Abstract
Free full text

Suppression of autophagy impedes glioblastoma development and induces senescence
ABSTRACT
The function of macroautophagy/autophagy during tumor initiation or in established tumors can be highly distinct and context-dependent. To investigate the role of autophagy in gliomagenesis, we utilized a KRAS-driven glioblastoma mouse model in which autophagy is specifically disrupted via RNAi against Atg7, Atg13 or Ulk1. Inhibition of autophagy strongly reduced glioblastoma development, demonstrating its critical role in promoting tumor formation. Further supporting this finding is the observation that tumors originating from Atg7-shRNA injections escaped the knockdown effect and thereby still underwent functional autophagy. In vitro, autophagy inhibition suppressed the capacity of KRAS-expressing glial cells to form oncogenic colonies or to survive low serum conditions. Molecular analyses revealed that autophagy-inhibited glial cells were unable to maintain active growth signaling under growth-restrictive conditions and were prone to undergo senescence. Overall, these results demonstrate that autophagy is crucial for glioma initiation and growth, and is a promising therapeutic target for glioblastoma treatment.
Introduction
Glioblastoma multiforme (GBM) is the most common adult brain tumor and one of the most aggressive cancers with a median survival of approximately one year.1 Understanding survival response processes in GBM that contribute to its resistance would be crucial for therapy. One such survival process is autophagy, a cellular catabolic pathway frequently activated upon various stresses encountered by the cell including nutrient deprivation and hypoxia.2
During autophagy, cytoplasmic material is targeted for lysosomal degradation by a finely orchestrated series of vesicle formation and fusion events. A number of autophagy-essential protein complexes are required for the formation of a double-membrane structure, the phagophore, that engulfs cytoplasmic materials, resulting in an enclosed vesicle called the autophagosome.3 At the heart of the autophagy pathway lies a family of ubiquitin-like proteins, such as MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3), essential for autophagosome maturation. Cytosolic LC3 (termed LC3-I) is recruited to the growing phagophore upon its ubiquitin-like conjugation to phosphatidylethanolamine forming LC3-II. This reaction is catalyzed by the activity of E1-, E2- and E3-like enzymes, which are ATG7, ATG3 and ATG12–ATG5-ATG16L1, respectively. Autophagy activation can be sensed by the ULK (unc-51 like kinase) signaling complex comprised of the protein kinase ULK1 and several regulatory components including ATG13 and RB1CC1/FIP200 (RB1-inducible coiled-coil 1).
Increasing evidence has demonstrated that autophagy is closely associated with cancer.2,4 Autophagic degradation of cytoplasmic components can promote tumor cell survival by providing nutrient supply, or can suppress tumorigenesis by clearing toxic cellular materials that can otherwise be carcinogenic. Such dual effects of autophagy on cancer progression are most likely dependent on the tumor type, genetic composition and stage. Genomic studies of GBM patient samples revealed mutations in multiple signaling pathways, including gain-of-function mutations in the receptor tyrosine kinase (RTK)-RAS-class I phosphoinositide 3-kinase oncogenic pathway,5 which modulate autophagy.6 Furthermore, in cultured GBM cells, autophagy is frequently activated as a stress response upon treatment with therapeutic agents.7 However, how autophagy affects GBM development in vivo has not been addressed.
Cellular and xenograft experiments suggest an important role of autophagy in GBM.8,9 However, these experiments utilize established GBM cell lines, and thus do not reflect the process of tumor initiation. For this reason, we aimed to inhibit autophagy along with oncogene activation in a genetically engineered mouse model. We used a Replication-Competent Avian Sarcoma-leukosis virus LTR splice acceptor/Tumor Virus A (RCAS/TVA) mouse model, which allows specific manipulation of genes of interest in glial progenitors. In this model, gliomagenesis is driven by oncogenic KRAS, and autophagy is specifically disrupted using shRNA sequences targeting Atg7, Atg13 and Ulk1. Combined expression of the shRNA sequences and oncogene ensured inactivation of autophagy in tumor-forming cells. Unlike genetic deletion, which results in complete autophagy disruption, RNAi-mediated gene knockdown can more closely model the therapeutic situation by retaining residual activity of the target gene. Our results indicate that the autophagy inhibition strongly reduced GBM development in mice and abrogated the oncogenic potential of mutant KRAS-expressing glial cells ex vivo.
Results and discussion
RCAS/TVA mouse model for gliomagenesis
In order to test the role of autophagy in GBM, we utilized the RCAS/TVA mouse model for gliomagenesis.10 GBM is induced in cdkn2a/ink4a-arf−/- mice using RCAS viruses carrying oncogenic KRAS (KRASG12D). Intracranial injection of the virus-producing chicken fibroblast DF-1 cells into neonatal mice results in somatic gene transfer from the RCAS viruses to glial cells expressing the viral receptor, TVA, under the control of the Nes/nestin promoter (Fig. 1A). Importantly, tumors driven by KRAS expression using this model show a histopathology that closely mimics high-grade human tumors, including features such as pseudopalisading necrosis and microvascular proliferation.11 The RCAS vectors were further modified to co-express shRNA sequences targeting autophagy-related (Atg) gene products along with KRAS expression to inhibit autophagy in tumor-initiating cells (such as KRAS:shAtg7, KRAS:shAtg13 or KRAS:shUlk1) or control shRNA (KRAS:shLacZ). Using this system, efficient gene knockdown (Fig. 1B) and oncogene overexpression (Fig. 1C) can be achieved upon the infection of primary TVA-expressing glial cells (termed XFM cells) derived from uninjected mice.12 Furthermore, shRNA-mediated inhibition of Atg7, Atg13 or Ulk1 resulted in autophagy inhibition as assessed by endogenous LC3 lipidation, but maintained residual autophagic activity (Fig. 1D).
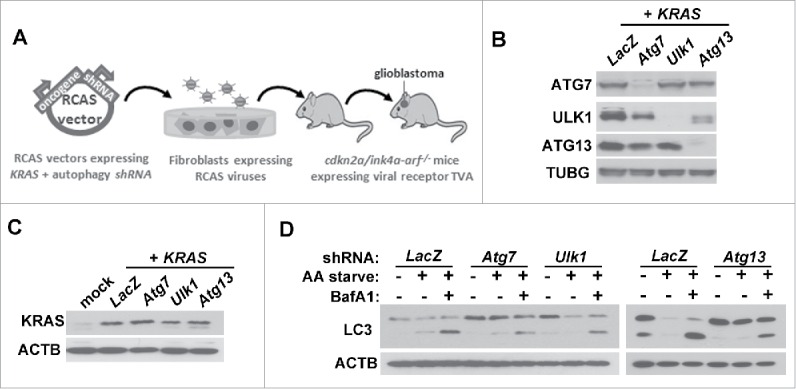
RCAS mouse model for gliomagenesis. (A) Outline of the RCAS mouse model. RCAS viruses harboring an oncogenic KRAS gene and autophagy shRNA are amplified in chicken fibroblasts. Intracranial injection of the fibroblasts in neonatal mice expressing the viral receptor, TVA, under the control of the Nes/nestin promoter results in gene transfer from the RCAS vectors to glial progenitors. (B-C) Validation of the RCAS-mediated gene knockdown (B) and KRAS overexpression (C). Primary glial cells (XFM) expressing the viral receptor TVA harvested from uninjected mice were infected in culture with RCAS viruses expressing KRAS and the indicated shRNA. (D) Inhibition of autophagy in glial cells expressing KRAS in the presence of shRNA against Atg7, Atg13 or Ulk1. XFM cells were amino acid starved (AA starve) for 2 h in the presence or absence of bafilomycin A1 (BafA1; 20 nM) to inhibit lysosomal degradation.
Autophagy is required for GBM development
To test the effect of autophagy inhibition on KRAS-mediated gliomagenesis, we performed intracranial injection of DF-1 cells expressing RCAS viruses into neonate mice. Injection of DF-1 expressing KRAS:shLacZ RCAS viruses resulted in tumor formation in approximately 60% of the mice within 6 wk after injection (Fig. 2A–B) as reported previously.13 In contrast, the expression of shRNA targeting autophagy genes resulted in a dramatic reduction in KRAS-mediated tumor development (KRAS:shAtg7, ~10% of the injected mice developed tumors) or complete inhibition of tumor formation (KRAS:shAtg13 and KRAS:shUlk1). Overall, these results indicate that autophagy is required for gliomagenesis induced by KRAS expression.
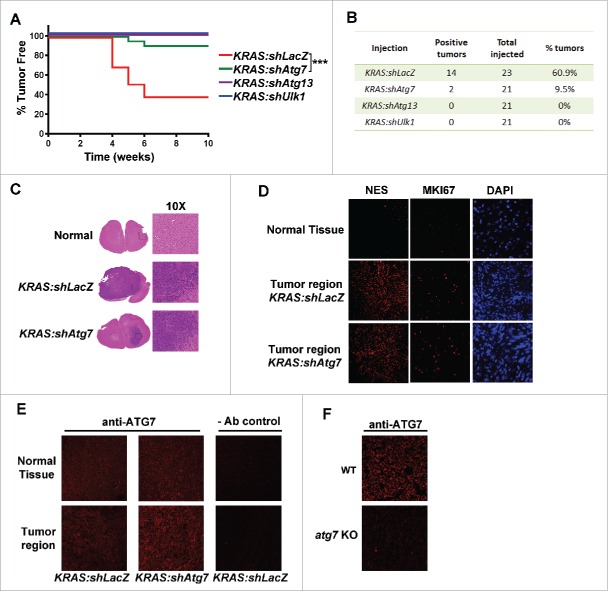
Autophagy is required for GBM development. Mice were injected with RCAS vectors expressing KRAS in the presence of shRNA against LacZ, Atg7, Atg13 or Ulk1. (A) Kaplan Meier curve measuring tumor-free animals monitored up to 10 wk after injection with the indicated RCAS expressing cells. ***, p < 0 .001 (Log-rank Mantel-Cox test). (B) Animal numbers of those plotted in (A). (C) Representative H&E images of mouse brain sections showing GBM development in the indicated injections. (D) Tissue sections of normal or GBM brains were stained with antibodies to NES/nestin, or MKI67/Ki67 or with DAPI. (E) Immunocytochemical staining of brain sections using an antibody against ATG7. (F) Similar staining procedures using atg7 knockout (KO) or wild-type (WT) MEF cells to confirmed antibody specificity.
Having observed that some tumors did form in KRAS:shAtg7-injected mice (2 out of 21 mice), we assessed whether these tumors differed from the control (KRAS:shLacZ)-injected mice. We analyzed tissue sections by hematoxylin and eosin (H&E) staining and observed no difference in the tumor morphology between KRAS:shLacZ and KRAS:shAtg7 tumors (Fig. 2C). Similarly, there was no difference in NES/nestin (marker of GBM) or MKI67/Ki67 (proliferation marker) staining (Fig. 2D). Next we examined whether KRAS:shAtg7-derived tumors exhibited efficient reduction of ATG7 protein levels. Interestingly, immunohistological staining showed comparable ATG7 levels between shAtg7- and shLacZ-expressing tumors, suggesting that the tumor cells bypassed the Atg7 shRNA effect (Fig. 2E). We further confirmed the specificity of the anti-ATG7 antibody by staining mouse embryonic fibroblasts (MEFs) derived from wild-type or atg7 knockout mice (Fig. 2F). Overall, these observations further support the conclusion that the expression of autophagy-essential genes is required for GBM formation in the KRAS-RCAS/TVA model.
Autophagy is required for KRAS-driven oncogenic growth of glial cells
The lack of tumor formation upon RNAi-mediated inhibition of Atg7, Atg13 or Ulk1 suggests that autophagy is required for KRAS-driven gliomagenesis in vivo. However, these results do not distinguish whether the absence of autophagy impedes KRAS-induced transformation or growth of transformed cells. In order to understand the role of autophagy in gliomagenesis, we infected XFM glial cells (cdkn2a/ink4a-arf−/-) with RCAS viruses co-expressing KRAS and shRNA against LacZ, Atg7, Atg13 or Ulk1 (as in Fig. 1B). We observed no growth defects upon autophagy inhibition in cells grown under normal culture conditions in monolayer. Interestingly, when cells were seeded in anchorage-independent conditions in soft agar, colony formation was strongly reduced in KRAS cells co-expressing shRNA targeting Atg7, Atg13 or Ulk1 compared to KRAS:shLacZ expressing cells (Fig. 3A). Therefore, autophagy appears to be critical for KRAS-driven glial transformation. Furthermore, we tested whether autophagy is required for clonogenic growth under suboptimal conditions by culturing cells in low serum (0.1% fetal bovine serum [FBS]), a condition mimicking poor angiogenic state in tumors. Under this condition, autophagy inhibition by Atg7-shRNA resulted in failure of clonogenic cell growth when cells were allowed to recover in full-growth medium containing 10% FBS compared to control cells (Fig. 3B–C). Overall, these studies indicate that autophagy is required for both oncogenic transformation of KRAS-expressing glial cells and their sustained viability under stressful conditions associated with gliomagenesis.
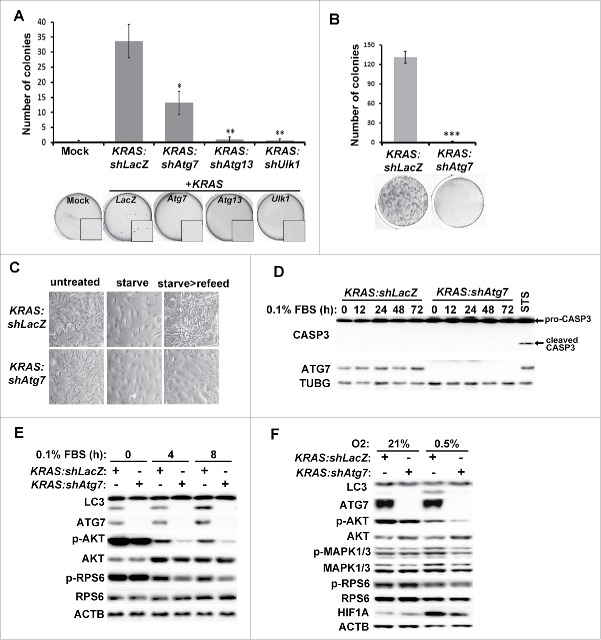
Inhibition of autophagy suppresses oncogenic growth and growth signaling pathways in KRAS-expressing glial cells. Primary glial cells (XFM) were transduced with RCAS viruses co-expressing KRAS along with the indicated shRNA and grown under the indicated conditions. (A-B) Transduced XFM cells were grown in (A) soft agar or (B) low serum (0.1% FBS) for 2 wk followed by incubation in full growth medium for 1 week. (C) Representative images of cell morphology for cells treated as in (B). (D) No induction of apoptosis was observed (measured by cleaved CASP3) in cells cultured in 0.1% FBS for the indicated time points. Staurosporine (STS) treatment in KRAS:shLacZ cells was used as a positive control for CASP3 activation. (E) Cells grown in low serum (0.1% FBS) for the indicated times and cell lysates were analyzed by western blot using antibodies against the indicated proteins. (F) KRAS:shLacZ- or KRAS:shAtg7-expressing XFM cells were grown under hypoxic conditions (0.5% O2) for 72 h and cell lysates analyzed. Statistical analyses are shown of 3 independent experiments performed in triplicates including error bar (SEM values). *, p < 0 .05; **, p < 0 .01, ***, p < 0 .001 (Student t test, unpaired 2-tailed).
We further tested the underlying mechanism for the lack of clonogenic growth in autophagy-inhibited cells by analyzing growth signaling and survival pathways. We observed neither morphological changes indicative of cell death induction in cells cultured under low-serum condition (0.1% FBS) nor CASP3/caspase-3 activation, indicating that the difference in growth capacity was not due to apoptotic cell death (Fig. 3D). Next, we tested whether growth signaling pathways were impeded by Atg7 knockdown. When grown in normal culture medium, the phosphorylation status of signaling factors AKT, MAPK1/ERK2-MAPK3/ERK1, and RPS6 was comparable in KRAS:shLacZ- and KRAS:shAtg7-expressing cells, indicating that autophagy was dispensable under this condition. Interestingly, under low-serum conditions, while KRAS:shLacZ-expressing cells could maintain modest levels of AKT, MAPK1/3 and RPS6 phosphorylation, KRAS:shAtg7-expressing cells were more susceptible to serum withdrawal (Fig. 3E). We further confirmed the role of autophagy in maintaining growth signaling by culturing cells in hypoxic conditions (a critical aspect of GBM). Autophagy was significantly induced in KRAS:shLacZ cells grown under low oxygen conditions (0.5% O2, Fig. 3F). Importantly, both AKT and MAPK1/3 phosphorylation were significantly lower in KRAS:shAtg7 cells compared to control cells, indicating that growth signaling under hypoxia was compromised in the absence of autophagy. In contrast, no significant effects were observed on RPS6 phosphorylation, suggesting that the MTOR (mechanistic target of rapamycin [serine/threonine kinase]) complex 1 (MTORC1) signaling pathway may be dispensable under these conditions. Overall, the above results strongly suggest that autophagy is required to maintain growth signaling pathways in cells deprived of growth factors or grown under hypoxia.
Autophagy inhibition induces senescence and reduces metabolic activity under low serum conditions
We further sought to characterize the consequences of autophagy inhibition in cells grown under suboptimal culture conditions. Having observed that in the absence of autophagy cells failed to recover after being cultured in low-serum media for 14 d (Fig. 3B), we examined whether cells underwent irreversible growth arrest. To do so, we measured senescence-associated β-galactosidase (SA-GLB1) activity as an assay for cellular senescence. As shown in Figure 4A, autophagy inhibition resulted in a significant increase of senescent cells in KRAS-expressing XFM cells compared to control cells when grown in low serum for 7 d and, to a lesser extent, 14 d. Similarly, BrdU incorporation assay showed a reduced proliferation rate in Atg7-knockdown cells compared to control cells after 7 d of serum starvation, but the rate became comparable at later time points (14 d, Fig. 4B). Consistent with these observations, we found that at various time points following serum starvation the dephosphorylation and thereby activation of RB1/p105 (retinoblastoma 1)14,15 and cellular levels of CDKN1B/p27 as well as markers of the senescence-associated secretory phenotype including IL1B/IL-1β and IL6 were elevated in KRAS:shAtg7 cells compared to KRAS:shLacZ cells (Fig. 4C–D). These results suggest that when autophagy is defective, cells are more prone to undergo senescence. Interestingly, under low serum conditions, activation of TRP53/p53 was not detected, suggesting that in this context senescence may occur independently of TRP5316,17 (Fig. 4C). These observations highlight the importance of autophagy in maintaining cell cycle progression in culture following growth factors withdrawal in KRAS-transformed cells.
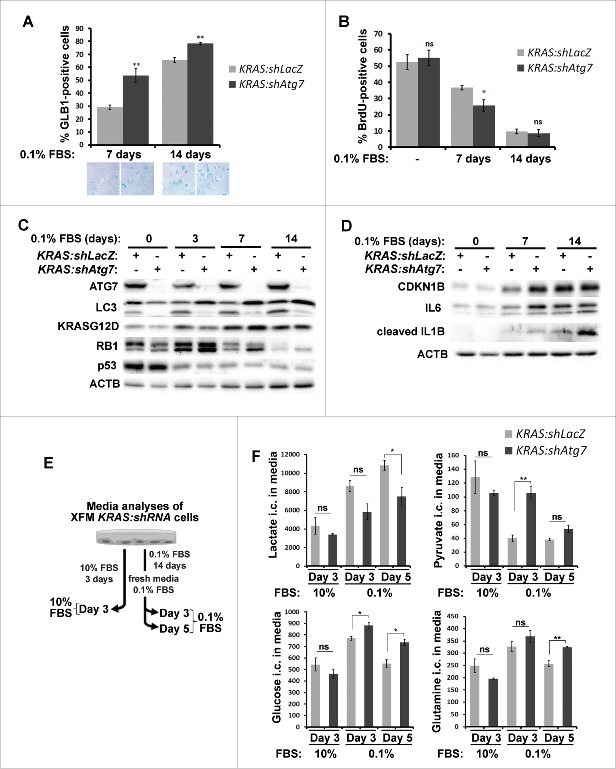
Autophagy is required for suppressing senescence and maintaining metabolism under low-serum conditions. (A) KRAS:shLacZ- or KRAS:shAtg7-expressing XFM cells were incubated in 0.1% serum for 7 or 14 d. Cells were fixed and SA-GLB1 activity was measured as an indicator of cellular senescence. Quantification of approximately 200 cells per condition of 3 independent experiments is shown with SEM values. Representative images are shown below. (B) BrdU incorporation assay of cells treated as in (A). Statistical analyses are shown of 3 independent experiments performed in triplicates including error bar (SEM values). (C-D) Western blot analyses of XFM cell lysates cultured in 10% FBS (d 0) or 0.1% FBS (3, 7 or 14 d). (E) Schematic presentation of the experimental outline used to analyze metabolites in tissue culture medium. Cells were cultured in 10% FBS and medium collected after 3 d (10%, d 3). Alternatively, cells were cultured in 0.1% FBS for 14 d, fresh medium containing 0.1% FBS was added and medium was collected following a further 3 or 5 d of culture (0.1%, d 3 or d 5). (F) Ion count (i.c.) measurements of the indicated metabolites normalized to cell numbers in tissue culture medium using mass spectrometry of XFM cells grown as described in (E). *, p < 0 .05; **, p < 0 .01; ***, p < 0 .001, ns, nonspecific (p > 0 .05; Student t test, unpaired 2-tailed).
We further sought to measure whether the metabolic activity of KRAS-expressing cells differed in the absence of autophagy in growth-arrested cells during prolonged serum starvation (Fig. 4E). When cells were grown under normal growth conditions (10% FBS), there was no significant difference in glucose, glutamine and pyruvate consumption or lactate production between the 2 cell types (Fig. 4F). Under conditions of serum starvation, however, uptake of these nutrients was markedly reduced in autophagy-deficient cells indicating that overall biosynthetic activity in these cells was lower. Lactate production was also lower in KRAS:shAtg7-expressing cells, indicating reduced aerobic glycolysis. Overall, these results indicate that autophagy acts as a stress response pathway and is required to bypass senescence and maintain metabolic activity in growth-arrested conditions.
In this study, we demonstrate that RNAi-mediated suppression of autophagy impedes tumor incidence in a KRAS-driven, RCAS/TVA GBM model. This is in agreement with previous studies where suppression of autophagy by genetic deletion restricts growth of KRAS- and BRAF-driven tumors of the lung, pancreas and skin.18-20 Further supporting our findings is the observation that tumors developing from KRAS:shAtg7 injections retain Atg7 expression. Loss of RNAi repression effect may be due to either silencing of the shRNA expression or additional mutations that render resistance to the shRNA.
The RCAS/TVA mouse model is a unique system to study gliomagenesis based on somatic gene transfer (using RCAS viruses) into a specific cell type (engineered to express viral TVA receptor).10 Unlike xenograft models that utilize established cancer cells, this genetically engineered mouse model allows for the assessment of tumor initiation in the presence of a functional immune system. In addition, xenograft experiments rely on the injection of a large number of cells into the recipient animals which differ considerably from the process of tumor development generally involving the transformation of a single or few cells.21 Indeed, considerable differences are observed when assessing the role of autophagy in GBM using the RCAS/TVA mouse model presented here and previously published xenograft studies. Xenograft studies using GBM cell lines show that autophagy inhibition cooperates with drug treatment to restrict cell growth but has no effect on tumor growth in the absence of treatment.8,9 However, our studies indicate that autophagy inhibition in the absence of drug treatment can affect tumor development (Fig. 2). The differences between these results may be due to the presence of an intact immune response in the RCAS/TVA model. Furthermore, GBM cell lines may have acquired additional mutations during prolonged in vitro culture thereby altering their response to autophagy inhibition. Alternatively, it is possible that inhibition of autophagy after tumor formation (xenograft models) may impose different effects compared to autophagy inhibition in a tumor initiation model (RCAS/TVA model). Further studies are required to elucidate these differences.
How does autophagy inhibition suppress gliomagenesis? Autophagy may facilitate cellular changes that are required for KRAS-mediated transformation, including genomic instability, metabolic stress and lipid homeostasis resulting from KRAS expression.19,22 Our cell culture-based studies indicate that autophagy can support the growth of KRAS-expressing glial cells in unfavorable conditions such as the absence of cell attachment or reduced growth factors, key hallmarks of cellular transformation. Molecularly, we found that when grown under growth restrictive conditions, autophagy-deficient cells were unable to maintain activated growth signaling and were prone to senescence, characterized by the activation of RB1 and increased levels of CDKN1B/p27 and markers of the senescence-associated secretory phenotype.23-25 Interestingly, our results show that autophagy is also required to maintain metabolic activity in growth-arrested cells during prolonged growth factor withdrawal, consistent with previous publications.18,26 A recent study suggests that maintaining glycolysis enhances cell survival during mitotic arrest induced by Cdc20 deletion.27 This may imply that, in a similar fashion, the absence of glycolysis in autophagy-defective cells may contribute to their inability to proliferate when restimulated with full growth medium as observed in our system. It remains to be defined whether the role of autophagy in maintaining cell growth and metabolic activity is required for gliomagenesis in vivo. Mouse models allowing conditional autophagy inhibition after tumor establishment will be useful for this purpose.
Our study is the first to demonstrate the effect of autophagy inhibition by RNAi on gliomagenesis in a genetically engineered mouse model. A recent study showed that adult mice with conditional whole body deletion of Atg7 live for 2–3 mo due to selective tissue damage.28 However, in this model, complete inhibition of autophagy in mice with lung tumors induces tumor regression within 5 wk, suggesting that a therapeutic window exists and can be exploited.28 Additionally, when autophagy is significantly inhibited but not completely ablated, an anti-tumorigenic effect could already be achieved, as demonstrated by RNAi-based autophagy inhibition in our GBM mouse models. A partially inhibited autophagy pathway is likely to maintain the homeostatic function of autophagy in normal tissues during prolonged treatment, thus avoiding the lethal effect of complete, whole body elimination of autophagy function.
Materials and methods
Cell culture and treatment
DF-1 chicken fibroblasts (ATCC, CRL-12203) and glial cells derived from the RCAS mouse model expressing TVA receptor XFM,12 were cultured in DMEM (Life Technologies, 41966029 supplemented with 10% FBS, L-glutamine (2 mM), penicillin (10 units/mL) and streptomycin (0.1 mg/ml).
For amino acid starvation experiments, cells were grown in DMEM lacking amino acids and serum for 2 h prior to harvest, as described previously.29 The lysosomal inhibitor, bafilomycin A1 (Sigma, B1793), was added as indicated at a final concentration of 20 nM for 2 h. Staurosporine (STS; Sigma, S5921) was added to cells for a final concentration of 1 µM for 16 h.
Western blot and antibodies
For western blot analyses, cell lysates were prepared in RIPA buffer (10 mM Tris pH 7.4, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1 mM ethylene glycol tetraacetic acid, 0.1% sodium dodecyl sulfate, 1% Triton-X100 [Sigma, T9284], 1 mM 2-mercaptoethanol, 0.5% sodium deoxycholate and 10% glycerol) and analyzed by SDS-PAGE as described previously.7,29 The following antibodies were used: anti-LC3 (Sigma, L7543); anti-ACTB/β-actin (Sigma, A5316); anti-ULK1 (Sigma, A7481); anti-ATG7 (Santa Cruz Biotechnology, clone H300, sc-33211; or Sigma, A2856); anti-pan-RAS (EMD Millipore, OP40); anti-RAS G12D mutant specific (Cell Signaling Technology, 14429); anti-ATG13 (Sigma, SAB4200100); anti-TUBG/γ-tubulin (Sigma, GTU-88); anti-HIF1A/HIF1α (R&D Systems, MAB1536); anti-phosphorylated (p)-AKT (Ser473; Cell Signaling Technology, 4060); anti-total AKT (Cell Signaling Technology, 9272); anti-p-MAPK1/ERK2-MAPK3/ERK1 (Thr202/Tyr204; Cell Signaling Technology, 4370); anti-total MAPK1/ERK2-MAPK3/ERK1 (Cell Signaling Technology, 9102); anti-pRPS6/S6 (Ser235/236; Cell Signaling Technology, 4858); anti-total RPS6/S6 (Cell Signaling Technology, 2217); anti-CASP3/caspase-3 (Cell Signaling Technology, 9665); anti-RB1/p105 (BD Biosciences, 554136); anti-TRP53/p53 (Cell Signaling Technology, 2524); anti-CDKN1B/p27 (Cell Signaling Technology, 2552); anti-IL6/interleukin 6 (R&D Systems, BAF-406); anti-IL1B/interleukin 1 β (R&D Systems, AF-301-NA).
Injections
All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committees of Memorial Sloan Kettering Cancer Center and followed NIH guidelines for animal welfare. The RCAS/TVA system used in this work to induce gliomas in vivo in immunocompetent mice has been described previously.30 N/TVA;cdkn2a/ink4a-arf)−/-;Ptenfl/fl mice were used for the RCAS mediated gliomagenesis in this study.13 Briefly, DF-1 cells were transfected with the relevant RCAS viral plasmid using Lipofectamine2000 (Invitrogen, 11668019) according to the manufacturer's protocol. The cells were regularly maintained for at least 3 passages for propagation of the RCAS viruses to entire cells. Cells were then used for injection into murine brain. Newborn pups were injected intracranially with 1 μL of ~1×105 DF-1 cells. Then mice were monitored until they developed symptoms of GBM such as lethargy, poor grooming, weight loss, dehydration or macrocephaly. GBM incidence or absence was confirmed by H&E staining of brain sections in all injected animals. Kaplan-Meier analysis demonstrating symptom-free survival in murine gliomas was performed using the Prism software (GraphPad).
Vector constructs
For the generation of the RCAS-shRNA vector, shRNAs were initially assembled in the pSUPER.retro vector (OligoEngine, VEC-PRT-0002). The shRNAs containing the H1 promoter were PCR-amplified and then inserted into the RCAS-Y vector (generated and provided by Dr Yi Li, Baylor College of Medicine, Texas)31 using NotI and PacI restriction sites. KRAS was subsequently cloned using NotI restriction sites. The shRNA target sequences are as follows: Atg7 5′ CACATAGCATCATCTTTGA; Atg13 5′ GAGAAGAATGTCCGAGAAT; Ulk1 5′GAGCAAGAGCACACGGAAA.
Immunocytochemistry
Mouse brains harvested from animals were fixed in 10% formalin for 24–72 h and then transferred to 70% ethanol. Samples were then paraffin-embedded, sectioned and stained with H&E by Histoserv Inc., Maryland. Similarly, MEFs derived from wild-type or atg7 knockout cells were pelleted and processed for paraffin embedding. For immunocytochemistry staining, paraffin-embedded sections were treated with xylene twice for 10 min each before being sequentially hydrated in decreasing concentrations of ethanol. Epitope retrieval was performed by incubating slides in 10 mM sodium citrate buffer, pH 6 for 15 min at 95°C then allowed to cool for 20–45 min. Subsequently, slides were blocked with TBST (150 mM NaCl and 10 mM Tris pH 7.5, 0.1% Tween 20 [Sigma, P5927] + 5% BSA [Thermo Fisher Scientific, BP1605-100]) for 45 min, and incubated with primary antibodies overnight at 4°C. Subsequently, slides were incubated with Alexa fluorescent secondary antibodies (Life Technologies, A-11012 and A-11032) and images acquired using a Nikon Eclipse Ti-U Confocal Microscope. The antibodies used in this study are anti-ATG7 (Sigma, A2856), anti-MKI67/Ki67 (Vector Labs, VP-K451), and anti-NES/nestin (BD Biosciences, 556309).
Colony formation assay
XFM cells were seeded in 6-cm dishes; 24 h later cells were cultured in 0.1% FBS for 2 wk (with medium replenished after 1 wk) followed by growth in 10% FBS for a further 1 wk. Cells were then fixed with 10% formaldehyde and stained with Giemsa (Sigma, GS500) to visualize colonies.
Hypoxia
For hypoxia experiments, cells were incubated in a Whitley H35 Hypoxystation set to 37°C and 0.5% O2. 72 h later, cells were lysed inside the Hypoxystation and analyzed by western blotting.
Soft agar assay32
XFM cells were seeded at 10,000 cells/well in a 6-well dish. Cell suspension (final volume 1.5 mL) containing 0.4% low gelling agarose (Sigma, A4018) in full growth DMEM was overlaid with a layer of 0.5% agarose and left to solidify at 4°C. Cells were then incubated at 37°C, fed weekly with 0.4% agarose-containing DMEM and analyzed after ~3 wk by staining with 0.02% Iodonitrotetrazolium chloride (Sigma, I10406). Colonies were quantified using an Optronix Gelcount (Oxford Optronix). All assays were conducted in triplicates in 3 independent experiments.
Senescence-associated β-galactosidase assay (SA-GLB1)23
Cellular senescence was measured using an SA-GLB1 assay. Briefly, cells were fixed with 0.5% glutaraldehyde then incubated with X-gal staining solution, pH 6.0 (1mM MgCl2 phosphate-buffered saline [Fisher BioReagents, BP399], X-gal [Thermo Fisher Scientific, R0941], 0.12 mM K3Fe[CN]6 [Sigma, 60299], 0.12 mM K4Fe[CN]6 [Sigma, 60279]) overnight at 37°C. Images of cells were taken using a Nikon Digital Sight DS-L3.
BrdU incorporation assay
For the BrdU incorporation assay, cells were pulsed with 50 μM BrdU (Sigma, B5002) for 18 h followed by fixation with 3.7% paraformaldehyde and permeabilization with 0.2% Triton-X100. Cells were blocked with 0.2% gelatin-fish (Sigma, G7765) in 5% BSA-phosphate-buffered saline and incubated with anti-BrdU primary antibody (1:2000; BD Biosciences, 555627), 0.5 U/L DNase (Sigma, D4527), and 1 mM MgCl2 in blocking solution. Subsequently, slides were incubated with Alexa secondary antibodies (Life Technologies, A-11001) and 1 μg/ml DAPI; images were acquired using ImageXpress and analyzed using MetaXpress software.
Metabolite measurement
XFM cells were grown in 0.1% FBS for 2 wk and fresh 0.1% FBS medium was added and cells grown for a further 3 or 5 d. Alternatively, cells grown in 10% FBS were analyzed after 3 d of culture. Metabolites were extracted from the medium using ice-cold extraction buffer (50% methanol, 30% acetonitrile). Extracted metabolites were separated on a Zic-pHILIC column (Merck Millipore) using a Thermo Ultimate BioRS HPLC with a single step linear gradient of 1095%– A over 20 min (mobile phases were [A] 20 mM ammonium carbonate [B] acetonitrile). Metabolites were eluted into a Q-Exactive mass spectrometer (Thermo Fisher Scientific) using a flow rate of 300 µl/min. Metabolite masses were acquired in negative mode within the range of 78–250 m/z. Relative metabolite abundance was then determined by integrating the ion peak area (Quan Thermo Xcalibur) and normalized to cell number.
Statistical analyses
All data are presented as the mean ± SEM. The statistical significance was evaluated using two-tailed, unpaired Student t test. Percentage tumor-free survival Kaplan Meier curve was analyzed using Log-rank Mantel-Cox test (Graphpad Prism software). The data were considered significant when the P value was less than 0.05 (*).
Abbreviations
- ATG
- autophagy related
- FBS
- fetal bovine serum
- GBM
- glioblastoma multiforme
- H&E
- hematoxylin and eosin
- KRAS
- Kirsten rat sarcoma viral oncogene homolog
- MAP1LC3/LC3
- microtubule-associated protein 1 light chain 3 (MAP1LC3)
- MEFs
- mouse embryonic fibroblasts
- RCAS
- replication-competent avian sarcoma-leukosis virus LTR splice acceptor/tumor virus A
- SA-GLB1
- senescence-associated galactosidase, β 1
- ULK1
- unc-51 like kinase 1
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank members of the Jiang and Gammoh laboratories for helpful discussions.
Funding
This work was supported in part by a Goodwin Experimental Therapeutic Centerss fund, a Cycle for Survival fund, and NIH R01CA166413 and R01GM113013 (to X.J.) as well as an NIH fellowship 1F32CA162691 and a Carnegie Trust for the Universities of Scotland Grant (to N.G.).
References
Articles from Autophagy are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1080/15548627.2016.1190053
Read article for free, from open access legal sources, via Unpaywall:
https://www.tandfonline.com/doi/pdf/10.1080/15548627.2016.1190053?needAccess=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Therapeutic Relevance of Inducing Autophagy in β-Thalassemia.
Cells, 13(11):918, 25 May 2024
Cited by: 0 articles | PMID: 38891049 | PMCID: PMC11171814
Review Free full text in Europe PMC
IGF2BP2 modulates autophagy and serves as a prognostic marker in glioma.
Ibrain, 10(1):19-33, 11 Mar 2024
Cited by: 1 article | PMID: 38682020 | PMCID: PMC11045200
Autophagy Modulation and Its Implications on Glioblastoma Treatment.
Curr Issues Mol Biol, 45(11):8687-8703, 29 Oct 2023
Cited by: 0 articles | PMID: 37998723 | PMCID: PMC10670099
Review Free full text in Europe PMC
Microorganism-regulated autophagy in gastrointestinal cancer.
PeerJ, 11:e16130, 27 Sep 2023
Cited by: 0 articles | PMID: 37786582 | PMCID: PMC10541808
Compensatory cross-talk between autophagy and glycolysis regulates senescence and stemness in heterogeneous glioblastoma tumor subpopulations.
Acta Neuropathol Commun, 11(1):110, 07 Jul 2023
Cited by: 7 articles | PMID: 37420311 | PMCID: PMC10327182
Go to all (69) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Autophagy activation, lipotoxicity and lysosomal membrane permeabilization synergize to promote pimozide- and loperamide-induced glioma cell death.
Autophagy, 17(11):3424-3443, 19 Jan 2021
Cited by: 39 articles | PMID: 33461384 | PMCID: PMC8632287
Inhibition of autophagy enhances apoptosis induced by proteasome inhibitor bortezomib in human glioblastoma U87 and U251 cells.
Mol Cell Biochem, 385(1-2):265-275, 09 Oct 2013
Cited by: 37 articles | PMID: 24104452 | PMCID: PMC3840293
A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: Hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway.
Autophagy, 12(7):1129-1152, 10 May 2016
Cited by: 80 articles | PMID: 27163161 | PMCID: PMC4990999
Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth.
J Biol Chem, 289(32):22306-22318, 12 Jun 2014
Cited by: 54 articles | PMID: 24923441 | PMCID: PMC4139240
Funding
Funders who supported this work.
Cancer Research UK (1)
Characterization of the Senescence Associated Extracellular Matrix (SA-ECM) and its role in cancer progression
Dr Juan Carlos Acosta, University of Edinburgh
Grant ID: 16243
NCI NIH HHS (3)
Grant ID: F32 CA162691
Grant ID: R01 CA166413
Grant ID: P30 CA008748
NIGMS NIH HHS (1)
Grant ID: R01 GM113013





