Abstract
Free full text

Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia
Associated Data
Abstract
The 8.1 haplotype of the HLA complex has been reproducibly associated with several autoimmune diseases and traits, notably with thymus hyperplasia in patients with acquired generalized myasthenia gravis, an autoantibody-mediated disease directed at the muscle acetylcholine receptor. However, the strong linkage disequilibrium across this haplotype has prevented the identification of the causative locus, termed MYAS1. Here, we localized MYAS1 to a 1.2-Mb genome segment by reconstructing haplotypes and assessing their transmission in 73 simplex families. This segment encompasses the class III and proximal class I regions, between the BAT3 and C3-2-11 markers, therefore unambiguously excluding the class II loci. In addition, a case-control study revealed a very strong association with a core haplotype in this same region following an additive model (P = 7 × 10-11, odds ratio 6.5 for one copy and 42 for two copies of the core haplotype). Finally, we showed that this region is associated with a marked increase in serum titers of anti-acetylcholine receptor autoantibodies (P = 8 × 10-6). Remarkably, this effect was suppressed by a second locus in cis on the 8.1 haplotype and located toward the class II region. Altogether, these data demonstrate the highly significant but complex effects of the 8.1 haplotype on the phenotype of myasthenia gravis patients and might shed light on its role in other autoimmune diseases.
Since the discovery of the HLAs (1), a considerable amount of work, which has recently culminated in the sequencing of the entire HLA region (2), has established its central role in the biology of the immune system and in predisposition to a large number of inflammatory and autoimmune diseases. Although HLA class I and class II loci have long been held responsible for the disease associations (3), there is now evidence that additional nonclass I and nonclass II HLA loci play a major role in controlling many immune phenotypes (4). The precise mapping and identification of the causative loci, however, have been hampered by the strong linkage disequilibrium across the HLA region plus the fact that most genes in this highly gene-rich region are expressed in the immune system and are therefore candidates. In this study, we focused on the 8.1 haplotype, which associates not only the HLA DR3, B8, and A1 antigens but also alleles of many other HLA loci and extends over 3 Mb stably across generations in Caucasians (5). This haplotype is associated with highly prevalent autoimmune diseases such as rheumatoid arthritis (RA), Graves' disease, type I diabetes, and celiac disease and also systemic lupus, myasthenia gravis (MG), IgA deficiency, and low antibody response to hepatitis B vaccine (5).
Acquired generalized MG is an autoimmune disease of the neuromuscular junction (6-8) that affects ≈400,000 individuals in the United States (9). It provides an ideal model for studying autoimmunity in humans because the self-antigen, the muscle nicotinic acetylcholine receptor (AChR), is well identified in most patients (10). It is targeted by pathogenic and highly specific autoantibodies, causing a functional defect in neuromuscular transmission by different mechanisms, including disappearance of the AChR, blockade of acetylcholine binding, and complement-mediated lysis of the postsynaptic membrane. The disease is life-threatening when respiratory muscles are involved. In its most frequent and typical form, the thymus is abnormal, showing hyperplasia and the presence of germinal centers containing B lymphocytes that produce anti-AChR autoantibodies (11). High levels of these autoantibodies are often detected in patient sera (12).
The inheritance of MG is complex, involving both genetic and environmental factors (13, 14). The form of MG with thymus hyperplasia is known to be associated with the 8.1 haplotype (15) and was recently shown to be genetically linked to HLA, defining the MYAS1 locus (16). Two previous studies suggested that the disease locus is located closer to HLA-B than to DRB1 (17, 18). However, no definitive conclusions could be drawn because of the relatively low density and informativity of the markers available and the use of unphased genotypes. Here, we considerably narrowed down the location of MYAS1 and notably excluded the class II region. We also investigated whether and how the 8.1 haplotype influences anti-AChR autoantibody titers in MG patients. Our data show that the 8.1 haplotype has complex and unexpected, yet highly significant, effects on the phenotype of MG patients.
Subjects and Methods
MG Patients. We recruited 136 unrelated French Caucasian patients (17 men and 119 women) from the participating centers. Their records were reviewed by one of the authors (P.G., B.E., or C.T.). Of these patients, 73 were included in a family study, including 62 nuclear families (42 with both parents and 20 with one parent) and 11 families with siblings only. No parents or other relatives were available for the other 63 patients. Written informed consent was obtained from the 488 participants in accordance with the Helsinki Declaration.
All patients fulfilled the clinical and electromyographical criteria for acquired generalized MG (7, 8) and underwent thymectomy. Thymus hyperplasia was defined by the presence of multiple lymphoid follicles with germinal centers in an otherwise normal organ (19, 20). Anti-AChR autoantibody titers were quantified for all 136 cases by radioimmunoprecipitation of serial dilutions of sera as described (21). Fourteen (10.3%) patients repeatedly gave measurements below the positivity threshold (0.6 nM). These “seronegative” MG patients were included in the study of the MYAS1 locus but not in the quantitative trait analysis of autoantibody titers against the AChR.
Genotyping and Haplotype Inference. Genotyping was conducted blindly to the clinical data. Generic genotyping of HLA-DRB1 had previously been performed in all subjects (16). The presence or the absence of the generic HLA-B8 allele was determined by PCR-restriction fragment length polymorphism, using the TaqI restriction endonuclease. The single nucleotide polymorphism at position -308 (A/G) in the tumor necrosis factor (TNF) α gene promoter was genotyped by PCR and dot-blot hybridization. Microsatellites spanning the HLA complex were genotyped by using standard procedures. Primer sequences and methods are given in Table 3 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site. All genotypes were confirmed by double-blind reading. Errors in family data were checked with pedcheck 1.1 (22). Aberrant data were corrected after repeating the genotyping or deleted if the inconsistency remained.
The haplotypes of the family probands were reconstructed by using pedigree information with the assistance of simwalk 2.83 (23). One simple recombinant of 201 informative meioses (0.5%) was observed between the 82-1 and 82-3 markers. Knowledge of the alleles associated with the 8.1 haplotype was used to infer an ambiguous phase at certain loci in some individuals. All available family members were used to infer missing genotypes. Inferred genotypes were not used for single-point transmission disequilibrium tests or ANOVA. At each marker, alleles that were in-phase with HLA-DR3 and HLA-B8 were assigned to the 8.1 ancestral haplotype (see Table 3) and were confirmed by typing the CEPH ws13 and ws15 cell lines (24). The criterion for assignment of the 8.1 haplotype was at least three consecutive markers with a reference allele. Alleles differing from the reference allele by one repeat (25) were considered to be associated with the 8.1 haplotype. Risk haplotypes were then subdivided into groups on the basis of the length and composition of consensus alleles. Transmitted and nontransmitted haplotypes of 46 heterozygous parents were counted manually and tested exactly by using the binomial law. For the patients who were not family probands, only haplotypes related to the 8.1 ancestral haplotype were reconstructed.
The control group consisted of 105 unrelated French Caucasian blood donors. Four-locus haplotypes were reconstructed by using phase 2.0 (26, 27). The surrogate 8.1 haplotype, defined by the associated alleles of the 8.1 haplotype (9N-I*310, 82-3*169, C1-2-5*207, and C2-4-5*278), was observed in 13 controls (6%) with probabilities of >0.95 (eight cases), 0.94, 0.82, 0.73, 0.59, and 0.46. The overall frequency of this haplotype was estimated at 0.048 ± 0.004. Two- and three-locus 8.1-related haplotypes were deduced from the four-locus data.
The case-control association of the surrogate 8.1 haplotypes under the additive model was tested with the Cochran-Armitage trend test. Exact calculations of odds ratios and two-sided P values were performed with statxact software (Cytel Software, Cambridge, MA).
Quantitative Analysis of Anti-AChR Autoantibody Titers. The distribution of serum levels of anti-AChR autoantibodies in the 122 seropositive MG patients was normalized by logarithmic transformation. Genetic markers were coded as binary variables, depending on the presence or absence of the 8.1-associated alleles. Their effect on autoantibody titers was tested by a one-way ANOVA with statistica 6.0 software (Statsoft, Tulsa, OK).
Serum titers of the patients carrying no 8.1-associated allele (baseline) were also compared to those of three other groups of patients. The first group included patients carrying an extended 8.1 haplotype. The second one included patients harboring a haplotype recombinant on the centromeric side, at the DRB1 locus or more distally. The third group pooled patients with a haplotype recombinant on the telomeric side, at the C2-4-5 locus or more proximally. Patients harboring a double recombinant haplotype or two different kinds of 8.1 haplotype were not considered. The mean autoantibody titers in each group were compared to the baseline with the post hoc Tukey test after a one-way ANOVA.
The qtphase program of the unphased suite (28) was used to evaluate the additive haplotypic effects of unphased genotype data on normalized anti-AChR antibody titers. Moreover, cisphase interactions could be investigated with conditional tests. Uncertain haplotypes were estimated with an expectation-maximization algorithm. Only haplotypes with a frequency >0.05 were tested, using the droprare option. With the individual option, each haplotype was compared to all others grouped together. In a first step, a three-locus window was moved across all HLA markers, with or without conditioning on the DRB1 marker. In a second step, the eight markers associated in the single-locus ANOVA were selected and reinvestigated by using a moving three-locus window or a single eight-locus window, with conditioning on DRB1.
Results
Mapping the MYAS1 Locus to the HLA Central Region. An association analysis was performed in 73 single-case families with probands affected with generalized MG and thymus hyperplasia. A panel of 27 markers encompassing the HLA complex between the class II and distal class I regions (Fig. 1) was selected for genotyping. This panel included 24 informative microsatellite markers plus the DRB1 locus, the HLA-B8 allele, and a single nucleotide polymorphism in the promoter of the TNF-α gene that were previously associated with MG (29). The number of markers was highest in the class III and proximal class I regions, which show the highest density of genes in the HLA complex (2).
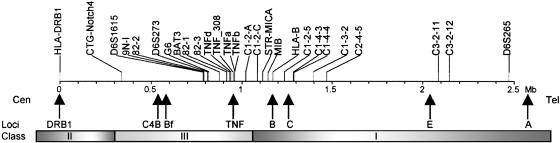
Map of the HLA complex showing the 27 markers used in the study. Distances (in Mb) are relative to the most centromeric marker, HLA-DRB1, and are based on the consensus HLA sequence available at The Sanger Institute (www.sanger.ac.uk) and from the University of California Santa Cruz Genome Browser (www.genome.ucsc.edu). Arrows below the map locate some important known genes in the HLA complex. Shaded bars depict the extent of the three main classes.
Single- and two-locus analysis of allele transmission showed that the strongest associations were in the class III and proximal class I regions, with local peaks at the 9N-I, 82-3, and C1-2-5 markers (Fig. 4, which is published as supporting information on the PNAS web site), whereas the class II markers did not reach the significance level after correction for the number of alleles tested. However, as previously reported with the 8.1 ancestral haplotype (5, 30), most markers showed strong linkage disequilibrium over the entire region in this sample of MG patients (Fig. 5, which is published as supporting information on the PNAS web site). To overcome this major obstacle, we reconstructed disease-associated haplotypes by using family information. In total, 46 patients among the 73 (63%) included in the family-based study carried the 8.1 haplotype or a fragment of it on at least one chromosome. Fig. 2A shows a graphical representation of these chromosomes. The extended 8.1 haplotype, from DRB1*03 (DR3) to D6S265, was found in 25 patients. In 16 of these 25 patients, it was in the heterozygous state. In the other nine patients, it was associated with a recombinant 8.1 haplotype on the other chromosome. No patients showed a homozygous extended 8.1 haplotype. Another 20 patients carried a recombinant 8.1 haplotype in the heterozygous state. One patient (no. 38 on Fig. 2 A) carried a recombinant 8.1 haplotype (of a different kind) on both chromosomes. The comparison of transmitted (T) and nontransmitted (NT) 8.1 haplotypes (56 versus 10, P = 6 × 10-10), whether complete or recombinant, strongly suggested that this haplotype played a role in MG predisposition. Remarkably, all untransmitted 8.1 haplotypes occurred in families in which the MG child received another 8.1 haplotype, either from the same parent or the other parent (data not shown).
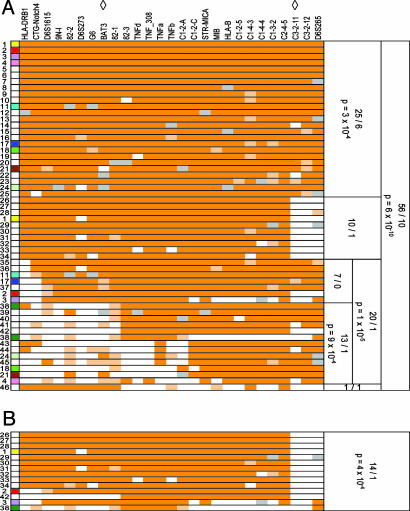
Graphical representation of the 8.1 haplotypes in the family probands and their transmission using the transmission disequilibrium test. Tested loci are shown at the top. Diamonds indicate the current boundaries of the MYAS1 interval. Each row shows a haplotype. Individual alleles are color-coded: alleles of the 8.1 haplotype are dark orange; for microsatellite, alleles differing from the 8.1 reference allele by one repeat are pale orange; all other alleles are white; missing data are gray. The numbers on the left refer to the patient carrying the haplotype. Next to them, a color code helps to associate both haplotypes belonging to a given patient. White indicates patients carrying only one 8.1 haplotype. Haplotypes were grouped by length. For each group, the counts of transmitted and untransmitted haplotypes and the exact P values are shown on the right. (A) All 8.1 haplotypes are displayed and sorted by centromeric length. (B) Only recombinant haplotypes with the breakpoint on the telomeric side are shown.
Although the transmission of the extended 8.1 haplotype was also markedly distorted (T/NT: 25/6, P = 3 × 10-4), visual inspection of recombinant haplotypes indicated that almost all of them shared the central part of the 8.1 haplotype, including the class III and proximal class I regions, suggesting that MYAS1 was located centrally. To refine the localization of MYAS1, we investigated the parental transmission of ancestrally recombinant haplotypes depending on the location of the breakpoint. On the proximal side (Fig. 2 A), counts of haplotypes that had lost at least the DR3 class II allele provided definitive evidence that DRB1 was not the MYAS1 locus in the 8.1 haplotype (T/NT: 20/1, P = 1 × 10-5). When more distal breakpoints (at the BAT3 microsatellite or beyond) were considered, the transmission distortion (T/NT: 13/1) was significant (P = 9 × 10-4). On the distal side of HLA, all recombinant haplotypes had a breakpoint proximal to the C3-2-11 marker (Fig. 2B). Their transmission was also significantly distorted (T/NT: 14/1, P = 4 × 10-4). Altogether, these data showed that the MYAS1 locus on the 8.1 haplotype was located in a 1.2-Mb region, which is the distance between BAT3 and C3-2-11.
Population Study: Evidence for an Additive Model. We also carried out a case-control study to assess the association of the 8.1 haplotype with MG and thymus hyperplasia. The group of 73 family probands was combined with a second group of 63 unrelated patients who were genotyped for the same markers. There were no phenotypic or genotypic differences between the two subgroups (Figs. 6 and 7, which are published as supporting information on the PNAS web site). The control group consisted of 105 unrelated French Caucasian healthy individuals. They were typed for the microsatellite markers 9N-I, 82-3, and C1-2-5, which were most strongly associated with MYAS1 in the family study, and for C2-4-5, located at the distal boundary of the MYAS1 interval. To minimize bias, no missing genotypes were allowed at any of the four test loci. This led to the exclusion of six patients (each with one missing genotype). Therefore, the comparison concerned 130 patients and 105 controls. Table 1 shows the numbers of patients and controls who carried zero, one, or two copies of the 8.1 haplotype or of its surrogate, based on two, three, or all four loci. There was very strong evidence for an additive model: odds ratios of 6.5 for one copy and 42.6 for two copies of the three-locus haplotype (P = 7.1 × 10-11). The frequency of the surrogate 8.1 haplotype among the controls was estimated to be ≈0.06. The parent-child transmission of the surrogate 8.1 haplotype in single-case families also showed a major distortion of transmission (Table 4, which is published as supporting information on the PNAS web site).
Table 1.
| No. of (%)§
| Additive
| ||||||
|---|---|---|---|---|---|---|---|
| Haplotypes† | Population‡ | -/- | -/+ | +/+ | P¶ | OR (95% CI), one copy one copy | OR (95% CI), two copies two copies |
| 9N-I/82-3/C1-2-5/C2-4-5 | P | 68 (52.3) | 55 (42.3) | 7 (5.4) | 1.9 × 10-9 | 6.0 (3.0-12.7) | 36 (9.30-161) |
| C | 92 (87.6) | 13 (12.4) | 0 (0.0) | ||||
| 82-3/C1-2-5/C2-4-5 | P | 62 (47.7) | 59 (45.4) | 9 (6.9) | 7.1 × 10-11 | 6.52 (3.37-13.5) | 42.6 (11.4-182) |
| C | 91 (86.7) | 14 (13.3) | 0 (0.0) | ||||
| C1-2-5/C2-4-5 | P | 53 (40.8) | 65 (50.0) | 12 (9.2) | 7.5 × 10-11 | 5.48 (3.05-10.3) | 30 (9.28-106) |
| C | 86 (81.9) | 18 (17.1) | 1 (1.0) | ||||
| 82-3/C1-2-5 | P | 61 (46.9) | 60 (46.2) | 9 (6.9) | 1.1 × 10-10 | 6.24 (3.3-12.7) | 39 (10.7-160) |
| C | 90 (85.7) | 15 (14.3) | 0 (0.0) | ||||
 Exact odds ratios (OR) and their 95% confidence intervals (CI) for one or two copies of the surrogate 8.1 haplotype relative to its absence
Exact odds ratios (OR) and their 95% confidence intervals (CI) for one or two copies of the surrogate 8.1 haplotype relative to its absenceOpposing Cis Effects of the 8.1 Ancestral Haplotype on Anti-AChR Autoantibody Expression. Patients with thymus hyperplasia have higher autoantibody titers than other nonthymoma patients on average (21). We investigated the effect of the 8.1 haplotype on autoantibody titers in the 122 seropositive patients with thymus hyperplasia. A one-way ANOVA comparing titers in patients positive or negative for 8.1-associated alleles revealed an association between the markers in the class III and proximal class I regions and increased autoantibody titers (Fig. 3A). In contrast, a haplotype-based comparison showed no difference between the titers in patients carrying the extended 8.1 haplotype (one or two copies) and patients who did not harbor this haplotype (baseline) (Fig. 3B). This discrepancy may be caused by the existence of two loci, one increasing autoantibody titers and one (in cis) decreasing titers. Consistent with this hypothesis, patients with a haplotype recombinant on the proximal side, between DRB1 and CTG-NOTCH4 or more distally, therefore excluding at least DR3, had significantly higher titers than baseline (Fig. 3B). This finding suggests that the autoantibody-increasing locus is located in the central region and that its modifier maps toward the proximal end of the HLA complex. Conversely, patients with a haplotype recombinant on the distal side (distal to C2-4-5) also appeared to have higher titers than baseline, but the difference was not significant.
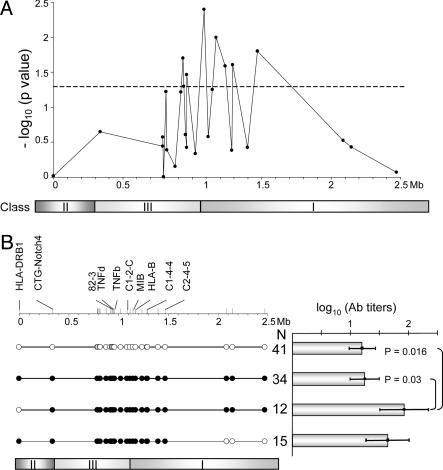
HLA control of anti-AChR autoantibody titers. The titer distribution was normalized by a logarithmic transformation. (A) Single-locus scan of the HLA region. One-way ANOVA was used to compare titers in patients carrying an 8.1-associated allele and in those not carrying this allele at each locus. The -log10 P value of the F test is plotted on the y axis. The dashed line indicates the uncorrected level of significance (0.05). (B) Haplotype-based analysis. (Left) Four groups of patients were defined on the basis of characteristic allelic associations; •, 8.1-associated alleles and ○, 8.1-nonassociated alleles at each site (see also Subjects and Methods). Marker positions are shown at the top. (Right) The bar chart shows the autoantibody titers (log10 of the means and 95% confidence intervals) in the four groups of patients with their size indicated. The P value of the ANOVA F test was 0.0085. Significant P values obtained with the post hoc Tukey test are shown.
The recently available qtphase program allows testing for associations between a quantitative trait and multilocus haplotypes from unphased genotype data, with optional conditioning on loci in linkage disequilibrium. A qtphase scan of the HLA complex using a window of three markers revealed associations in the class III and proximal class I regions (Fig. 8, which is published as supporting information on the PNAS web site). Conditioning the analysis on the DR3 allele considerably increased the significance of the findings (P = 4.5 × 10-5). We then carried out a similar three-point qtphase scan (Table 2) focusing on the set of eight markers found to be associated by the one-way ANOVA (Fig. 3A). The recombinant haplotypes consistently showed significantly higher titers of autoantibodies than the nonrecombinant 8.1 and the non-8.1 haplotypes. The most significant window associated C1-2-C, MIB, and HLA-B8 (P = 8.8 × 10-6); titers were ≈7-fold higher in patients with a recombinant 8.1 haplotype than at the baseline. Simultaneous testing of all eight loci yielded similar results. These data support the existence of a locus that increases autoantibody titers in the class III region or proximal class I region and suppressed in cis by a proximal locus.
Table 2.
| Haplotypes†
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-DR | 82-3 | TNFd | TNFb | C1-2-C | MIB | HLA-B8 | C1-4-4 | C2-4-5 | Frequencies | Mean of log10 titers | Individual P | Overall P |
| 1 | 1 | 1 | 1 | 0.49 | 1.378 | 0.10 | 0.0028 | |||||
| 1 | 2 | 2 | 2 | 0.08 | 2.051 | 0.0016 | ||||||
| 2 | 2 | 2 | 2 | 0.24 | 1.482 | 0.81 | ||||||
| 1 | 1 | 1 | 1 | 0.52 | 1.247 | 0.0035 | 0.00080 | |||||
| 1 | 2 | 2 | 2 | 0.07 | 1.984 | 0.0013 | ||||||
| 2 | 2 | 2 | 2 | 0.27 | 1.448 | 0.62 | ||||||
| 1 | 1 | 1 | 1 | 0.49 | 1.317 | 0.023 | 0.00035 | |||||
| 1 | 2 | 2 | 2 | 0.08 | 2.168 | 0.00010 | ||||||
| 2 | 2 | 2 | 2 | 0.26 | 1.433 | 0.79 | ||||||
| 1 | 1 | 1 | 1 | 0.49 | 1.354 | 0.13 | 0.0000088 | |||||
| 1 | 2 | 2 | 2 | 0.09 | 2.221 | 0.0000047 | ||||||
| 2 | 2 | 2 | 2 | 0.28 | 1.429 | 0.96 | ||||||
| 1 | 1 | 1 | 1 | 0.53 | 1.394 | 0.094 | 0.000020 | |||||
| 1 | 2 | 2 | 2 | 0.08 | 2.357 | 0.000018 | ||||||
| 2 | 2 | 2 | 2 | 0.27 | 1.461 | 0.81 | ||||||
| 1 | 1 | 1 | 1 | 0.29 | 1.242 | 0.016 | 0.000063 | |||||
| 1 | 2 | 2 | 2 | 0.09 | 2.218 | 0.000050 | ||||||
| 2 | 2 | 2 | 2 | 0.26 | 1.451 | 0.92 | ||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.20 | 1.210 | 0.036 | 0.00042 |
| 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0.06 | 2.443 | 0.000094 | |
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0.22 | 1.416 | 0.75 | |
Discussion
Our study demonstrates that the 8.1 haplotype has very significant but complex and antagonizing effects on MG patients. We show that the MYAS1 interval does not overlap with the class II region. It mapped to the class III and proximal class I regions. Very interestingly, it overlaps with the locus recently identified in patients with RA, which is also associated with the 8.1 haplotype (31, 32). This finding is consistent with the frequent association of RA in MG patients (33) and, conversely, with the observation of thymic hyperplasia in RA patients (34). This region notably contains the cluster of the TNF/lymphotoxin genes, which are major candidates given their role in the formation of germinal centers (35, 36), and the IκB-L gene, recently shown to be associated with RA (37). In contrast, the class I HLA-B8 allele was weakly associated and the class I-like locus MICA, previously associated with type I diabetes (38), was not associated at all; these loci are therefore not primary candidates.
Although the class II loci on the 8.1 haplotype showed no proper association, we have previously shown that a different class II gene allele, DQA1*0101, not associated with the 8.1 haplotype, is associated with MG and interacts with a polymorphism of CHRNA1, one of the AChR-encoding genes (39, 40). Therefore, class II loci do contribute to MG predisposition as they do in many autoimmune diseases (41). However, their lack of association with the 8.1 haplotype in MG should facilitate the identification of MYAS1, unlike in type I diabetes and celiac disease in which the additional nonclass II HLA-linked disease loci are in strong linkage disequilibrium with the predisposing class II loci. Hence, MG provides an invaluable model.
The population study confirmed the results of the family-based analysis and provided very strong evidence for an additive model for MYAS1. The odds ratios, 6.5 for a single copy of the core 8.1 haplotype and 42 for a double copy, were higher than those measured in most previous studies, demonstrating the importance of appropriate modeling and marker selection. The frequency (<6%) of the disease-associated core haplotype in our control group was similar to that reported in Caucasian populations (42). The actual frequency of the disease allele at the MYAS1 locus is likely to be even lower if the causative polymorphism appeared after the constitution of the 8.1 ancestral haplotype. This observation challenges the widespread assumption of the common variant for a complex disease and has important implications for the future identification of MYAS1.
As firmly established by passive transfer experiments and the observation of neonatal MG, anti-AChR autoantibodies are primary effectors of MG, even though other factors are necessary for its clinical expression (43), which explains why their serum titers correlate poorly with disease severity. Understanding the mechanisms of their production and regulation is therefore of both research and clinical importance. Early studies suggested HLA-dependent control in nonthymoma MG patients (12, 44). This group of patients, however, is heterogeneous. The age at onset of the disease and the thymus histopathology are confounding factors that must be taken into account as they are correlated with one another and autoantibody titers (21). Therefore, it was critical to restrict the genetic analysis to the subset of patients with thymus hyperplasia to remove the variation in autoantibody titers caused by thymus histopathology. A quantitative trait locus (QTL) controlling anti-AChR autoantibody titers could be mapped to an interval that overlaps with MYAS1, although the peak of association mapped closer to the class I loci and involved the HLA-B8 allele. Therefore, whether this QTL is MYAS1 itself or a different locus remains to be clarified. The variation in the titers of autoantibodies may also reflect their heterogeneity, i.e., their ability to react with different epitopes of the AChR. These differences could be explained by the class I alleles. In addition, the QTL was suppressed by an allele acting in cis at a proximal locus, in the class II region or proximal class III region. This finding is reminiscent of a previous report of gene interactions within the MHC in experimental models of autoimmunity (45). It remains to be determined whether these loci specifically modulate anti-AChR autoantibodies or influence humoral responses more broadly, notably those previously reported to be associated with the 8.1 haplotype including low antibody response to hepatitis B vaccine (46), allergen-specific IgE production (47), and IgA deficiency (48).
In summary, three loci influencing the phenotype of MG patients and associated with the 8.1 haplotype can now be added to the list of class II genes, which are dissociated from the 8.1 haplotype. These pleiotropic effects of the 8.1 haplotype are consistent with its involvement in multiple autoimmune traits and diseases (5). Future studies, combining very high-resolution genotyping, in situ analysis of candidate gene expression, and investigation of endophenotypes such as cytokine production, should further elucidate the importance of this haplotype in MG and other autoimmune diseases.
Acknowledgments
We thank Dr. Elizabeth Dulmet (Service d'Anatomie Pathologique, Hôpital Marie-Lannelongue, Le Plessis-Robinson, France) for providing full access to histopathology records and Drs. Matthieu Levi-Strauss and Saoussen Karray for critical reading of the manuscript. This study was funded by the Institut National de la Santé et de la Recherche Médicale, the Association Française Contre les Myopathies, and the Fondation pour la Recherche Médicale. C.V. was supported by the Ministère de la Recherche et de la Technologie and is now an Association Française Contre les Myopathies Fellow.
Notes
Author contributions: H.-J.G. designed research; C.V., G.B., M.G., C.H.-B., B.E., C.T., and P.G. performed research; C.V. and J.D. contributed new reagents/analytic tools; C.V. and H.-J.G. analyzed data; C.V. and H.-J.G. wrote the paper; and H.-J.G. coordinated the research.
Abbreviations: MG, myasthenia gravis; AChR, acetylcholine receptor; T/NT, transmitted/nontransmitted; TNF, tumor necrosis factor; RA, rheumatoid arthritis.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0406756101
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc524438?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0406756101
Article citations
Genome-wide meta-analysis of myasthenia gravis uncovers new loci and provides insights into polygenic prediction.
Nat Commun, 15(1):9839, 13 Nov 2024
Cited by: 0 articles | PMID: 39537604 | PMCID: PMC11560923
Pathophysiology of Childhood-Onset Myasthenia: Abnormalities of Neuromuscular Junction and Autoimmunity and Its Background.
Pathophysiology, 30(4):599-617, 02 Dec 2023
Cited by: 0 articles | PMID: 38133144 | PMCID: PMC10747330
Review Free full text in Europe PMC
Sodium butyrate alleviates R97-116 peptide-induced myasthenia gravis in mice by improving the gut microbiota and modulating immune response.
J Inflamm (Lond), 20(1):37, 03 Nov 2023
Cited by: 2 articles | PMID: 37924056 | PMCID: PMC10625296
Commentary: Conserved polymorphic sequences protect themselves for future challenges.
Front Genet, 13:993944, 11 Nov 2022
Cited by: 1 article | PMID: 36437948 | PMCID: PMC9691882
Next-Generation Sequencing Identifies Extended HLA Class I and II Haplotypes Associated With Early-Onset and Late-Onset Myasthenia Gravis in Italian, Norwegian, and Swedish Populations.
Front Immunol, 12:667336, 07 Jun 2021
Cited by: 2 articles | PMID: 34163474 | PMCID: PMC8215161
Go to all (55) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Linkage of HLA to myasthenia gravis and genetic heterogeneity depending on anti-titin antibodies.
Neurology, 57(9):1555-1560, 01 Nov 2001
Cited by: 59 articles | PMID: 11706089
[Thymic lymphoid hyperplasia of myasthenia gravis patients: correlation with clinical features and efficacy of thymectomy].
Kyobu Geka, 48(6):447-451, 01 Jun 1995
Cited by: 1 article | PMID: 7602854
Genetic control of autoantibody expression in autoimmune myasthenia gravis: role of the self-antigen and of HLA-linked loci.
Genes Immun, 5(5):398-404, 01 Aug 2004
Cited by: 17 articles | PMID: 15201863
[Antibodies in myasthenia gravis].
Rev Neurol (Paris), 165(2):137-143, 21 Jan 2009
Cited by: 10 articles | PMID: 19162288
Review




