Abstract
Free full text

Genetic Deletion of the Repressor of Estrogen Receptor Activity (REA) Enhances the Response to Estrogen in Target Tissues In Vivo
Abstract
We previously identified a coregulator, repressor of estrogen receptor activity (REA), that directly interacts with estrogen receptor (ER) and represses ER transcriptional activity. Decreasing the intracellular level of REA by using small interfering RNA knockdown or antisense RNA approaches in cells in culture resulted in a significant increase in the level of up-regulation of estrogen-stimulated genes. To elucidate the functional activities of REA in vivo, we have used targeted disruption to delete the REA gene in mice. The targeting vector eliminated, by homologous recombination, the REA exon sequences encoding amino acids 12 to 201, which are required for REA repressive activity and for interaction with ER. The viability of heterozygous animals was similar to that of the wild type, whereas homozygous animals did not develop, suggesting a crucial role for REA in early development. Female, but not male, heterozygous animals had an increased body weight relative to age-matched wild-type animals beginning after puberty. REA mRNA and protein levels in uteri of heterozygous animals were half that of the wild type, and studies with heterozygous animals revealed a greater uterine weight gain and epithelial hyperproliferation in response to estradiol (E2) and a substantially greater stimulation by E2 of a number of estrogen up-regulated genes in the uterus. Even more dramatic in REA heterozygous animals was the loss of down regulation by E2 of genes in the uterus that are normally repressed by estrogen in wild-type animals. Mouse embryo fibroblasts derived from heterozygous embryos also displayed a greater transcriptional response to E2. These studies demonstrate that REA is a significant modulator of estrogen responsiveness in vivo: it normally restrains estrogen actions, moderating ER stimulation and enhancing ER repression of E2-regulated genes.
The biological activity of estrogens, acting through estrogen receptors (ERs), is critically dependent on coregulator proteins (coactivators or corepressors) that are recruited to the ligand-receptor complex at various gene regulatory sites. Recent work indicates that this is a highly dynamic situation, with the equilibrium between corepressors and coactivators in a cell and the nature of the hormonal ligand determining the state of nuclear receptor activation or inhibition (4, 8, 11, 17, 21, 30, 32, 34, 35, 40, 43, 44, 47).
Coactivator and corepressor proteins assemble into distinct, dynamic multiprotein complexes. In the case of coactivators, these complexes are constituted of the SRC/p160 family of proteins, CREB binding protein (CBP) and/or p300, and other factors that are recruited in a temporally ordered fashion (4, 44) and up-regulate nuclear receptor activity, at least in part, through enhanced histone acetyltransferase activity (8, 30, 32). ATP-dependent chromatin remodeling complexes, such as SWI/SNF, and the TRAP-DRIP-ARC (mediator) complex, which act sequentially or combinatorially, also enhance gene transcription by facilitating RNA polymerase II recruitment to promoters and its activation (43).
While many coactivator proteins have been identified, far fewer corepressors are known. Among the corepressors, the most fully characterized are NCoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoic acid and thyroid hormone receptors), which function as major negative regulators of several members of the nuclear receptor family through recruitment of large multicomponent complexes containing histone deacetylases to promote a repressive chromatin state (6, 11, 17, 43, 58). In addition, we have identified the coregulator, named repressor of estrogen receptor activity (REA), which binds to ER alpha and ER beta, effectively suppressing transcriptional activity of both ERs, that was shown to play an important role in determining the responsiveness of estrogen target cells in culture to estrogen (5, 27, 36). Other corepressors include a novel DEAD box RNA helicase, DP97 (41), and a metastasis-associated protein corepressor, MTA1 (28), that serve as corepressors for all of the steroid hormone receptors, a small unique nuclear corepressor, SUN-CoR (61), and the scaffold attachment factor, SAF-B (51). Among the nuclear hormone receptors investigated, REA was unique in being ER selective and in markedly repressing ER transcriptional activity but not that of other steroid (progesterone receptor, glucocorticoid receptor, and androgen receptor) and nonsteroid (thyroid hormone and retinoic acid) nuclear receptors (36).
Mapping of domains in the 37-kDa REA protein showed that distinct regions were required for its interaction with ER and for its repression of ER activity. REA was shown to compete with p160 coregulators, such as SRC-1, for binding to the ER, thereby functioning as an anticoactivator that eliminated the functional coactivation of ER transcriptional activity (5, 27, 36). In addition, REA repressive activity results from its recruitment of class I and II histone deacetylases (20). The action of REA was also found to be modulated by its binding to the chromatin remodeling protein prothymosin-α (PTα), an interaction that is competitive with REA binding to ER. Intriguingly, because PTα, which binds to REA but not ER, is known to be a chromatin-remodeling protein associated with cell proliferation and we observed that PTα is itself up-regulated by estrogen, this protein could participate in a feed forward loop, supporting estrogen-induced cell proliferation by progressively removing REA from ER (26, 27).
Supporting the importance of REA in ER action was our demonstration that decreasing the intracellular level of REA by using small interfering RNA knockdown or antisense RNA approaches in cells resulted in a significant increase in the activity of estrogens (5, 27), suggesting that REA plays a significant role in restraining the stimulatory effect of estrogens. Further, there is a strong positive correlation between REA and ER positivity in breast cancers, and higher levels of REA in low-grade versus high-grade more-advanced breast tumors (37, 45). These observations support the proposal that REA activity may be a restraint on estrogen-driven activities. Therefore, to elucidate the functional activities of REA in estrogen target tissues in vivo and its role in regulating hormone activities, we have generated an REA knockout mouse, the study of which is detailed in this report.
MATERIALS AND METHODS
Animal care and treatment of animals with estrogen.
All animals were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and all procedures described here were approved by the University of Illinois and Baylor College of Medicine Institutional Animal Care and Use Committees. Adult female C57BL mice and 129Sv mice were purchased from Harlan Co. (Indianapolis, Ind.). Immature female mice were injected subcutaneously (s.c.) daily with estradiol (E2) (0.5 μg/animal ·
· day) for 4 days. Injections consisted of compound dissolved in dimethyl sulfoxide and then diluted 1:10 in corn oil. At 24 h after the last injection, animals were killed by CO2 sedation and cervical dislocation. Uteri were removed, washed in cold phosphate-buffered saline (PBS), and weighed after removal of associated fat and expression of any luminal fluid. One uterine horn was then stored in RNA later (Ambion, Austin, Tex.) for RNA isolation, and the other was fixed for immunocytochemistry. The experiments were repeated three times with 3 to 5 animals per group per replicate.
day) for 4 days. Injections consisted of compound dissolved in dimethyl sulfoxide and then diluted 1:10 in corn oil. At 24 h after the last injection, animals were killed by CO2 sedation and cervical dislocation. Uteri were removed, washed in cold phosphate-buffered saline (PBS), and weighed after removal of associated fat and expression of any luminal fluid. One uterine horn was then stored in RNA later (Ambion, Austin, Tex.) for RNA isolation, and the other was fixed for immunocytochemistry. The experiments were repeated three times with 3 to 5 animals per group per replicate.
Preparation of mouse REA genomic DNA.
The mouse bacterial artificial chromosome (BAC) library clone 284H12 (Research Genetics, Inc.) was positive for the REA gene and confirmed by Southern blot analysis with a radiolabeled probe specific for the REA gene. After restriction enzyme-treated DNA was separated on a 0.8% agarose gel, the DNA fragments were transferred to a nitrocellulose membrane and probed with a 32P-labeled REA probe, followed by several stringent washing steps. Then the membrane was exposed at −80°C and developed. The BAC clone 284H12 DNA was prepared by the alkaline lysis method, and this clone DNA was used as the template for mouse REA genomic DNA to generate a targeting vector.
Targeting vector and ES cell culture.
The targeting vector, designed to eliminate the REA exon sequence encoding amino acids 12 to 201, which are required for REA repressive activity and for interaction with ER, contained 5.2 kb (5′ targeting arm) and 6.9 kb (3′ targeting arm) of mouse REA and neighboring sequences flanking a neomycin-resistance gene and loxP sequences (loxP-PGK-neo). In addition, the herpes simplex virus thymidine kinase (HSV-TK) gene was located outside the REA sequence and served as a negative drug selection marker. The pBluescript II SK(+) plasmid (Stratagene) was used as the backbone vector (Fig. (Fig.1).1). First, the HSV-TK gene, the negative selection marker gene, was fused next to the 5′ targeting arm through the ClaI site. The 5′ targeting arm, a 5.2-kb genomic DNA fragment, contained part of REA spanning from the end of exon 5 to the end of the gene plus an additional genomic sequence after the REA gene. As the 3′ targeting arm, we used 6.9 kb of genomic DNA containing the 5′ flanking region of REA and part of the adjacent C2f gene. A loxP-flanked pPGK-neo gene was fused upstream of the 3′ targeting arm and used as a positive selection marker for screening of transfected mouse embryonic stem (ES) cells. Finally, the fragment containing the HSV-TK gene and 5′ targeting arm was inserted upstream of the loxP-flanking pPGK-neo through the NotI site. The sequences of all constructs were verified. (We incorporated the loxP sites so that we would have been able to delete the neo gene had we encountered problems with expression of an aberrant fragment of REA or of neighboring genes driven by the PGK promoter of the neo gene.) The targeting vector was linearized by ClaI digestion and purified with the QIAGEN plasmid maxi kit.
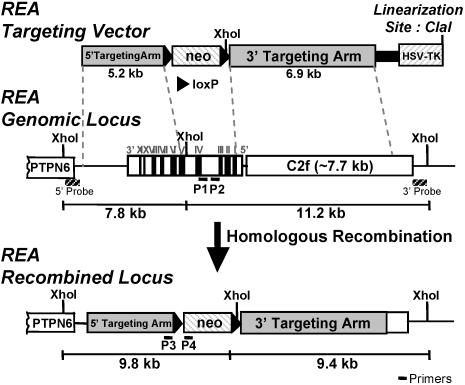
Design of targeting vector and targeting strategy for disruption of the REA gene. The targeting vector designed for disruption of the REA gene contained 5.2-kb (5′ targeting arm) and 6.9-kb (3′ targeting arm) mouse REA and neighboring genomic sequences. The neomycin resistance gene flanking loxP sequences (loxP-PGK-neo) was used as a positive selection marker, and the HSV-TK gene was used as a negative drug selection marker. The pBluescript II SK (+) plasmid (Stratagene) was used as the backbone vector. After homologous recombination on the REA genomic locus, the XhoI restriction enzyme sites within the REA gene were deleted, causing a different fragmentation pattern with XhoI treatment. 3′ and 5′ probes for Southern hybridization can detect the difference in fragmentation. Primer set P1 and P2 and primer set P3 and P4 were designed for genotyping analysis.
ES cell work was carried out as previously described (18). Briefly, the TC-1 ES cell line with a 129Sv/Ev strain background was cultured on the feeder layer of G418-resistant mouse embryonic fibroblasts in ES cell medium containing 750 U of leukemia inhibitory factor/ml. ES cells were electroporated in PBS containing 20 μg of targeting vector DNA per ml. Growth selection was carried out in ES cell medium containing 300 μg of G418/ml and 0.2 μM FIAU [1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil]. A total of 576 G418- and FIAU-resistant ES clones were isolated, and their genomic DNA samples were prepared for Southern blot analysis.
Four targeted ES cell lines including 1E7, 1G1, 1G9, and 2H8 were microinjected, which produced 6, 5, 6, and 2 male chimeric mice, respectively. All four independent chimeric lines carried REA germ line mutations. Two lines, 1E7 and 1G9, were expanded for experimental analysis and maintenance.
Preparation of DNA for genotype analysis.
Genomic DNA extraction from mouse tails for genotypic analysis was performed by using the Wizard kit (Promega, Madison, Wis.) according to the manufacturer's protocol. DNA from embryos was extracted by using proteinase K digestion solution (100 mM NaCl, 50 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.5% sodium dodecyl sulfate, 0.4 μg of proteinase K/μl) (Invitrogen, Carlsbad, Calif.) and purified with phenol and phenol-chloroform-isoamyl alcohol (Invitrogen).
Genotype analysis by Southern blotting and PCR.
REA genotypic analyses by Southern blotting and PCR were used to identify REA heterozygous and homozygous mutants. The 5′ probe for Southern screening was generated by PCR by using the BAC 284H12 clone DNA as a template. The sequences of the primers are 5′-CGGAATTCCTATGGACTTGTGTGAGAAGCCG-3′ and 5′-CGGAATTCAAGGTTGTCTGGGAAGGGGATAC-3, and the product of the PCR is 475 bp covering upstream of the 5′ targeting arm. In the same way, a 3′ probe, 560 bp, was generated by using primers 5′-CGGAATTCGGCGGTATCGGCACATATACAAT-3′ and 5′-CGGAATTCATGTCTTCTTCTTGAGCCTACTA-3′. Both 3′ and 5′ probes were designed to detect the external region of the targeted locus for selection of correct insertion. A Neo probe was also used to confirm that homologous recombination occurred correctly and was produced from the neo gene-containing plasmid.
To perform PCR for the genotyping, two primer sets were designed: one set (GT-reaF3/GT-reaR4) is for detecting the deleted part of REA and another set (GT-5′ arm-2F/GT-neo-5R) is for detecting the 5′ arm and neo gene. The sequences of the primers are as follows: GT-reaF3 (P1), 5′-GCTGCGAGGCATTGAACTTG-3′; GT-reaR4 (P2), 5′-TGTGCTTCTCTGTTGCTGTTTGTG-3′; GT-5′ arm-2F (P3), 5′-GCACAATCTTCTGTCGCTG-3′; GT-neo-5R (P4), 5′-AGACTGCCTTGGGAAAAGC-3′.
RNA isolation and real-time PCR.
Total RNA was isolated from whole uterine tissue by using Trizol reagent (Invitrogen) according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed in a total volume of 20 μl with 200 U of reverse transcriptase, 50 pmol of random hexamer, and 1 mM deoxynucleotide triphosphate (New England Biolabs, Inc., Beverly, Mass.). The resulting cDNA was then diluted to a total volume of 100 μl with sterile H2O. Each real-time PCR consisted of 1 μl of diluted reverse transcriptase product, 1× SYBR Green PCR master mix (Applied Biosystems, Foster City, Calif.), and 50 nM forward and reverse primer. Reactions were carried out in an ABI Prism 7700 sequence detection system (Applied Biosystems) for 40 cycles (95°C for 15 s, 60°C for 1 min) following an initial 10-min incubation at 95°C. Primers used for real-time PCR are as follows: complement component 3 (C3) forward, 5′-TCATCCTCATTGAGACCCCC; complement C3 reverse, 5′-CTGCCCCATGTTGACCAGTT; lactoferrin (LF) forward, 5′-CCCTTGAGGAAGCGGTATCC; LF reverse, 5′-ACACGAGCTACACAGGTTGGG; glucose-6-phosphate dehydrogenase (G6PDH) forward, 5′-CCCATCTGGAATCGAGACAAC; G6PDH reverse, 5′-CCCCACGACCCTCAGTACC; androgen receptor (AR) forward, 5′-AGGTCTTCCCCTGGACGAA; AR reverse, 5′-TTTGGCCTAACCTCCCTTGA; progesterone receptor (PR) forward, 5′-TTCATCCAATCCCGGACACT; PR reverse, 5′-AGGATCTTGGGCAACTGGG; 36B4 forward, 5′-CGACCTGGAAGTCCAACTAC; 36B4 reverse, 5′-ATCTGCTGCATCTGCTTG. The relative change in expression was calculated by using the ΔΔCt (cycle threshold) method (23) with the ribosomal protein 36B4 mRNA as an internal control.
MEF cell culture and transfection.
Nine- to eleven-day-postcoitum embryos were separated from maternal tissues, and the yolk sac and internal organs, such as the liver, were removed. Then the embryos were incubated at 4°C for 16 h in 0.25% (wt/vol) trypsin-EDTA solution (Invitrogen), followed by incubation at 37°C for 1 h. The embryo tissues were then broken down by vigorous pipetting and were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in a 5% CO2 atmosphere. Mouse embryo fibroblasts (MEFs) were grown to confluence in tissue culture flasks at 37°C in a 5% CO2 atmosphere (considered here to be passage zero). MEFs from the first or second passage at about 90% confluence were used in transfection experiments.
MEF cells were plated at 5 × 104 cells per well in 24-well plates in Dulbecco's modified Eagle's medium supplemented with 10% charcoal dextran-treated FCS and transfected 24 h later with 1 μg of (ERE)2-pS2-Luc, 10 ng of pCMV5-ER expression vector, and 0.3 μg of pCMV-β-galactosidase internal control plasmid. The transfection utilized FuGene 6 reagent (Roche Applied Science, Indianapolis, Ind.) and was done by following the manufacturer's protocol. Cells were harvested 24 h after hormone treatment and were lysed with three cycles of freezing on dry ice and thawing at 37°C. Cell extracts were prepared after centrifugation at 20,800 × g (14,000 rpm in an Eppendorf 5417R microcentrifuge) at 4°C for 10 min. β-Galactosidase activity, which was measured to normalize for transfection efficiency, and luciferase activity were assayed by using whole-cell lysates.
Western blot analysis.
MEF cultures growing at confluence after 4 days were washed twice with PBS and lysed in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, 1 mM sodium orthovanadate, 1 mM dithiothreitol, 10 mM β-glycerophosphate, and 4 μg of Complete protease inhibitor cocktail [Roche Applied Science]/ml). The lysate was centrifuged for 10 min at 15,000 × g to remove cell debris. Protein concentrations of lysates were determined by the BCA protein assay system (Pierce, Rockford, Ill.) with bovine serum albumin as the standard.
Protein from the uterus was prepared by using Trizol reagent (Invitrogen) according to the manufacturer's instructions. For protein analysis by immunoblotting, 15 μg of protein was mixed with 2× Laemmli sample buffer (Bio-Rad, Hercules, Calif.), denatured at 95°C for 5 min, and electrophoresed in sodium dodecyl sulfate-4 to 15% polyacrylamide gel electrophoresis gels. Gels were transferred to nitrocellulose membranes that were blocked for 2 h at room temperature in TBS-T (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat milk. Blots were sequentially incubated with anti-REA antibody (5) at a concentration of 2 μg/ml or with anti-β-actin primary antibody at a 1:5,000 dilution for 16 h at 4°C. After washing the filters with TBS-T, the filters were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Zymed Laboratory, Inc., South San Francisco, Calif.) at a dilution of 1:8,000 or rabbit anti-mouse immunoglobulin G (Zymed Laboratory, Inc.) at a dilution of 1:4,000 for 2 h in TBS-T buffer. The REA protein was visualized with X-ray film by using a chemiluminescence system (Pierce) after washing the filters again under the same conditions with the horseradish peroxidase-bound secondary antibody. β-Actin protein was used as a loading control. The intensities of the bands were analyzed with ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, Calif.)
Histological analysis.
For hematoxylin-eosin staining, one uterine horn from each animal was fixed in 10% neutral buffered formalin for 24 h at 4°C, transferred to 70% ethanol, and then embedded in paraffin. Four-micrometer sections were deparaffinized in xylene and rehydrated in ethanol. Endogenous peroxidase was inactivated by using 0.3% H2O2 in methanol for 30 min at room temperature.
Statistical analysis.
All experiments were repeated a minimum of three times, and results are expressed as means ± standard errors of the means (SEM). Statistical differences were determined by Student's t test, and chi-square analysis was employed to determine whether hypothesized results were verified experimentally.
RESULTS
REA targeting vector and genotype analysis of REA knockout ES cells.
To disrupt the endogenous REA gene in ES cells derived from the 129Sv/Ev mouse strain, gene targeting by homologous recombination was used. The targeting vector was designed to eliminate the REA exon sequence encoding amino acids 12 to 201, which are required for REA repressive activity and for interaction with ER (5). The mouse REA gene and the targeting vector used for REA deletion through homologous recombination are shown in Fig. Fig.1.1. The Southern analysis and PCR screening used for distinguishing and selecting positive colonies and identifying REA heterozygous and homozygous animals are shown in Fig. Fig.2.2. After correct recombination, the targeting event deleted 2,545 bp of REA genomic sequence encoding 190 amino acids (Pro12 to Val201) and replaced it with the neo gene sequence. Because there is a XhoI site within the deleted 2,545 bp region, the XhoI fragment from genomic DNA is found at 7.8 kb by the 5′ probe in Southern analysis, whereas after correct recombination, the XhoI fragment is found at 9.8 kb. Likewise, by using a 3′ probe, an 11.2-kb fragment is detected from genomic DNA and a 9.4-kb fragment is detected after recombination. To select against random insertion, in addition to using the TK-negative selection marker, we used external 3′ and 5′ probes, which can recognize the external region of the targeted locus, and also a neo probe, as detailed in Materials and Methods. In this manner, we identified correctly targeted ES cell lines (Fig. (Fig.22).

Isolation of ES cell clones and genotyping analysis by Southern hybridization (A) and PCR (B). The ES cells were selected by resistance to G418 (positive selection) and by growth on FIAU-containing media (negative selection) that excluded the random integration of the plasmid DNA. Isolated ES cell clones were analyzed by Southern hybridization to establish that a homologous recombination event that interrupted the REA gene had occurred. Mouse genomic DNA extracted from tail samples of all pups was genotyped by both PCR and Southern blotting and digested with XhoI. Two primer sets were designed: one is for detecting the deleted part of REA and another set is for detecting the 5′ arm and neo gene. 3′ and 5′ probes are identical to probes used for ES cell Southern screening. WT, wild type; KO, knockout.
Breeding of chimeric REA mice to generate heterozygous and homozygous progeny.
ES cells positive for incorporation of the neo gene after correct homologous recombination were injected into blastocysts donated from a C57BL/6 strain and implanted into pseudopregnant female FVB mice to generate independent chimeric founder lines. Chimeric animals, identified by their agouti coat color, were used for outbreeding with the wild-type C57BL/6 mouse strain. Because there can be mouse strain-dependent phenotype variability, we also bred our chimeras with the mouse strain from which our ES cell line was derived (129Sv), so we could analyze the offspring phenotype also on an inbred background. In both cases, fertile heterozygous REA+/− male and female mice were born and bred further to produce REA−/− mice. All animals were genotyped by PCR with genomic DNA extracted from tail samples and digested with XhoI (Fig. (Fig.2B2B).
Genotypic analysis of live-born progeny from mating of heterozygous animals revealed that no homozygous REA mutant pups were recovered from 260 progeny in the outbred strain and 97 progeny in the inbred strain (Table (Table1).1). The numbers of heterozygous mutant animals in the outbred genetic background were underrepresented among weaned pups (animals at 19 to 21 days of age) compared to the expected number assuming normal Mendelian inheritance (P < 0.01). However, we did not observe any death of REA heterozygous animals after birth, and genotyping of embryos even at early gestational stages showed that the ratio between wild-type and heterozygous animals followed the Mendelian ratio of 1:2 (wild type/heterozygous). In addition, wild-type and heterozygous REA mutant embryos were not morphologically distinguishable at the age of embryonic day 9 (E9.0), E10.0 and E11.0. Based on our findings, there is the possibility that a fraction of the REA heterozygous embryos may have died in utero during a late gestational stage. This did not appear to occur in the inbred genetic background, where the numbers of wild-type and REA heterozygous pups at weaning (Table (Table1)1) followed the Mendelian ratio (P > 0.25), although the number of animals analyzed was lower.
TABLE 1.
Genotyping analysis of F2 offspring from REA heterozygous mouse intercrossesa
| Genetic background and gender | No. of F2 offspring | No. (%) of mice of genotype:
| ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Outbred | 260 | 106 (41) | 154 (59) | 0 (0) |
    Male Male | 136 | 58 | 78 | |
    Female Female | 124 | 48 | 76 | |
| Inbred | 97 | 37 (38) | 60 (62) | 0 (0) |
    Male Male | 44 | 16 | 28 | |
    Female Female | 53 | 21 | 32 | |
The analysis of offspring showed that the REA mutation is recessive embryonic lethal. To investigate when lethality develops, embryos at different ages of gestation were obtained from heterozygous animal intercrosses and genotyped (Table (Table2).2). Homozygous REA mutant embryos could not be identified during the middle stages of development (from E9.0 to E11.0), and there was no evidence of resorption during these stages of development. These facts indicate that the complete absence of REA is lethal at a stage before E9.0, implying that REA has an important role in early stages of mouse development.
TABLE 2.
Genotyping analysis of embryos from REA heterozygous mouse intercrossesa
| Age | No. of embryos | No. (%) of embryos of genotype:
| ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E9.0 | 31 | 11 (35) | 20 (65) | 0 (0) |
| E10.0 | 33 | 11 (33) | 22 (67) | 0 (0) |
| E11.0 | 20 | 10 (50) | 10 (50) | 0 (0) |
REA heterozygous male and female mice survived and were fertile. Outbred intercrosses (× C57BL) exhibited larger litter sizes than inbred crosses (× 129Sv), and this phenomenon was observed in both first-generation (F1, pure) and second-generation (F2, mixed) intercrosses, so litter size was dependent on mouse strain. However, in both the outbred (× C57BL) intercrosses and the inbred (× 129Sv) crosses, litter sizes of second-generation (F2, mixed) mice were reduced significantly from those of the first-generation (F1) mice, indicating reduced embryonic survival during gestation (C57BL litter size, 7.5 ± 0.4 [F1] versus 5.9 ± 0.3 [F2], P < 0.05; 129Sv litter size, 6.1 ± 0.3 [F1] versus 4.1 ± 0.3 [F2], P < 0.05).
Growth of REA heterozygous mice.
The body weights of female and male REA+/+ and REA+/− mice were measured at various ages (Fig. (Fig.3).3). The growth curves revealed similar growth rates of age-matched REA wild-type and heterozygous males over time, while the growth of age-matched REA wild-type and heterozygous females showed a statistically significant difference, with REA heterozygous females exhibiting a greater gain in body weight from age 9 weeks to 26 weeks (P < 0.05). This increase in body weight may indicate that REA is involved in regulating female growth-related biological processes in response to estrogen, since the increase in body weight is seen only after the onset of puberty. Heterozygous REA female mice showed no obvious morphological abnormalities other than larger size.

Analysis of growth in wild-type and REA heterozygous mice. (A) Growth curve of females. REA+/− female mice grow significantly faster than the wild-type female mice. Values are means ± SEM of at least 13 mice each. *, significantly different from age-matched wild-type females (P < 0.05). (B) Growth curve of males. The growth of REA+/− male mice is similar to that of wild-type males. These mice have an outbred genetic background. Values are means ± SEM of at least 13 mice each.
Determination of the effect of REA haploinsufficiency on the responsiveness of the uterus to estrogen.
We examined uterine response to estrogen stimulation to gain insight into the effects of REA on female reproductive function. Wild-type and REA heterozygous female animals were treated for 4 days with E2 or control vehicle, and estrogen responsiveness of the uterus was analyzed by measuring uterine weight. In the absence of added estrogen, uterine weight was the same in wild-type and REA heterozygous animals, but the uterine weight increase in response to estrogen was 40% greater in REA heterozygous animals (Fig. (Fig.4A).4A). As expected, the REA mRNA level in uteri of REA heterozygous animals was reduced to 50% of that in uteri of wild-type animals, and Western blot analysis with REA antibody confirmed that heterozygous uteri had a similarly reduced level of REA protein (Fig. 4B and C).
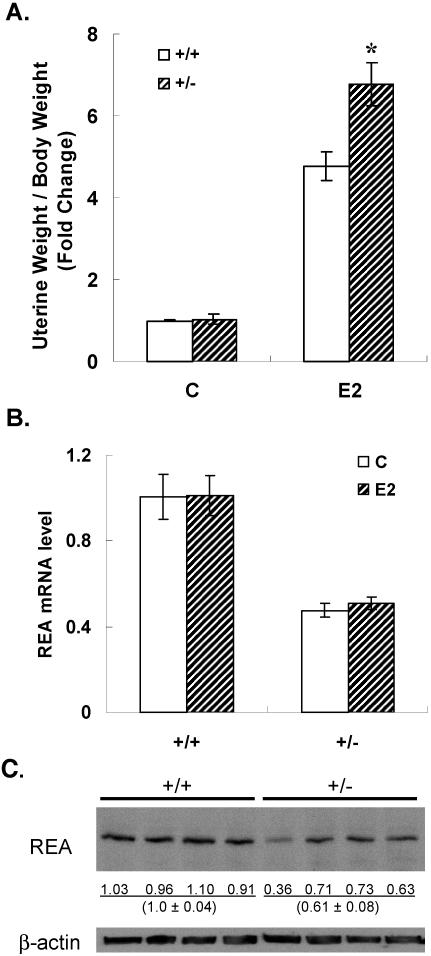
E2 responsiveness of the uterus (A) and REA mRNA (B) and REA protein levels (C) in wild type (+/+) and REA heterozygous (+/−) mice. Immature (day 21) mice were injected s.c. daily with control vehicle or 0.5 μg of E2 for 4 days. Uterine weight (A) was measured 24 h after the last injection and adjusted to each animal's body weight. *, P < 0.05. RNA was extracted from the uteri with Trizol, and the REA expression level was measured by real-time PCR. (B) The REA mRNA level was calculated relative to that of the vehicle-treated +/+ control, which was set at 1. (C) REA protein level in uteri from four mice of each genotype. Each lane contains uterine protein from a separate mouse of the indicated genotype. REA was detected by Western immunoblotting with REA polyclonal antibody. The numbers indicate the relative intensities of bands analyzed with ImageQuant software. β-Actin was used as a loading control.
Uterine morphology in the presence and absence of E2 was also examined (Fig. (Fig.5).5). Uterine weight and histology were similar in control (vehicle treated) wild-type and REA heterozygous uteri (Fig. (Fig.5,5, top panels). With E2 treatment, uterine luminal epithelial height and stromal and myometrium thickness were increased in both REA+/+ and REA+/− uteri, and epithelial cells changed from a single layer of low columnar cells to pseudostratified columnar cells. Of note, hyperproliferation was evident in the REA+/− luminal epithelium (Fig. (Fig.5,5, bottom right), and in addition, the apical surface of the luminal epithelium in the uteri of E2-treated REA heterozygous mice was more convoluted than the smooth apical surface of the wild-type luminal epithelium (Fig. (Fig.5,5, bottom panels).

Histological analysis of control and E2-treated uteri of REA heterozygous (+/−) and wild type (+/+) mice. Immature REA +/+ and +/− mice (21 days old) were injected s.c. daily with control vehicle or 0.5 μg of E2 for 4 days. At 24 h after the last hormone injection, tissue samples were collected, fixed, and stained with hematoxylin and eosin as described in Materials and Methods. (A) Control vehicle-treated uteri of wild type (+/+) mice; (B) control vehicle-treated uteri of REA heterozygous (+/−) mice; (C) E2-treated uteri of wild-type (+/+) mice; (D) E2-treated uteri of REA heterozygous (+/−) mice. Magnification, ×40.
Gene expression in uteri of REA+/+ and REA+/− females.
In the immature mouse uterus, several genes are known to be up-regulated by estrogen, namely complement C3, LF, G6PDH, and PTα (22, 26), whereas others, such as AR, PR, and ERα (7, 19, 50, 54), are known to be down-regulated by E2. Because REA is an ER coregulator that restrains estrogen activity in cells in culture (5, 27, 36), we wished to determine whether changes of REA levels in heterozygous REA animals would affect the expression of genes in the uterus. To investigate this, RNA was extracted from uteri of wild-type and heterozygous REA animals and analyzed for the modulation of gene expression by estradiol by real-time PCR.
As shown in Fig. Fig.6A,6A, the expression of C3 and LF genes was stimulated in the presence of estrogen by 21- and 30-fold, respectively, in uteri of wild-type animals. By contrast, the stimulation of these genes by estrogen was much greater (71- or 101-fold, respectively, for C3 and LF) in uteri of REA heterozygous animals. With two other genes, G6PDH and PTα, where the magnitude of increase by E2 was much lower, there appeared to be a weak trend toward increased expression in REA heterozygous versus wild-type uteri, but the differences were not significant (Fig. (Fig.6A6A).

Modulation of E2-up-regulated and E2-downregulated gene expression in uteri of REA wild-type (+/+) and heterozygous (+/−) mice. RNA was isolated from uteri of animals that received control vehicle or E2 treatments as described in the legend to Fig. Fig.5.5. Levels of complement component C3, lactoferrin, G6PDH, and PTα were examined by real-time PCR, and changes were calculated relative to the vehicle-treated controls. Levels of the downregulated genes PR, AR, and ERα were also examined by real-time PCR, and changes were calculated relative to the vehicle-treated controls.
REA also had a striking effect on E2 down-regulated genes (PR, AR, and ERα). In uteri of heterozygous REA animals, E2 failed to elicit the down-regulation of expression of these genes (Fig. (Fig.6B),6B), whereas as expected in REA wild-type uteri, E2 elicited a decrease in expression of these genes.
Therefore, the data shown in Fig. Fig.66 support the hypothesis that REA normally suppresses ER activity such that the reduction of REA level in the REA heterozygous mice enhances E2 stimulation of some up-regulated genes and dampens or eliminates the suppression of E2 down-regulated genes. The findings also imply that the effect of REA, as a coregulator of ER-mediated gene transcription, may be dependent on the particular target gene.
Studies in REA wild-type and heterozygous MEFs.
To examine the effect of REA on gene transcription in MEFs, REA wild-type and heterozygous MEFs were transfected with an ERα expression vector and an E2-responsive luciferase reporter gene containing two estrogen response elements (EREs). A strong increase in E2-responsive reporter gene expression (25-fold) was observed in wild-type MEFs; however, the response to E2 was significantly greater in REA heterozygous MEFs (Fig. (Fig.7A).7A). This enhanced response to E2 was observed over a wide range of E2 concentrations and was particularly strong at the lower, physiological concentrations of E2. These results show that the heterozygous MEFs, with half the wild-type level of REA mRNA and protein (Fig. 7B and C), give a more robust transcriptional response to the E2-ER complex, consistent with the results of gene expression stimulation by E2 observed in the uteri of REA heterozygous animals.
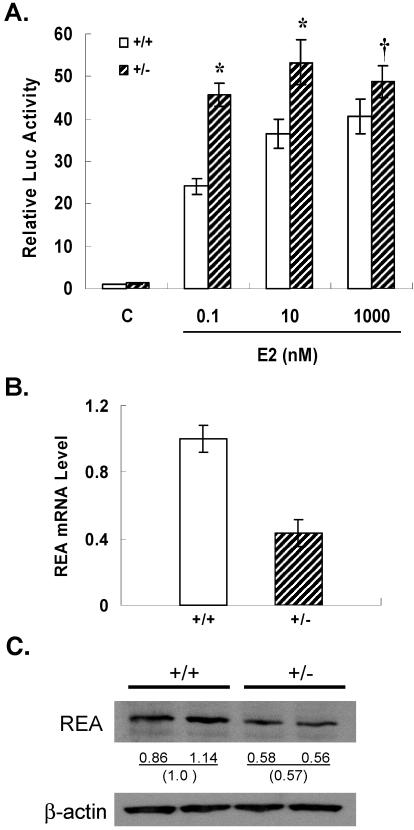
Enhancement of ER-driven transcriptional activation in REA heterozygous MEFs and examination of REA mRNA and protein levels in wild-type and heterozygous MEFs. (A) MEFs of each genotype were transfected with ERα (10 ng), (ERE)2-pS2-Luc reporter (1 μg), and β-galactosidase (300 ng) and treated with control vehicle (C) or the indicated concentration of E2 for 24 h, and luciferase activities were measured. *, significantly different from wild-type (+/+) MEFs at the same concentration of E2 (P < 0.05); †, P = 0.2. All values are means ± SEM and have been normalized for β-galactosidase activity. (B) MEFs were derived from either wild-type or REA heterozygous embryos as described in Materials and Methods. Total mRNA was isolated by using Trizol, and the REA mRNA level was measured by real-time PCR. (C) REA protein level was monitored with REA polyclonal antibody. β-Actin, used as a loading control, was monitored by using β-actin antibody. Numbers indicate the relative intensities of bands analyzed with ImageQuant software.
DISCUSSION
Coregulators play critical roles in nuclear receptor regulation of gene expression and physiological activities, determining responsiveness of target tissues to diverse ligands. Our functional studies in vivo with REA gene targeting have provided insight into gene regulatory mechanisms and biological processes in which REA plays a role, including early development, growth of female mice, and response of the reproductive system to estrogen.
REA and early development, viability, and fertility.
We have found that homozygous REA gene knockout animals do not develop, implying a critical role for REA in early development and, further, that no compensatory factors take over to rescue this lethality. For example, up-regulation of other corepressor proteins, such as NCoR, SMRT, or DP97, may have counterbalanced at least some of the results of REA knockout, but any such changes did not overcome the embryonic lethality in REA−/− mice. Such compensatory changes were noted in the SRC-1 knockout, where SRC-2 up-regulation was observed in the uterus, perhaps accounting for the only partial steroid resistance that was observed in this organ in SRC-1−/− animals (57).
Homozygous REA knockout embryos died before E9.0. This embryonic lethality is earlier than reported for most mammalian transcriptional mutants. Although estrogen has an apparent role in blastocyst activation (39) and ER expression is detected in early development (10), REA mutation resulted in a severe developmental defect, embryonic lethality, suggesting that REA has a significant role in regulating biological processes during development. The early lethality occurred in two different genetic backgrounds, the inbred 129Sv strain and the outbred C57BL strain, implying that the lethality of REA homozygotes is intrinsic to the embryo and not due to the genetic makeup of the mothers.
In addition to being a recessive lethal, the numbers of wild-type and REA heterozygous pups from REA heterozygous intercrosses in the outbred genetic background (129Sv × C57BL) were somewhat lower than expected from a Mendelian ratio (1:2). Since no neonatal lethality of REA outbred heterozygous animals was observed, no distinct differences were observed in the morphology between wild-type and REA heterozygous animals during early gestation, and the Mendelian ratio was normal in early gestation, some REA heterozygotes may die in the uterus during late gestation. However, this was not observed with REA heterozygous crosses in the inbred genetic background (129Sv × 129Sv) where the numbers of wild-type and REA heterozygous pups were within the statistically normal range and followed the expected 1:2 Mendelian ratio, although the total numbers of pups were much lower than in the outbred genetic background. This indicates that there may be a factor(s) or modifier gene(s) in the C57BL strain that affects the viability of REA heterozygous embryos. The near-Mendelian ratio of 1:2 for REA+/+ and REA+/− mice indicates that REA deletion has no major effect on gamete formation or function.
Although heterozygous mutant animals for most genes do not usually have a phenotype different from the wild type, there is evidence for a relationship between gene dosage and viability and phenotypic alteration of heterozygotes for several factors important in transcription. For example, null mutation of Brg1, a component of mammalian SWI/SNF complexes, resulted in very early embryonic lethality and in exencephaly in a small portion of Brg1 heterozygotes. Haploinsufficient Brg1 gene dosage particularly influenced the development of the central nervous system, where Brg1 is highly expressed (3, 42). The transcriptional integrator p300 null mutation exhibited embryonic lethality and dosage-dependent heterozygous lethality, which was also affected by genetic background (60). As observed for REA, the transcriptional regulator Trap220 null mutation elicited early embryonic lethality, while Trap220 heterozygous mutants were viable and exhibited altered growth (retardation in the case of Trap220+/− mice) and altered (i.e., impaired) transcription (14). Hence, genes responsible for transcriptional regulation are often essential for mouse development, and their dosage is frequently important in regulating important biological processes.
REA effects on body size.
Because both inbred and outbred strains of REA homozygous knockout mice were not viable, we examined REA heterozygous (+/−) animals. Of note was the faster growth and larger body size of REA heterozygous females that was first observed at 9 weeks of age (i.e., postpuberty) and continued to persist throughout the 26-week period of monitoring. The increased body weight reflects an overall increase in body size. No obvious increase in adiposity was evident, although this aspect needs further examination. Because estrogen is known to increase levels of insulin-like growth factor 1 and many other hormones and growth factors (12, 13, 38), this increased growth, which was observed in female +/− but not in male +/− mice, may be due to a greater estrogenic stimulation of blood levels of these growth-promoting factors, and these aspects will be assessed in future studies.
REA effects on estrogen response in the uterus and MEFs.
Estrogen is well known to elicit a marked increase in uterine weight in female animals (24). In this study, wild-type animals responded normally with a fivefold increase in uterine weight, and REA heterozygous animals, expressing half the wild-type level of this repressor protein, responded even better to estrogen with a much greater increase in uterine weight. In addition, the REA heterozygous mice showed hyperproliferation of the luminal epithelium, indicating that REA normally serves as a brake on estrogen-driven proliferation. Since growth factors such as insulin-like growth factor 1 and transforming growth factor alpha have been linked to estrogen action in the uterus (12, 13, 38), REA reduction in the heterozygous mutant mice may impact the production and/or actions of these growth factors.
REA mutation resulting in REA depletion to ca. 50% in heterozygous animals also resulted in marked changes in the magnitude of gene regulation by E2. Stimulation of the E2-up-regulated genes, complement C3 and LF, was markedly enhanced in uteri of REA heterozygous females. Also of note was the finding that the suppressive effect of E2 on down-regulated genes, AR, PR, and ERα (7, 19, 50, 54), was completely lost in REA heterozygous animals. These findings are consistent with the reports that REA competes with coactivators for binding to ER and also recruits histone deacetylases (5, 20, 27, 36), suppressing ER stimulatory activity and potentiating ER inhibitory effects. Thus, these data reveal that REA plays a significant role in transcriptional regulation of estrogen-regulated genes in vivo and that its effects appear to be gene specific, since regulation of PTα and G6PDH by E2 was similar in REA+/+ and REA+/− uteri. Gene-selective effects of another coregulator, CITED, have been noted also (59). In a tissue such as the uterus that consists of several cell types where the biological responses may differ, it must be borne in mind that increased gene responses may be due to gene-specific as well as cell type-specific effects.
Transfection studies with MEFs isolated from wild-type and REA heterozygous embryos revealed that the 50% reduction in REA significantly increased the magnitude of response to E2. These results in the uterus and in MEFs indicate that REA is important in determining target tissue sensitivity to estrogen.
Although coregulators, including REA, are widely distributed in many tissues, it is of note that knockout of specific coregulators has revealed that these regulators have remarkably distinct, tissue-selective functions. For example, although SRC-1, SRC-3, and E6-AP are all expressed in reproductive tissues, only SRC-3 and E6-AP knockout females exhibit fertility defects (46, 56, 57). Likewise, genetic elimination of the three highly homologous p160 coactivators (SRC-1, SRC-2, and SRC-3) result in distinct reproductive phenotypes (9). The phenotypes provide compelling evidence that coregulators play distinctive, largely nonredundant physiological roles and that multiple coregulators are required to modulate the full range of steroid hormone activities in vivo (25, 29-31).
Of note, knockout of only two other corepressors has been reported. Deletion of RIP-140/Nrip-1 leads to infertility and anovulation, characterized by failure to release mature oocytes at ovulation (55). In contrast, knockout of the broadly acting corepressor NCoR (15) or the broadly acting coactivator proteins CBP, p300, and Trap220, results in embryonic lethality (14, 48, 60). Because of the lethality observed with REA knockout mice, the use of conditional knockout approaches targeted to specific tissues such as the mammary gland, ovary, and uterus should allow its possibly tissue-specific effects to be more fully examined in the future.
REA as a repressor of estrogen receptor transcriptional activity.
REA is a member of the prohibitin family of proteins, showing about 50% amino acid identity with prohibitin. The prohibitins share a high degree of sequence homology between species and are highly conserved through evolution, implying important roles for these proteins. The REA gene is located in chromosomal region 12p13 in humans and chromosome 6 in mice (1, 2). Although REA was initially identified in B cells and named B-cell receptor-associated protein, BAP 37 (16, 49), this protein was shown to be present in many other cells and tissues examined, clearly indicating that it functions in cells that do not contain the B-cell receptor (49). We find that REA is present in many human tissues but is at its highest levels in the liver, ovary, colon, skeletal muscle, thyroid, and adrenal gland; at moderate levels in the uterus, bladder, prostate, and mammary gland; and at lower levels in most regions of the brain. The important and possibly diverse function of prohibitin is supported by recent observations that this protein represses E2F transcriptional activity and cell proliferation by recruiting HDAC1 (52, 53) and the chromatin remodeling proteins Brg-1/Brm to E2F-responsive promoters for the repression of E2F-mediated transcription.
Among the nuclear hormone receptors, REA was shown to interact selectively with the estrogen receptor (27, 36). A recent report that REA interacts also with the orphan nuclear receptors, COUP-TFI and COUP-TFII, implies that REA may affect a subset of receptors within the large nuclear receptor superfamily (20). This interaction of REA with both ER and COUP-TFI and COUP-TFII is of particular interest because ER and COUP-TF have been shown to form a complex in cells, with this association increasing ER phosphorylation by ERK2/p42 mitogen-activated protein kinase and enhancing ER transcriptional activity (33). Hence, the basis for the lethality observed upon complete REA knockout is likely to be complex and may result from its influence on the actions of ER, COUP-TFs, histone deacetylases, and other partners with which REA interacts, including the chromatin remodeling protein prothymosin alpha.
Our study reveals that REA has essential roles in regulating physiological processes and that some are sensitive to REA gene dosage. Intriguingly, REA heterozygous female animals showed enhanced growth and modulation of transcriptional regulation by the estradiol-ER complex in both an estrogen target tissue, the uterus, and also in REA heterozygous MEFs. The studies provide evidence that REA is a significant modulator of estrogen action in vivo. Its presence normally serves as a brake on estrogen-driven gene expression and physiological actions.
Acknowledgments
We thank Surojeet Sengupta for contributions to some of these studies.
We gratefully acknowledge the support of this work by grants from the NIH, CA18119 to B.S.K., DK58242 to J.X., and HD07857 and DK59820 to B.W.O.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.25.5.1989-1999.2005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc549370?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Inhibition of Prolyl Oligopeptidase Restores Prohibitin 2 Levels in Psychosis Models: Relationship to Cognitive Deficits in Schizophrenia.
Int J Mol Sci, 24(7):6016, 23 Mar 2023
Cited by: 0 articles | PMID: 37046989 | PMCID: PMC10093989
Activation of AMP-activated protein kinase (AMPK) through inhibiting interaction with prohibitins.
iScience, 26(4):106293, 28 Feb 2023
Cited by: 4 articles | PMID: 36950117 | PMCID: PMC10025096
Role of Prohibitins in Aging and Therapeutic Potential Against Age-Related Diseases.
Front Genet, 12:714228, 29 Oct 2021
Cited by: 8 articles | PMID: 34868199 | PMCID: PMC8636131
Review Free full text in Europe PMC
Prohibitin S-Nitrosylation Is Required for the Neuroprotective Effect of Nitric Oxide in Neuronal Cultures.
J Neurosci, 40(16):3142-3151, 09 Mar 2020
Cited by: 9 articles | PMID: 32152200 | PMCID: PMC7159891
Prohibitin 2 deficiency impairs cardiac fatty acid oxidation and causes heart failure.
Cell Death Dis, 11(3):181, 12 Mar 2020
Cited by: 27 articles | PMID: 32165613 | PMCID: PMC7067801
Go to all (61) article citations
Other citations
Wikipedia
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Analysis of transcription and estrogen insensitivity in the female mouse after targeted disruption of the estrogen receptor gene.
Mol Endocrinol, 9(11):1441-1454, 01 Nov 1995
Cited by: 274 articles | PMID: 8584021
Methoxychlor stimulates estrogen-responsive messenger ribonucleic acids in mouse uterus through a non-estrogen receptor (non-ER) alpha and non-ER beta mechanism.
Endocrinology, 140(8):3526-3533, 01 Aug 1999
Cited by: 47 articles | PMID: 10433208
Haploinsufficiency of the corepressor of estrogen receptor activity (REA) enhances estrogen receptor function in the mammary gland.
Proc Natl Acad Sci U S A, 103(45):16716-16721, 25 Oct 2006
Cited by: 28 articles | PMID: 17065319 | PMCID: PMC1636521
Estrogen receptor gene disruption: molecular characterization and experimental and clinical phenotypes.
Recent Prog Horm Res, 51:159-86; discussion 186-8, 01 Jan 1996
Cited by: 173 articles | PMID: 8701078
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA018119
Grant ID: CA18119
NICHD NIH HHS (2)
Grant ID: R01 HD007857
Grant ID: HD07857
NIDDK NIH HHS (4)
Grant ID: DK59820
Grant ID: DK58242
Grant ID: P01 DK059820
Grant ID: R01 DK058242





