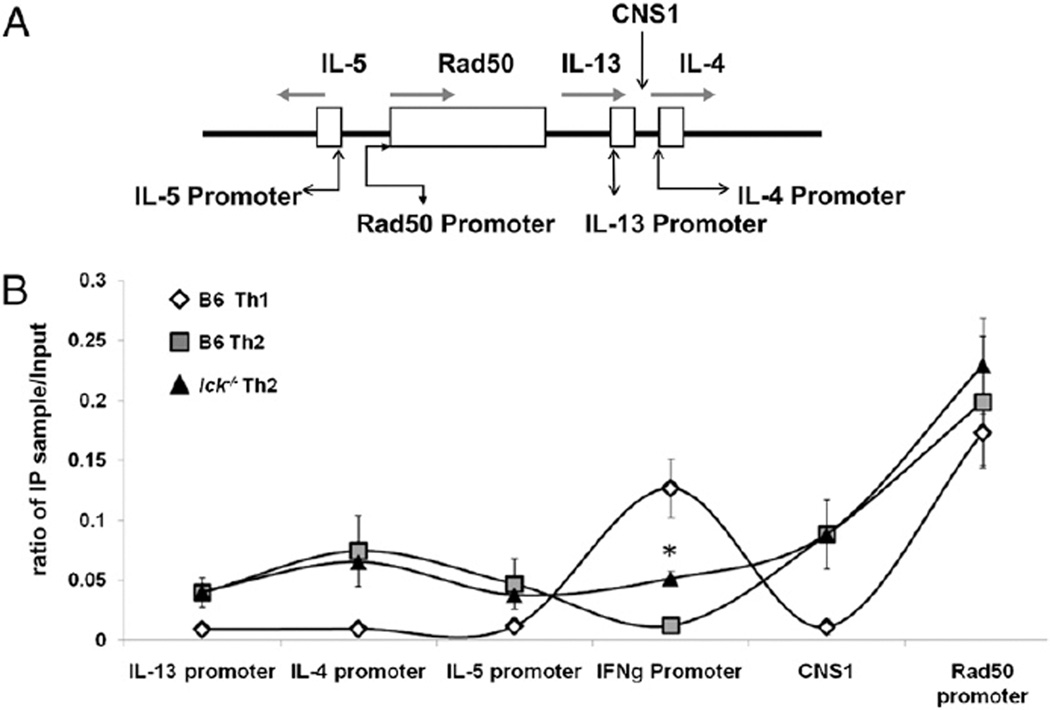
lck−/− Th2 cells have increased H3 acetylation at the Ifng locus but normal H3 acetylation at the Th2 cytokine gene locus. A, A schematic of the Th2 cytokine gene locus showing regions that were analyzed for H3 acetylation. B, H3 acetylation patterns of skewed cells. CD4+ cells were skewed for 7 d under Th1 and Th2 conditions, and H3 acetylation in nucleosomes associated with the Th2 cytokine gene locus and Ifng gene promoter were analyzed by ChIP. Analysis of the Rad50 gene promoter was included as a positive control for general H3 acetylation, whereas B6 Th1 cells served as a negative control. The data are graphed as the ratio of the immunoprecipitated DNA over input DNA. The asterisk indicates that the data are statistically significant (Student unpaired t test; p < 0.05). These results are the average of three experiments. The sequences used to analyze the ChIP are listed in Table II.
Discussion
Lck plays a role in T cell development via activation of downstream signaling pathways (33–36). However, the function of Lck in peripheral T cell responses is not well explored. We present evidence that Lck is required for proper Th2 differentiation but appears not to be required for Th1, Th17, and Treg differentiation (Fig. 1A, 1B, and data not shown). The loss of Lck in established Th2 clones affected only the kinetics of IL-4 cytokine expression but did not lead to IFN-γ expression (18). This indicates that in established Th2 cells, Lck is not required for Th2 cytokine expression or for the silencing of IFN-γ. In contrast, expression of a dominant-negative form of Lck in CD4+ cells placed under Th2 polarizing conditions resulted in highly reduced IL-4 expression and gain of IFN-γ expression (17). In our model, IL-4 levels are not reduced to the same degree (Fig. 1A, 1B). However, as mice age, the global Th2 defect becomes more pronounced (Supplemental Fig. 1). Overall, the differences in results may reflect differences in mouse models and/or skewing protocols. It is also possible that the presence of the dominant-negative form of Lck could inhibit functions of other signaling molecules involved in Th differentiation, and this could lead to a more profound defect in Th2 skewed cells. For example, in our model, Fyn may be able to partially compensate for the absence of Lck; in the dominant-negative model, Fyn could be displaced by the kinase defective Lck and, therefore, is unable to provide substitute signals.
We do not believe the decrease in IL-4 expression found in lck−/− Th2 skewed cells is due to inhibition of IL-4R signaling, because this pathway is intact when Lck is inhibited (17). Furthermore, IL-4R signaling leads to the activation of Stat6, which is involved in the transcription of GATA-3 (reviewed in Ref. 1). Although GATA-3 protein is decreased, we find that both wt and lck−/− Th2 skewed have similar levels of GATA-3 mRNA early in the differentiation process (day 4) and in fully differentiated cells (day 7) (Fig. 3A and data not shown). Furthermore, we find that IL-4R expression is normal in these cells (data not shown).
The Il4 and Ifng loci are nonselectively hyperacetylated in naive cells upon TCR stimulation; by 48 h, H3 acetylation is markedly reduced at the Ifng locus in Th2 skewed cells, and by day 7, it is virtually undetectable at the Ifng locus (4). These epigenetic changes allow for IL-4 production and inhibition of IFN-γ in Th2 skewed cells and are required for cells to appropriately differentiate into the Th2 lineage. Furthermore, epigenetic modifications at the Th2 cytokine gene locus are dependent, in part, on the transcription factor GATA-3 (reviewed in Ref. 26). By day 3, Th2 cells should express GATA-3 and trace amounts of T-bet (7, 8, 10). The phenotypic reduction of IL-4 and ectopic expression of IFN-γ in fully differentiated lck−/− Th2 skewed cells is consistent with the observed decreased GATA-3 and increased T-bet protein expression (Figs. 1, ,3B).3B). Furthermore, the aberrant T-bet expression in lck−/− Th2 skewed cells correlates with a 4-fold increase in H3 acetylation at the Ifng locus compared with wt Th2 cells (Fig. 4). Even though Lck-deficient Th2 cells have reduced IL-4 expression, H3 acetylation at the Th2 cytokine gene locus is normal. This indicates that the level of GATA-3 expressed is sufficient to mediate H3 acetylation at the Th2 cytokine gene locus but not inhibit T-bet expression. Overall, these data indicate that in the absence of Lck, cells fail to appropriately differentiate into the Th2 lineage.
Studies have shown that T-bet and GATA-3 directly and indirectly antagonize each other’s expression and function (6, 8, 9). We observe decreased GATA-3 and elevated T-bet in lck−/− Th2 skewed cells, and this suggests that inhibition of GATA-3, possibly by T-bet, may contribute to the reduction in IL-4 expression (Figs. 1, 3A, 3B). This hypothesis is consistent with our observation that overexpression of GATA-3 restores IL-4 production in lck−/− Th2 skewed cells (Fig. 3C).
The GATA-3 retrovirus appears to increase the percentage of IL-4+ cells in the GFP− (nontransduced) population. This may be an anomalous observation, reflecting the fact that fixation and subsequent intracellular staining results in a large loss of the GFP signal. Therefore, many of the GFP− cells actually represent successfully transduced cells that have difficult-to-detect levels of GFP. Because these fall into the GFP− gate, this could account for the apparent increase in IL-4+ cells in the uninfected population.
GATA-3 has been shown to be important for mediating epigenetic changes to the Th2 cytokine gene loci. In differentiated Th2 cells, GATA-3 is then required for maintenance of IL-5 and IL-13 expression but not for IL-4 or IL-10 (37). Although GATA-3 can exhibit haploinsufficiency (38), GATA-3+/− Th2 cells do not appear to have an appreciable defect in cytokine production (37). In the lck−/− Th2 cells, the GATA-3 levels appear reduced ~50%, which may be sufficient for production of IL-13 and IL-5, and may explain why we do not observe a significant defect in synthesis of the Th2 cytokines other than IL-4.
In addition, the selective defect for IL-4 in lck−/− Th2 cells suggests that there may be impaired expression of other transcription factors specific for IL-4 expression. We find that c-Maf, a transcription factor required for IL-4 expression, but not other Th2 cytokines is actually upregulated 2-fold in lck−/− Th2 skewed cells (Fig. 3D). Significantly, we find Runx3 mRNA to be elevated in the mutant Th2 cultures, and this expression level is similar to what is observed in wt Th1 cells (Fig. 3D and data not shown). Runx3 is upregulated by T-bet in Th1 cells, where it can then form a complex with T-bet and bind to the Il4 silencer region (also known as HSIV) to prevent IL-4 production (28). Because T-bet is ectopically expressed in lck−/− Th2 skewed cells, the diminished IL-4 production could be due to a similar mechanism. Moreover, GATA-3 can inhibit Runx complexes from binding to the Il4 silencer (39). Because GATA-3 is reduced in lck−/− Th2 skewed cells, it may only partially interfere with these complexes and thus allow for some IL-4 transcription. Alternatively, overexpression of GATA-3 in lck−/− Th2 skewed cells could alleviate the inhibition of IL-4 by removal of Runx3/T-bet complexes from the Il4 locus. Therefore, interference of GATA-3 expression and function, mediated by T-bet, may allow for Runx3 and T-bet to bind to the Il4 silencer and mediate repression of IL-4 (Fig. 5).
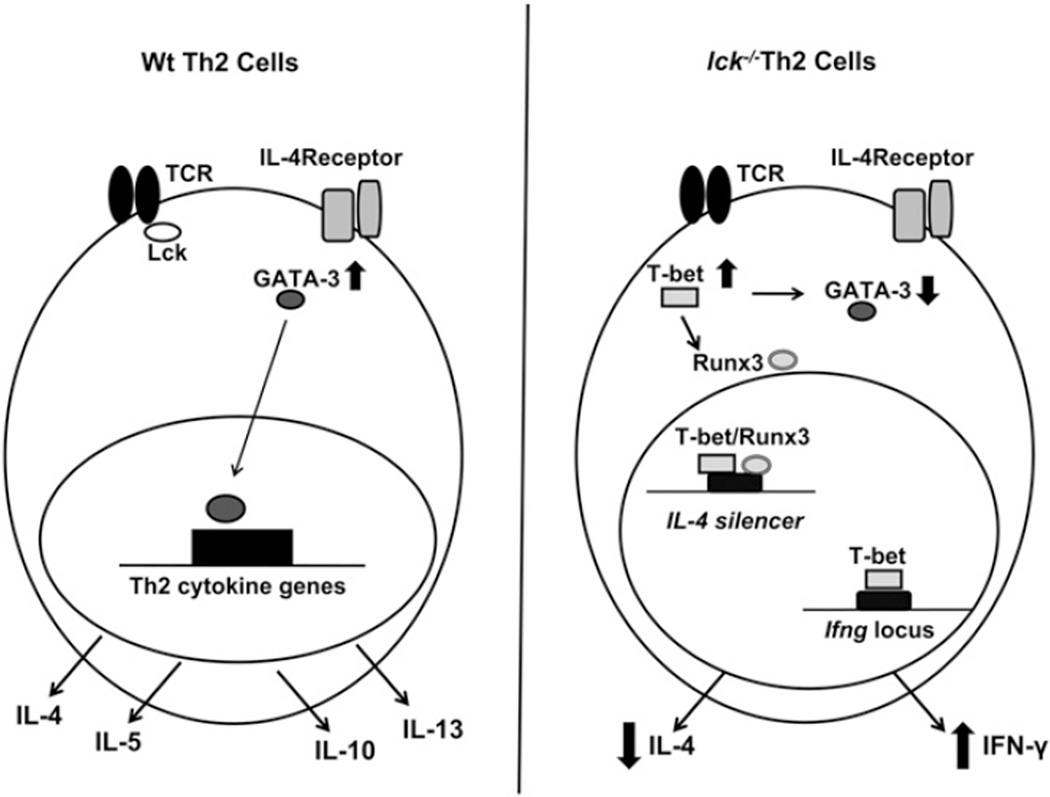
Model of how Lck may influence Th2 differentiation. The left panel illustrates what typically occurs during Th2 differentiation, and the right panel represents what may occur in the absence of Lck. Under Th2 conditions, GATA-3 is upregulated and is then able to mediate epigenetic modifications and/or transcription of Th2 cytokines. This leads to the expression of IL-4, IL-5, IL-13, and IL-10. In the absence of Lck (right panel), T-bet is ectopically expressed. T-bet interferes with GATA-3 protein expression and upregulates IFN-γ and Runx3. T-bet and Runx3 then complex and bind to the Il4 silencer, which leads to decreased IL-4 expression. The reduction of GATA-3 protein results in a smaller pool of GATA-3 capable of removing the Runx3/T-bet complexes from the Il4 silencer.
Runx1 is suspected of controlling IL-4 expression in naive cells, and it has been shown to be capable of binding to the Il4 silencer (39). In addition, overexpression of Runx1 in Th2 skewed cells inhibits GATA-3 expression and Th2 cytokine expression (40). We find that Runx1 is elevated >2-fold at day 4 in lck−/− Th2 skewed cells but returns to wt levels by day 7 (data not shown). It is possible that Runx1, in combination with Runx3 and T-bet, inhibits GATA-3 during early differentiation, leading to more substantial defects by day 7.
There is evidence that Fyn, another Src family kinase found in T cells, is involved in mediating Th differentiation. It has been reported that unpolarized fyn−/− cells have elevated IL-4 and IL-5 in response to TCR stimulation compared with wt (41), and Fyn levels are elevated in Th1 cells compared with Th2 cells (42). We find that Fyn expression is equivalent in wt and lck mutant Th2 cells, indicating that Fyn is not increased in the absence of Lck (data not shown). However, Fyn has been shown to phosphorylate p38 in vitro (43), and the p38 MAPK pathway has been implicated in activating T-bet and IFN-γ expression (14, 44–46). It is possible that in the absence of Lck, the p38 MAPK pathway is activated inappropriately, and this leads to elevated IFN-γ and T-bet expression in lck−/− Th2 skewed cells.
In summary, our results demonstrate that Lck is important in appropriately regulating expression of IL-4 and IFN-γ, potentially through affecting the normal regulation of T-bet and GATA-3.
Acknowledgments
We thank Sheila Chari, Dr. William Karpus, Mary Quirion, Dr. Steve Reiner, Aki Ueda, Sarah Umetsu, and Dr. Susan Winandy for technological expertise and support.
This work was supported in part by National Institutes of Health Grants R01CA086867 (to P.L.S.) and F31AI077322 (to K.L.K.) and National Institutes of Health, National Cancer Institute Training Grant T32CA09560 (to K.L.K.).
Abbreviations used in this paper
| ChIP | chromatin immunoprecipitation |
| CNS | conserved noncoding sequence |
| H3 | histone 3 |
| HPRT | hypoxanthine phosphoribosyltransferase |
| ICCS | intracellular cytokine staining |
| qRT-PCR | quantitative RT-PCR |
| Treg | T regulatory |
| wt | wild-type |
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.4049/jimmunol.0901282
Read article for free, from open access legal sources, via Unpaywall:
https://www.jimmunol.org/content/jimmunol/184/8/4178.full.pdf
Free to read at www.jimmunol.org
http://www.jimmunol.org/cgi/content/abstract/184/8/4178
Free after 12 months at www.jimmunol.org
http://www.jimmunol.org/cgi/content/full/184/8/4178
Free after 12 months at www.jimmunol.org
http://www.jimmunol.org/cgi/reprint/184/8/4178
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4049/jimmunol.0901282
Article citations
A Novel Biallelic LCK Variant Resulting in Profound T-Cell Immune Deficiency and Review of the Literature.
J Clin Immunol, 44(1):1, 15 Dec 2023
Cited by: 2 articles | PMID: 38100037 | PMCID: PMC10724324
Review Free full text in Europe PMC
Blockade of Tyrosine Kinase, LCK Leads to Reduction in Airway Inflammation through Regulation of Pulmonary Th2/Treg Balance and Oxidative Stress in Cockroach Extract-Induced Mouse Model of Allergic Asthma.
Metabolites, 12(9):793, 25 Aug 2022
Cited by: 7 articles | PMID: 36144198 | PMCID: PMC9506330
New horizons in drug discovery of lymphocyte-specific protein tyrosine kinase (Lck) inhibitors: a decade review (2011-2021) focussing on structure-activity relationship (SAR) and docking insights.
J Enzyme Inhib Med Chem, 36(1):1574-1602, 01 Dec 2021
Cited by: 8 articles | PMID: 34233563 | PMCID: PMC8274522
Review Free full text in Europe PMC
Naturally Occurring Genetic Alterations in Proximal TCR Signaling and Implications for Cancer Immunotherapy.
Front Immunol, 12:658611, 03 May 2021
Cited by: 3 articles | PMID: 34012443 | PMCID: PMC8126620
Review Free full text in Europe PMC
MicroRNA-155: Regulation of Immune Cells in Sepsis.
Mediators Inflamm, 2021:8874854, 08 Jan 2021
Cited by: 21 articles | PMID: 33505221 | PMCID: PMC7810547
Review Free full text in Europe PMC
Go to all (23) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription.
J Exp Med, 203(3):755-766, 06 Mar 2006
Cited by: 208 articles | PMID: 16520391 | PMCID: PMC2118252
Lck regulates IL-10 expression in memory-like Th1 cells.
Eur J Immunol, 40(11):3210-3219, 27 Oct 2010
Cited by: 9 articles | PMID: 21061443 | PMCID: PMC3517127
Elevated gene expression of Th1/Th2 associated transcription factors is correlated with disease activity in patients with systemic lupus erythematosus.
J Rheumatol, 34(1):89-96, 15 Nov 2006
Cited by: 56 articles | PMID: 17117487
Tissue-specific regulation of cytokine gene expression.
Cold Spring Harb Symp Quant Biol, 64:573-584, 01 Jan 1999
Cited by: 14 articles | PMID: 11232334
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R01CA086867
Grant ID: R01 CA086867
Grant ID: T32 CA009560
Grant ID: T32CA09560
NIAID NIH HHS (2)
Grant ID: F31 AI077322
Grant ID: F31AI077322





