Abstract
Free full text

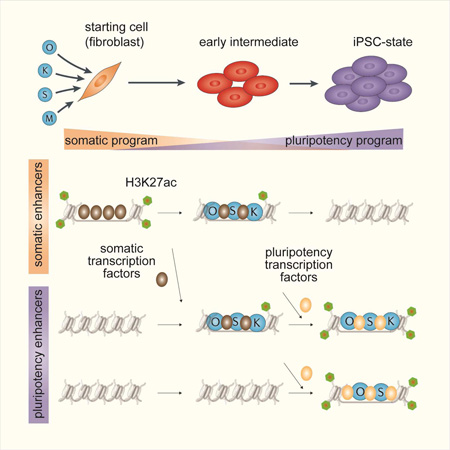
Cooperative binding of transcription factors orchestrates reprogramming
Summary
Oct4, Sox2, Klf4, and cMyc (OSKM) reprogram somatic cells to pluripotency. To gain a mechanistic understanding of their function, we mapped OSKM-binding, stage-specific transcription-factors (TFs), and chromatin-states in discrete reprogramming stages and performed loss- and gain-of-function experiments. We found that OSK predominantly bind active somatic-enhancers early in reprogramming and immediately initiate their inactivation genome-wide by inducing the redistribution of somatic TFs away from somatic-enhancers to sites elsewhere engaged by OSK, recruiting Hdac1, and repressing the somatic-TF Fra1. Pluripotency-enhancer selection is a step-wise process that also begins early in reprogramming through collaborative binding of OSK at sites with high OSK-motif density. Most pluripotency-enhancers are selected later and require OS and other pluripotency TFs. Somatic and pluripotency-TFs modulate reprogramming efficiency when overexpressed by altering OSK-targeting, somatic-enhancer inactivation, and pluripotency-enhancer selection. Together, our data indicate that collaborative interactions among OSK and with stage-specific-TFs direct both somatic-enhancer inactivation and pluripotency-enhancer selection to drive reprogramming.
Introduction
Differentiated cells can be reprogrammed to pluripotency by overexpression of the four transcription factors (TFs) Oct4, Sox2, Klf4, and cMyc (OSKM) (Takahashi and Yamanaka, 2006). Successful reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) leads to the faithful shutdown of the somatic and activation of the target program. Conversely, in TF-induced conversions of one somatic cell type to another incomplete extinction of the starting cell program represents a major barrier (Cahan et al., 2014). Hence, understanding the mechanisms by which OSKM inactivate the starting cell program and induce the pluripotency network will provide insights into the principles by which cell identity can be effectively manipulated.
The interaction of OSKM with chromatin has been primarily studied in ESCs, where O, S, and K preferentially bind enhancers and M primarily associates with promoters (Chen et al., 2008; Kim et al., 2008). In ESCs, enhancers are often occupied by additional pluripotency TFs including Nanog and Esrrb (Chen et al., 2008; Kim et al., 2008; Whyte et al., 2013) suggesting that complex regulatory interactions perpetuate the pluripotent state. Among the pluripotency TFs, O, S, and Nanog are thought to form a pivotal circuitry as they co-occupy enhancers with a higher frequency than other TFs (Chen et al., 2008), raising the questions of why K is an effective reprogramming factor when combined with O and S and how these factors interact during reprogramming. Moreover, it is unclear how and when pluripotency enhancer selection happens during reprogramming given that most pluripotency TFs are only available late in the process (Polo et al., 2012; Samavarchi-Tehrani et al., 2010). Since enhancers play a central role in driving cell-type specific gene expression (Heinz et al., 2015), defining how the reprogramming factors control the reorganization of the enhancer landscape is critical for a mechanistic understanding of reprogramming.
A few studies reported that the target sites of the reprogramming factors change during reprogramming (Chen et al., 2016; Sridharan et al., 2009). In addition, it has been shown that O, S, and K each can act as pioneer factor since they can engage nucleosome-occluded sites in human fibroblasts and nucleosomal templates in vitro (Soufi et al., 2012; Soufi et al., 2015). Whether these properties are relevant for their binding to pluripotency enhancers during the reprogramming process, however, remains elusive. Moreover, the pioneer factor model does not provide a mechanistic explanation for the silencing of the somatic program, and, therefore, it has remained unclear how the reprogramming factors would induce this process.
In our study, we delineated the interaction of the reprogramming factors with somatic and pluripotency enhancers. We uncovered that OSK mediate both somatic enhancer silencing and pluripotency enhancer selection through collaborative interactions among themselves and with stage-specific TFs.
Results
Comprehensive mapping of TFs, chromatin features and expression at defined reprogramming stages
To characterize the role of OSKM in reprogramming, we carried out chromatin immunoprecipitation for each reprogramming factor coupled to high-throughput sequencing (ChIP-seq) at four distinct stages of mouse embryonic fibroblast (MEF) reprogramming (Fig 1A). These stages included (i) MEFs carrying a tetracycline-inducible polycistronic OSKM expression cassette to capture the starting state; (ii) the same MEFs induced for OSKM expression with doxycycline (dox) for 48 hours (48h); (iii) two independently generated pre-iPSC lines (pre-i#1 and pre-i#2); and (iv) the pluripotent state represented by mouse ESCs for the end state (Fig S1A-C). The 48h time point represents an early reprogramming stage and was chosen to examine the initial interaction of OSKM with MEF chromatin. Importantly, within the first 48h, fibroblasts respond to OSKM activation in a homogeneous manner and with limited expression changes (Buganim et al., 2012; Koche et al., 2011; Polo et al., 2012). Since reprogramming cultures are heterogeneous at later time points (Pasque et al., 2014; Polo et al., 2012), we turned to pre-iPSC lines with closely related transcriptional, epigenetic, and OSKM binding profiles (Fig S1D/E, S2E/F) that were isolated clonally from reprogramming cultures infected with OSKM-encoding retroviruses (Sridharan et al., 2009) for a proxy of a late intermediate stage. Since M and K are expressed endogenously in starting MEFs (Fig S1A-C), we mapped both in all four reprogramming stages, whereas O and S were profiled at 48h, in pre-iPSCs and ESCs.
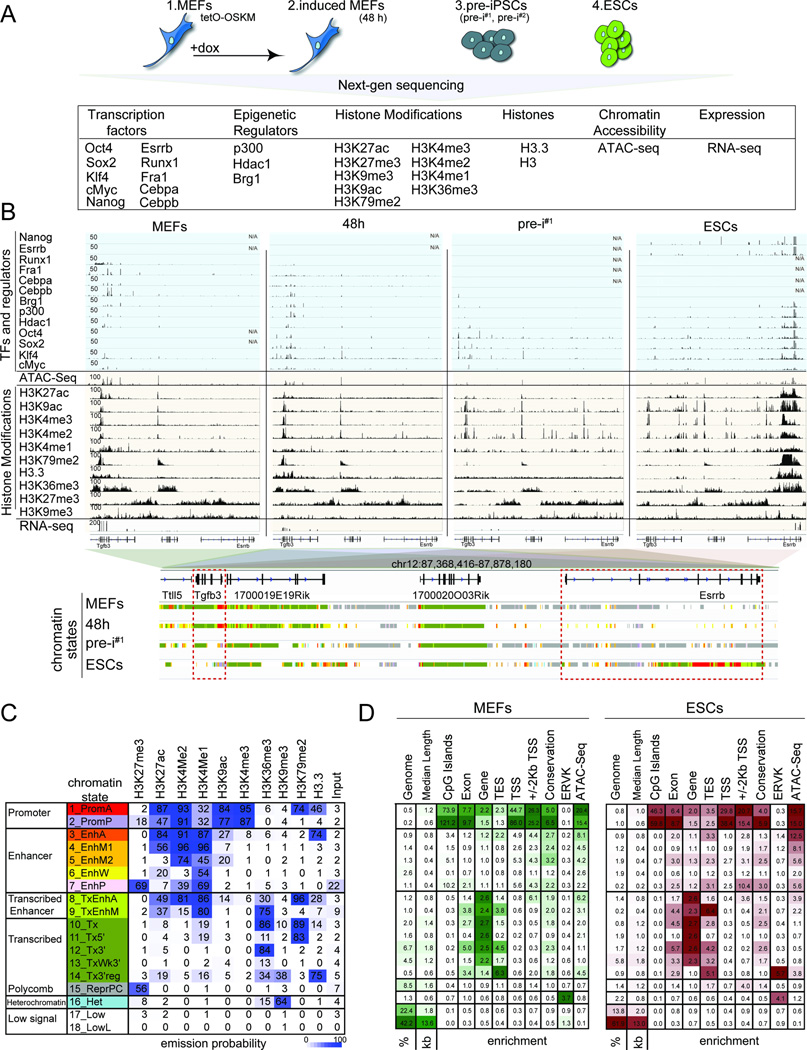
A) Summary of reprogramming stages and data sets produced.
B) Snapshot of indicated genomics data at a candidate genomic locus. N/A = no data. The color code represents the stage-specific chromatin states defined in (C). Red boxes mark the somatic gene Tgfb3 and the pluripotency gene Esrrb.
C) Rows represent chromatin states and their representative mnemonics, color-coded and grouped based on their putative annotation. Cells show the frequency of each histone marks, H3.3, and input for each state (ChromHMM emission probabilities).
D) Columns give % genome occupancy, median length in kilo bases (kb), and fold-enrichment of indicated features (transcription end sites, TES; transcription start sites, TSS; conservation phastCons elements; endogenous retrovirus K elements, ERVK) for each chromatin state described in (C) for MEFs and ESCs. Color-code per column from highest to lowest value.
See also Fig S1.
Additionally, we determined the targets of endogenously expressed TFs (Cebpa, Cebpb, Fra1, Runx1, Esrrb and Nanog) and chromatin regulators (p300, Hdac1 and Brg1) in relevant reprogramming stages to determine their interplay with OSKM, mapped histone H3 to assess nucleosome occupancy, and measured chromatin accessibility by ATAC-seq and gene expression by RNA-sequencing (Fig 1A) (Tables S1/S2). We also generated maps for nine histone modifications and the histone variant H3.3 for each reprogramming stage. The histone modifications included H3K4me3 and H3K9ac primarily associated with promoters; H3K4me1, H3K4me2 and H3K27ac characteristic of active promoters and enhancers; H3K79me2 and H3K36me3 associated with transcription, and the repressive marks H3K9me3 and H3K27me3 (Fig 1A) (Ernst et al., 2011). A snapshot of the various data sets is shown in Figure 1B. Data reproducibility was confirmed by correlating replicate experiments, experimental and imputed data (Ernst and Kellis, 2015), and through comparisons with published data sets (Table S3), leading to the merging of replicate data sets for downstream analyses. Additionally, for TFs their known motifs were identified at occupied sites (Fig S1F), validating our data sets.
Identification of cis-regulatory elements at each reprogramming stage
To enable a characterization of the chromatin environment at sites engaged by OSKM, we summarized the combinatorial and spatial patterns of histone modifications and H3.3 for each reprogramming stage by building a chromatin state model with 18 states using ChromHMM, and assigned candidate functional annotations to each state on the basis of present marks (Fig 1C) (Ernst and Kellis, 2012). The 18 states defined active and poised promoters, inter- and intragenic enhancers of varying activity levels, various transcribed regions, repressed regions, and genomic regions with minimal or no signal of any histone mark (Fig 1C/D). These chromatin state annotations were supported by associations with genomic landmarks such as CpG islands and transcriptional start sites (TSSs) of genes, chromatin accessibility, expression of nearby genes (Fig 1D, S1G/H), and captured epigenetic states expected to occur at somatic and pluripotency loci during reprogramming (Fig 1B, S1I).
OSKM predominantly occupy active and poised promoters and enhancers at each reprogramming stage
First, we investigated the characteristics of OSKM binding sites at each reprogramming stage. Regardless of reprogramming stage, O, S, and K predominantly bound in distal regions >2kb away from the TSS, whereas M binding occurred more often in close proximity to the TSS (Fig 2A, S2A). Intersection of binding sites with chromatin states revealed that at each reprogramming stage, all four reprogramming factors bound both active and poised promoters and that the distal binding sites of OSK were predominantly located within active enhancers (Fig 2B, S2B). These binding preferences also applied when considering co-binding between the reprogramming factors, such that M in combinations with O, S, or K displayed strong promoter bias, whereas combinations of O, S, or K binding without M, preferentially targeted active enhancers (Fig S2C). Sites occupied by O, S, K, or M displayed pronounced nucleosome depletion and chromatin accessibility at each stage (Fig 2C, S2D). Together, these results demonstrated that OSKM prefer to bind active and poised promoters and enhancers regardless of reprogramming stage.
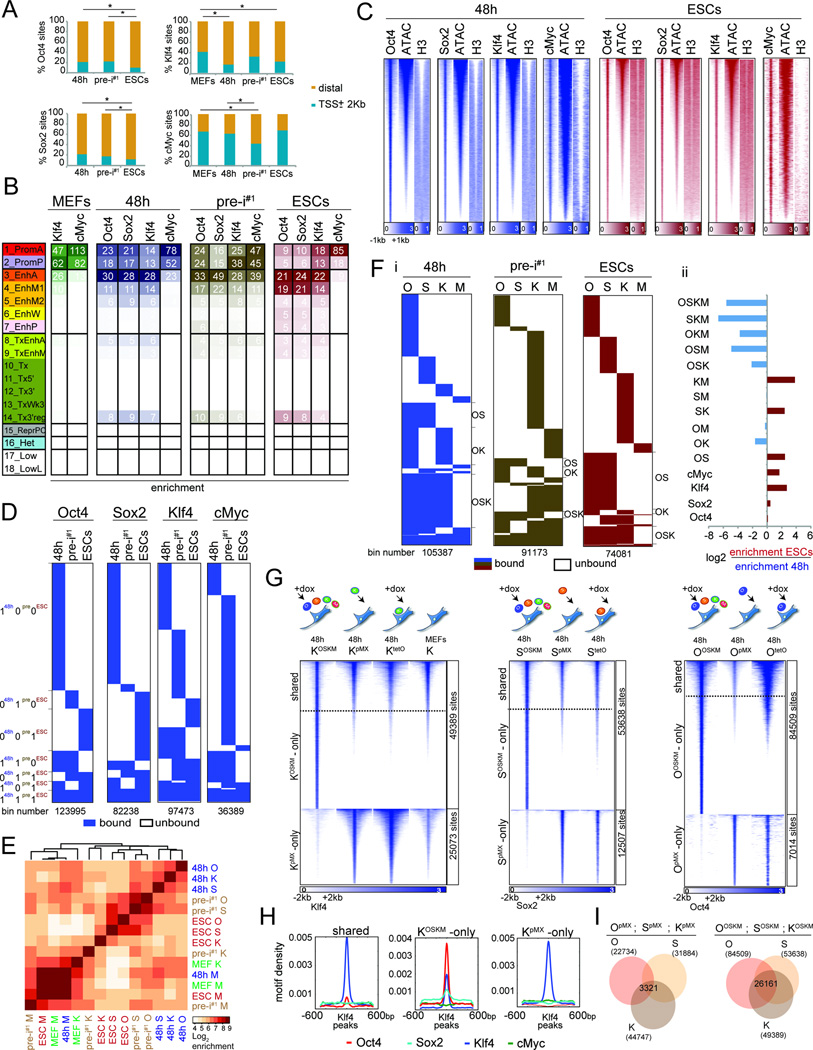
A) Fraction of TF binding sites within promoter (TSS+/−2kb) and distal (>2Kb from TSS) promoter regions. *p-val < 0.0001, chi-square test.
B) Fold-enrichment of TF binding sites per chromatin state (Fig 1C) in the corresponding reprogramming stage, colored per column from highest to lowest.
C) Heatmap of O, S, K, and M ChIP-seq signal for 48h and ESCs peaks and corresponding signals for ATAC-seq and histone H3, ranked by ATAC-seq signal strength.
D) Comparison of binding events of each reprogramming factor between 48h, pre-i#1, and ESCs (0/white=unbound, 1/blue=bound), at 500bp resolution (bin).
E) Hierarchical clustering of pairwise enrichments of O, S, K, and M binding events.
F) (i) Clustering of O, S, K, and M binding events at 500bp resolution (bin). OS, OK, and OSK co-binding events are marked. (ii) Differential enrichments of co-binding groups between ESCs and 48h.
G) Heatmaps of ChIP-seq signal for K, S, or O peaks at 48h of OSKM or individual reprogramming factor expression (retrovirally (pMX) or inducibly (tetO)). Peaks were grouped based on presence/absence of peak calls comparing the OSKM and single TF expressing (pMX) samples. For K, binding events in MEFs were also plotted.
H) Density plots of O, S, M, and K motifs in sets of K peaks defined in (G).
I) Overlap of O, S, and K sites (number given) obtained from MEFs individually expressing O, S, and K for 48h (pMX, left) and MEFs co-expressing OSKM for 48h (right).
OSKM redistribution and binding partner switch during reprogramming
A comparison of binding sites between 48h, pre-iPSCs, and ESCs revealed that the genomic locations of each reprogramming factor differed dramatically between stages and that the majority was stage-specific (coined ‘100’; ‘010’; and ‘001’; where 1 represents presence and 0 absence of binding, and the digits from left to right binding at 48h, pre-i#1, and ESCs) (Fig 2D, S2E/F). For instance, 48% of all Oct4 binding events occurred exclusively at 48h (‘100’ sites) and 16% were specific for the pluripotent stage (‘001’ sites). 48h-specific Oct4 binding events (‘100’ sites) occurred close to genes with fibroblast functions based on gene ontology (GO) analysis, whereas pluripotency-specific sites (‘001’ sites) were linked to genes that control stem cell function and early developmental decisions (Fig S2G; Table S4), suggesting that stage-specific binding events are associated with stage-specific gene functions. Together, these data indicated the interaction of OSKM with somatic sites early in reprogramming and the redistribution to pluripotency-associated sites at later stages.
The remaining binding events were transient (‘110’ and ‘011’), absent in pre-iPSCs (‘101’), or constitutive (‘111’) (Fig 2D, S2E). Constitutively-bound Oct4 sites, for instance, represented 8% of all 48h-bound sites and occurred in the vicinity of genes implicated in blastocyst formation, chromosome organization and inhibition of MAPK signaling, which is closely tied to the maintenance of pluripotency (Ying et al., 2008) (Fig S2G; Table S4). Thus, the majority of sites associated with the pluripotent state become engaged by the reprogramming factors only late in the process, but certain sites are targeted within the first 48h. Motif analysis revealed lower densities of OSKM DNA binding sequences at ‘100’ sites compared to ‘001’ and ‘111’ sites (Fig S2H), suggesting that temporal binding events differ in their regulation.
In addition, we found that M binding differed strongly from that of O, S, and K throughout reprogramming, and, more surprisingly, that K sites coincided more with those of O and S at 48h but diverged from these in pre-iPSCs and the pluripotent state (Fig 2E, S2I/J). Consequently, we observed significantly more co-binding events of OSK and OK at 48h than in ESCs and, conversely, an increase in OS co-occupancy in ESCs relative to 48h (Fig 2F). Thus, co-binding preferences change from OSK/OK to OS during reprogramming, consistent with O and S composing the core pluripotency network in ESCs alongside Nanog instead of Klf4 (Chen et al., 2008).
OSK co-occupancy at 48h depends on their co-expression
By comparing K binding between MEFs and 48h, we found that many binding sites were gained whereas others were lost at 48h (Fig 2G, S3A). Upon overexpression of only Klf4 in MEFs for 48h, either retrovirally (KpMX) or inducibly (KtetO), without the other reprogramming factors, K predominantly engaged sites that were targeted by it in MEFs and not those newly accessible in the context of OSKM co-expression (Fig 2G), despite its higher expression level (Fig S3B). We conclude that O and S availability, and not the expression level of K per se, is responsible for the differential binding at 48h compared to MEFs. Moreover, whereas sites targeted by endogenous K in MEFs or upon individual overexpression of K carried only the K motif (Fig 2G/H ‘KpMX-only’ and ‘shared’ sites, S3A), new locations bound by K at 48h of OSKM-reprogramming were co-occupied by O and S and enriched for the motifs of all three factors (Fig 2G/H, ‘KOSKM-only’ sites; S3A), revealing an unexpected dependence of K occupancy on OS early in reprogramming. Conversely, the targeting of O and S at 48h also strongly depended on the presence of the other reprogramming factors (Fig 2G). Specifically, when individually expressed, O and S bound many sites in open MEF chromatin that carried the motif of the respective reprogramming factor (Fig S3B-E), which did not overlap substantially between the factors (Fig 2I). Yet, when co-expressed in the context of OSKM for 48h, O and S co-occupied a many new sites with K carrying the motifs of all three factors (Fig 2G/I, Fig S3B-E). M was largely dispensable for the redistribution of K and OSK co-binding at 48h as co-expression of OSK, without M, led to engagement of largely the same sites at 48h as in OSKM-induced reprogramming (Fig S3F). We conclude that cooperative binding of O, S, and K is critical for the targeting of a vast number of genomic sites early in reprogramming and additionally restrict access to locations that carry the motif of only one reprogramming factor.
Enhancers are sites of most dramatic chromatin changes in reprogramming
To examine the association between temporal OSKM binding events and chromatin changes during reprogramming, we derived another chromatin state model that took into consideration the combination of histone marks and H3.3 at any given genomic location within each reprogramming stage as well as the changes of these histone marks/H3.3 between the stages (Fig 3A), and defined 35 chromatin states that will be referred to as chromatin trajectories (tr.) hereafter. Based on the histone mark/H3.3 composition of each trajectory, we annotated genomic regions as candidate promoters (tr. 1–4), enhancers (tr. 5–18), units of transcription (tr. 19–27), repressed (tr. 28–32), transcribed repeats (tr. 33), and devoid of histone marks (tr. 34, 35). These annotations were consistent with enrichments for genomic landmarks and expression of neighboring genes (Fig S4A/B). Differences in temporal histone marks/H3.3 composition between the reprogramming stages defined the stage-specific or constitutive chromatin character of each trajectory. We noted that the promoter states (tr. 1–4) did not carry a strong stage-specific identity (Fig 3A, S4B) consistent with promoter states being more conserved across cell types (Heintzman et al., 2009). Around 16% of the genome represented enhancers, and, in contrast to promoters, the enhancer trajectories strongly differed in their histone mark composition between reprogramming stages and therefore likely in their activity and regulation (Fig 3A, tr. 5–18).
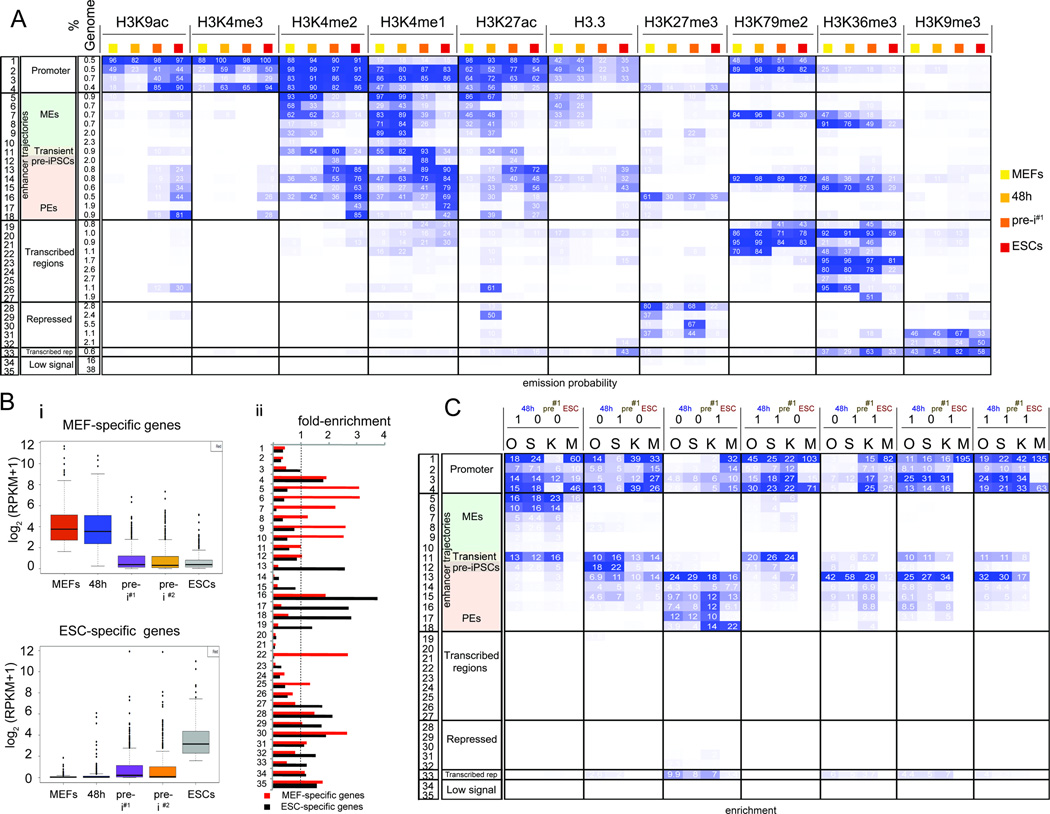
A) Definition of the 35 chromatin trajectories that capture the major chromatin changes during reprogramming. The first three columns give the number, functional annotation, and genome fraction of each trajectory. Following columns are organized by histone mark and sub-ordered by reprogramming stage displaying the frequency of each mark per reprogramming stage and trajectory, colored from 0 (white) to 100 (blue).
B) (i) Boxplots of expression levels of MEF- and ESC-specific genes per reprogramming stage. (ii) Relative enrichment of each trajectory defined in (A) within +/− 20kb of the TSS of MEF- and ESC-specific genes compared to +/− 20kb of the TSS of all active genes. Values above the dashed line indicate higher enrichment in MEF- and ESC-specific genes, respectively.
C) Fold-enrichment of stage-specific and constitutive O, S, K, and M binding events defined in Fig 2D for each trajectory in (A), colored per column from highest to lowest.
See also Fig S4.
Based on the presence of the active enhancer mark H3K27ac in MEFs and its absence in ESCs, we defined MEF enhancers (MEs) (tr. 5, 6, 9, 10 and 7, 8, respectively) (Fig 3A, S4A/B). MEs were either inter- or intragenic and typically located in the vicinity of genes with fibroblast-specific functions that tended to be expressed specifically early in reprogramming (Fig 3B, S4B/C). Pluripotency enhancers (PEs) were defined based on the presence of H3K27ac in ESCs and near absence in MEFs (tr. 13–18). PEs of tr. 13 and 17 were intergenic, neighboring genes highly expressed in ESCs and implicated in stem cell maintenance, blastocyst formation, and developmental programs based on GO analysis (Fig 3A/B, S4A-C). PEs associated with tr. 14, 15, 16, and 18 were predominantly intragenic or poised (carrying H3K27me3) and close to or within genes that tended to be either constitutively expressed or repressed across during reprogramming (Fig 3A, Fig 4B) and implicated in chromatin regulation and cell fate specification.
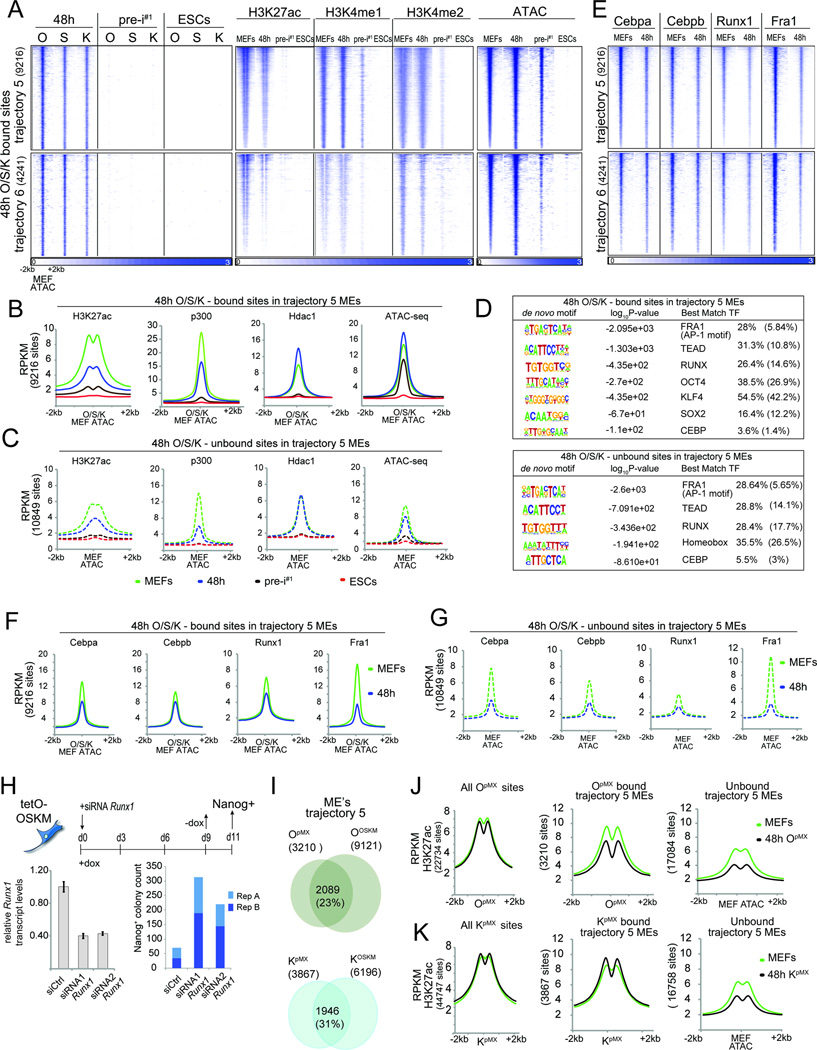
A) Heatmaps of O, S, K, H3K27ac, and H3K4me1/2 ChIP-seq signal and the ATAC-seq signal at O, S, and K binding events in tr. 5 and 6 MEs at 48h, ordered by the ATAC-seq signal strength. The total number of peaks is given in brackets.
B) Metaplots of signal intensities for H3K27ac, p300, Hdac1, and ATAC-seq data in MEFs, 48h, pre-i#1, and ESCs at tr. 5 MEs occupied by O, S, or K at 48h, centered on ATAC-seq summits in MEFs.
C) As in (B), except for tr. 5 MEs not bound by O, S, or K at 48h.
D) De novo motifs identified under 48h O, S, or K- bound or unbound tr. 5 MEs. Last column: observed and expected motif frequencies (in parentheses).
E) Heatmaps of somatic TF ChIP-seq signal at sites defined in (A).
F) As in (B), except for somatic TFs in MEFs and at 48h.
G) As in (C), except for somatic TFs in MEFs and at 48h.
H) Schematic of the reprogramming experiment with Runx1 knockdown. Runx1 transcript levels were determined at 48h (error bars represent standard deviation) and Nanog-positive colonies were counted from two technical replicates (A, B).
I) Comparison of O or K binding events at tr. 5 MEs in MEFs individually expressing the respective reprogramming factor (OpMX or KpMX) and MEFs co-expressing OSKM for 48h (OOSKM or KOSKM). Number of sites is given in brackets.
J) Metaplots of signal densities for H3K27ac in starting MEFs and MEFs expressing only O for 48h (OpMX) at all OpMX bound sites and tr. 5 MEs bound or unbound by OpMX at 48h.
K) As in (J), but for KpMX.
See also Fig S5.
One group of intergenic enhancers was marked by H3K4me1/2 at all four stages but displayed activity, defined by H3K27ac presence, in a transient manner at 48h and in pre-iPSCs (tr, 11, transient enhancers) (Fig 3A). These enhancers were linked to transiently expressed genes involved in various signaling pathways, most notably those acting in the BMP pathway (Fig S4B/C). Since BMPs have a positive role early in reprogramming (Samavarchi-Tehrani et al., 2010), activation of these enhancers may be critical for reprogramming progression. Other enhancers were active exclusively in pre-iPSCs (tr. 12) (Fig 3A) and their neighboring genes enriched for neuronal ontologies (Fig S4C) consistent with the observations that neuronal genes are ectopically induced during reprogramming (Ho et al., 2013). In summary, we identified enhancers as the most dynamic part of the epigenome during reprogramming and defined groups of enhancers that are selectively used at different reprogramming stages.
Changes in OSK binding mirror enhancer re-organization
To investigate how the redistribution of OSKM relates to the chromatin rearrangement during reprogramming, we intersected the genomic coordinates of temporal OSKM binding events (Fig 2D) with the chromatin trajectories (Fig 3A) and made four key observations (Fig 3C, S4D): First, we confirmed that OSK binding predominantly occurred in promoters and enhancers whereas M preferred promoters throughout reprogramming. Second, the majority of O, S, and K binding events at 48h (‘100’, ’110’) occurred in promoters, MEs, and transient enhancers, indicating that early in reprogramming, O, S, and K predominantly target sites with open chromatin character in starting MEFs, unlike what has been reported for human cell reprogramming (Soufi et al., 2012). Third, O, S, and K binding at enhancers was typically observed when they were active (based on H3K27ac). For instance, pluripotency-specific O, S, K binding events (‘001’) were enriched specifically within PEs (tr. 13–18). Conversely, 48hspecific binding events (‘100’) enriched most in active MEs (tr. 5/6) and transient enhancers (tr, 11). These observations identified a dramatic shift of O, S, and K binding from MEs to PEs during reprogramming that accompanies their inactivation and selection/activation, respectively, and suggested that the reprogramming factors may directly control these two opposing processes. Fourth, we noted that a specific subset of PEs was targeted by O, S and K early in reprogramming. Among all enhancers, constitutive binding by O, S, and K (‘111’) was most enriched in tr. 13 PEs, and located proximal to genes involved in stem cell maintenance, blastocyst formation (Nanog, Lif, Esrrb, Stat3, Nodal etc) and negative regulation of MAP kinase signaling (Fig S4E), indicating that PE selection starts early in reprogramming and is finished in a step-wise manner throughout the process.
Since promoters displayed relatively little stage-specificity with respect to chromatin state and temporal reprogramming factor binding events whereas enhancers were often stagespecific for both (Fig 3C), we focused the rest of our study on the targeting and action of OSK at MEs and PEs to understand the regulation of ME silencing and PE selection as well as the regulation of distinct temporal binding patterns of OSK at enhancers.
MEs are suppressed genome-wide early in reprogramming
Since it has remained unexplored how MEs become silenced during reprogramming and how the reprogramming factors contribute to this process, we examined active intergenic MEs captured by tr. 5/6 in more detail, approximately half of which were bound by O, S, or K at 48h. Considering tr. 5/6 MEs engaged by O, S, or K, we found extensive co-occupancy of these TFs at 48h, which was accompanied by an increased ATAC-seq signal (Fig 4A/B, S5A/B). Later in reprogramming, in pre-iPSCs and ESCs, these MEs were depleted of active enhancer marks, OSK binding, and presented diminished chromatin accessibility defined by ATAC-seq (Fig 4A/B, S5A/B), consistent with a predominant ‘100’ OSK binding pattern at these enhancers.
Surprisingly, OSK-bound MEs displayed a lower level of the active enhancer mark H3K27ac at 48h compared to MEFs, which was corroborated by a decrease of the H3K27 acetyltransferase p300 (Fig 4A/B, S5B), indicating that somatic enhancer inactivation is initiated quite extensively very early in reprogramming. The enhancer marks H3K4me1/2 displayed smaller or no changes at 48h (Fig 4A, Fig S5A/B). The observation that H3K27ac levels were maintained or increased at other genomic locations (tr. 4, 9, and 11) at 48h (Fig S5C-E) argued against a global loss of p300 activity and H3K27ac. The histone deacetylase Hdac1 was also present at OSK- targeted MEs and, unlike p300, increased at 48h (Fig 4B), which was also seen in independent replicates, and, as for p300, occurred without alteration in its expression level (Fig S5F). We conclude that the change in balance of both p300 and Hdac1 observed at OSKbound MEs at 48h likely accounts for the reduction in H3K27ac at these enhancers in the earliest phase of reprogramming. The completion of silencing of these enhancers occurred later indicating that ME inactivation is a step-wise process.
Unexpectedly, we observed that MEs of tr. 5/6 that were not engaged by OSK also had strongly reduced H3K27ac and p300 levels at 48h (Fig 4C, S5A/B). These findings suggested that the disruption of the most active ME network takes place genome-wide early in reprogramming and extends beyond direct OSK targets. Interestingly, the increase in Hdac1 was specific to OSK-bound MEs and not observed at MEs that were not targeted by OSK (Fig 4B/C), potentially as a consequence of a direct action of O, S, or K.
Loss of somatic TFs from OSK-bound and unbound MEs at 48h
To investigate how MEs could be globally affected, we performed de novo motif scanning in OSK-bound and unbound tr. 5 MEs and identified DNA motifs of the Fra1 (AP-1 family), Tead, Runx, and Cebp families of TFs in both sets (Fig 4D). O, S, and K motifs were enriched specifically in the bound set (Fig 4D). We then performed ChIP-seq for the corresponding TFs Fra1, Cebpa, Cebpb, and Runx1, all highly expressed in MEFs (Fig S6J), and found that these TFs indeed occupied both OSK-bound and-unbound MEs in MEFs (Fig 4E-G). At 48h, all four TFs displayed reduced binding at OSK-bound and unbound MEs (Fig 4E-G), which was independently supported by a reduction in ATAC-seq signal at MEs not targeted by OSK (Fig 4C, S5A/B). These results suggested that the loss of somatic TFs from active MEs causes the reduction of p300 and H3K27ac at MEs at 48h.
To test the functional significance of somatic TF loss, we performed siRNA-mediated knockdown of Runx1 (Fig 4H) and Cebpa/b (Fig S5G) during reprogramming. Both increased the number of Nanog-positive colonies indicating that the depletion of ME-bound somatic TFs represents a mechanism for improving reprogramming efficiency, likely by augmenting ME inactivation.
Reprogramming factors can individually induce ME silencing
To determine whether OSK co-expression is required for global ME silencing, we analyzed O and K binding and H3K27ac levels at MEs in MEFs expressing only Oct4 or Klf4 for 48h. Only 23% and 31% of tr. 5 MEs bound by O and K, respectively, in the context of OSKM co-expression (OOSKM and KOSKM) were engaged by the single factors (OpMX and KpMX) (Fig 4I), emphasizing the importance of co-operative binding for the engagement of MEs. H3K27ac levels were reduced at tr. 5 MEs, but maintained over all binding sites of the individually expressed reprogramming factor (Fig 4J/K). Notably, individual reprogramming factors induced an H3K27ac drop at tr. 5 MEs comparable to that observed for OSKM-induced reprogramming (Fig S5H). Interestingly, for O, we observed a reduction of H3K27ac at tr. 5 MEs irrespective of its binding, but for K only at MEs not targeted by this reprogramming factor at 48h (Fig 4J/K), suggesting that Oct4, but not Klf4, may enhance silencing at its target MEs directly by increasing Hdac1 levels.
Somatic TF redistribution at 48h is guided by OSK
A comparison of peak locations for Cebpa, Cebpb, Runx1 and Fra1 revealed a loss and gain of binding events between MEFs and 48h as well as sites that were maintained (Fig 5A/B, S6A/B). Gain and loss of binding occurred predominantly at sites occupied by only one of the somatic TFs (Fig 5C, clusters I-III and IV-VII), whereas binding sites maintained at 48h were more often co-occupied by the somatic TFs (Fig 5C, cluster VIII). Binding events lost or maintained at 48h were located predominantly in MEs (tr. 5–10), promoters (tr. 1–4), as well as transient enhancers (tr. 11) (Fig 5D, MEF-only and shared sites). Conversely, new binding sites of the TFs at 48h were primarily enriched within promoters (tr. 1–4), transient enhancers (tr. 11) and tr. 13 PEs (Fig 5D, 48h-only sites). Together, these data revealed an unexpected redistribution of somatic TFs away from sites that include MEs towards new sites that include PEs.
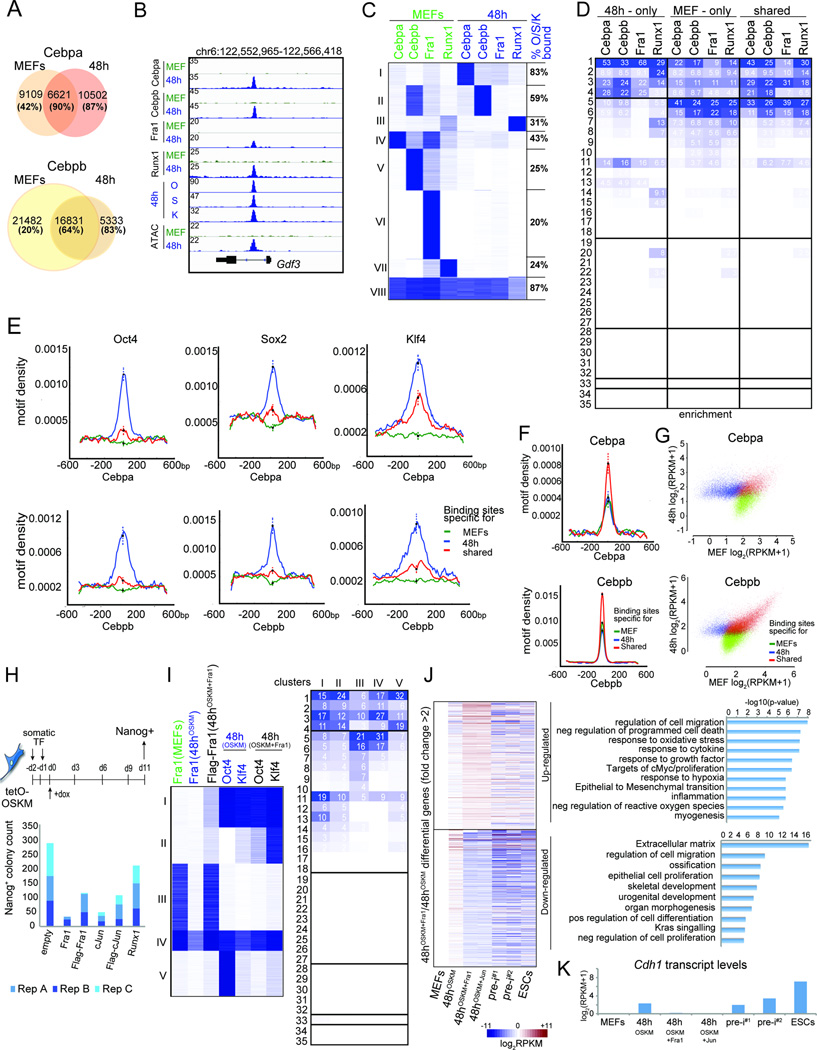
A) Intersection of Cebpa or Cebpb binding sites between MEFs and 48h. The fraction of sites also bound by O, S, or K is given in brackets for each group.
B) Genome browser view at the Gdf3 locus of OSK, somatic TF binding and ATAC-seq data in MEFs and at 48h.
C) K-means clustering of somatic TF binding events in MEFs and at 48h. The fraction of sites in each cluster also bound by O, S, or K is provided on the right.
D) Fold-enrichment of MEF-only, 48h-only, and shared binding sites of somatic TFs from (A) in chromatin trajectories defined in Fig 3A, colored per column from highest to lowest.
E) Density of O, S, or K motifs at MEF-only, 48h-only, and shared Cebpa (top) and Cebpb (bottom) sites from (A). Error bars = 95% confidence interval at summits.
F) As in (E), but for Cebpa (top) and Cebpb (bottom) motifs.
G) MEF and 48h input-normalized ChIP-seq signal for MEF-only, 48h-only, and shared binding events of Cebpa and Cebpb from (A).
H) Schematic of the reprogramming experiment with retroviral overexpression of somatic TFs. Nanog-positive colony counts from three biological replicates are shown.
I) K-means clustering of Fra1 peaks in MEFs and Fra1, O and K peaks at 48h of OSKM or OSKM+Fra1 co-expression. Right: Fold-enrichments of each cluster on the left in chromatin trajectories defined in Fig 3A, colored per column from highest to lowest.
J) Heatmap of differential gene expression between reprogramming stages indicated at the bottom and MEFs, for genes with 2-fold differentially expressed between 48hOSKM+Fra1 and 48hOSKM. Right: GO ontologies of these genes.
K) E-cadherin (Cdh1) transcript levels for indicated samples based on RNA-seq data.
See also Fig S6.
48h-specific sites of Cepba, Cebpb, and Fra1, respectively, were extensively co-occupied by O, S, or K at 48h (>80%) and had a high density of OSK motifs, whereas MEF-specific sites displayed lower reprogramming factor occupancy (<42%) and lacked OSK motifs (Fig 5A/C/E, S6B). Thus, somatic TFs relocate from MEs towards new sites that become available by binding of the reprogramming factors early in reprogramming, suggesting that OSK directly guide this process, which in turn leads to the global destabilization of MEs. In support of an inter-dependency of somatic TFs and OSK, we found that somatic TF bindings sites maintained at 48h were also targets of OSK early in reprogramming (Fig 5A/C, S6A/B) and that Cebpb co-occupied many sites with OSK in pre-iPSCs (Fig S6C).
We also noted that somatic TF binding sites maintained at 48h (shared sites) exhibited higher normalized tag counts and motif density of the respective TF than either MEF- or 48h-specific peaks (Fig 5F/G, S6D/E). These results suggested that binding events maintained early in reprogramming display higher affinity for the somatic TF compared to those lost or gained and that OSK induce the relocation of somatic TFs from one set of lower affinity binding sites to another.
Runx1 also relocated early in reprogramming but the new sites at 48h occurred often in transcribed units and did not overlap as extensively with OSK binding as Fra1 and Cebpa/b (Fig 5C/D, S6A) suggesting that a different mechanism controls the redistribution of this TF. In addition, we noticed that more sites were lost and fewer sites gained for Fra1 compared to Cebpa/b or Runx1 at 48h (Fig 5A/C, S6B, also seen in independent replicates), which raised the question of whether the level of Fra1 was altered. RNA-seq revealed limited transcriptional changes early in reprogramming (Fig S6F/G) (Koche et al., 2011) with Cebpa, Cepbb, and Runx1 transcript levels remaining largely unchanged whereas Fra1 transcript levels decreased substantially (2.7-fold) (Fig S6J/K, Table S2). Hence, repression of Fra1 appears to be an additional mechanism that contributes to the loss of somatic TFs from MEs. Loss of Fra1 binding at its own locus at 48h (Fig S6L) could enhance the down-regulation of this TF via its known auto-regulation (Verde et al., 2007). Of note, genes up-regulated early in reprogramming were enriched for 48h-specific somatic TF binding and down-regulated genes for MEF-specific somatic TF peaks (Fig S6H/I), suggesting that the redistribution of somatic TFs contributes to the few expression changes detected at 48h.
Fra1 repression is critical for somatic program silencing and reprogramming
To test if Fra1 repression is critical for ME silencing, we ectopically expressed Fra1 or a Flag-tagged version together with OSKM, which dramatically lowered the efficiency of reprogramming (Fig 5H, S6M). In comparison, Runx1 overexpression had a limited inhibitory effect on iPSC formation (Fig 5H, S6M), again hinting at differential control of reprogramming by Runx1. Ectopic expression of Flag-tagged Fra1 for 48h abrogated the loss of Fra1 from MEs that occurred early in OSKM-mediated reprogramming (Fig 5I, clusters III, IV, Fig S6L). Upon 48h overexpression, Fra1 also engaged new sites in promoters, PEs and transient enhancers that were co-occupied by O and K (Fig 5I, cluster I, S was not tested here), and induced the targeting of these reprogramming factors to new sites (Fig 5I, clusters II and V) emphasizing the co-dependency of somatic TF and reprogramming factor binding events. Fra1 overexpression also reversed expression changes observed under standard reprogramming conditions at 48h and prevented the upregulation of the epithelial signature gene E-cadherin (Fig 5J/K). These data suggested that Fra1 loss from MEs is critical for their silencing and iPSC production. Overexpression of cJun, the binding partner of Fra1, was also detrimental for reprogramming (Fig 5H, S6M) (Liu et al., 2015) and produced similar expression changes as Fra1 overexpression (Fig 5J/K), suggesting that cJun may block reprogramming in synergy with Fra1.
Step-wise PE selection is not explained by starting chromatin state
Besides ME silencing, the selection of PEs is critical for reprogramming. The temporal differences in PE engagement, with a large number of PEs targeted by OSK only late in reprogramming and others first engaged at 48h or in pre-iPSCs (Fig 3C), prompted us to ask what distinguishes temporally different reprogramming factor binding at PEs. We focused this analysis on ‘111’ and ‘001’ O binding events in intergenic PEs of tr. 13 and 17 because of their association with genes involved in stem cell-related functions and high expression in ESCs (Fig S4B/C/E).
We first analyzed enhancer-associated histone marks at ‘111’ and ‘001’ O binding events in tr. 13 PEs and ‘001’ O sites in tr. 17 PEs (Fig 6A). All sites existed in a closed chromatin conformation in MEFs lacking active histone marks and ATAC-seq signal (Fig 6A, S7A). For ‘111’ sites in tr. 13 and ‘001’ sites in tr. 17, O binding correlated with the gain of active enhancer marks H3K4me1/2 and H3K27ac and ATAC-seq signal (Fig 6A, S7A) suggesting that chromatin opening and selection of these sites is linked to reprogramming factor binding. At ‘001’ sites in tr. 13 PEs, the gain of enhancer marks and chromatin accessibility preceded O binding (Fig 6A) implying a role for non-reprogramming TFs in the opening of these sites. Regardless, the transition of PEs from ‘closed’ to ‘open’ chromatin was associated with the recruitment of Brg1 (Fig 6B).
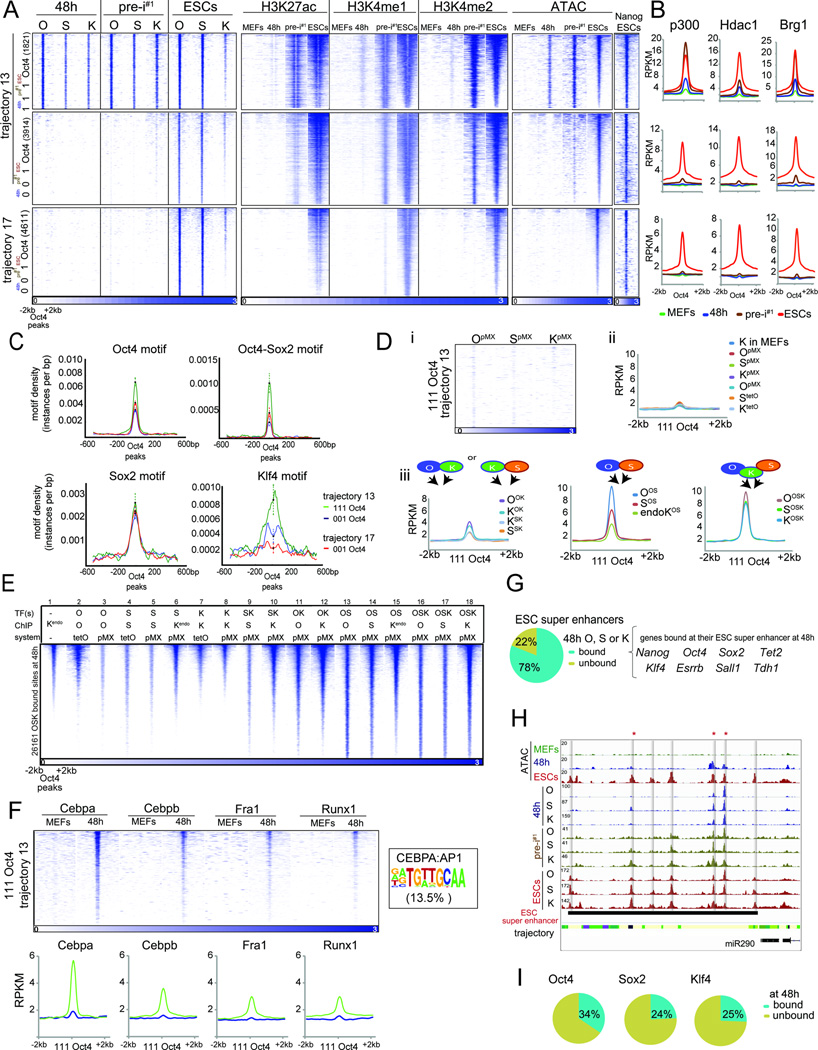
A) Heatmaps of O, S, K, H3K27ac, H3K4me1/2, and Nanog ChIP-seq signal and ATAC-seq data in indicated reprogramming stages at ‘111’ or ‘001’ Oct4 binding sites within tr. 13 and 17 PEs, sorted by ESC ATAC-seq signal intensity. Number of peaks in each set is given in brackets.
B) Metaplots of signal intensities of p300, Hdac1 and Brg1 for sites in (A).
C) Motif density for sites in (A), with 95% confidence interval at the summits.
D) (i) Heatmaps of O, S, and K ChIP-Seq signal at ‘111’ Oct4 sites in tr. 13 PEs, in MEFs individually expressing O, S, or K for 48h. (ii) Metaplots of signal intensities of the indicated reprogramming factor individually expressed (pMX or tetO) in MEFs for 48h and of Klf4 in MEFs in tr. 13 ‘111’ Oct4 sites. (iii) As in (ii), except for MEFs expressing OK, SK, OS, or OSK for 48h. Binding of endogenous K in OS-expressing MEFs is also given.
E) Heatmap of ChIP-seq signal for the factor indicated by ‘ChIP’, in MEFs ectopically expressing one or combinations of reprogramming factor(s) for 48h (‘TF’) using retroviral (pMX) or inducible (tetO) expression (‘system’), for sites co-bound by OSK at 48h of OSKM-induced reprogramming, sorted by Klf4 signal in MEFs. Kendo refers to targets of endogenously expressed K.
F) As in (D), except for binding of somatic TFs in MEFs and at 48h. The given CEBPA:AP1 composite motif was identified in 13.5% of tr. 13 ‘111’ Oct4 sites.
G) Fraction of ESC super enhancers occupied by O, S or K at 48h and associated genes.
H) Genome browser view of OSK ChIP-seq, ATAC-seq data and chromatin trajectories (color-coded as in Fig S4A) at the mir290 ESC super enhancer. Grey bars indicate seven sub-elements engaged by OSK in ESCs and the asterisks mark those bound at 48h.
I) Fraction of ESC-bound O, S or K locations within ESC super enhancers engaged by the respective TF at 48h.
See also Fig S7.
Interestingly, at ‘111’ sites in tr. 13 PEs the levels of H3K4me1/2 and H3K27ac were much lower at 48h than in pre-iPSCs and ESCs (Fig 6A, S7A). This pattern was recapitulated by p300 and Hdac1 binding (Fig 6B) demonstrating that reprogramming factor binding at 48h induced the selection of these PEs but not their full activation, which occurred at a later reprogramming stage (Fig 6A/B). One intriguing hypothesis is that reprogramming factor binding to PEs early in the process allows for the binding of additional TFs at later reprogramming stages, which is required for full enhancer activation. Regardless, these data showed that the step-wise selection and activation of PEs is largely controlled by parameters beyond chromatin state.
Motif density and OSK co-occupancy distinguish early- and late-engaged PEs
Since the chromatin state in MEFs did not distinguish early- (‘111’) and late- (‘001’) engaged PEs, we examined properties of the underlying DNA sequence. Early-bound O sites in tr. 13 PEs carried significantly more Oct4, Oct4/Sox2 composite, and Klf4 consensus motifs compared to late-bound sites in tr. 13 and 17 (Fig 6C). De novo motif scanning also revealed a stronger enrichment of the Klf4 motif in ‘111’ O sites in tr. 13 PEs compared to ‘001’ sites in tr. 17 PEs (Fig S7B). Consistent with these differences in motif occurrence, ‘111’ Oct4 sites in tr. 13 PEs were co-bound by S and K when they were first engaged at 48h, whereas tr. 13 and 17 PEs bound by O late (‘001’) were predominantly co-occupied with S but not K in ESCs (Fig 6A, S7C). These data demonstrated that OSK co-occupancy is associated with PE selection early and OS co-binding with PE engagement late in reprogramming, which is driven by motif presence. Despite co-binding by OSK at 48h, ‘111’ O sites in tr. 13 PEs were mostly bound by OS in ESCs (Fig 6A, S7C) in agreement with K binding being more distinct to O and S binding in ESCs (Fig 2E/F).
PE engagement early in reprogramming requires collaborative binding by OSK
To test mechanistically how the selection of PEs occurs early in reprogramming, we determined the independent ability of O, S, and K to engage these sites at 48h. We found that tr. 13 PEs were not targeted when O, S, and K were individually expressed (Fig 6Di,ii, S3B/C/G) indicating that the ability of these reprogramming factors to act as pioneer factors is not at play for the opening of these sites. Retroviral co-expression of combinations of reprogramming factors and mapping of binding sites at 48h further demonstrated that OSK co-expression was sufficient for the selection of tr. 13 PEs at 48h, showing that ectopic M is not essential for PE selection, and additionally revealed lower occupancy when two reprogramming factors were expressed (OS, SK, OK) compared to three (OSK) (Fig 6Diii). Though these data were consistent with PE section requiring a collaborative mode of action of OSK, one exception was that OS co-expression resulted in binding levels close to those seen with OSK co-expression, particularly for O, despite the lack of ectopic K (Fig 6Diii). This result likely can be explained by the relocation of endogenously expressed K to these sites in OS-expressing MEFs (Fig 6Diii). Thus, we conclude that the selection of PEs early in reprogramming requires the collaborative action of O, S, and K, and suggest that the necessity of OSK for reprogramming is linked to their ability to open a subset of PEs together.
We made similar observations when considering all sites that were co-occupied by O, S, and K at 48h of OSKM-induced reprogramming, only ~30% were accessible to individually expressed O, S, or K, mainly at locations representing endogenous Klf4 binding in MEFs (Fig 6E, columns 1–8). The number of accessible sites increased when double combinations of reprogramming factors were expressed, rising from SK, OK to OS (Fig 6E, columns 9–15).
We also noted that 13.5% of ‘111’ O sites in tr. 13 PEs carried the CEBPA:AP1 composite motif, and that Cebpa, and to lesser extent Cebpb, Fra1, and Runx1, occupied these PEs with OSK at 48h (Fig 6F, ,5B).5B). Additionally, Fra1 extensively engaged tr. 13 PEs early in reprogramming upon overexpression together with the reprogramming factors (Fig 5I, clusters I/II/V), supporting a link between somatic TFs and OSK at PEs. At PEs, somatic TFs may be required for their selection, prevent their activation early in reprogramming, or may simply bind due to the open chromatin character, which will need to be studied further.
Early-engaged PEs are close to core pluripotency genes
Recently, super enhancers, defined as dense clusters of enhancers with high activity, received attention as cis-regulatory elements of genes that control cell identity (Whyte et al., 2013). We found that ESC super enhancers gained enhancer marks gradually during reprogramming and that their neighboring genes were activated progressively (Fig S7D/E). ESC super enhancers were enriched most strongly in tr. 13 PEs (Fig S7F) and typically engaged by O, S, and K early in reprogramming (of 231 ESC super enhancers 78% were bound by O; 61% by S; 66% by K at 48h) including those at the mir290, Pou5f1, Sox2, Klf4, Tdh1, and Nanog loci (Fig 6G, S7G/H). Thus, critical regulatory sites of the pluripotent state are among the PEs that are selected early in reprogramming. Interestingly, only a subset of sites within ESC super enhancers bound by O, S, and K in ESCs was engaged at 48h (Fig 6H/I, S7I) suggesting that super enhancers do not act as a single entity and that the opening at specific sites early in reprogramming may be critical for full selection/activation later in the process. Notably, ESC super enhancers represented only a small fraction of all early-engaged PEs as only ~4% of the ‘111’ O sites in tr. 13 were located within them.
Esrrb enhances both ME inactivation and PE selection
The data described above argue in favor of a model where cooperative binding of TFs, including both reprogramming factors and endogenously expressed TFs, dictates their genomic targeting and thereby enhancer selection. For late-engaged PEs (‘001’ sites), we therefore hypothesized that additional TFs that become available progressively during reprogramming are required for selection either prior to or together with OS. In support of this idea, we observed that in ESCs PEs were occupied by additional TFs that become expressed later in reprogramming, such as Nanog and Esrrb (Fig 6A, Table S5). Moreover, ‘001’ O sites in PEs could be distinguished from ‘111’ sites by the presence of the Esrrb motif (Fig 7A), establishing Esrrb, which is turned on very late in reprograming (Buganim et al., 2012; Pasque et al., 2014; Polo et al., 2012) (S7J/K), as a unique candidate to test our hypothesis.
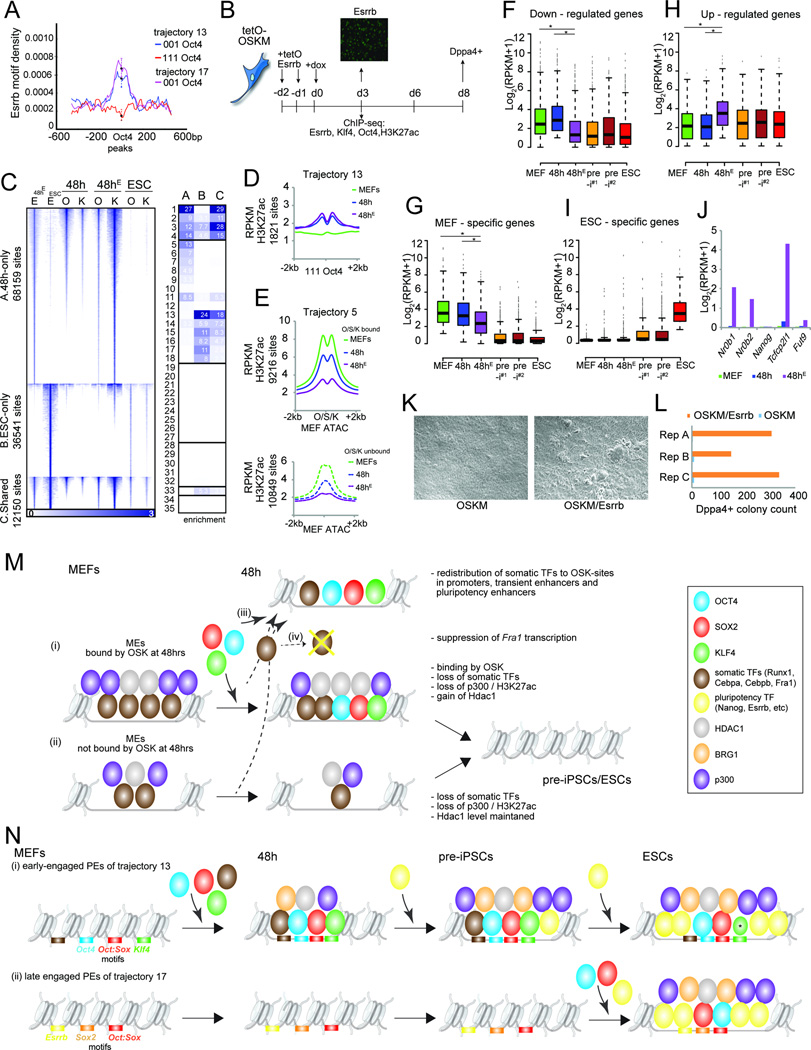
A) Esrrb motif density in ‘111’ and ‘001’ Oct4 peaks in tr. 13 and 17 PEs. Error bars = 95% confidence interval at summits.
B) Schematic of the reprogramming experiment with lentiviral overexpression of Esrrb (tetOEsrrb). Image: Esrrb expression was confirmed by immunostaining at day 3.
C) Heatmap of Esrrb (E) ChIP-seq signal for Esrrb peaks identified in ESCs and at 48h of coexpression of OSKM and Esrrb (48hE). Peaks were divided into three groups (A-C) based on their reprogramming stage specificity. The O and K signal at 48h of OSKM (48h) or OSKM/Esrrb expression (48hE) and in ESCs for the same sites are also shown. Right: Fold-enrichments of sites in sets A-C in chromatin trajectories defined in Fig 3A, colored per column from highest to lowest.
D) Metaplot of signal intensity of H3K27ac at ‘111’ Oct4 sites in tr. 13 PEs for MEFs, 48h and 48hE (OSKM/Esrrb).
E) As in (D), except for OSK-bound and unbound tr. 5 MEs, centered on ATAC-seq summits in MEFs.
F) Boxplots of expression levels of genes down-regulated at 48h of reprogramming with OSKM/Esrrb (48hE) relative to OSKM alone (48h). Asterisks mark any significant differences between MEFs, 48h, and 48hE samples (Wilcoxon test, adj. p-val<0.05).
G) As in (F), for MEF-specific genes.
H) As in (F), for up-regulated genes.
I) As in (F), for ESC-specific genes.
J) Expression of pluripotency genes known to be regulated by Esrrb.
K) Bright-field image at day 6 of reprogramming with OSKM or OSKM/Esrrb.
L) Count of DPPA4-positive colonies at day 8 of OSKM or OSKM/Esrrb expression, from three biological replicates.
M) Model for the functions of OSK at MEs.
N) Model for the functions of OSK at PEs. Asterisk indicates reduced K binding in ESCs.
See also Fig S7.
To this end, we expressed Esrrb alongside OSKM from an inducible lentivirus and profiled binding of Esrrb, O, K, and H3K27ac at 48h (48hE samples) (Fig 7B, S7K). As for OSKM, most Esrrb binding sites at 48h differed from those in ESCs (Fig 7C). 48h-specific binding occurred predominantly in promoters, MEs, and transient enhancers (Fig 7C-group A), whereas ESC-specific sites enriched in PEs (Fig 7C-group B). 25% of ESC targets of Esrrb became engaged at 48h, many of which were located in promoters and, as seen for OSK, in tr. 13 PEs (Fig 7C-group C). The sites in group C were targeted by O and K in ESCs as well as upon OSKM/Esrrb co-expression at 48h, but only a third were engaged by O and K at 48h when merely OSKM were overexpressed (Fig 7C). Similarly, 2291 sites in PEs of tr. 13–18 normally engaged by O, S, or K only late (‘001’ sites) (882 in tr. 13 and 415 in tr. 17), were targeted by O or K at 48h upon OSKM/Esrrb over-expression. Thus, PEs not accessible to the reprogramming factors early in reprogramming became accessible early when Esrrb was co-expressed. These data provide evidence for the cooperation of a pluripotency TF with OSK in the selection of PEs and highlight the need of additional pluripotency TFs for the reconstitution of the pluripotency network.
Interestingly, at 48h, tr. 13 PEs reached a similar level of H3K27ac in the presence of Esrrb as reprogramming cells not exposed to Esrrb (Fig 7D). However, the average level of H3K27ac was much lower at tr. 5 MEs at 48h upon Esrrb expression indicating even more pronounced ME silencing (Fig 7E). Consistent with this observation, MEF signature genes were more strongly repressed at 48h with Esrrb (48hE versus 48h; Fig 7F/G, S7L). With the exception of a few pluripotency genes such as Nr0b1/2, Tcfcp2l1, and Fut9, Esrrb did not induce precocious expression of ESC-specific genes at 48h, but instead induced genes involved in metabolic pathways ectopically (Fig 7H-J, S7L). Lastly, we found that the molecular changes induced by Esrrb early in reprogramming correlated with a more than 100-fold increase in the number of Dppa4+ colonies and shortened kinetics of iPSC-like colony formation (Fig 7K/L). Together, these data demonstrate a dramatic effect of the pluripotency TF Esrrb on MEF identity, binding and inactivating MEs, and, in parallel, on the induction of the pluripotency program by enhancing PE selection.
Discussion
Our study provides a comprehensive analysis of OSKM occupancy at four reprogramming stages. Among the four reprogramming factors, M is distinct as it primarily targets promoters throughout reprogramming, whereas O, S, and K favor enhancers. We found that OSK switch from somatic to pluripotency enhancers during reprogramming and, unexpectedly, orchestrate both the inactivation of MEs and PE selection. Most importantly, our work revealed that the selection of genomic target sites of OSK and the opposing effects on MEs and PEs are controlled by the combinatorial interplay of O, S, and K with endogenously expressed TFs, many with a stage-specific expression.
The extensive silencing of MEs at 48h, affecting MEs bound by OSK and those not targeted by the reprogramming factors, was a particularly surprising finding and indicated the widespread interference with the somatic epigenetic network very early in reprogramming. We determined that OSK initiate the silencing of MEs by at least three distinct mechanisms: First, OSK bind to approximately 50% of the most active MEs and increase their Hdac1 levels, potentially attributable to the interaction of Hdac1 with Oct4 (Pardo et al., 2010) (Fig 7Mi). Second, OSK induce the removal of somatic TFs from OSK-bound and unbound MEs (Fig 7Mi/ii). Mechanistically, the loss of somatic TFs from MEs is accomplished by their relocation away from MEs to new sites including PEs that become bound by OSK at 48h and carry the motifs for the reprogramming factors and the somatic TFs (Fig 7Miii). Third, OSK expression leads to a decrease in Fra1 transcript levels, which contributes to the extensive loss of this TF from MEs (Fig 7Miv).
Binding sites in MEs contained fewer consensus DNA binding motifs of each reprogramming factor than those at PEs suggesting that the interaction of OSK with MEs differs from that at PEs. Therefore, we propose that the targeting of MEs may involve non-consensus motifs that are accessible to the reprogramming factors due to the open chromatin state or protein-protein interactions with endogenously expressed factors. For instance, protein-protein interactions with Cebpa/b, Fra1, or Runx1 may contribute to the recruitment of OSK to MEs. Reciprocally, the same interactions could facilitate the redistribution of somatic TFs to new sites with OSK binding at 48h and contribute to the combinatorial binding between OSK and somatic TFs. The presence of fewer cognate OSK motifs at MEs could mediate a generally weaker binding that in turn facilitates the disengagement of OSK from these sites when somatic TFs become unavailable.
In addition to MEs, OSK engage a substantial number of PEs at 48h including sites neighboring critical pluripotency-associated genes (Fig 7Ni). However, the majority of PEs are bound only at later stages (Fig 7Nii) revealing that PEs selection is a step-wise process. The different kinetics of PE selection was not explainable by differences in the chromatin state in starting MEFs. Instead, we propose that the timing of PE selection is dictated by (i) the collaborative binding among O, S, and K, and with additional, endogenously expressed TFs, and (ii) cis-encoded properties, i.e. the presence and combination of motifs at these sites (Fig 7N). O, S, and K together, potentially with somatic TFs, are required for the opening of PEs early in reprogramming (Fig 7Ni), which perhaps explains why this combination of reprogramming factors is so successful in establishing pluripotency. At early-engaged PEs, opening by OSK does not result in strong enhancer activation at 48h, which likely requires other TFs that become available later. Conversely, late-engaged PEs are targeted by OS, without K, indicating that OS alone are not sufficient to effectively compete with nucleosomes at these sites early in reprogramming (Fig 7Nii). Here, additional TFs that only become available later, such as Esrrb, are required for their selection in concert with OS (Fig 7Nii).
Ectopic Esrrb not only influenced OSK binding at PEs, but also bound MEs and facilitated their silencing. Equally, Fra1 acted on both MEs and PEs when overexpressed. Thus stage-specific TFs, including both somatic and pluripotency TFs, influence OSK binding, ME silencing, and PE selection, reinforcing the idea of the combinatorial control of TF binding during reprogramming. These observations and the fact that targeting of PEs in closed chromatin require binding of multiple TFs indicate that the pioneer factor model proposed for human somatic cell reprogramming (Soufi et al., 2012; Soufi et al., 2015) does not act at enhancers in mouse cell reprogramming. Additional work will be required to understand the differences between reprogramming processes in different species.
STAR Methods
Contact for Reagent and Resource Sharing
Please direct any requests for further information or reagents to the lead contact, Professor Kathrin Plath (ude.alcu.tendem@htalpk), Department of Biological Chemistry, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA 90095.
Experimental Model and Subject Details
Mouse embryonic fibroblasts (MEFs) harboring the M2rtTA construct in the R26 locus (heterozygously) together with a single dox-inducible polycistronic cassette coding for OSKM in the Col1A locus (tetO-OSKM) (Ho et al., 2013; Sridharan et al., 2013) or a dox-inducible cassette encoding a single reprogramming factor (tetO-Oct4, tetO-Sox2, or tetO–Klf4) in the Col1A locus, or wildtype MEFs were used for reprogramming experiments, ChIP-seq, ATAC-seq, and RNA-seq assays. In addition, pre-iPSCs derived by retroviral overexpression of OSKM in MEFs and the mouse ESC line V6.5 were used to study different stages of reprogramming. All cell lines are described in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-Nanog | eBioscience | Cat#14-5761-80 |
| Rabbit polyclonal anti-Nanog | cosmobio | Cat#REC-RCAB001P |
| Goat polyclonal anti-DPPA4 | R&D | Cat#AF3730 |
| Goat polyclonal anti-Oct4 | RnD | Cat#AF1759 |
| Goat polyclonal anti-Sox2 | RnD | Cat#AF2018 |
| Goat polyclonal anti-Klf4 | RnD | Cat#AF3158 |
| Goat polyclonal anti-cMyc | RnD | Cat#AF3158 |
| Mouse monoclonal anti-Esrrb | RnD | Cat#H6705 |
| Rabbit polyclonal anti-H3K9ac | Abcam | Cat#ab4441 |
| Rabbit polyclonal anti-H3K4me3 | Abcam | Cat#ab8580 |
| Rabbit polyclonal anti-H3K4me2 | Abcam | Cat#ab7766 |
| Rabbit polyclonal anti-H3K4me1 | Abcam | Cat#ab8895 |
| Rabbit polyclonal anti-H3K27me3 | Active Motif | Cat#39155 |
| Rabbit polyclonal anti-H3K27ac | Abcam | Cat#ab4729 |
| Rabbit polyclonal anti-H3K36me3 | Abcam | Cat#ab9050 |
| Rabbit polyclonal anti-H3K79me2 | Active Motif | Cat#39143 |
| Rabbit polyclonal anti-H3K9me3 | Abcam | Cat#ab8898 |
| Mouse monoclonal anti-H3K9me3 | Millipore | Cat#05-1242 |
| Rabbit polyclonal anti-H3 | abcam | Cat#ab1791 |
| Mouse monoclonal anti-H3.3 | Abnova | Cat#H00003021- M01 |
| Rabbit polyclonal anti-p300 | SantaCruz | Cat#sc-585 |
| Rabbit polyclonal anti-Runx1 | Novus Biologicals | Cat#NBP1-61277 |
| Rabbit polyclonal anti-Fra1 | SantaCruz | Cat#sc-183X |
| Rabbit polyclonal anti-Cebpa | SantaCruz | Cat#sc-61X |
| Rabbit polyclonal anti-Cebpb | SantaCruz | Cat#sc-150X |
| Rabbit polyclonal anti-Hdac1 | abcam | Cat#ab7028 |
| Rabbit monoclonal anti-Brg1 | abcam | Cat#ab110641 |
| Mouse monoclonal anti-Gapdh | Fitzgerald | Cat#10R-G109A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Micrococcal nuclease | Roche | Cat#10107921001 |
| Formaldehyde | Fisher Scientific | Cat#F79-500 |
| DSG | ThermoFisher Scientific | Cat#201593 |
| Critical Commercial Assays | ||
| TruSeq ChIP Sample Prep Kit | Illumina | Cat#IP-202-1012 |
| TruSeq stranded mRNA sample preparation kit | Illumina | Cat#RS-122-2101 |
| Nextera DNA library preparation kit | Illumina | Cat#FC-121-1030 |
| RNeasy MiniElute Cleanup kit | Qiagen | Cat#74204 |
| Qiagen MinElute reaction clean up kit | Qiagen | Cat#28204 |
| Deposited Data | ||
| ChIP-seq data, RNA-seq, ATAC-seq data: | This study | GEO accession: GSE90895 |
| Experimental Models: Cell Lines | ||
| Mouse embryonic fibroblasts isolated from 129SV/Jae mice | Laboratory of K.Plath Sridharan et al., 2009 | N/A |
| Mouse embryonic fibroblasts isolated from 129SV/Jae/C57BL6J mice carrying Col1A:tetO- OSKM/wt Rosa26:M2rtTA/wt | Laboratory of K.Plath Sridharan et al., 2013 | N/A |
| Mouse embryonic fibroblasts isolated from 129SV/Jae/C57BL6J mice carrying Col1A:tetO-Oct4/wt Rosa26:M2rtTA/wt | This paper | N/A |
| Mouse embryonic fibroblasts isolated from 129SV/Jae/C57BL6J mice carrying Col1A:tetO-Sox2/wt Rosa26:M2rtTA/wt | This paper | N/A |
| Mouse embryonic fibroblasts isolated from 129SV/Jae/C57BL6J mice carrying Col1A:tetO-Klf4/wt Rosa26:M2rtTA/wt | This paper | N/A |
| Pre-iPSC line 1 (12.1) | Laboratory of K.Plath Sridharan et al., 2013 | N/A |
| Pre-iPSC line 2 (1A2) | Laboratory of K.Plath Sridharan et al., 2009 | N/A |
| Mouse embryonic stem cell line V6.5 | Laboratory of R. Jaenisch | N/A |
| PlatE cell line; 293T based | Laboratory of T. Kitamura Morita et al., 2000 | N/A |
| 293T cells | ATCC | Cat#CRL3216 |
| Experimental Models: Organisms/Strains | ||
| Mouse: 129SV/Jae/C57BL6J, Col1A: OSKMtetO/wt R26: M2rtTA/wt | Laboratory of K.Plath Sridharan et al., 2013 | N/A |
| Mouse: 129SV/Jae/C57BL6J, Col1A: OtetO/wt R26: M2rtTA/wt | This paper | N/A |
| Mouse: 129SV/Jae/C57BL6J, Col1A: StetO/wt R26: M2rtTA/wt | This paper | N/A |
| Mouse: 129SV/Jae/C57BL6J, Col1A: KtetO/wt R26: M2rtTA/wt | This paper | N/A |
| Recombinant DNA | ||
| FUW-tetO Esrrb | Buganim et al., 2012 | Addgene:#40798 |
| pMX-RUNX1 | This paper | N/A |
| pMX-Fra1 | This paper | N/A |
| pMX-Flag-Fra1 | This paper | N/A |
| pMX-cJun | This paper | N/A |
| pMX-Flag-cJun | This paper | N/A |
| Sequence-Based Reagents | ||
| siRNA for Runx1-A | Dharmacon | Cat#D-048982-01 |
| siRNA for Runx1-B | Dharmacon | Cat#D-048982-03 |
| siRNA for Cebpa-A | Dharmacon | Cat#D-040561-03 |
| siRNA for Cebpa-B | Dharmacon | Cat#D-040561-04 |
| siRNA for Cebpb-A | Dharmacon | Cat#D-043110-06 |
| siRNA for Cebpb-B | Dharmacon | Cat#D-043110-22 |
| siRNA for Luciferase | Dharmacon | Cat#D-001210-02 |
| Software and Algorithms | ||
| ChromHMM v1.1.0 | Ernst and Kellis, 2012 | http://compbio.mit.edu/ChromHMM/ |
| ChromImpute v1.0.0 | Ernst and Kellis, 2015 | http://www.biolchem.ucla.edu/labs/ernst/ChromImpute/ |
| DESeq2 | Love et al., 2014 | https://bioc.ism.ac.jp/packages/3.1/bioc/html/DESeq2.html |
| Metascape | Tripathi et al., 2015 | http://metascape.org/gp/index.html#/main/step1 |
| GREAT | McLean et al., 2010 | http://bejerano.stanford.edu/great/public/html/ |
| Bowtiev2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Tophat | Trapnell et al., 2009 | https://ccb.jhu.edu/software/tophat/index.shtml |
| MACS2 2.1.0 | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| Other | ||
Method Details
Cell lines, culture conditions, and reprogramming experiments
The following cell lines were used for the comprehensive genomics analysis of the reprogramming process at discrete stages: primary MEFs harboring a heterozygous R26- M2rtTA allele and a single dox-inducible polycistronic cassette coding for OSKM in the Col1A locus (tetO-OSKM) (Ho et al., 2013; Sridharan et al., 2013), derived from day 14.5 embryos of timed mouse pregnancies; two independently generated male pre-iPSC lines (line 12–1 (pre-i#1) and 1A2 (pre-i#2) obtained upon retroviral (pMX-based) expression of Oct4, Sox2, Klf4, and cMyc in Nanog-GFP reporter MEFs (Sridharan et al., 2013; Sridharan et al., 2009); and the male ESC line V6.5 from the Jaenisch laboratory. All cell types were grown in standard mouse ESC media containing KO DMEM, 15% fetal bovine serum (FBS), recombinant leukemia inhibitory factor (Lif), β-mercaptoethanol, 1× penicillin/streptomycin, L-glutamine, and non-essential amino acids. Pre-iPSCs and ESCs were grown on irradiated MEFs (feeders), but feeder-depleted and grown overnight on gelatin for genomics experiment. For the 48h reprogramming time point, tetO-OSKM MEFs were cultured in ESC media containing 2µg/ml doxycycline for 48 hours to induce the expression of OSKM. For all ChIP-seq, ATAC-seq and RNA-seq experiments, cells were grown in ESC medium.
For single reprogramming factor overexpression ChIP-seq experiments (Figures 2G, 4J/K, 6D/E, S3, and S5H), MEFs containing a dox-inducible cassette encoding a single reprogramming factor (tetO-Oct4, tetO-Sox2, or tetO–Klf4) in the Col1A locus and the tet-transactivator M2rtTA in the R26 locus (heterozygous) were generated as described (Beard et al., 2006) by targeting V6.5 ESCs carrying a FRT site in the Col1A locus, generating chimeric mice upon blastocyst injection, and breeding for germline transmission. These MEFs were induced with 2ug/ml doxycycline for 48h to assess the binding events of individually expressed reprogramming factors. Alternatively, wild-type 129SVJae MEFs were infected with a pMX retrovirus encoding an individual reprogramming factor (either pMX-Oct4, pMX-Sox2, or pMX-Klf4) for single factor overexpression, or with a combination of reprogramming factor baring retroviruses for double or triple reprogramming factor combinations (OK, SK, OS, or OSK). Briefly, the cDNAs of the three factors (Oct4, Sox2 or Klf4) were cloned into the pMX retroviral vectors and individually transfected into PlatE packaging cells (Maherali et al., 2007). Viral supernatants were harvested 48 hours post-infection and used to infect MEFs twice, for 8hrs continuously in the presence of 10 mg/ml polybrene. MEFs were harvested for genomics analyses 48 hours post infection.
The role of somatic TFs in the reprogramming process was tested via overexpression by infecting tetO-OSKM MEFs with pMX-retroviruses encoding the Fra1, cJun, or Runx1 cDNA. N-terminally Flag-tagged versions of cJun and Fra1 were also cloned into pMX vectors and tested for reprogramming efficiency. Viral supernatants were produced in 293T cells as described above. Subsequently, tetO-OSKM MEFs were infected twice for a span of 8 hours each time, followed by dox-induction of OSKM expression. For Esrrb overexpression, a lentiviral construct encoding the tet-inducible Essrb cDNA, obtained from (Buganim et al., 2014), was transfected alongside viral packaging vectors (pMDLg, pRSV-REV, pCMV-VSVG) into 293T cells using the CalPhos mammalian transfection kit (Clontech 062013) as per manufacturer’s instructions. Lentiviral production was performed for 48h, and the harvested supernatant used to infect tetOOSKM MEFs containing M2rtTA twice. To initiate reprogramming and Esrrb expression, the cells ESC medium containing 2mg/ml doxycycline was added. The viral packaging vectors were a generous gift from Dr Zack laboratory in UCLA.
For the Runx1, Cebpa and Cebpb siRNA experiments, a set of four different siRNAs was purchased from Dharmacon and initially transfected into MEFs using lipofectamine–RNAi max (Life Technologies) according to manufacturer’s instructions to assess knockdown efficiency. Of the four siRNAs, the two producing the most efficient knockdown were used in reprogramming experiments at a final concentration of 20uM. For Runx1 these were D-048982-01 and D- 048982-03, for Cebpa D-040561-03 and D-040561-04, and for Cebpb D-043110-06 and D- 043110-22. For control siRNA treatment, we used the non-targeting Luciferase control (D-001210-02). siRNAs against Runx1 were transfected into tetO-OSKM MEFs two times: first 12 hours before reprogramming was started by doxycycline addition, and second at the time of doxycycline addition, to induce depletion of Runx1 only early in reprogramming. siRNAs against Cebpa/b were first transfected 12 hours before reprogramming was started by doxycycline addition and were re-transfected every three days to maintain knockdown throughout reprogramming.
In all experiments that assessed reprogramming efficiency, reprogramming cultures were shifted to reprogramming media, which is similar to ESC medium but contains 15% KSR instead of FBS, at day 3 of reprogramming. Reprogramming efficiency was scored by counting Nanog-positive colonies after immunostaining cultures with an anti-Nanog antibody (eBioscience 14-5761-80), 11 days post doxycycline induction. For the Runx1 siRNA experiment, doxycyline was withdrawn from the cultures at day 9, for the last 48 hours, before fixation of the reprogramming cultures at day 11. For OSKM/Esrrb-induced reprogramming cultures reprogramming efficiency was calculated by counting DPPA4-positive colonies after immunostaining with an antibody directed against DPPA4 (R&D AF3730), 8 days post-OSKM/E induction with doxycycline. We have shown previously that DPPA4 is induced after Nanog expression during the final steps of reprogramming (Pasque et al., 2014).
Immunofluorescence
Cells were grown on coverslips pretreated with 0.3% porcine gelatin (Sigma G2500) in ESC medium for 48h. After fixation with 4% paraformaldehyde the cells were washed with 1×PBS- 0.05% Tween, permeabilized with 1×PBS-0.5% Triton-X, and blocked with 5% donkey serum in 1×PBS-0.05% Tween. Primary antibody incubation was carried out at 4°C overnight, secondary antibody incubation was carried out at RT for 30min, each in blocking buffer. Between each incubation, cells were washed with 1×PBS-0.05% Tween for three times. Cells were then mounted using a mounting medium with DAPI (Vector Labs H-1200). Antibodies used for Nanog and DPPA4 to detect reprogrammed colonies are listed above. Antibodies for the detection of O,S,K,M or Esrrb were: anti-Oct4 (RnD; AF1759), anti-Sox2 (RnD AF2018), anti-Klf4 (RnD; AF3158), anti-cMyc (RnD; AF3158) and anti-Esrrb (RnD; H6705).
Native ChIP-seq (N-ChIP)
Native ChIP-seq was performed for as described in (Wagschal et al., 2007) for all histone modification except H3K79me2 and H3K9me3. Briefly, 50×106 Nuclei were isolated from noncrosslinked cells (MEFs, 48h, pre-i#1 and ESC) by incubation in 2 ml of a hypotonic solution (0.3M sucrose, 60mM KCl, 15mM NaCl, 5mM MgCl2, 15mM Tris-HCl pH 7.5, 0.5mM DTT, 0.1% NP40, and protease inhibitor cocktail) followed by centrifugation through a sucrose cushion (1.2M sucrose, 60mM KCL, 15mM NaCl, 5mM MgCl2, 0.1mM EGTA, 15 mM Tris-HCl pH 7.5, 0.5mM DTT, and protease inhibitor cocktail). Nuclei were then re-suspended in MNAse-digestion buffer (0.32M sucrose, 50mM Tris-HCl pH 7.5, 4mM MgCl2, 1mM CaCl2, and protease inhibitor cocktail) and digested with 3 units of MNase (Roche 10107921001) for 10 minutes at 37°C. The first soluble fraction (S1) was recovered by centrifugation for 10 min at 10,000 rpm. The pellet containing nuclei was then dialyzed overnight in 1l of dialysis buffer (1mM Tris-HCl pH7.5, 0.2mM EDTA, protease inhibitors) to more completely release the chromatin fraction (S2) from nuclei. 10 ug of soluble chromatin (S1 and S2) were then incubated with 5 ug of antibody targeting histone modifications-conjugated to magnetic beads (Active Motif; 53014) under constant stirring at 4°C for 16 hrs. The antibodies used were: anti-H3K9ac (Abcam; ab4441), anti-H3K4me3 (Abcam; ab8580), anti-H3K4me2 (Abcam ab7766), anti-H3K4me1 (Abcam; ab8895), anti-H3K27me3 (Active Motif; 39155), antiH3K27ac (Abcam; ab4729), and anti-H3K36me3 (Abcam; ab9050). Beads were washed twice with wash buffer A (50mM Tris-HCl pH 7.5, 10mM EDTA, 75mM NaCl), wash buffer B (50mM Tris-HCl pH 7.5, 10mM EDTA, 125mM NaCl), and wash buffer C (50mM Tris-HCl pH 7.5, 10mM EDTA, 175 mM NaCl). DNA was extracted using phenol:chloroform:iso-amylacohol and used for downstream library construction. DNA from fractions S1 and S2 was also isolated directly using phenol:chloroform:iso-amylacohol extraction and used as an whole genome input control (native Input). All protocols for Illumina/Solexa sequencing library preparation, sequencing, and quality control were performed as recommended by Illumina, with the minor modification of limiting the PCR amplification step to 10 cycles. All constructed libraries were sequenced using single-end 50 bp reactions.
Cross-linked ChIP-seq (X-ChIP)
Transcription factor and epigenetic regulator occupancy data generated in this study were acquired using ChIP after crosslinking cells (X-Chip). X-ChIP was also employed for mapping H3K79me2 (Active Motif, 39143), H3K9me3 (abcam, ab8898 or Millipore, 05-1242), H3 (abcam, ab1791) and H3.3 (Abnova, H00003021-M01). Briefly, cells were grown to a final concentration of 5×107 cells for each ChIP-seq experiment. To stabilize HATs/HDACs (p300, Hdac1) and Brg1 on chromatin, cells were treated with 2 mM disuccinimidyl glutarate (DSG) for 10 minutes prior to formaldehyde crosslinking. For all other targets, cells were chemically cross-linked at room temperature by the addition of formaldehyde to 1% final concentration for 10 minutes and quenched with 0.125 M final concentration glycine. Cross-linked cells were re-suspended in sonication buffer (50mM Hepes-KOH pH 7.5, 140mM NaCl, 1mM EDTA, 1% TritonX-100, 0.1% Na-deoxycholate, 0.1% SDS) and sonicated using a Diagenode Bioruptor for three 10-minute rounds using pulsing settings (30 sec ON; 1 min OFF). 10 ug of sonicated chromatin was then incubated overnight at 4°C with 5 ug of antibody conjugated to magnetic beads. The antibodies used were: anti-Esrrb (RnD; H6705), anti-Klf4 (RnD; AF3158), anti-cMyc (RnD; AF3696), anti- Nanog (cosmobio REC-RCAB001P), anti-Oct4 (RnD; AF1759), anti-Sox2 (RnD AF2018), antip300 (SantaCruz;sc-585), anti-Runx1 (Novus Biologicals NBP1-61277), anti-Fra1 (SantaCruz;sc-183X), anti-Cebpa (SantaCruz; sc-61X), anti-Cebpb (SantaCruz;sc-150X), anti- Hdac1(abcam; ab7028) and anti-Brg1(abcam; ab110641). Following the IP, beads were washed twice with RIPA buffer (50mM Tris-HCl pH8, 150 mM NaCl, 2mM EDTA, 1% NP-40, 0.1% Na-deocycholate, 0.1% SDS), low salt buffer (20mM Tris pH 8.1, 150mM NaCl, 2mM EDTA, 1% Triton X-100, 0.1% SDS), high salt buffer (20mM Tris pH 8.1, 500mM NaCl, 2mM EDTA, 1% Triton X-100, 0.1% SDS), LiCl buffer (10mM Tris pH 8.1, 250mM LiCl, 1mM EDTA, 1% Nadeoxycholate, 1% NP-40), and 1×TE. Finally, DNA was extracted by reverse crosslinking at 60°C overnight with proteinase K (20ug/ul) and 1% SDS followed by phenol:chloroform:isoamylacohol purification. Libraries were constructed as indicated above and sequenced using single-end 50 bp reactions.
ATAC-seq library construction and sequencing
ATAC-seq was done as previously described (Buenrostro et al., 2013). Briefly, 50000 cells (129SVJae MEFs, un-induced tetO-OSKM MEFs, 48hOSKM, pre-i#1, pre-i#2 or ESCs) were resuspended in 50µl lysis buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40, and 1 × Complete Protease inhibitor (Roche)) and spun at 500g for 10 min at 4 °C to collect nuclei. Nuclei were washed in 1× PBS and subsequently re-suspended in 50 µl Transposase reaction (25 µl 2 × tagmentation buffer, 22.5 µl water, 2.5 µl Tn5 Transposase, following instructions by Illumina). Reactions were incubated for 30 min at 37 °C and DNA purified using Qiagen MinElute columns (Qiagen). The transposed DNA was subsequently amplified with custom primers as described (Buenrostro et al., 2013) for 7–9 cycles and libraries were visualized on a 2% TBE gel prior to sequencing with a single-end-sequencing length of 50 nucleotides.
RNA-seq
RNA from independent biological replicates of each un-induced MEFs, induced MEFs at 48hrs, pre-iPSCs (pre-i#1& pre-i#2 ) and ESCs, was isolated using the RNeasy Mini kit. RNA was treated on column with 0.5 kunitz units of DNAse prior to elution according to manufacturers instructions. RNA from MEF cultures induced for 48h to express OSKM/Esrrb, OSKM/Fra1, OSKM/cJun or a single reprogramming factor (tetO-Oct4, tetO-Sox2, or tetO–Klf4, tetO–Myc) was also isolated. In all cases, messenger RNA was captured using oligodT Dynabeads (Life Technologies). Strand-specific RNA-seq libraries were constructed as described in (Parkhomchuk et al., 2009).
Quantification and Statistical Analysis
Data Analysis and Visualization
Reads from ChIP-seq experiments were mapped to the mouse genome (mm9) using Bowtie software (Langmead et al., 2009) and only those reads that aligned to a unique position with no more than two sequence mismatches were retained for further analysis. Multiple reads mapping to the exact same location and strand in the genome were collapsed to a single read to account for clonal amplification effects. For ChIP-seq of TFs and ATAC-seq, peaks were called using MACS2 software (Zhang et al., 2008) using a bandwidth parameter of 150bp. Peaks with q-val cut-off < 0.005 and fold >= 4-fold were retained. Identified peak locations can be found in Table S1.
Reads from RNA-seq experiments were mapped to the mouse genome (mm9) using TopHat software (Trapnell et al., 2009) and only those reads that aligned with no more than two sequence mismatches were retained. Replicates were merged and RPKM values of mm9 RefSeq genes were calculated as described (Mortazavi et al., 2008) (Table S2). Prior to log2 transformation of RPKM values, a pseudo-count of 1 was added to all RPKM values (log2(RPKM+1).
Genome signal tracks of features (TFs, histone marks, ATAC-seq and RNA-seq) were calculated by partitioning the genome into non-overlapping bins of fixed size (100b for TFs, ATAC-seq and RNA-seq, and 25bp for the histone marks). RPKM values were calculated for each bin using the number of sequencing reads that overlap with the corresponding bin. For histone marks, each read was extended by 200 bp in the direction of the alignment. Tracks were visualized in the IGV genome browser (Thorvaldsdottir et al., 2013).
To produce the heatmaps, in Figures 2C/G, S2D/E, S3A/D/F, 4A/E, 6A/D/E/F, S6C, and 7C, we aligned the given feature (such as peaks of a TF) at their summit and tiled the flanking up- and downstream regions within +/−2kb in 100bp bins. For each location, we calculated RPKM values over all 100bp bins by using the number of sequencing reads that overlap each bin after extension by 200bp in the direction of the alignment. To control for input, we computed at each bin a log2 input-normalized RPKM value as log2(RPKMFOREGROUND) - log2(RPKMInput), where RPKMFOREGROUND denotes the RPKM of the corresponding TF, ATAC or histone data set and RPKMInput denotes the RPKM value of the corresponding whole genome ‘Input’. For visualization in figures, each 100 bp bin was displayed with JavaTreeview (Eisen et al., 1998). All metaplots were produced by computing the average input-normalized RPKM value for each 100bp bin across all locations in the given set.
The scatter plots in Figures 5G and S6D/E were produced by first computing log2(RPKM+1) values over 200bp windows centered at each binding site for the TF signal in MEFs and 48h. To control for the input, we computed log2(RPKM+1) for the input signal in MEFs at each 200bp window and subtracted it from the values in MEFs and 48h to obtain an input-normalized log2 RPKM value for each cell type: log2(RPKMTF in X+1) - log2(RPKMMEF_Input+1), where RPKMTF in X is the RPKM value in MEF or 48h.
Figures S7D and S7I were generated with ngs.plot (Shen et al., 2014).
ChIP-seq and ATAC-seq data validation
Several external (published) data sets were used to validate our ChIP-seq data (Table S3). Moreover, the majority of ChIP-seq data sets in this study were generated in biological replicates (Table S3), and the correlation of replicate data sets demonstrated a high reproducibility of our data. Furthermore, to ensure that un-induced (starting) tetO-OSKM MEFs were not already representing a ‘leaky’ expression state for the reprogramming factors (already partially reprogrammed), we also profiled wildtype MEFs not carrying any reprogramming factor transgene for ATAC-seq. These ATAC-seq data sets correlated most closely with those of the un-induced tetO-OSKM MEFs.
Correlation of our data sets with imputated data sets
We created an imputed version of the H3, H3.3, H3K27ac, H3K27me3, H3K36me3, H3K4me1, H3K4me2, H3K4me3, H3K79me2, H3K9ac, H3K9me3, p300, ATAC-seq and INPUT (Native Input) data for MEFs, 48h, pre-i#1, and ESCs, using ChromImpute v1.0.0 (Ernst and Kellis, 2015). In creating the imputed version of a data set, we used all other data sets but the data set being imputed. The imputed version of each data set can be viewed as a pseudo-replicate for each data set and can be used to assess reproducibility. The data put into ChromImpute were the RPKM normalized signals at 25bp resolution after removing reads that map to blacklisted regions in the mouse genome (https://sites.google.com/site/anshulkundaje/projects/blacklists, ENCODE Project Consortium, 2012) and excluding chrM. The signal files for all histone marks, H3, H3.3, and INPUT were generated by extending reads by 200bp in the direction of the alignment. The signal for p300 and ATAC-seq was generated without extension of the reads. ChromImpute was run with default options except the flag ‘-b 20 -tieglobal’ was added to the GenerateTrainData command, the flag ‘-b 20’ to the Train command, and the flag ‘-b 20 -tieglobal’ to the Apply command. The imputed data were converted to a 1000bp resolution by averaging the signal for each 25bp within it. Signal tracks for the observed data were produced at 1000bp resolution in the same way as the signal at 25bp used as input for ChromImpute. Pairwise Pearson correlations were then computed based on the 1000-bp resolution data (Table S3). For each observed data set, we also reported the maximum correlation with any of the other three observed data sets (for the other reprogramming stages) for the mark based on the 1000-bp resolution data (Table S3).
Differential gene expression analysis
HTSeq (Anders et al., 2015) was used to determine gene counts from replicate experiments, and DESeq2 (Anders and Huber, 2010) for differential analysis. Our quadruplicate data sets were used to identify differential genes between MEFs and OSKM-induced MEFs at 48h (48h), using an adjusted p-val < 0.05. DESeq2 was also used to identify differential genes from the following comparisons 1) OSKM-induced MEFs at 48h against and OSKM/Esrrb-induced MEFs at 48h; 2) OSKM-induced MEFs at 48h against and OSKM/Fra1-induced MEFs at 48h; and 3) OSKM-induced MEFs at 48h against and OSKM/cJun-induced MEFs at 48h. In addition, genes were called MEF- or ESC- specific using the following criteria: 1) DESeq2 differential calls with an adjusted p-val < 0.05 between ESCs and MEFs; 2) Fold-change of >=5× between transcript levels in MEFs and ESCs; 3) low RPKM value of in the non-expressing type (typically <1 RPKM).
Defining combinatorial OSKM binding groups per reprogramming stage
We generated sets of sites co-bound by the reprogramming factors at a given reprogramming stage by extending TF summits produced by MACS2 by 100 bp in each direction and intersecting the extended summits between Oct4, Sox2, Klf4, and cMyc per reprogramming stage (Figure 2F). In this case, we first defined sites bound by all four TFs by intersecting the extended summits of all four factors in any possible order and merging overlapping intersections. Analogously, we defined triply bound sites, and, subsequently, removed those regions that overlapped with the quadruply bound sites from them. Next, we defined doubly bound sites by intersecting the extended summits of every pair of TFs and removing regions that overlapped triply and quadruply bound sites. Finally, we defined solo bound sites as all sites that were not doubly, triply, or quadruply bound. To calculate the enrichment scores of the co-bound groups in Figure 2Fii, we used the middle point between the start and the end coordinates of quadruply-, triply-, and doubly- bound sites. For solo sites, the coordinates of the original summits were used.
Determination of temporal OSKM binding groups
Seven co-binding groups (Figure 2D: ‘100’, ‘010’, ‘001’, ‘110’, ‘011’, ‘101’, ‘111’) were generated in a similar manner as the combinatorial OSKM binding groups described above, by intersecting the extended TF summits (100bp) of a given TF among the three reprogramming stages: 48h, pre-i#1, and ESCs.
Transcription factor clusters
K-means clustering was employed to identify coherent groups of TF binding in Figures S2J, 5C and 5I. To define these TF clusters, the genome was tiled into 500bp windows and the presence of TF peaks in each bin was determined. This procedure resulted in a vector of binary data for each TF reflecting its absence or presence within 500bp windows across the genome. The windows represented by these vectors were then clustered using R’s k-means function applying the Hartigan-Wong method to obtain groups of windows exhibiting common combinatorial binding patterns across the genome. The number of clusters was chosen to reduce the number of potential combinatorial TF groups, while ensuring that each cluster was represented by a significant number of windows.
Ontology Annotation
To associate transcription factor peaks with the closest gene for Ontology analysis (Figures S2G, S4C/E; Table S4) we used the GREAT tool (McLean et al., 2010) with default parameters. Differentially-regulated genes defined by DESeq2 (Figures 5J, S6FG and S7L) were assigned to relevant GO ontology groups using the Metascape software (Tripathi et al., 2015).
ChromHMM modeling parameters
To derive chromatin state segmentations for each reprogramming stage (Figure 1C), we used ChromHMM (version V1.1.0) (Ernst and Kellis, 2012) with default parameters. First, we binarized the mapped reads for all chromatin marks and the native ‘Input’ indicated in Figure 1C with the ChromHMM’s BinarizeBed procedure, using a p-value cutoff of 1e-4. To reduce effects of artifacts, we removed redundancy in the input data by keeping only one sequencing read in cases where multiple reads mapped to the same genomic position and strand orientation. We examined models with different numbers of states ranging from 2 to 30 and chose a model with 18 chromatin states that is both interpretable and able to capture the combinatorial complexity of chromatin marks in each reprogramming stage.
In addition to the 18 chromatin state model shown in Figure 1C, which captures chromatin states per reprogramming stage, we used ChromHMM in the “stacked” mode to capture the chromatin changes between MEFs, 48h, pre-iPSCs (pre-i#1), and the pluripotent state, which yielded the 35 chromatin trajectories defined in Figure 3A. In particular, we constructed a single virtual cell type that has all datasets from MEFs, 48h, pre-i#1, and ESCs as individual marks by setting the label of each original dataset in the input file for ChromHMM to contain both the source cell type and the histone mark name. Then, we used ChromHMM to discover and annotate the genome for chromatin states in the virtual cell type. The rest of the preprocessing and ChromHMM parameters were the same as for the 18 state model described above. We considered models with different numbers of states ranging from 25 to 100 and chose 35 states, because it was the model with the minimum number of states that captured unique biological events. We termed the resulting 35 chromatin states (Figure 3A) “chromatin trajectories” to distinguish them from the chromatin states specific to each reprogramming stage (Figure 1C).
TF enrichment in the vicinity of differentially expressed genes early in reprogramming (Figures S6H/I)
OSKM binding combinations and groups of MEF-only, 48h-only, and shared somatic TF binding events were intersected with the genomic intervals encompassing TSS+/− 20kb regions of differentially expressed genes at 48h. To compute the fraction of bound up-regulated genes for each type of TF set, we counted the number of up-regulated genes between MEFs and 48h that have at least one such binding event within 20kb of their TSS and divided this number by the total number of up-regulated genes between MEFs and 48h. Analogously, we computed the fraction of bound down-regulated genes between MEF and 48h for each TF combination. We then divided the two fractions and plotted the ratio on log2 scale. The statistical significance of each log2 ratio was assessed by a chi-squared test that compares the number of TF bound genes between the two groups given the total number of genes in each group.
Positional expression plots (Figures S1H, S4B)
For each chromatin state, we calculated average gene expression levels in MEFs, 48h, pre-i#1, and ESCs, conditioned on the state’s distance from annotated transcription start sites. We restricted this analysis to 50kb up- or downstream of transcriptional start sites (TSS). We partitioned this region into non-overlapping bins of 200 bp. For each bin, we computed the average log2 (RPKM+1) value of genes that have a particular chromatin state at this distance relative to their TSS.
Calculations of fold-enrichment
Using the ChromHMM OverlapEnrichment function (Ernst and Kellis, 2012), we calculated enrichment scores for genomic features (TF binding events, conserved elements, repeats, exon, gene-bodies, TSS, TES, ESC super enhancers, etc) in the chromatin state of each corresponding reprogramming stage (18-state chromatin model) and for the 35 chromatin trajectories capturing the chromatin changes during reprogramming, respectively. The enrichment scores were calculated as the ratio between the observed and the expected overlap for each feature and chromatin state based on their sizes and the size of the mouse genome:
- where F is the number of base pairs annotated for the feature F, S is the size of chromatin state S and G is the total length of the mouse genome.
To calculate log2 differential enrichments in Figure 2Fii, we used the following formula:
- where each enrichment is calculated based on a binomial background model that treats the corresponding TFs as independent in each cell type (48h and ESCs).
Coordinates for TSS, TES, CpG islands, Exon and Gene Body features used were part of the mm9 annotation included in the ChromHMM software (Ernst and Kellis, 2012). For the calculation of enrichments of conserved genomic regions in Figures 1D and S1G, we downloaded the 30-way Euarch phastCons elements from the UCSC genome browser for the mm9 genome that represent 30 vertebrate species (euarchontoglires) including human and mouse (Siepel et al., 2005).
In Figures 2E and S2I, we applied complete linkage hierarchical clustering with optimal leaf ordering to cluster the enrichments of all pairs of TFs (Bar-Joseph et al., 2001). The pairwise enrichments at base-pair resolution were calculated as the observed overlap divided by the expected overlap based on the binomial background model that treats both transcription factors as independent:
- where the numerator is the size of the overlap between peaks of TFA and TFB and the denominator is the product between the total number of bp occupied by peaks of TFA and TFB divided by the size of the genome (G). A pseudo-count of 100 was added to both the numerator and the denominator to avoid instabilities due to division of small numbers and the maximum enrichment was set to 500.
In Figure 3Bii, the fold-enrichment of the 35 chromatin trajectories in the vicinity of MEF-and ESC-specific genes was calculated with the following formula:
- where the numerator is the percentage of genes (MEF- or ESC- specific; as described above and Table S2) carrying each trajectory i within 20kb from their TSS. As control, we divided by the percentage of all active genes in the same cell type (>1 RPKM) carrying that trajectory.
Assigning peaks to TSS (+/−2Kb) regions
We computed the proportion of transcription factor binding summits of Oct4, Sox2, Klf4, and cMyc that are located within 2 kb of annotated transcription start sites (mm9 RefSeq TSS) and the rest (distal). P-values in Figures 2A and S2A were calculated based on an exact two-sided Binomial test of the null hypothesis that the probability of TSS in one of the samples is given by the frequency in the other sample.
Motif analyses
We calculated motif densities at 10 bp resolution within 500 bp around ChIP-seq summits by using the annotatePeaks procedure from HOMER (Heinz et al., 2010) with the following command line arguments:
We used the positional weight matrices for the corresponding transcription factor binding motifs provided by HOMER with their default thresholds.
When we scanned regions that were bound by multiple transcription factors, we centered each region at the summit of the corresponding TF. For example, regions co-bound by OSKM were centered at the corresponding Oct4 summits when we scanned them for the Oct4 motif, then centered at the Sox2 summits for the Sox2 motif, then centered at the Klf4 summits for Klf4 motif, and, finally, centered at the cMyc summits for the cMyc motif.
We subsequently smoothed the motif densities by applying a box kernel of length 5 bins centered at each bin. To calculate confidence intervals at the summit bin, we generated 1000 bootstrap samples within each group and calculated the 95% percentile bootstrap confidence intervals (Efron and Tibshirani, 1991).
For de novo motif discovery we used the findMotifsGenome.pl procedure from Homer using the following arguments
Data and Software availability
The data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession number GEO: GSE90895.
Peak locations derived from Chip-seq and ATAC-seq experiments are given in Table S1, and normalized expression measurements based on RNA-seq are given in Table S2.
Supplementary Material
1
Table S1. ChIP-Seq peak locations for TFs and epigenetic regulators and ATAC-seq peak locations produced in this study (related to Figure 1)This table contains genomic coordinates for peaks (based on the mm9 genome-build) of:
Klf4, cMyc, p300, Hdac1, Brg1, Cebpa, Cebpb, Fra1, and Runx1 in MEFs (sheet 1, MEFpeaks).
Klf4, Oct4, p300, Sox2, cMyc, Hdac1, Brg1, Cebpa, Cebpb, Fra1, Runx1 at 48h of OSKM-induced reprogramming (48h_OSKM) as well as the peak coordinates for Esrrb, Oct4, and Klf4 at 48h of reprogramming with OSKM/Esrrb overexpression (48h_OSKMEsrrb), and the peak coordinates for Fra1(FlagFra1), Oct4, and Klf4 at 48h of reprogramming with OSKM/Fra1 overexpression (sheet 2, 48h_peaks).
Klf4, Oct4, p300, Sox2, cMyc, Hdac1, Brg1, and Cebpb for pre-i#1 and Klf4, Oct4, Sox2, and cMyc for pre-i#2 (sheet 3, pre-iPSC_peaks). (iv) Esrrb, Klf4, Nanog, Oct4, p300, Sox2, cMyc, Hdac1, and Brg1 in ESCs (sheet 4, ESC_peaks).
single, double and triple combinations of the reprogramming factors expressed retrovirally in MEFs for 48hrs (nomenclature: pMX_O_Oct4 = pMX (retroviral), O (only Oct4 overexpressed), Oct4 (peaks for Oct4)
ATAC-seq peaks in MEFs, 48h, pre-i#1, pre-i#2, and ESCs (sheet 6, ATAC-seq).
7
Figure S2. Additional characterization of OSKM binding sites at each reprogramming stage and OSKM redistribution during reprogramming (related to Figure 2)(A) Percentage of O, S, K, and M binding events in promoter-proximal (TSS +/− 2Kb) and distal genomic locations for pre-i#2. This figure accompanies Figure 2A.
(B) Percentage of O, S, K, and M binding events in each of the 18 chromatin states from Figure 1C, per reprogramming stage. Specifically, peaks of O, S, K, and M, respectively, in MEFs were analyzed with respect to the chromatin state in MEFs, 48h peaks to the chromatin state at 48h, pre-i#1 peaks against the chromatin state in these cells, and ESC targets to ESC chromatin state. This figure accompanies Figure 2B that shows the fold-enrichment for the same data.
(C) Fold-enrichment of OSKM co-binding groups defined in Figure 2Fi per chromatin state as defined in Figure 1C, for each reprogramming stage. Specifically, co-binding events of O, S, M, and K, respectively, at 48h were analyzed with respect to the chromatin state at 48h, those in pre-i#1 to the chromatin state in pre-i#1, etc.
(D) Heatmap of normalized tag densities (log2RPKM) for O, S, K, and M binding events and the corresponding ATAC-seq and histone H3 signals at the same sites for MEFs and the two pre-iPSC lines pre-i#1 and pre-i#2. For each bound site, the signal is displayed within a 2 kb window centered on the peak summit for the respective reprogramming factor and peaks were ranked based on ATAC-seq signal strength.
(E) Heatmap of normalized tag densities for O binding events (log2RPKM) for 48h, pre-i#1, and ESCs, for Oct4 binding groups shown in Figure 2D, depicting the actual signal at regions surrounding 2kb in either direction of the peak calls. In addition, the figure displays the normalized tag densities for O binding events for the same genomic locations in the independently derived pre-iPSC line pre-i#2.
(F) Venn diagram depicting the overlap of O, S, K, and M binding events, respectively, between the pre-i#1 and pre-i#2 lines. The total number of binding events and the number of overlapping sites and their percentage (against the pre-i#1 events) are given.
(G) Ontology of genes associated with ‘111’, ‘001’, and ‘100’ Oct4 sites defined in Figure 2D.
(H) Densities of the Oct4 and Oct4:Sox2 composite motifs at 48h-specific (‘100’), constitutive (‘111’), and ESC-specific (‘001’) binding events of Oct4, of the Sox2 motif within Sox2 peaks, the cMyc motif in cMyc peaks, and the Klf4 motif in Klf4 peaks. 95% confidence intervals at peak summits are indicated by the error bars
(I) Hierarchical clustering with optimal leaf ordering of the pairwise enrichment of O, S, K, and M binding events in the four reprogramming stages and pre-i#2, at base pair resolution. Black boxes emphasize clusters of TFs. O and S bind similar targets in pre-i#1, pre-i#2 and ESC, and Klf4 binding events are more distinct at these stages, clustering away from OS and closer to Myc. At 48h, binding events of O, S, and K cluster together. Myc peaks are more similar to each other than to those of the other reprogramming factors.
(J) K-means clustering of O, S, K, and M peaks across MEFs, 48h, pre-i#1, pre-i#2, and ESCs. Extensive OSK and OK co-binding was observed at 48h, whereas OS co-binding was more prevalent in ESCs. Notably, a subset of sites co-bound by OSK at 48h remained bound throughout reprogramming (second cluster from left). This clustering approach of binding events supports the conclusions made in Figures 2E/F.
8
Figure S3. Additional characterization of binding sites of individually and co-expressed reprogramming factors at 48h (related to Figure 2)(A) Klf4 has relocated to new sites that are co-bound by Oct4 and Sox2 at 48h of reprogramming. (i) A comparison of Klf4 peaks in MEFs (endogenously expressed Klf4) and at 48h of reprogramming revealed sites bound at both stages (shared), sites that were bound in MEFs but not at 48h (lost sites), and sites that were targeted at 48h but not in MEFs (de novo sites). The heatmap shows normalized Klf4 ChIP-seq signal (log2RPKM) at these sites. Each row shows the +/− 2kb region around each Klf4 summit. The number of sites in each category is given. The normalized signal for Oct4, Sox2 and cMyc binding at 48h and in MEFs were added for the same genomic sites. (ii) The metaplots present the average normalized signal of Klf4 in MEFs and at 48h for the three binding groups defined in (i) demonstrating that shared sites have higher Klf4 signal strength than ‘lost’ and ‘de novo’ sites. Density plots of the Oct4, Sox2, cMyc, and Klf4 motifs for the three groups of Klf4 binding events defined in (i) are given in (iii). Oct4, Sox2, and Klf4 motifs can be found at de novo Klf4 sites, while only the Klf4 motif is present at lost and shared sites.
(B) Transcript levels (log2(RPKM+1)) of the reprogramming factors in MEFs, at 48h of reprogramming with OSKM, and at 48h in MEFs over-expressing individual reprogramming factors from a dox-inducible cassette, based on RNA-seq data. Individually expressed reprogramming factors are 50× (Oct4), 2.5× (Sox2) and 8.8× (Klf4) up-regulated compared to the corresponding factor at 48h of OSKM-induced reprogramming.
(C) Western blot for Oct4 on starting MEFs and MEFs expressing the indicated individual reprogramming factor or combinations thereof either retrovirally (pMX) or inducibly (tetO) for 48h, pre-i#1, and ESCs. Whole cell extracts of equal cell numbers were used.
(D) Heatmap of normalized tag density for ATAC-seq data (log2RPKM) at sites bound by the indicated reprogramming factor at 48h of individual overexpression in MEFs (OpMX, SpMX, or KpMX). The MEF ATAC-seq signal at the same sites is also shown in each heatmap and the number of sites per reprogramming factor is given. Metaplots of the averaged normalized signal intensities of the ATAC-seq data are presented at the bottom.
(E) Density plots of Oct4, Sox2, Klf4, and cMyc motifs in Sox2 and Oct4 binding groups defined in Figure 2G (shared, OSKM-only, pMX-only). These data show that motif presence discriminates OSKM-only from shared and pMX-only sites.
(F) Heatmaps of normalized log2RPKM signals for all Oct4, Sox2, and Klf4 binding events, respectively, at 48h of reprogramming with MEFs carrying all four reprogramming factors (OSKM). In addition, the figure displays the normalized tag densities for the binding events of the same reprogramming factor when only OSK were expressed together retrovirally for 48h in MEFs (OSKpMX), without cMyc, for the same genomic locations. The number of peaks per reprogramming factor is given. These heatmaps demonstrate that the sites targeted by O, S, and K early in reprogramming in the context of OSKM co-expression are also largely targeted when only OSK are co-expressed in MEFs (without ectopic cMyc).
(G) Fold-enrichment for O, S, and K binding groups, defined in Figure 2G against the 35 chromatin trajectories described in Figure 3A, colored within each column from high (blue) to low (white) (left table). Percentage of binding events in each of the 35 chromatin trajectories is also given (right table; each column totals 100%) with each column colored from high (blue) to low (white).
9
Figure S4. Additional characterization of the 35 chromatin trajectories describing the major chromatin changes that occur during reprogramming (related to Figure 3)(A) Fold-enrichment of various genomic features for each of the 35 chromatin trajectories defined in Figure 3A. Columns represent fold-enrichment for CpG islands, exons, gene bodies, transcription end sites (TES), transcription start sites (TSS), promoters (defined as TSS +/−2kb), conserved elements (phastCons), satellite repeats as defined by RepeatMasker (RepeatMasker Open-4.0) and endogenous retrovirus 1 elements (ERV1). Enrichment scores were calculated as the ratio between the observed overlap and the expected overlap based on the state size, and colored within each column from high (blue) to low (white).
(B) Relationship between the 35 chromatin trajectories and the expression level of associated genes. The average expression level of genes is plotted as a function of the position of the chromatin state relative to RefSeq-TSS up to 50 kb in both directions. Each larger row corresponds to one of the 35 chromatin trajectories. Within each larger row are smaller rows corresponding to each of our four reprogramming stages (MEFs, 48h, pre-i#1, and ESCs). Each small row shows for the presence of the given chromatin trajectory at each position relative to the TSS, the average expression level of those corresponding genes at the given reprogramming stage. Red indicates higher expression, yellow intermediate expression, and blue low or no expression based on log2(RPKM+1) values from RNA-seq data. For instance one can observe that the pluripotency enhancer trajectory 13 is associated with a gradual increase in expression of associated genes from 48h to ESCs, while enhancer trajectory 17 is associated more clearly with ESC-specific gene expression. Conversely, the MEF enhancer states (trajectories 5 to 10) display higher expression in MEFs and at 48h than in pre-iPSCs and ESCs.
(C) Gene ontology analysis for enriched biological processes for the indicated chromatin trajectories based on the 35 chromatin states defined in Figure 3A.
(D) Percentage of stage-specific and constitutive O, S, K, and M binding events as defined in Figure 2D (‘100’, ‘001’, ‘111’ sites etc) for each of the 35 chromatin trajectories defined in Figure 3A. The total number of binding sites observed for each of the seven binding groups of O, S, K, and M, respectively, is given at the bottom of each column. Color scale within each column ranges from the highest (blue) to lowest (white).
(E) Gene ontology analysis for ‘111’ Oct4 sites in trajectory 13.
10
Figure S5. Additional characterization of changes occurring at MEs during reprogramming (related to Figure 4)(A) Metaplots of averaged normalized signal intensities of ATAC-seq data and ChIP-seq data for H3K4me1 and H3K4me2 at trajectory 5 MEs bound by O, S, or K (solid lines) and those not bound by any of the three reprogramming factors (dotted lines) in MEFs (green), 48h (blue), pre-i#1 (brown), and ESCs (red). The plots are centered on the summits of ATAC-seq peaks in MEFs.
(B) As in (A), except for trajectory 6 MEs and additional metaplots for H3K27ac.
(C) As in (B), except for trajectory 9 MEs.
(D) As in (B), except for trajectory 11 elements (transient enhancers).
(E) As in (B), except for trajectory 4 (promoters).
(F) Normalized transcript levels of p300 and Hdac1 for the reprogramming stages indicated, based on RNA-seq data.
(G) Schematic of the reprogramming experiment testing the role of Cebpa/b in reprogramming. OSKM-inducible MEFs were transfected with siRNAs targeting Cebpa or Cebpb or with siCtrl every 3 days during the course of reprogramming. Cebpa/b transcript levels were determined 48h post dox-addition (error bars indicate standard deviation of duplicate qPCR measurements) and Nanog-positive colonies counted at day 11 post OSKM induction for two replicates. Each replicate was generated using different siRNA reagents (siRNA 1 and 2).
(H) Metaplots of averaged normalized tag densities (RPKM) of the enhancer mark H3K27ac at trajectory 5 MEs engaged by O, S, or K (left) and those not engaged by either O, S, or K (right) at 48h post OSKM induction (blue). For the same two sets of trajectory 5 MEs, H3K27ac levels in starting MEFs (green) and in MEFs individually expressing Oct4 (top panels) or Klf4 (bottom panels) for 48h (black) were plotted.
11
Figure S6. Additional characterization of the role of somatic TFs in reprogramming (related to Figure 5)(A) Venn diagrams representing the overlap of Runx1 binding sites in MEFs and at 48h of reprogramming. The number of MEF-only, 48h-only and shared sites is given as well as the fractions of each set also bound by O, S, or K (in brackets).
(B) As in (A), for binding sites of Fra1.
(C) Heatmaps of normalized tag densities (log2RPKM) of the Cebpb ChIP-seq signal at the 12927 Cebpb binding sites obtained in pre-i#1. In addition, the data for O, S, and K occupancy in pre-i#1 and the independent pre-iPSC line pre-i#2 are shown for the same sites, indicating extensive co-binding of O, S, and K with Cebpb in pre-iPSC lines.
(D) Scatterplot of input normalized ChIP-seq signal (log2(RPKM+1) of Runx1 for MEF-only (green), 48h-only (blue), and shared (red) Runx1 binding events defined in (A).
(E) As in (D), except for Fra1 at sites defined in (B).
(F) Expression changes early in reprogramming. 609 genes (adjusted p-val <0.05) were differentially regulated within the first 48h of OSKM induction based on RNA-seq, with 372 genes induced and 237 genes down-regulated (Table S2). Transcription levels of these up- and downregulated genes in MEFs and at 48h of reprogramming are represented as boxplots.
(G) Gene ontology groups associated with up and down-regulated genes defined in (F).
(H) Differential enrichment of OSKM co-binding events in genes up- and down-regulated early in reprogramming as defined in (F). We computed the log2 ratio between the fraction of bound down-regulated genes out of all down-regulated genes and the fraction of bound up-regulated genes out of all up-regulated genes for different combinations of OSKM binding. Bound genes were defined as genes that have at least one binding site of the corresponding combination within 20kb of their TSS. Blue and red coloring represent higher fractions in down- and upregulated genes, respectively. Only the enrichment of the OSK co-binding event was significant (*; p<0.01 Chi-squared test) indicating that sites co-occupied by O, S, and K are enriched in upregulated genes.
(I) As in (H), but showing the differential enrichment of MEF-only, 48h-only, and shared binding events of Cebpa, Cebpb, Fra1, and Runx1 as defined in Figures 5A and S6A/B in genes up-and down-regulated early in reprogramming. These data demonstrate that up-regulated genes carry more 48h-only somatic TF binding events compared to down-regulated genes whereas higher fractions of down-regulated genes are occupied by MEF-only somatic TF binding relative to up-regulated genes. * denotes significance (p<0.01 Chi-squared test).
(J) Transcript levels of the somatic TFs Fra1, Cebpa, Cepbb, and Runx1 in MEFs, 48h, pre-i#1, pre-i#2, and ESCs, based on RNA-seq data.
(K) Fra1 transcript level in MEFs, iPSCs, and days 3, 6, 9 and 12 sorted SSEA1+ reprogramming populations, which are considered to be enriched for cells with higher reprogramming potential, as defined in (Polo et al., 2012).
(L) Genome browser view of the Fra1 locus. RNA-seq reads and Fra1 ChIP-seq data (both in RPKM) in MEFs (green), at 48h of OSKM-induced reprogramming (48h; blue) and at 48h of reprogramming with OSKM in the presence of Fra1 over-expression (black, 48hF) are shown. O and K binding in the locus for 48h and 48hF reprogramming samples are also depicted. Shaded areas represent regions within the Fra1 locus that lose Fra1 binding within the first 48h of reprogramming but have re-gained Fra-1 upon Fra1 overexpression in the context of OSKM/Fra1 (48hF). The asterisk (*) denotes an intronic enhancer that is known to auto-regulate Fra1 expression (Verde et al., 2007)
(M) Fold-increase of Fra1, cJun, and Runx1 transcript levels determined by RT-PCR at 48h of reprogramming with OSKM in combination with Fra1, cJun, or Runx1 overexpression, respectively, relative to 48h of reprogramming with OSKM only.
12
Figure S7. Additional characterization of reprogramming factor binding at PEs and the Esrrb overexpression effect (related to Figures 6 and and77)(A) Metaplots of averaged normalized signal intensities (RPKM) of H3K27Ac, H3K4me1, and H3K4me2 at ‘111’ Oct4 binding events within trajectory 13 PEs for MEFs (green), 48h (blue), pre-i#1 (brown), and ESCs (red).
(B) De novo scanning for motif identification in ‘001’ and ‘111’ Oct4 binding events in PEs of trajectories 13 and 17. The top enriched motifs per indicated set of peaks, the log10(P-value) for each motif and the best matching TF are given.
(C) (top) Percentage of ‘111’ Oct4 binding sites in trajectory 13 PEs that are also bound by Klf4 at 48h (left) or in ESCs (right), demonstrating prominent co-binding of Klf4 with Oct4 at these sites particularly early in reprogramming. (bottom) Percentage of ‘001’ Oct4 sites in PEs of trajectories 13 or 17 also bound by Klf4 in ESCs.
(D) Metaplot of normalized signal intensities of H3K27ac, H3K4me1, and H3K4me2 for all ESC super enhancers defined by (Whyte et al., 2013), for each of the four reprogramming stages. 5’ and 3’ denote the start and stop coordinates for the super enhancers, and the shading represents one standard deviation from the mean.
(E) Boxplots of transcript levels for genes neighboring ESC super enhancer in each of our four reprogramming stages. Asterisks (*) mark significant change (p-val < 0.007 and < 8.2e-12 for the MEF to pre-i#1 and MEF to ESC comparison, respectively, based on Wilcoxon test).
(F) Fold-enrichment of the 35 chromatin trajectory described in Figure 3A within ESC super enhancers colored within the column from highest (blue) to lowest (white).
(G) Snapshot of 48h and ESC O, S, and K ChIP-seq data (RPKM) at the ESC super enhancer regions associated with the Nanog, Sox2, Oct4, and Klf4 genes. In addition, the chromatin changes of these region are given by the trajectory annotation (from the 35 chromatin state model) based on the color-code in Figure S4A. Sites bound by O, S, or K already at 48h are highlighted by the grey shading.
(H) Genome browser view of O, S, and K ChIP-seq data and ATAC-seq data at the Tdh1 ESC super enhancer (RPKM) at the indicated reprogramming stages. In addition, the chromatin changes of this region are given by the trajectory annotation (from the 35 chromatin state model) based on the color code in Figure S4A. Of the five major sites in this super enhancer bound by O, S, or K in ESCs (highlighted by grey bars), one is engaged already at 48h (labeled with 1) and the others are bound only at later reprogramming stages (labeled with asterisks).
(I) Metaplot for normalized Klf4 (top) and Oct4 (bottom) ChIP-seq signal (RPKM) averaged across all ESC super enhancers for our four reprogramming stages. Oct4 data for MEFs were not available since it is not expressed in these cells. 5’ and 3’ denote the start and stop coordinates for ESC super enhancers and the shading indicates one standard deviation from the mean. Based on the comparison of Klf4 binding in MEFs and at 48hrs, we conclude that Klf4 already significantly binds ESC super enhancers at 48h.
(J) Transcript levels of Esrrb in MEFs, iPSCs, and days 3, 6, 9 and 12 sorted SSEA1+ reprogramming populations, which are thought to enrich for cells with higher reprogramming potential, as defined in (Polo et al., 2012).
(K) Transcript levels of Esrrb in our reprogramming stages (MEFs, 48h of OSKM expression (48h), 48h of OSKM and Esrrb co-expression (48hE), pre-iPSCs (pre-i#1,pre-i#2), and ESCs, based on RNA-seq.
(L) Gene ontology analysis for enriched biological processes for down- and up-regulated genes defined comparing MEFs expressing OSKM/Esrrb for 48h (48hE) versus MEFs expressing only OSKM for 48h (48h).
2
Table S2. Transcript levels of different gene sets for different reprogramming stages (related to Figures 1, ,2,2, ,5,5, ,77)This table contains information on the transcript levels (RPKM) of different sets of genes generated in this study:
for all genes for different reprogramming stages (MEFs, 48h of OSKM expression, pre-i#1, pre-i#2, and ESCs), for MEFs expressing OSKM and Fra1 for 48h (48h_OSKMFra1), for MEFs expressing OSKM and cJun for 48h (48h_OSKMcJun), for MEFs expressing OSKM and Esrrb for 48h (48h_OSKMEsrrb), and for MEFs individually expressing either Oct4, Sox2, or Klf4 (sheet 1, All_genes_RPKM)
for those genes determined to be significantly up-regulated (adjusted p-val <0.05) at 48h compared to MEFs, in MEFs, 48h of OSKM expression, pre-i#1, pre-i#2, and ESCs (sheet 2, up-regulated genes 48h)
for those genes determined to be significantly down-regulated at 48h compared to MEFs, for MEFs, 48h of OSKM expression, pre-i#1, pre-i#2, and ESCs (sheet 3, down-regulated genes 48h)
MEF-specific genes, for MEFs, 48h of OSKM expression, pre-i#1, pre-i#2, and ESCs (sheet 4, MEF specific genes)
ESC-specific genes, for MEFs, 48h of OSKM expression, pre-i#1, pre-i#2, and ESCs (sheet 5, ESC specific genes)
genes associated with ESC super enhancers, for MEFs, 48h of OSKM expression, prei#1, pre-i#2, and ESCs (sheet 6, ESC-super enh_associated genes)
genes significantly up-regulated at 48h when OSKM was co-expressed with Esrrb compared to 48h OSKM only (sheet 7, 48h_OSKMEsrrb_Upregulated)
genes down-regulated at 48h when OSKM was co-expressed with Esrrb compared to 48h OSKM only (sheet 8, 48h_OSKMEsrrb_Downregulated)
genes up-regulated at 48h when OSKM was co-expressed with Fra1 compared to 48h OSKM only (sheet 9, 48h_OSKMFRa1_Upregulated)
genes down-regulated at 48h when OSKM was co-expressed with Fra1 compared to 48h OSKM only (sheet 8, 48h_OSKMFra1_Downregulated)
3
Table S3. Summary of datasets generated in this study and correlation analysis (related to Figure 1)This table contains information on the number of replicates for the various genomic approaches and the correlations of replicate data sets, of merged data sets with published findings, and of experimental and imputed data. The table contains:
a summary of all genomics data sets generated in this study and information on replicate number and uniquely aligned reads (sheet 1, All_datasets_description).
correlation scores at 1Kb resolution between biological ChIP-seq replicates for TFs and chromatin regulators (sheet 2, TF replicate cor).
correlation scores at 1Kb resolution between replicate data sets of the enhancer marks H3K27ac, H3K4me1, and H3K4me2 obtained from MEFs and 48h, and for 48h of OSKM and Esrrb co-expression (sheet 3, MEF-48h enhancer mark rep cor).
correlation scores at 1Kb resolution between ATAC-seq replicate data sets for each reprogramming stage (sheet 4, ATAC-Seq replicate cor).
correlation scores at 1Kb resolution between experimental data sets (mostly histone modifications) and imputed data (see Extended Experimental Procedures for the description of the imputation) (sheet 5, imputed_dataset_correlation).
correlation scores at 1Kb resolution between histone modification data sets generated in this study and published data sets (sheet 6, Published histone data_cor).
4
Table S4. Gene ontology analysis for temporal binding events of Sox2, Klf4, and cMyc (related to Figure 2)This table contains ontology information for genes associated with ‘100’, ‘001’, and ‘111’ Sox2, Klf4, and cMyc binding sites defined in Figure 2D. This table is also associated with Figure S2G.
5
Table S5. Enrichment of pluripotency TF binding sites in ESCs in the 35 chromatin trajectories (related to Figure 6)This table contains the fold-enrichment of ESC-binding sites of pluripotency-related transcription factors and regulators in the 35 chromatin trajectories described in Figure 3A.
6
Figure S1. Validation of genomics data and characterization of stage- specific chromatin states (related to Figure 1)(A) Immunostaining for Oct4, Sox2, Klf4, and cMyc (green) in MEFs and at 48h of dox addition to MEFs carrying the polycistronic OSKM cassette, demonstrating endogenous expression of cMyc and Klf4 in MEFs and homogeneous induction of each of the four reprogramming factors across all cells upon dox treatment for 48h.
(B) Western blot for Oct4, Sox2, Klf4 and cMyc in MEFs, at 48h, pre-i#1, and ESCs. Whole cell extracts of equal cell numbers were used and Gapdh protein levels served as a loading control.
(C) Transcript levels of the reprogramming factors in the four reprogramming stages (MEFs, 48h of dox-induction, pre-iPSCs and ESCs) based on RNA-seq data. Transcripts of Oct4 and Sox2, unlike those of cMyc and Klf4, are not present in MEFs prior to induction of transgenic expression.
(D) Unsupervised hierarchical clustering of the top 10000 genes with most variant gene expression across MEFs, 48h, pre-i#1, pre-i#2, and ESCs. Scale is in log2RPKM. This heatmap demonstrates that the independently generated pre-iPSC lines pre-i#1 and pre-i#2 clustered together and that both lines are more similar to ESCs than to the early reprogramming states.
(E) Hierarchical clustering with optimal leaf ordering of the pairwise enrichment of ATAC-seq peaks in MEFs, 48h, pre-i#1, pre-i#2, and ESCs, at base pair resolution. The pre-iPSCs lines were more similar to each other followed by ESCs, while MEFs and 48h formed a separate node.
(F) Motif analysis of binding sites of OSKM, somatic TFs, and pluripotency TFs in MEFs, at 48h, in pre-i#1, and ESCs, as indicated. At 48h and in pre-iPSCs Oct4, Sox2, Klf4 and cMyc were ectopically expressed. Esrrb was ectopically expressed at 48h in OSKM-induced MEFs. N/A indicates that ChIP-seq data were not generated for the given TF at the indicated reprogramming stage. The Homer tool was used to scan for motif presence under the peaks of the corresponding TF. We scanned these peaks for all known motifs present in the Homer database and reported the top-scoring motif (canonical motif), which in all cases identified the respective known canonical motif. The same motifs were identified as the top represented by de novo motif analysis, with the exception of Oct4 and Sox2 in ESCs and Sox2 in pre-iPSCs, where the composite Oct4:Sox2 motif was most over-represented. For Cebpa and Cebpb similar motifs were identified.
(G) Genomic enrichments of chromatin states defined in Figure 1C at 48h of reprogramming and in pre-i#1. Columns represent percentage (%) of genome occupancy, median length of each state in kilo bases (kb), and fold-enrichments for CpG islands, exons, gene bodies, transcription end sites (TES), transcription start sites (TSS), promoters (defined as TSS +/−2kb), conserved elements (phastCons), ATAC-seq peaks, and endogenous retrovirus K elements (ERVK), colored within each column from highest (darkest) to lowest (white).
(H) Relationship between chromatin states and expression level of nearby genes. The average expression level of genes was plotted as a function of the position of the chromatin state relative to RefSeq-TSS up to 50 kb in both directions. Each larger row corresponds to a chromatin state (1–18) defined in Figure 1C. Within each larger row, smaller rows corresponding to each of our four reprogramming stages (MEFs, 48h, pre-i#1, and ESCs). Each small row shows for the presence of the given chromatin state at each position relative to the TSS, the average expression level of those corresponding genes at the given reprogramming stage. Red indicates higher expression, yellow intermediate expression, and blue low or no expression based on log2(RPKM+1) values from RNA-seq data. For instance, one can observe that the active promoter state (state 1) is present at the TSS of highly expressed genes, whereas the presence of the inactive/poised promoter state (state 2) around the TSS corresponds to a low or no expression. Also the strong enhancer state (state 3) is proximal to genes with higher expression than the weaker enhancer states (states 4–7).
(I) Validation of reprogramming stage-specific chromatin state annotations defined in Figure 1C by visualization of expected chromatin changes in reprogramming. A comparison of the chromatin states for each of the four reprogramming stages for genes known to be repressed during reprogramming (Col3a1 and Col5a2), induced (Dppa4/Dppa2 and mir290 clusters), and constitutively expressed (Hprt and Phf6). The color code of chromatin states is given in Figure 1C. Notably, the Dppa2/Dppa4 cluster is embedded in low signal chromatin states until the pluripotent state. Conversely, the genomic regions upstream the ESC-specific miR290 cluster gains enhancer marks (orange/yellow) as early as 48h post OSKM induction and forms a large enhancer domain in pre-iPSCs and ESCs.
Acknowledgments
We are grateful to Drs. N. S. Thomas, K. Zaret, S. Smale, M. Pellegrini, S. Kurdistani, N. Davidson, W. Lowry, and the Plath lab for discussions and reading of the manuscript. C.C. was supported by CIRM (TG2-01169) and Leukemia and Lymphoma Research (#10040) fellowships; P.F. by CIRM (TG2-01169) and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research (BSCRC) at UCLA; G.B. by Whitcome, Dissertation Year and QCB fellowships from UCLA; J.E. by NIH (R01ES024995; U01HG007912), NSF (CAREER Award 1254200), a Sloan Fellowship, and KP by the BSCRC, David Geffen School of Medicine, and Jonnson Comprehensive Cancer Center at UCLA, CIRM, and NIH (GM099134).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, C.C. and K.P.; Methodology, C.C. and K.P; Software, C.C., P.F., S.S., G.,B. and J.E.; Validation, C.C. and P.F.; Investigation, C.C., B.P. and S.B.; Formal Analysis, C.C., P.F., S.S., G.,B. J.E. and K.P. ; Data Curation, C.C., P.F., S.S., and G.,B.; Writing – Original Draft, C.C. and K.P.; Writing – Review & Editing, C.C., P.F., B.P., G.B., J.E. and K.P.; Resources, J.E. and K.P.; Visualization, C.C., P.F., J.E. and K.P.; Supervision, J.E. and K.P.; Project Administration, K.P.; Funding Acquisition, J.E. and K.P.References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [Europe PMC free article] [Abstract] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [Europe PMC free article] [Abstract] [Google Scholar]
- Bar-Joseph Z, Gifford DK, Jaakkola TS. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics. 2001;17(Suppl 1):S22–S29. [Abstract] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. [Abstract] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. [Europe PMC free article] [Abstract] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. [Europe PMC free article] [Abstract] [Google Scholar]
- Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. [Europe PMC free article] [Abstract] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen J, Chen X, Li M, Liu X, Gao Y, Kou X, Zhao Y, Zheng W, Zhang X, Huo Y, et al. Hierarchical Oct4 Binding in Concert with Primed Epigenetic Rearrangements during Somatic Cell Reprogramming. Cell Rep. 2016;14:1540–1554. [Abstract] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. [Abstract] [Google Scholar]
- Efron B, Tibshirani R. Statistical data analysis in the computer age. Science. 1991;253:390–395. [Abstract] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. [Europe PMC free article] [Abstract] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature Methods. 2012;9:215–216. [Europe PMC free article] [Abstract] [Google Scholar]
- Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol. 2015;33:364–376. [Europe PMC free article] [Abstract] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-typespecific gene expression. Nature. 2009;459:108–112. [Europe PMC free article] [Abstract] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. [Europe PMC free article] [Abstract] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. [Europe PMC free article] [Abstract] [Google Scholar]
- Ho R, Papp B, Hoffman JA, Merrill BJ, Plath K. Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep. 2013;3:2113–2126. [Europe PMC free article] [Abstract] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. [Europe PMC free article] [Abstract] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell stem cell. 2011;8:96–105. [Europe PMC free article] [Abstract] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu J, Han Q, Peng T, Peng M, Wei B, Li D, Wang X, Yu S, Yang J, Cao S, et al. The oncogene c-Jun impedes somatic cell reprogramming. Nat Cell Biol. 2015;17:856–867. [Abstract] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. [Abstract] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. [Europe PMC free article] [Abstract] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. [Abstract] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. [Europe PMC free article] [Abstract] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. [Europe PMC free article] [Abstract] [Google Scholar]
- Pasque V, Tchieu J, Karnik R, Uyeda M, Sadhu Dimashkie A, Case D, Papp B, Bonora G, Patel S, Ho R, et al. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. [Europe PMC free article] [Abstract] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. [Europe PMC free article] [Abstract] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. [Abstract] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. [Europe PMC free article] [Abstract] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. [Europe PMC free article] [Abstract] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. [Europe PMC free article] [Abstract] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. [Europe PMC free article] [Abstract] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. [Europe PMC free article] [Abstract] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. [Europe PMC free article] [Abstract] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. [Abstract] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): highperformance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [Europe PMC free article] [Abstract] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. [Europe PMC free article] [Abstract] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. Meta- and Orthogonal Integration of Influenza"OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. [Europe PMC free article] [Abstract] [Google Scholar]
- Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. Deciphering AP-1 function in tumorigenesis: fra-ternizing on target promoters. Cell Cycle. 2007;6:2633–2639. [Abstract] [Google Scholar]
- Wagschal A, Delaval K, Pannetier M, Arnaud P, Feil R. Chromatin Immunoprecipitation (ChIP) on Unfixed Chromatin from Cells and Tissues to Analyze Histone Modifications. CSH Protoc. 2007 2007, pdb prot4767. [Abstract] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. [Europe PMC free article] [Abstract] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2016.12.016
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867416317305/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The emergence of Sox and POU transcription factors predates the origins of animal stem cells.
Nat Commun, 15(1):9868, 14 Nov 2024
Cited by: 0 articles | PMID: 39543096 | PMCID: PMC11564870
Manipulating cell fate through reprogramming: approaches and applications.
Development, 151(19):dev203090, 30 Sep 2024
Cited by: 0 articles | PMID: 39348466
Review
Genome access is transcription factor-specific and defined by nucleosome position.
Mol Cell, 84(18):3455-3468.e6, 28 Aug 2024
Cited by: 0 articles | PMID: 39208807 | PMCID: PMC11420395
The yin and yang of pioneer transcription factors: Dual roles in repression and activation.
Bioessays, 46(10):e2400138, 26 Jul 2024
Cited by: 0 articles | PMID: 39058903
Engineering mouse cell fate controller by rational design.
Nat Commun, 15(1):6200, 23 Jul 2024
Cited by: 1 article | PMID: 39043686 | PMCID: PMC11266670
Go to all (289) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE90895
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Understanding impediments to cellular conversion to pluripotency by assessing the earliest events in ectopic transcription factor binding to the genome.
Cell Cycle, 12(10):1487-1491, 19 Apr 2013
Cited by: 11 articles | PMID: 23603987 | PMCID: PMC3680528
Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome.
Cell, 151(5):994-1004, 15 Nov 2012
Cited by: 579 articles | PMID: 23159369 | PMCID: PMC3508134
Spermatogonial stem cells and progenitors are refractory to reprogramming to pluripotency by the transcription factors Oct3/4, c-Myc, Sox2 and Klf4.
Oncotarget, 8(6):10050-10063, 01 Feb 2017
Cited by: 9 articles | PMID: 28052023 | PMCID: PMC5354640
Directing reprogramming to pluripotency by transcription factors.
Curr Opin Genet Dev, 22(5):416-422, 03 Aug 2012
Cited by: 24 articles | PMID: 22868173
Review
Funding
Funders who supported this work.
BSCRC
CAREER Award (1)
Grant ID: 1254200
CIRM (1)
Grant ID: TG2-01169
David Geffen School of Medicine
Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research (BSCRC)
Jonnson Comprehensive Cancer Center at UCLA
Leukemia and Lymphoma Research (1)
Grant ID: #10040
NHGRI NIH HHS (1)
Grant ID: U01 HG007912
NIEHS NIH HHS (1)
Grant ID: R01 ES024995
NIGMS NIH HHS (1)
Grant ID: P01 GM099134
NIH (3)
Grant ID: U01HG007912
Grant ID: R01ES024995
Grant ID: P01-GM099134