Abstract
Free full text

Reducing ribosome biosynthesis promotes translation during low Mg2+ stress
Associated Data
Summary
The synthesis of ribosomes is regulated by both amino acid abundance and the availability of adenosine triphosphate (ATP), which regenerates guanosine triphosphate (GTP), powers ribosomes, and promotes transcription of ribosomal RNA genes. We now report that bacteria supersede both of these controls when experiencing low cytosolic magnesium (Mg2+), a divalent cation essential for ribosome stabilization and for neutralization of ATP’s negative charge. We uncover a regulatory circuit that responds to low cytosolic Mg2+ by promoting expression of proteins that import Mg2+ and that lower ATP amounts. This response reduces the levels of ATP and ribosomes, making Mg2+ ions available for translation. Mutants defective in Mg2+ uptake and unable to reduce ATP levels accumulate non-functional ribosomal components and undergo translational arrest. Our findings establish a new paradigm whereby cells reduce the amounts of translating ribosomes to carry out protein synthesis.
Introduction
The ribosome is the primary site of protein synthesis. The amount of ribosomes determines the rate or protein synthesis because the majority of ribosomes in a cell are actively translating (Forchhammer and Lindahl, 1971), and the rate of translation per ribosome is constant (Maaløe and Kjeldgaard, 1966). Ribosome activity controls ribosome production. That is, substrates consumed in translation also function as signals that regulate the synthesis of ribosomal components (Jinks-Robertson et al., 1983; Cole et al., 1987; Condon et al., 1993; Schneider and Gourse 2003a) (Figure 1A). For instance, adenosine triphosphate (ATP) both promotes the initiation of ribosomal RNA (rRNA) transcription, the rate-limiting step in ribosome biosynthesis, and is consumed by translating ribosomes (Gaal et al., 1997; Schneider et al., 2002). A rise in cellular ATP levels leads to a surge in rRNA transcription and a subsequent increase in ribosome numbers (Gaal et al., 1997; Schneider et al., 2002; Murray et al., 2003). Therefore, a cell maximizes its translation capacity by matching the rate of ribosome synthesis to the levels of ATP available for consumption. Here, we uncover a stress response pathway that supersedes this regulation and, unexpectedly, aids translation by decreasing ribosome production.
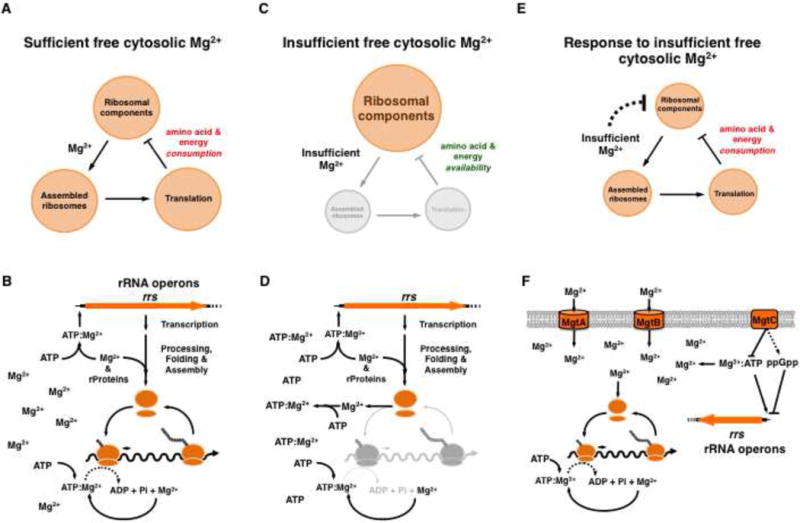
(A) Schematic depicting the regulatory circuit controlling ribosome production when Mg2+ is abundant. Mg2+ mediates the assembly of ribosomal components into functional ribosomes. Translation consumes energy and amino acids. A decrease in energy or amino acids decreases expression of ribosomal components. (B) Cartoon representation showing the role of Mg2+ and ATP on the synthesis of rRNA, assembly of ribosomes and translation when free cytosolic Mg2+ is abundant. (C) Schematic depicting the effect of disturbances on the regulatory circuit depicted on (A) due to insufficient free cytosolic Mg2+. This condition impairs ribosome assembly and, therefore, translation, creating an abundance of energy and amino acids, which promotes expression of ribosomal components. (D) Cartoon representation highlighting the effects of insufficient free cytosolic Mg2+ on ribosome synthesis, assembly and translation, resulting in the accumulation of non-functional ribosomes (in grey). (E) Schematic depicting a regulatory circuit that enables cells to repress the synthesis of ribosomal components when free cytosolic Mg2+ is limiting. This circuit integrates information about the availability of Mg2+ ions for ribosomal stabilization into the regulatory circuit governing ribosome biosynthesis. (F) Cartoon representation illustrating the proteins comprising the circuit depicted on (E) in S. enterica serovar Typhimurium. Mg2+ uptake into the cytosol by the Mg2+ transporters MgtA and MgtB, and a decrease in ATP promoted by the MgtC protein enables formation of functional ribosomes that are lower in number than when bacteria have abundant Mg2+ (A and B).
Mg2+ is the most abundant divalent cation in living cells (Maguire and Cowan, 2002; Wacker, 1969). While Mg2+ participates in a multitude of essential processes, the majority of cytosolic Mg2+ is involved in various aspects of protein synthesis (Pontes et al., 2015a). First, Mg2+ is required in several enzymatic reactions that generate biosynthetic precursors for translation (Wacker, 1969). Second, Mg2+ functions as a counter ion for ATP, guanosine triphosphate (GTP), and other nucleotide triphosphates (NTPs) (Maguire and Cowan, 2002; Wacker, 1969). And third, Mg2+ plays an essential role in the assembly of ribosomes by neutralizing negative charges from phosphates present in the rRNA backbone and, thus, allowing folding and compaction of the rRNA (Gesteland, 1966; Klein et al., 2004).
A single ribosome in the bacterium Escherichia coli contains at least 170 Mg2+ ions (Schuwirth et al., 2005). Removal of Mg2+ from purified ribosomes leads to loss of peptidyl-transferase activity and subsequent ribosome disassembly into its individual constituents (Gesteland, 1966). Mg2+ cannot be replaced by other cations without compromising the structural stability and/or the catalytic activity of the ribosome (Weiss and Morris, 1973; Weiss et al., 1973; Fagerbakke et al., 1999). Notably, Mg2+ exerts a regulatory effect on the cellular translation status: removal of Mg2+ from the growth medium leads to a reduction of translation both in mammalian and bacterial cells (McCarthy, 1962; Terasaki and Rubin, 1985). Furthermore, in bacteria, prolonged periods of Mg2+ deprivation leads to the depletion of ribosomes and cessation of protein synthesis, despite the fact that cells are able to maintain viability (McCarthy, 1962). These findings indicate that cells deprived of environmental Mg2+ are able to maintain sufficient amounts of this cation in their cytosol to carry out vital enzymatic reactions. Moreover, they imply that cells harbor genetic programs that specifically repress the synthesis of ribosomes during Mg2+ starvation.
We now report the identification of such a program that, counter intuitively, favors translation by reducing ribosome synthesis. We establish that the bacterium Salmonella enterica serovar Typhimurium induces expression of Mg2+ transporters and of an inhibitor of the F1FO ATP synthase, the machine responsible for the synthesis of the majority of cellular ATP (Okuno et al., 2011), when experiencing cytosolic Mg2+ limitation. Mutants lacking these proteins overproduce ribosomal components but fail to carry out protein synthesis because they are incapable of assembling functional ribosomes. The identified adaptation is also displayed by E. coli and likely to be widespread because all ribosomes are stabilized by Mg2+ and powered by ATP.
Results and Discussion
Low Mg2+ Represses rRNA Transcription
We hypothesized that Mg2+ regulates ribosome production because it plays crucial roles in protein synthesis by stabilizing the ribosome and participating in various biochemical reactions. To investigate the possible connection between ribosome production and Mg2+, we monitored the levels of the rrs-encoded 16S rRNA produced by wild-type Salmonella grown in different Mg2+ concentrations. Because rRNA synthesis is the rate-limiting step in ribosome production (Gaal et al., 1997), rRNA transcription can serve as a proxy for ribosome synthesis. As hypothesized, rRNA transcription (measured as the ratio between the leader rRNA to the structural rRNA; see Experimental Procedures for rationale) was 10-fold lower following growth in low (10 μM) than in high (10 mM) Mg2+ (Figure S1A).
We reasoned that the transcription factor PhoP (Shin and Groisman, 2005) might be responsible for the reduction in rrs transcription taking place in low Mg2+ because PhoP is activated in low extracytoplasmic Mg2+ detected by its cognate sensor PhoQ (Soncini et al., 1996; Véscovi et al., 1996). As hypothesized, a phoP null mutant displayed higher rRNA than wild-type Salmonella (Figure S1A). That Salmonella represses rrs transcription when grown in low Mg2+ in a phoP dependent manner was verified by primer extension analysis (Figure 2A), a technique often used to examine rrs promoter activity (Zhi et al., 2003; Gaal et al., 1997; Schneider et al., 2002; Murray et al., 2003). The mRNA levels of the control ompA gene were unaffected by changes in the Mg2+ concentration or by mutation of the phoP gene (Figure 2A). PhoP governs rRNA synthesis rather than degradation because the rrs RNA decreased with similar kinetics in phoP and wild-type Salmonella (Figure S1B). Taken together, these results established that low Mg2+ represses rrs transcription in a PhoP-dependent manner.
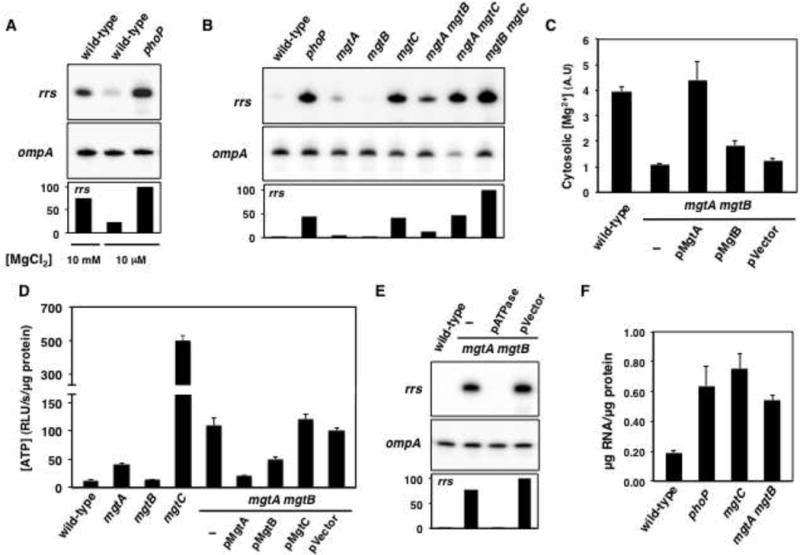
(A) Primer extension analyses of wild-type Salmonella (14028s) grown in high (10 mM) Mg2+, and wild-type (14028s) and phoP (MS7953) Salmonella grown in low (10 μM) Mg2+. (B) Primer extension analyses of wild-type (14028s), phoP (MS7953), mgtA (EG16735), mgtB (EL5), mgtC (EL4), mgtA mgtB (EG16741), mgtA mgtC (EG16740), and mgtB mgtC (EL6) Salmonella following growth in low Mg2+. (C) Cytosolic Mg2+ concentration in wild-type (14028s), mgtA mgtB (EG17048) and mgtA mgtB (EG17048) Salmonella harboring the mgtA-expressing plasmid (pMgtA), the mgtB-expressing plasmid (pMgtB) or the plasmid vector (pUHE-21-2-lacIq) during growth in low Mg2+. Cytosolic Mg2+ concentrations were obtained using a genetic reporter (Figure S4A and B; Cromie et al., 2006). Values represent the normalized activities of this Mg2+ genetic reporter (i.e., β-galactosidase activity values from Figure S4C were normalized as 1/[pPlac1-6-mgtAleader-lacZ/pPlac1-6r-lacZ]), and are depicted in arbitrary units (A.U.). (D) ATP levels of wild-type (14028s), mgtA (EG16735), mgtB (EL5), mgtC (EL4), mgtA mgtB (EG16741), and mgtA mgtB (EG16741) Salmonella harboring pMgtA, pMgtB or the plasmid vector (pUHE-21-2-lacIq). (E) Primer extension analyses of wild-type (14028s), mgtA mgtB (EG17048) and mgtA mgtB (EG17048) Salmonella harboring the ATPase expressing plasmid (pATPase) or the plasmid vector (pCP44). Bacteria were grown in N-minimal medium containing high (10 mM) or low (10 μM) Mg2+. Measurements were carried following 3 h (high Mg2+) and 6 h (low Mg2+) growth (OD600 ≈ 0.4–0.6). Error bars represent standard deviations. Graphs are the average of 3 to 4 independent experiments. Primer extension analyses are representative of 2 to 4 independent experiments. (F) RNA/protein ratio of wild-type (14028s) and phoP (MS7953) Salmonella grown in low Mg2+. See also Figures S1, S2 and S4.
Mg2+ Transporters and F1FO ATP Synthase Inhibitor Repress rRNA Transcription Initiation
PhoP appears to repress rrs transcription indirectly, via a PhoP-regulated gene(s). This is because the purified PhoP protein did not alter the mobility of DNA fragments corresponding to the rrsC P1 and P2 promoters (Figure S1C), which lack sequences resembling a PhoP box (Kato et al., 1999; Zwir et al., 2012). The PhoP protein shifted a DNA fragment corresponding to the phoP promoter (Figure S1C), which was used as a positive control (Soncini et al., 1995; Shin and Groisman, 2005), but not one harboring the PhoP-independent hisL promoter, which was used as a negative control (Figure S1C).
We hypothesized that the PhoP-activated mgtA gene and mgtCBR operon might be responsible for the regulation of rrs transcription taking place in low Mg2+. The mgtA and mgtB genes encode distinct Mg2+ transporters (Snavely et al., 1989a and 1989b), and mgtC encodes an inhibitor of the F1FO ATP synthase (Lee et al., 2013) (MgtR is a regulator of MgtC proteolysis; Alix and Blanc-Potard, 2008). Transcription of the mgtA and mgtCBR coding regions is promoted when cytosolic Mg2+ levels decrease below a certain threshold (Cromie et al., 2006; Spinelli et al., 2008). Together, MgtA, MgtB and MgtC maintain cytosolic Mg2+ homeostasis by importing Mg2+ into the cytosol and by freeing Mg2+ from high affinity complexes with ATP (Groisman et al., 2013; Pontes et al., 2015a).
Both an mgtC mutant and an mgtA mgtB double mutant exhibited elevated rrs transcription during growth in low Mg2+ (Figure 2B). This was also true for mgtA mgtC and mgtB mgtC double mutants (Figure 2B), but not for mgtA or mgtB single mutants, which displayed minor or no alterations in rrs transcription (Figure 2B). The observed effects are specific to rrs because ompA mRNA levels were the same in all examined strains (Figure 2B). The elevated rrs transcription in the mgtC and mgtA mgtB mutants was reduced to the level observed in wild-type cells by plasmids expressing MgtC and MgtA, respectively, but not by the plasmid vector (Figures S1D and S1E). Moreover, the joint expression of the mgtC and mgtB genes from a heterologous promoter lowered rrs transcription in the phoP mutant to near wild-type levels (Figure S1F and S7C), but isogenic plasmids expressing mgtC or mgtB, or the plasmid vector did not (Figure S1F and S7C).
In rapidly growing cells, the majority of rRNA is transcribed from the rrs P1 promoters (Sarmientos and Cashel, 1983). To determine whether the MgtA, MgtB and MgtC proteins exert their effects at the transcription initiation level, we constructed a transcriptional fusion between the Salmonella rrsC P1 promoter and a promoterless gfp gene, and measured the fluorescence produced in three isogenic strains. rrsC P1 promoter activity was 12- and 7-fold higher in mgtC and mgtA mgtB mutants than in wild-type Salmonella, respectively (Figure S2A and S2B). Cumulatively, the data presented in this section indicate that a decrease in cytosolic Mg2+ triggers the production of the MgtA, MgtB and MgtC proteins, which, in turn, repress rrs transcription initiation.
MgtC Represses rRNA Transcription by Reducing ATP Levels and by Increasing Guanosine Tetraphosphate (ppGpp) Amounts
Transcription initiation at the rrs P1 promoter is regulated by the concentration of the initiating nucleotide triphosphate (iNTP) (Gourse, 1988; Gaal et al., 1997; Schneider et al., 2002) and by the levels of (p)ppGpp, a second messenger mediating the response to nutrient limitation often referred to as the stringent response (Cashel and Gallant, 1969; Sarmientos and Cashel, 1983; Sarmientos et al., 1983; Xiao et al., 1991; Ross et al., 2013). The iNTP and (p)ppGpp regulate P1 promoters by controlling the stability of the RNA polymerase promoter open complex (Gaal et al., 1997; Barker et al., 2001).
ATP is the iNTP in all Salmonella rrs P1 promoters (Figure S2C), and MgtC lowers intracellular ATP levels (Lee et al., 2013). We therefore reasoned that the abnormal rRNA accumulation displayed by the mgtC mutant might be due to heightened ATP levels stimulating rrs P1 transcription. As proposed, ATP hydrolysis promoted by the ATPase-expressing plasmid pATPase (Koebmann et al., 2002; Pontes et al., 2015b) drastically reduced rrs levels in the mgtC mutant, whereas the plasmid vector had no effect (Figure 3A and Figure S7A). By contrast, plasmid pATPase did not alter the mRNA levels of the control ompA gene (Figure 3A). Given that pATPase did not lower rrs transcription of the mgtC mutant to the levels displayed by wild-type Salmonella (Figure 3A and Figure S7A), we hypothesized that MgtC might also increase (p)ppGpp amounts. Such a dual mode of rrs regulation by ATP and (p)ppGpp would be analogous to that exerted by the FbaA protein (Schneider and Gourse, 2003b).
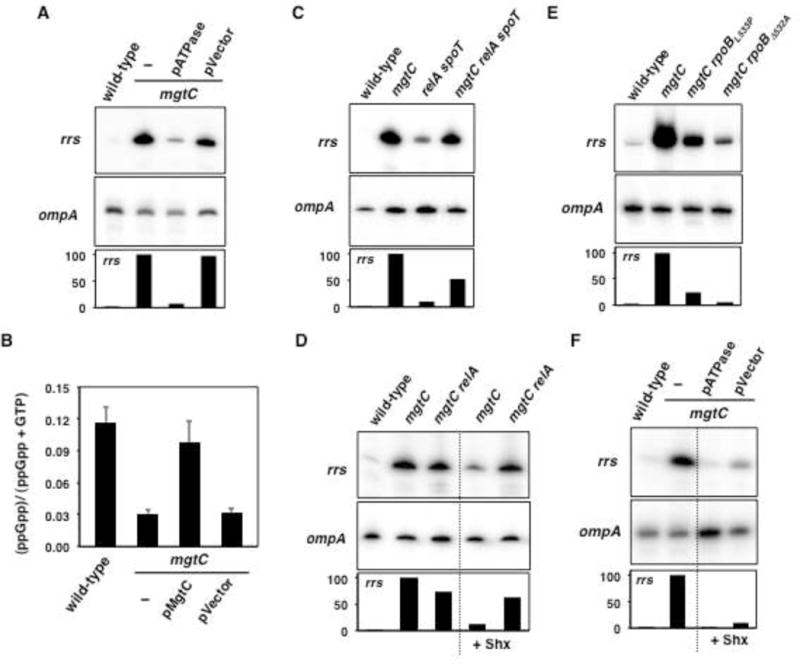
(A) Primer extension analyses of wild-type (14028s), mgtC (EL4) and mgtC (EL4) Salmonella harboring the ATPase expressing plasmid pATPase or the plasmid vector (pCP44) during growth in low (10 μM) Mg2+. For rrs primer extension: lanes 1 and 2, and 3 and 4 correspond to lanes 1 and 2, and 7 and 8 of the gel image displayed on Figure S7A, respectively. (B) Quantification of ppGpp levels in wild-type (14028s), mgtC (EL4) and mgtC (EL4) strains of Salmonella harboring the mgtC-expressing plasmid (pMgtC) or the plasmid vector (pUHE-21-2-lacIq) during growth in low Mg2+. Error bars represent standard deviations. Graph shows the average of four independent experiments. (C) Primer extension analyses of wild-type (14028s), mgtC (EL4), relA spoT (MP342) and mgtC relA spoT (MP343) Salmonella growth in MOPS glucose medium containing low (10 μM) Mg2+. For rrs primer extension: lanes 1 and 2, and 3 and 4 correspond to lanes 1 and 2, and 5 and 6 of the gel image displayed on Figure S7B, respectively. (D) Primer extension analyses of wild-type (14028s), mgtC (EL4), and mgtC relA (MP335) Salmonella grown in low Mg2+, in the presence/absence of serine hydroxamate (Shx, 50 μg/ml). (E) Primer extension analyses of wild-type (14028s), mgtC (EL4), mgtC rpoBL533P (MP571) and mgtC rpoBΔ532A (MP570) Salmonella during growth in low Mg2+. (F) Primer extension analyses of wild-type (14028s), mgtC (EL4) and mgtC (EL4) Salmonella with pATPase or the plasmid vector (pCP44) during growth in low (10 μM) Mg2+, in the presence/absence of 50 μg/ml Shx. Unless stated otherwise, strains were grown in N-minimal medium. All measurements were carried following 6 h of growth in low Mg2+ (OD600 ≈ 0.4–0.6). Primer extension analyses are representative of 3–4 independent experiments. See also Figures S1, S2, S3 and S7.
The mgtC mutant displayed lower ppGpp levels than wild-type Salmonella (Figures 3B and S3A). This defect was corrected by heterologous expression of the mgtC gene, but not by the plasmid vector (Figures 3B and S3A). A number of experiments indicated that the MgtC-mediated repression of rRNA transcription results from its effects on the cellular pools of both ppGpp and ATP. First, a relA spoT double mutant, which is unable to synthesize ppGpp (Xiao et al., 1991) (Figure S3A) but retains normal ATP levels during cytosolic Mg2+ starvation (Figure S3B), displayed more rrs transcription than wild-type Salmonella (Figure 3C and Figure S7B). Furthermore, rrs transcription was similar in mgtC single and mgtC relA spoT triple mutants, and higher than those in the relA spoT double mutant (Figure 3C and Figure S7B). Second, serine hydroxamate (Shx), a serine analog that triggers RelA mediated (p)ppGpp production (Tosa and Pizer, 1971; Haseltine and Block, 1973; Figure S3A), decreased rrs transcription in the mgtC mutant but not to wild-type levels (Figure 3D). Third, alleles of the RNA polymerase beta-subunit gene rpoB that shorten the half-life of promoter open complexes (Zhou and Jin, 1998) decreased rrs transcription in the mgtC mutant (Figure 3E). Despite making RNA polymerase behave stringently (Zhou and Jin, 1998), these rpoB alleles did not normalize rrs transcription to wild-type levels (Figure 3E). Fourth, Shx treatment restored rrs transcription to wild-type levels if the mgtC mutant carried plasmid pATPase but not when harboring the vector control (Figure 3F). As expected ompA mRNA levels were not affected by pATPase (Figure 3A), Shx treatment (Figure 3D) or the rpoB allele (Figure 3E). Together, these results demonstrated that MgtC represses rrs transcription both by reducing ATP levels and by accumulating ppGpp.
MgtA and MgtB Repress rRNA Transcription by Preventing ATP Accumulation
During growth in low Mg2+, PhoP promotes transcription of the mgtA and mgtB genes (Soncini et al., 1996) and muzzles the activity of the Mg2+ channel CorA (Alteri et al., 2011). This response indicates that Salmonella relies on MgtA and MgtB to maintain physiological cytosolic Mg2+ levels. As suggested, when experiencing low Mg2+ conditions, the concentration of free cytosolic Mg2+ was lower in the mgtA mgtB double mutant than in wild-type Salmonella (Figures 2C and S4A–C). The MgtA-expressing plasmid restored cytosolic Mg2+ to wild-type levels, an isogenic plasmid expressing mgtB increased cytosolic Mg2+ only partially, and the vector control had no effect (Figure 2C). These results indicate that MgtA functions as the main Mg2+ transporter under the investigated conditions.
Mg2+ is essential for ribosome assembly (Gesteland, 1966). Because most cellular ATP is consumed in translation (Stouthamer, 1973), a sudden decrease in protein synthesis resulting from insufficient cytosolic Mg2+ would increase ATP levels, thus stimulating rrs P1 promoters (Gaal et al., 1997; Schneider et al., 2002). In agreement with this notion, we established that ATP levels were ~10-fold higher in the mgtA mgtB double mutant than in the wild-type strain (Figure 2D), and ~4-fold higher in the mgtA single mutant than in the mgtB single mutant, which retained wild-type ATP levels (Figure 2D). ATP levels were ~50-fold higher in the mgtC mutant than in wild-type Salmonella (Figure 2D). Whereas the mgtA-expressing plasmid restored ATP levels of the mgtA mgtB double mutant to those of the wild-type strain, the isogenic mgtB-expressing plasmid lowered ATP levels of the mgtA mgtB double mutant to those of the mgtA single mutant, and the plasmid vector had no effect (Figure 2D). Notably, we established that a mgtC and mgtA mgtB mutations had an additive effect on ATP levels of cells grown in low Mg2+ (Figure S1G), indicating that MgtC and MgtA MgtB decrease ATP levels through distinct pathways. Thus, the rrs transcription profile (Figure 2B) mirrors the ATP levels of the Mg2+ transporter mutants.
Plasmid pATPase decreased rrs transcription in the mgtA mgtB double mutant to that of wild-type Salmonella (Figure 2E), whereas the vector control had no effect (Figure 2E). As expected, neither pATPase nor the plasmid vector altered ompA mRNA levels, which were the same in all strains (Figure 2E). Taken together, these results indicate that MgtA and MgtB inhibit rrs transcription by preventing a rise in ATP levels.
RNA/Protein Ratio, Growth Rate and the Efficiency of Translation
Ribosome synthesis is normally coupled to growth rate, such that rapidly growing cells have more ribosomes (Schaechter et al., 1958; Bremer and Dennis, 1996). The total RNA/protein ratio is also proportional to the amount of ribosomes and, thus, to the growth rate (Schaechter et al., 1958; Bremer and Dennis, 1996). Surprisingly, the RNA/protein ratio was higher in the phoP mutant than in wild-type Salmonella (Figure 2F) even though the phoP mutant grows slower than wild-type Salmonella in low Mg2+ (Soncini et at., 1996). Such uncoupling between the RNA/protein ratio and growth rate has been observed in mutants with slow translation rates, and in cells whose protein synthesis capability was compromised by pharmacological treatment (Nomura and Watson, 1959; Mikkola and Kurland, 1991). These findings suggested that the phoP mutant has decreased efficiency of protein synthesis during cytosolic Mg2+ starvation.
Cytosolic Mg2+ Homeostasis is Required for Ribosome Assembly
A decrease in the efficiency of protein synthesis in the phoP mutant could reflect a lower rate of peptide chain elongation, and/or a decrease in the fraction of ribosomes actively participating in protein synthesis. Because PhoP regulates rrs transcription via MgtA, MgtB and MgtC, and the mgtC single and mgtA mgtB double mutants also displayed elevated RNA/protein ratios (Figure 2F), we asked whether mutants lacking these proteins exhibited altered polysome profiles. Both the mgtC mutant and the mgtA mgtB double mutant accumulated 30S and 50S ribosomal subunits that did not efficiently assemble into 70S ribosomes (Figure 4A). This is in contrast to wild-type Salmonella, which assembled ribosomal subunits into 70S ribosomes that partake into mRNA translation as polysomes both in high (Figure S5A) and low Mg2+ (Figure 4A). The mgtC-expressing plasmid lowered the amounts of 30S and 50S subunits and increased polysome amounts in the mgtC mutant (Figure 4A). And the same was true for complementation of the mgtA mgtB double mutant with the mgtA-expressing plasmid (Figure 4A). The vector control rescued neither the mgtC nor the mgtA mgtB mutants (Figure 4A). These data established that MgtA, MgtB and MgtC are necessary for the assembly of functional ribosomes when bacteria experience low cytoplasmic Mg2+.
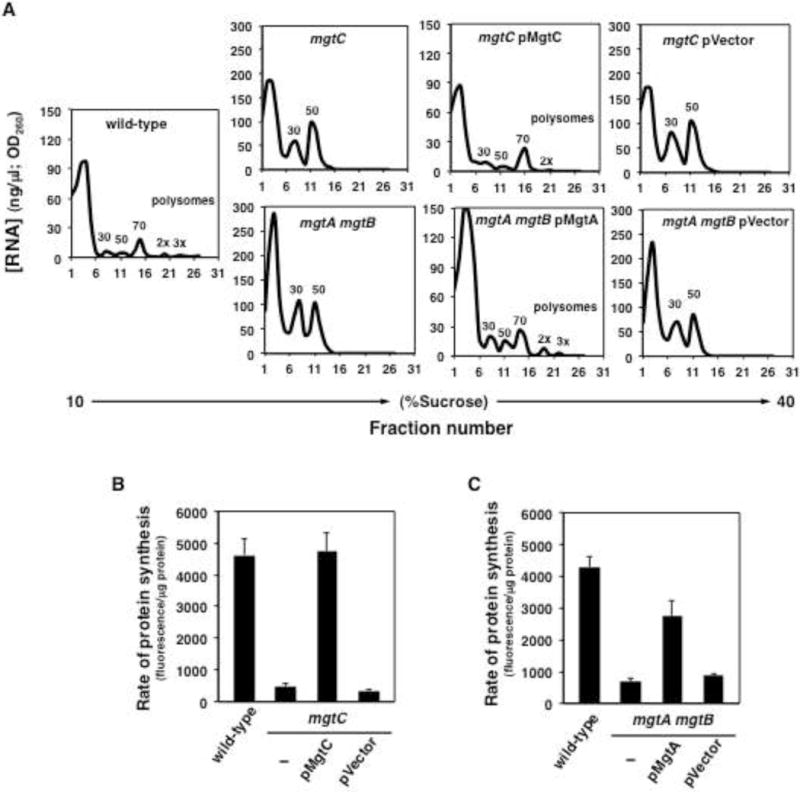
(A) Polysome analyses of wild-type (14028s), mgtC (EL4), and mgtC (EL4) Salmonella harboring the mgtC-expressing plasmid (pMgtC), or the plasmid vector (pUHE-21-2-lacIq), mgtA mgtB (EG16741) and mgtA mgtB (EG16741) Salmonella harboring the mgtA-expressing plasmid (pMgtA) or the plasmid vector (pUHE-21-2-lacIq) during growth in low (10 μM) Mg2+. Cells were collected for polysome analysis following 6 h (low Mg2+) of growth (OD600 ≈ 0.4–0.6). Polysome profiles are representative of four independent experiments. (B) Quantification of L-azidohomoalanine (AHA) labeling of wild-type (14028s), mgtC (EL4) and mgtC (EL4) Salmonella harboring pMgtC or the plasmid vector (pUHE-21-2-lacIq) following growth in low (10 μM) Mg2+. (C) Quantification of AHA labeling of wild-type (14028s), mgtA mgtB (EG16741) and mgtA mgtB (EG16741) Salmonella harboring pMgtA or the plasmid vector (pUHE-21-2-lacIq) during growth in low Mg2+. Measurements were carried following 6 h growth in low Mg2+ N-minimal medium lacking methionine (OD600 ≈ 0.4–0.6). Error bars represent standard deviations. Graphs are the average of 4 independent experiments. See also Figures S5 and S6.
Cytosolic Mg2+ Homeostasis Enables Translation
The data presented above suggest a model whereby maintenance of a free cytosolic Mg2+ pool supports ribosomal assembly, which makes translation possible (Figures 1A and and1B).1B). As the cytosolic concentration of free Mg2+ decreases, the assembly of 70S ribosomes is compromised and translation slows down (Figures 1C and and1D).1D). Wild-type Salmonella responds by increasing Mg2+ intake and by reducing ATP levels. The latter action alleviates low cytoplasmic Mg2+ stress both by releasing Mg2+ ions trapped in complexes with negatively charged ATP molecules and by reducing rRNA transcription (Figures 1E and and1F).1F). The inability to mount such a response results in a self-perpetuating cycle in which increased ATP levels stimulate the synthesis of ribosomal subunits not competent for translation (Figure 4A).
To test the proposed model, we examined protein synthesis in vivo by measuring the incorporation of the non-toxic methionine analog L-azidohomoalanine (AHA) into nascent proteins (Dieterich et al., 2007). The mgtC mutant displayed much lower AHA levels than wild-type Salmonella (Figures 4B and S5B). The protein synthesis defect of the mgtC mutant appears to result primarily from excess ATP because the mgtC mutant was rescued not only by the mgtC-expressing plasmid (Figure 4B and S5B) but also by pATPase (Figure S6A). By contrast, neither the mgtA-expressing plasmid nor the vector control corrected the mgtC mutant defect (Figures S6A and S6B). These results indicate that by inhibiting ATP synthesis, MgtC frees up Mg2+ ions required for translation.
The mgtA mgtB double mutant also incorporated less AHA than the wild-type strain (Figures 4C and S5C). The defect of the mgtA mgtB mutant was corrected by the increased cytosolic Mg2+ conferred by the mgtA-expressing plasmid, but not by the vector control (Figure 4C and S5C). In contrast to the rescue of the mgtC mutant (Figure S6A), pATPase-mediated ATP hydrolysis did not correct protein synthesis in the mgtA mgtB mutant (Figure S6C). These results likely reflect that the mgtA mgtB double mutant fails to import (as opposed to re-distribute) Mg2+ ions required for translation.
Wild-Type Salmonella Reduces the Rate of Protein Synthesis during Growth in Low Cytoplasmic Mg2+
Growth of wild-type Salmonella in low Mg2+ media is characterized by a logarithmic phase taking place in the first 4 h (OD600 ≤ 0.4), followed by a linear, slow-growing phase (Figure 5A). The onset of the slow-growing phase coincides with a decrease in cytosolic Mg2+ that triggers the synthesis of MgtA, MgtB and MgtC (Figure 5A) (Cromie et al. 2006; Cromie and Groisman, 2010; Alix and Blanc-Potard, 2008). By reducing the synthesis of ribosomal components (Figures 2B and and5A),5A), MgtA, MgtB and MgtC are expected to reduce the translation rate of the wild-type strain. As predicted, AHA incorporation decreased two-fold between 2.5 and 5.5 h of growth in low Mg2+ (Figures 5B and and5C),5C), which correspond to times before and after the synthesis of MgtA, MgtB and MgtC, respectively. A reduced translation rate may help Salmonella enter a semi-quiescent state characteristic of chronic infections and increased tolerance to antimicrobial agents. Notably, the MgtC protein, which plays a key role in this transition (Figure 5A), is required for Salmonella survival in a mouse model of chronic infection (Lawley et al., 2006).
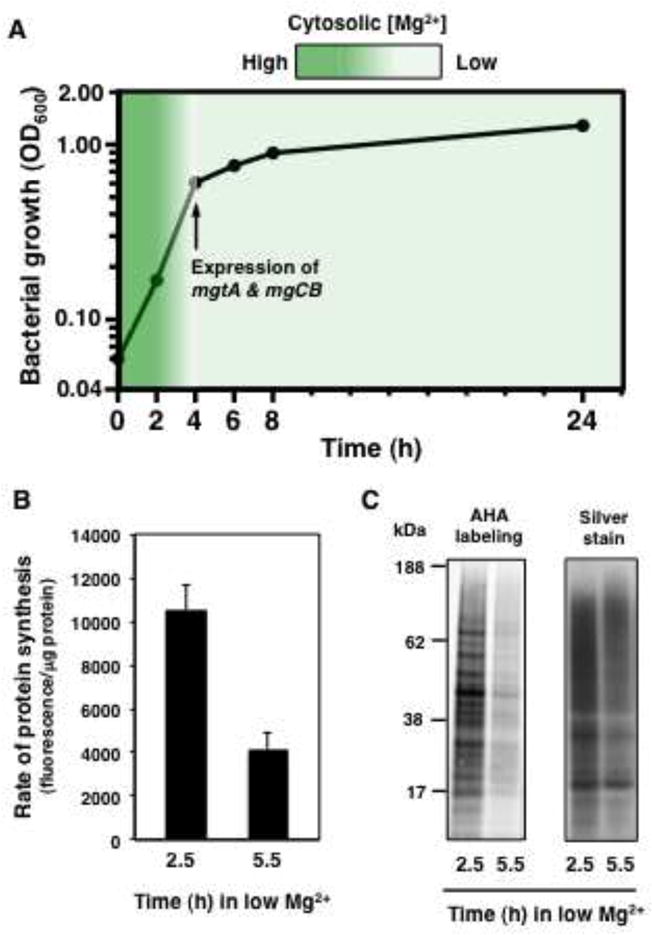
(A) Growth curve of wild-type Salmonella (14028s) in low (10 μM) Mg2+. The arrow indicates the time when MgtA, MgtB and MgtC are expressed. The color intensity of the legend depicts the concentration of Mg2+ in the cytosol. (B) Quantification of L-azidohomoalanine (AHA) labeling of wild-type Salmonella (14028s) prior to (2.5 h) and after (5.5 h) cytosolic Mg2+ starvation. Error bars represent standard deviations. Graphs show the average of four independent experiments. (C) SDS-PAGE analysis of AHA labeling of wild-type (14028s) prior to (2.5 h) and after (5.5 h) cytosolic Mg2+ starvation. The gel is representative of four independent experiments. All experiments were carried out in low Mg2+ (10 μM) Mg2+ N-minimal medium lacking methionine.
E. coli Buffers Translation During Cytosolic Mg2+ Starvation by Importing Mg2+ and Lowering ATP Levels
E. coli has homologs of the phoP, phoQ and mgtA genes (Kato et al., 1999) but lacks mgtC and mgtB (Blanc-Potard et al., 1999). We thus wondered whether E. coli is capable of a response analogous to that described above for Salmonella when facing low cytosolic Mg2+.
We determined that when Mg2+ is removed from the growth medium, wild-type E. coli experienced a decrease in both cytosolic Mg2+ (Figure 6A) and ATP levels (Figure 6B). By contrast, ATP levels actually increased in an E. coli phoP mutant, and to a lesser degree in an mgtA mutant (Figure 6B). In E. coli, PhoP promotes transcription of the yhiD gene, which specifies a product with low sequence similarity to MgtC and hypothesized to function as an Mg2+ transporter (Alteri et al., 2011; Hattori et al., 2009). However, in contrast to MgtC, YhiD does not interact with and inhibit the F1FO ATP synthase (Lee et al., 2013). In agreement with this notion, a yhiD mutant displayed wild-type ATP levels (Figure 6B).
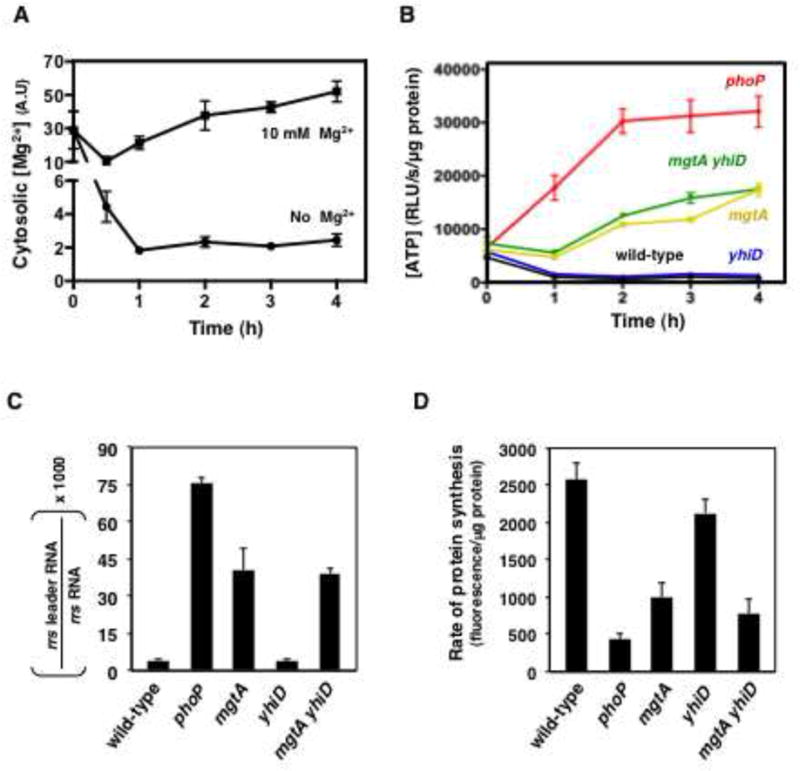
(A) Time-course cytosolic Mg2+ concentrations of wild-type E. coli grown in medium containing 0 or 10 mM Mg2+. Cytosolic Mg2+ concentrations reflect the normalized β-galactosidase activity values obtained with a genetic reporter for cytosolic Mg2+ and are expressed in arbitrary units (A.U.) (see Experimental Procedures). (B) Time-course quantification of ATP levels in wild-type (MG1655), phoP (EG12976), mgtA (EG16771), yhiD (MP751) and mgtA yhiD (MP752) E. coli grown in medium lacking Mg2+. (C) qPCR quantification of rrs transcriptional activity in wild-type (MG1655), phoP (EG12976), mgtA (EG16771), yhiD (MP751) and mgtA yhiD (MP752) E. coli 2 h following removal of Mg2+ from the growth medium. (D) Quantification of L-azidohomoalanine labeling of wild-type (MG1655), phoP (EG12976), mgtA (EG16771), yhiD (MP751) and mgtA yhiD (MP752) strains of E. coli 2 h following removal of Mg2+ from the growth medium. All experiments were carried out in MOPS glucose medium. Error bars represent standard deviations. Graphs show the average of four independent experiments.
We established that under cytosolic Mg2+ starvation, translation efficiency is inversely related to rrs transcription and cytosolic ATP concentrations. That is, wild-type and yhiD E. coli displayed low ATP (Figure 6B) and rrs transcription (Figure 6C), and synthesized proteins efficiently (Figure 6D). By contrast, mutants defective in phoP, mgtA or both mgtA and yhiD had elevated ATP (Figure 6B) and rrs transcription (Figure 6C), and were defective for translation (Figure 6D). The phoP mutant displayed the highest ATP levels (Figure 6B) and rrs transcriptional activity (Figure 6C), and exhibited the greatest translation defect (Figure 6D). Taken together, the results presented in this section indicate that E. coli utilizes the PhoP-activated MgtA and yet an unidentified gene(s) to maintain translation homeostasis when faced with low cytosolic Mg2+.
Concluding Remarks
We have now identified a genetic program that enables bacteria to coordinate the synthesis of ribosome precursors with the cytosolic Mg2+ available for their assembly. The existence of such program indicates that when cells are deprived of this cation and their cytosolic Mg2+ levels start to decrease, the assembly of ribosomal subunits will become compromised while the Mg2+-dependent enzymatic reactions responsible for the synthesis of ribosomal subunit themselves are still unaffected. The physiological adaptations implemented by this program enables cells to maintain the levels of free cytosolic Mg2+, allowing the use and recycling of this cation for various processes including the Mg2+-demanding association between the 30S and 50S ribosomal subunits (Zitomer and Flaks, 1972; Blaha et al., 2002) (Figure 7A–C).
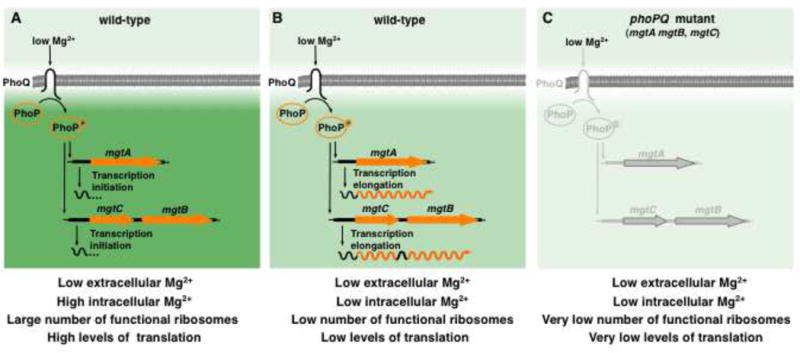
(A) Low extracytoplasmic Mg2+ activates the sensor PhoQ, which promotes the phosphorylation state of the regulator PhoP (PhoP-P). PhoP-P binds to the promoters of mgtA and mgtCB and recruit RNA polymerase. Transcription at these promoters is initiated but does not proceed into the coding regions because cytosolic Mg2+ levels are still high. There is a high abundance of functional ribosomes and translation is efficient. (B) As cells grow and consume the existing Mg2+ in the environment, levels of free cytosolic Mg2+ eventually drops bellow a certain threshold. This decrease in free cytosolic Mg2+ impairs ribosomal subunit association, causing a global slow down on translation that promotes transcription elongation into the coding regions of the mgtA, mgtC and mgtB genes. The Mg2+ transporters MgtA and MgtB import Mg2+ into the cytoplasm, and MgtC decreases ATP levels and promotes ppGpp accumulation. This response increases the levels of free cytosolic Mg2+, but decreases the number of functional of ribosomes, which enables cells to continue translation, albeit at a reduced rate. (C) Mutants unable to elicit the response presented in (B) (i.e., phoP, phoQ, mgtC and mgtA mgtB) are unable to maintain the pool of free cytosolic Mg2+ and thus, are defective to assemble their ribosomes and have much reduced translation.
The PhoP protein promotes transcription initiation of the mgtA gene and mgtCBR operon when extracytoplasmic Mg2+ is low (Véscovi et al., 1996; Soncini et al., 1996) (Figure 7A). However, transcription of the mgtA and mgtCBR mRNA coding regions takes place only at low cytosolic Mg2+ (Cromie et al., 2006; Spinelli et al., 2008), through mechanisms that rely on the inefficient translation of small open reading frames (ORFs) located in their respective leader mRNAs (Park et al., 2010; Zhao et al., 2011; Lee and Groisman 2012a; Lee and Groisman 2012b) (Figure 7B). These translation-dependent regulatory mechanisms enable Salmonella to link information about its cellular translation status with the expression of these proteins. On this note, it is interesting that the expression of the major vertebrate Mg2+ transporter TRMP7 is also triggered by inefficient translation of small leader ORFs (Nikonorova et al., 2014). The multilayered regulation of mgtA and mgtCBR transcription ensures that the PhoP-mediated repression of ribosome synthesis occurs only when cytosolic Mg2+ drop to levels that compromise the function of ribosomes.
From a broader perspective, our results help explain several findings pertaining to the relationship between ribosomes and Mg2+. First, the dramatic decrease in ribosomes manifested by E. coli cultures deprived of Mg2+ (McCarthy, 1962) results from a genetically encoded response. Second, the growth rate of an E. coli variant with engineered tethered ribosomal subunits increases upon mutations of the ribosomal subunit gene rpsA and the putative Mg2+ efflux gene ybeX (Orelle et al. 2015). Third, overexpression of a Mg2+ uptake system or inactivation of a Mg2+ efflux protein suppresses a defect in ribosomal subunit association in Bacillus subtilis (Akanuma et al., 2014; Nierhaus, 2014). And fourth, the cytosolic Mg2+ concentration decreases as the number of rrs genes in B. subtilis is reduced (Akanuma et al., 2014).
The synthesis of cellular machinery, such as enzymes and ribosomes, is typically responsive to the availability of the corresponding substrates and/or the energy requirements of the machine. For instance, ribosome production is promoted when ATP levels are high (Gaal et al., 1997; Schneider et al., 2002), and inhibited when organisms experience amino acid limitation (Sarmientos et al., 1983). Our findings now provide a singular example of a small molecule governing the synthesis of a cellular machine despite being neither a substrate nor an energy source. That is, cytosolic Mg2+ limitation supersedes amino acid availability and ATP abundance to shut down ribosome production because, without Mg2+, ribosomes would not assemble or function properly.
All living organisms translate mRNAs using ribosomes that are stabilized by Mg2+ ions and powered by ATP. Therefore, the principles governing translation homeostasis during cytosolic Mg2+ starvation are likely to be widely conserved, even if the proteins mediating the response may differ across species. That is, organisms respond to this stress by importing extracellular Mg2+ and repurposing cytosolic Mg2+ by reducing both ATP and ribosome biosynthesis (Pontes et al. 2015a). The repression of ATP and ribosome production serves to fulfill two physiological requirements: (i) a readjustment in the synthesis of ribosomes dictated by the amount of Mg2+ available in the cytosol, and (ii) a reallocation of Mg2+ ions bound to ATP molecules and idle ribosomes. Ultimately, this response allows translation to continue with a reduced ribosome pool.
Experimental Procedures
Bacterial Strains, Plasmid Constructs, Primers, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table S1. Oligonucleotide sequences are presented in Table S2. Single gene knockouts and deletions were carried out as described (Datsenko and Wanner, 2002). Mutations generated using this method were subsequently moved into clean genetic backgrounds via phage P22- (Salmonella) or P1vir- (E. coli) mediated transduction as described (Davis et al., 1980; Thomason et al., 2007). Bacterial strains used in recombination and transduction experiments were grown in LB medium at 30°C or 37°C (Datsenko and Wanner, 2002). When required, LB medium was supplemented with ampicillin (100 μg/mL), chloramphenicol (20 μg/mL), kanamycin (50 μg/mL), rifampicin (50 μg/mL), erythromycin (200 μg/mL), tetracycline (15 μg/mL) and/or L-arabinose (0.2% wt/vol). For Salmonella strains, growth under defined Mg2+ concentrations was carried out in N-minimal medium supplemented with 0.1% casamino acids (Soncini et al., 1996). However, in experiments involving relA spoT double mutants, all strains were grown in MOPS medium (Neidhardt et al., 1974) containing 30 mM glucose, 0.2% casamino acids and lacking CaCl2 to avoid repression of the PhoP/PhoQ system (Véscovi et al., 1996). These media were supplemented with either 10 mM (high) or 10 μM (low) MgCl2. For E. coli strains, growth under defined Mg2+ concentrations was carried out in the aforementioned MOPS medium containing 10 mM MgCl2 or lacking Mg2+. When required, defined media were supplemented with ampicillin (25 μg/mL), 200 μg/mL (overnight growth) or 80 μg/mL (physiological experiment) of erythromycin, 15 μg/mL chloramphenicol, 250 μM (pMgtC) or 100 μM (pMgtA or pMgtB) isopropyl β-D-1-thiogalactopyranoside (IPTG) and serine hydroxamate (50 μg/mL).
In Mg2+ starvation experiments, all strains were grown in a shaking water bath at 250 rpms and 37°C. For Salmonella strains, cultures were grown overnight in N-minimal liquid media containing high Mg2+. Cells were then washed 3x with Mg2+-free medium and inoculated 1:25 (strains harboring pATPase plasmid) or 1:50 (all other strains) into low Mg2+ N-minimal medium. For E. coli, following overnight growth in high Mg2+ MOPS medium, strains were subcultured (1:100) intro fresh high Mg2+ MOPS medium and grown for 2.5 h. Cells were then harvested by centrifugation, washed 3x with MOPS medium lacking Mg2+ and resuspended in MOPS medium containing either 10 mM Mg2+ or lacking Mg2+.
Construction of mgtC rpoBL533P and mgtC rpoBδ532a Strains
Introduction of point mutation in the rpoB gene was carried out via λ-red-mediated recombination (Datsenko and Wanner, 2002), as described in the Supplemental Experimental Procedures.
Construction of Plasmid Harboring rrsC P1-gfp
Wild-type Salmonella genomic DNA was used as template in a PCR reaction with primers pair W1480 and W1482. PCR product corresponding to the promoter region of rrsC P1 (14028s chromosomal base pairs 4,113,424–605) was digested with BamHI and EcoRI and ligated into pFPV25AAV(Choi and Groisman, 2013) digested with the same restriction enzymes. Ligation reactions were transformed into DH5α cells, and the identity of the inserts was verified by DNA sequencing.
Estimation of ATP Levels from Bacterial Samples
ATP measurements were carried out as described (Pontes et al., 2015b).
RNA Extraction and Primer Extension
RNA extraction and primer extension reactions were carried out using primers W2596 (rrs leader) and W2595 (ompA) as described (Prévost et al., 2007), with the exception that 20 μg of RNA were used in reverse transcription reactions.
RNA Extraction and Quantitative PCR
RNA extraction and quantification was carried out as described (Park et al., 2010), with the exception that cDNA was synthesized from RNA samples using the Superscript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Relative amounts of cDNA were determined using a standard curve obtained from qPCR with serially diluted wild-type E. coli genomic DNA, wild-type Salmonella genomic DNA or pFPV25AAV DNA. For quantification of the reporter fusion, gfp mRNA levels were normalized by mRNA levels derived from plasmid-borne bla gene. The following strategy was used to quantify rrs transcriptional activity: following rrs transcription, the structural portion of the 16S RNA is assembled into a stable 30S subunit whereas the leader region is quickly degraded (Shajani et al., 2011; Zhi et al., 2003). Thus, a decline in transcription initiation at the rrs promoters will result in a decrease in the ratio of the unstable leader to its corresponding stable structural 16S rRNA.
L-azidohomoalanine (AHA) Labeling and Quantification
Labeling of nascent protein synthesis was carried out using Click-iT Protein Reaction Buffer Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Detailed procedure is described in Supplemental Experimental Procedures.
Polysome Analysis
Following overnight growth in high Mg2+ N-minimal medium, cells were washed 3x in Mg2+-free medium and used to inoculate (1:50) fresh N-minimal medium containing high or low Mg2+. After 3 (high Mg2+) or 6 h (low Mg2+) of growth (OD600 ≈ 0.4–0.6), cells were collected, lysed and their polysomes were separated in a sucrose gradient as described (Qin and Fredrick, 2013). Sucrose gradients were separated into 30 fractions using a Gradient Station IP fractionator (BioComp), and the first 27 fractions were analyzed. The absorbance at 260 nm of the fractions was determined using a NanoDrop 8000 (Thermo Scientific). The identity of the 260 nm peaks were established by extracting fractions with phenol:chloroform, ethanol precipitating, and separating them by denaturing agarose gel electrophoresis.
Estimation of Protein/RNA Ratios
Following overnight growth in high Mg2+ N-minimal medium, Salmonella strains were washed 3x in Mg2+-free medium and used to inoculate (1:50) fresh low Mg2+ N-minimal medium. After 6 h of growth (OD600 ≈ 0.4–0.6), cells were collected, resuspended in dH2O and lysed by sonication. Crude cell extracts were used for protein and RNA quantification using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) and orcinol assay, respectively.
Estimation of Cytosolic Mg2+
Quantification of cytosolic Mg2+ concentration was carried out using a genetic reporter for intracellular Mg2+ (Cromie et al., 2006) and is detailed in Supplemental Experimental Procedures.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were performed as described (Choi and Groisman, 2013), with the minor modifications described in the Supplemental Experimental Procedures.
Nucleotide Labeling and Quantification
Salmonella strains were grown overnight in high Mg2+ N-minimal medium or MOPS (wild-type and relA spoT control strains). Strains were washed 3x with Mg2+-free media, used to inoculate (1:50) low Mg2+ N-minimal medium or MOPS (relA spoT strain) and grown at 37°C with aeration. Following 3 h of growth, 32P orthophosphoric acid (Perkin Elmer) was added to each culture at a final concentration of 150 μCi/mL. Labeling was allowed to proceed for 3 h at 37°C with aeration. Nucleotides were extracted and separated on a PEI-cellulose thin layer chromatography (Sigma-Aldrich) as described (Xiao et al., 1991). Wild-type and relA spoT control strains were treated with 50 μg/mL of serine hydroxamate 15 min prior to nucleotide extraction.
Supplementary Material
Figure S1. Repression of rrs Transcription by the PhoP-Activated mgtA, mgtB and mgtC Genes, Related to Figures 2 and and3.3. (A) rrs transcriptional activity of wild-type Salmonella (14028s) grown in high (10 mM), and wild-type (14028s) and phoP (MS7953) Salmonella grown in low (10 μM) Mg2+. Error bars represent standard deviations. Graph is the average of three independent qPCR experiments. (B) % of rrs leader rRNA region in wild-type (14028s) and phoP (MS7953) Salmonella following rifampicin (200 μg/ml) treatment to stop new transcription. Error bars represent standard deviations. Graph is the average of four independent experiments. (C) Electrophoretic mobility shift assays of DNA fragments harboring the Salmonella phoP, hisL, rrsC P1 and rrsC P2 promoters with phosphorylated PhoP protein. The gel is representative of three independent experiments. (D) Primer extension analyses of wild-type (14028s), mgtC (EL4) and mgtC (EL4) Salmonella harboring the mgtC-expressing plasmid (pMgtC) or the plasmid vector (pUHE-21-2-lacIq) following growth in low Mg2+. (E) Primer extension analyses of wild-type (14028s), mgtA mgtB (EG16741) and mgtA mgtB (EG16741) Salmonella harboring the mgtA-expressing plasmid (pMgtA) or the plasmid vector (pUHE-21-2-lacIq) during growth in low Mg2+. (F) Primer extension analyses of wild-type (14028s), phoP (MS7953) and phoP (MS7953) Salmonella harboring the plasmid vector (pUHE-21-2-lacIq), the mgtB-expressing plasmid (pMgtB), pMgtC and the mgtCB-expressing plasmid (pMgtCB). For rrs primer extension: lanes 1 and 2, and 3 to 5 correspond to lanes 1 and 2, and 9 to 12 of the gel displayed on Figure S7C, respectively. (G) ATP levels of wild-type (14028s), mgtA mgtB (EG16741), mgtC (EL4), mgtA mgtB mgtC (EG16738) and phoP (MS7953) Salmonella strains following 6 h growth in low Mg2+. All measurements were carried following 6 h growth in low Mg2+ N-minimal medium (OD600 ≈ 0.4–0.6). Primer extension analyses are representative of 2–4 independent experiments.
Figure S2. The PhoP-Activated mgtA, mgtB and mgtC Genes Repress rrs P1 Promoters, Related to Figures 2 and and3.3. (A) gfp mRNA levels in wild-type (14028s), mgtC (EL4) and mgtA mgtB (EG16741) Salmonella harboring pP1rrsC-gfpAAV following 6 h growth in low (10 μM) Mg2+ N-minimal medium (OD600 ≈ 0.4–0.6). Error bars represent standard deviations. Graphs show the average of four independent experiments analyzed by qPCR. Values are normalized to those of the constitutively transcribed bla gene, which is also in the plasmid. (B) Schematic illustration of plasmid pP1rrsC-gfpAAV, containing a transcriptional fusion between Salmonella’s rrsC P1 promoter and an unstable gfp reporter gene. (C) Sequence alignment of Salmonella rrs P1 promoters. Promoters’ UP-element is underlined, −35 and −10 regions are in bold and highlighted in gray, and transcription start site (+1) is depicted in red.
Figure S3. Effects of Mg2+ Starvation on Growth, ATP and ppGpp Levels, Related to Figure 3. (A) Detection of 32P-labeled ppGpp by thin layer chromatography (TLC). Leftmost: 32P-labeled nucleotides derived from wild-type (14028s) (positive control) and relA spoT (MP342) (negative control) Salmonella following 30 min treatment with serine hydroxamate (Shx, 50 μg/ml). Rightmost: nucleotides derived from wild-type (14028s), mgtC (EL4) and mgtC (EL4) Salmonella harboring the mgtC-expressing plasmid (pMgtC) or the plasmid vector (pUHE-21-2-lacIq). Nucleotides were extracted following 6 h of growth in low (10 μM) Mg2+ MOPS (leftmost strains) or N-minimal medium (rightmost strains) (OD600 ≈ 0.4–0.6). Arrows indicate signals corresponding to ATP, GTP, ppGpp and the origin. Top TLC shows signals from the origin to ATP. Bottom TLC shows high-contrasted region encompassing the origin to GTP. TLCs are representative of four independent experiments. (B) Quantification of ATP levels in wild-type (14028s) and mgtC (EL4), and relA spoT (MP342) and mgtC relA spoT (MP343) Salmonella following 6 h growth in MOPS glucose medium containing low Mg2+. Error bars represent standard deviations. Graphs show the average of four independent experiments. (C) Growth curves of wild-type (14028s), mgtC (EL4), relA spoT (MP342) and mgtC relA spoT (MP343) Salmonella in MOPS glucose medium containing low (10 μM) Mg2+. Dashed lines indicate the time where wild-type (14028s) and mgtC (EL4), and relA spoT (MP342) and mgtC relA spoT (MP343) Salmonella experience cytosolic Mg2+ starvation.
Figure S4. MgtA and MgtB Maintain Cytosolic Mg2+ Concentrations, Related to Figures 2 and and6.6. (A) Cartoon representation of the genetic reporter for cytosolic Mg2+ levels present in the pPlac1–6-mgtAleader-lacZ plasmid. This reporter consists of a constitutive promoter driving transcription of Salmonella’s mgtA Mg2+-sensing riboswitch fused to a promoterless lacZ gene. When cytosolic Mg2+ levels decrease, the riboswitch adopts a conformation that allows transcription to continue into lacZ, resulting in heighted β-galactosidase activity (Cromie et al., 2006). (B) Cartoon representation of the control pPlac1–6r-lacZ, consisting of an isogenic lacZ fusion lacking the Mg2+-sensing riboswitch (Cromie et al., 2006). β-galactosidase activities derived from this control fusion can serve as normalization factor, by accounting for potential differences in plac1–6 promoter activity or lacZ translation efficiency across strains. (C) β-galactosidase activities of wild-type (14028s), mgtA mgtB (EG17048) and mgtA mgtB (EG17048) Salmonella harboring the mgtA-expressing plasmid (pMgtA), the mgtB-expressing plasmid (pMgtB) or the plasmid vector (pUHE-21-2-lacIq) with pPlac1–6-mgtAleader-lacZ or pPlac1-6r-lacZ in following 6 h growth in low (10 μM) Mg2+ N-minimal medium (OD600 ≈ 0.4–0.6).
Figure S5. MgtA, MgtB and MgtC Enable Translation During Cytosolic Mg2+ Starvation, Related to Figure 4. (A) Leftmost panel: Polysome analysis of wild-type (14028s) Salmonella after 3 h growth in high (10 mM) Mg2+ N-minimal medium (OD600 ≈ 0.5–0.6). Rightmost panel: denaturing agarose gel electrophoretic analysis of polysomal fractions shown on the leftmost panel. Polysome profile is representative of four independent experiments. (B) SDS-PAGE analysis of L-azidohomoalanine (AHA) labeling of wild-type (14028s), mgtC (EL4) and mgtC (EL4) Salmonella harboring the mgtC-expressing plasmid (pMgtC) or the plasmid vector (pUHE-21-2-lacIq) during growth in low (10 μM) Mg2+. (C) SDS-PAGE analysis of AHA labeling of wild-type (14028s), mgtA mgtB (EG16741) and mgtA mgtB (EG16741) Salmonella harboring the mgtA-expressing plasmid (pMgtA) or the plasmid vector (pUHE-21-2-lacIq) during growth in low Mg2+. In (B) and (C), bacteria were grown in low (10 μM) Mg2+ N-minimal medium lacking methionine. AHA labeling was carried out from 5.5 to 6 h growth (final OD600 ≈ 0.4–0.5). Gels are representative of four independent experiments.
Figure S6. MgtA and MgtB Promote Translation by Importing Mg2+, Whereas MgtC Enables Translation by Redistributing Cytosolic Mg2+, Related to Figure 4. (A) Quantification and SDS-PAGE analysis of L-azidohomoalanine (AHA) labeling of wild-type (14028s) and mgtC (EL4) Salmonella harboring the ATPase expressing plasmid pATPase or the plasmid vector (pCP44) during growth in low (10 μM) Mg2+. (B) Quantification and SDS-PAGE analysis of AHA labeling of wild-type (14028s) and mgtC (EL4) strains of Salmonella harboring the mgtA-expressing plasmid (pMgtA) or the plasmid vector (pUHE-21-2-lacIq) during growth in low Mg2+. (C) Quantification and SDS-PAGE analysis of AHA labeling of wild-type (14028s) and mgtA mgtB (EG16741) Salmonella harboring pATPase or the plasmid vector (pCP44) during growth in low Mg2+. Strains were grown in low Mg2+ N-minimal medium lacking methionine. AHA labeling was carried out from 5.5 to 6 h growth (final OD600 ≈ 0.4–0.5). Error bars represent standard deviations. Graphs show the average of four independent experiments. Gels are representative of four independent experiments.
Figure S7. Ribosomal rrs primer extension analyses, Related to Figure 3. (A) Primer extension analyses of wild-type (14028s; lane 1), mgtC (EL4; lane 2), and mgtC (EL4) Salmonella harboring the ATPase expressing plasmid pATPase (lane 7) or the plasmid vector (pCP44; lane 8) during growth in low (10 μM) Mg2+. (B) Primer extension analyses of wild-type (14028s; lane 1), mgtC (EL4; lane 2), relA spoT (MP342; lane 5) and mgtC relA spoT (MP343; lane 6) Salmonella growth in MOPS glucose medium containing low (10 μM) Mg2+. (C) Primer extension analyses of wild-type (14028s; lane 1), phoP (MS7953; lane 2) and phoP (MS7953) Salmonella harboring the plasmid vector (pUHE-21-2-lacIq; lane 9), the mgtB-expressing plasmid (pMgtB; lane 10), pMgtC (lane 11) and the mgtCB-expressing plasmid (pMgtCB; lane12).
Table S1. Bacterial Strains and Plasmids Used in this Study, Related to Figures 2, ,3,3, ,44 and and55.
Table S2. Oligonucleotides Sequences Used in this Study, Related to Figures 2, ,3,3, ,44 and and55.
Acknowledgments
The authors would like to thank Dr. Peter R. Jensen (Technical University of Denmark) for kindly providing plasmids pCP44 and pCP41-AtpAGD, Anastasia Sevostiyanova for kindly providing purified PhoP*-His, Hubert Salvail for assistance with primer extension analyses, Anastasia Sevostiyanova, Richard L. Gourse and Michael Ibba for critical reading of this manuscript. This research was supported by grant AI49561 from the National Institutes of Health to EAG.
Footnotes
Author Contributions
E.A.G and M.H.P. designed the experiments and wrote the paper. M.H.P. conducted the vast majority of the experiments. J.Y. participated in the polysome profiling assays.References
- Akanuma G, Kobayashi A, Suzuki S, Kawamura F, Shiwa Y, Watanabe S, Yoshikawa H, Hanai R, Ishizuka M. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J Bacteriol. 2014;196:3820–3830. [Europe PMC free article] [Abstract] [Google Scholar]
- Alix E, Blanc-Potard AB. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008;27:546–557. [Europe PMC free article] [Abstract] [Google Scholar]
- Alteri CJ, Lindner JR, Reiss DJ, Smith SN, Mobley HL. The broadly conserved regulator PhoP links pathogen virulence and membrane potential in Escherichia col. Mol Microbiol. 2011;82:145–163. [Europe PMC free article] [Abstract] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitr. J Mol Biol. 2001;305:673–688. [Abstract] [Google Scholar]
- Blaha G, Burkhardt N, Nierhaus KH. Formation of 70S ribosomes: large activation energy is required for the adaptation of exclusively the small ribosomal subunit. Biophys Chem. 2002;96:153–161. [Abstract] [Google Scholar]
- Blanc-Potard AB, Solomon F, Kayser J, Groisman EA. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. [Europe PMC free article] [Abstract] [Google Scholar]
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Washington D.C.: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia col. Nature. 1969;221:838–841. [Abstract] [Google Scholar]
- Chamnongpol S, Groisman EA. Acetyl phosphate-dependent activation of a mutant PhoP response regulator that functions independently of its cognate sensor kinase. J Mol Biol. 2000;300:291–305. [Abstract] [Google Scholar]
- Choi J, Groisman EA. The lipopolysaccharide modification regulator PmrA limits Salmonella virulence by repressing the type three-secretion system Spi/Ssa. Proc Natl Acad Sci USA. 2013;110:9499–9504. [Europe PMC free article] [Abstract] [Google Scholar]
- Cole JR, Olsson CL, Hershey JW, Grunberg-Manago M, Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J Mol Biol. 1987;198:383–392. [Abstract] [Google Scholar]
- Condon C, French S, Squires C, Squires CL. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 1993;12:4305–4315. [Europe PMC free article] [Abstract] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. [Abstract] [Google Scholar]
- Cromie MJ, Groisman EA. Promoter and riboswitch control of the Mg2+ transporter MgtA from Salmonella enteric. J Bacteriol. 2010;192:604–607. [Europe PMC free article] [Abstract] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. [Europe PMC free article] [Abstract] [Google Scholar]
- Davis RW, Bolstein D, Roth JR. Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab; 1980. [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–540. [Abstract] [Google Scholar]
- Fagerbakke KM, Norland S, Heldal M. The inorganic ion content of native aquatic bacteria. Can J Microbiol. 1999;45:304–311. [Abstract] [Google Scholar]
- Forchhammer J, Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J Mol Biol. 1971;55:563–568. [Abstract] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. [Abstract] [Google Scholar]
- Gesteland RF. Unfolding of Escherichia coli ribosomes by removal of magnesium. J Mol Biol. 1966;18:356–371. [Abstract] [Google Scholar]
- Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitr. Nucleic Acids Res. 1988;16:9789–9809. [Europe PMC free article] [Abstract] [Google Scholar]
- Groisman EA, Hollands K, Kriner MA, Lee EJ, Park SY, Pontes MH. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet. 2013;47:625–646. [Europe PMC free article] [Abstract] [Google Scholar]
- Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. [Europe PMC free article] [Abstract] [Google Scholar]
- Hattori M, Iwase N, Furuya N, Tanaka Y, Tsukazaki T, Ishitani R, Maguire ME, Ito K, Maturana A, Nureki O. Mg2+-dependent gating of bacterial MgtE channel underlies Mg2+ homeostasis. EMBO J. 2009;28:3602–3612. [Europe PMC free article] [Abstract] [Google Scholar]
- Jinks-Robertson S, Gourse RL, Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983;33:865–876. [Abstract] [Google Scholar]
- Kato A, Tanabe H, Utsumi R. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol. 1999;181:5516–5520. [Europe PMC free article] [Abstract] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. [Europe PMC free article] [Abstract] [Google Scholar]
- Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J Bacteriol. 2002;184:3909–3916. [Europe PMC free article] [Abstract] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee EJ, Groisman EA. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature. 2012a;486:271–275. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee EJ, Groisman EA. Tandem attenuators control expression of the Salmonella mgtCBR virulence operon. Mol Microbiol. 2012b;86:212–224. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee EJ, Pontes MH, Groisman EA. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1FO ATP synthase. Cell. 2013;154:146–156. [Europe PMC free article] [Abstract] [Google Scholar]
- Maaløe O, Kjeldgaard NO. Control of Macromolecular Synthesis: A Study of DNA, RNA, and Protein Synthesis in Bacteria. New York: W. A. Benjamin; 1966. [Google Scholar]
- Maguire ME, Cowan JA. Magnesium chemistry and biochemistry. Biometals. 2002;15:203–210. [Abstract] [Google Scholar]
- McCarthy BJ. The effects of magnesium starvation on the ribosome content of Escherichia col. Biochim Biophys Acta. 1962;55:880–889. [Google Scholar]
- Mikkola R, Kurland CG. Evidence for demand-regulation of ribosome accumulation in E col. Biochimie. 1991;73:1551–1556. [Abstract] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. [Abstract] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. [Europe PMC free article] [Abstract] [Google Scholar]
- Nierhaus KH. Mg2+, K+, and the ribosome. J Bacteriol. 2014;196:3817–3819. [Europe PMC free article] [Abstract] [Google Scholar]
- Nikonorova IA, Kornakov NV, Dmitriev SE, Vassilenko KS, Ryazanov AG. Identification of a Mg2+-sensitive ORF in the 5′-leader of TRPM7 magnesium channel mRNA. Nucleic Acids Res. 2014;42:12779–12788. [Europe PMC free article] [Abstract] [Google Scholar]
- Nomura M, Watson JD. Ribonucleoprotein particles within chloromycetin-inhibited Escherichia col. J Mol Biol. 1959;1:204–217. [Google Scholar]
- Okuno D, Iino R, Noji H. Rotation and structure of FoF1-ATP synthase. J Biochem. 2011;149:655–664. [Abstract] [Google Scholar]
- Orelle C, Carlson ED, Szal T, Florin T, Jewett MC, Mankin AS. Protein synthesis by ribosomes with tethered subunits. Nature. 2015;254:119–124. [Abstract] [Google Scholar]
- Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. [Europe PMC free article] [Abstract] [Google Scholar]
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–1273. [Abstract] [Google Scholar]
- Qin D, Fredrick K. Analysis of polysomes from bacteria. Methods Enzymol. 2013;530:159–172. [Abstract] [Google Scholar]
- Pontes MH, Sevostyanova A, Groisman EA. When Too Much ATP Is Bad for Protein Synthesis. J Mol Biol. 2015a;427:2586–2594. [Europe PMC free article] [Abstract] [Google Scholar]
- Pontes MH, Lee EJ, Choi J, Groisman EA. Salmonella promotes virulence by repressing cellulose production. Proc Natl Acad Sci USA. 2015b;112:5183–5188. [Europe PMC free article] [Abstract] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarmientos P, Cashel M. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. Proc Natl Acad Sci USA. 1983;80:7010–7013. [Europe PMC free article] [Abstract] [Google Scholar]
- Sarmientos P, Sylvester JE, Contente S, Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983;32:1337–1346. [Abstract] [Google Scholar]
- Schaechter M, Maaløe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimuriu. J Gen Microbiol. 1958;19:592–606. [Abstract] [Google Scholar]
- Schneider DA, Gaal T, Gourse RL. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc Natl Acad Sci USA. 2002;99:8602–8607. [Europe PMC free article] [Abstract] [Google Scholar]
- Schneider DA, Gourse RL. Changes in Escherichia coli rRNA promoter activity correlate with changes in initiating nucleoside triphosphate and guanosine 5′ diphosphate 3′-diphosphate concentrations after induction of feedback control of ribosome synthesis. J Bacteriol. 2003a;185:6185–6191. [Europe PMC free article] [Abstract] [Google Scholar]
- Schneider DA, Gourse RL. Changes in the concentrations of guanosine 5′-diphosphate 3′-diphosphate and the initiating nucleoside triphosphate account for inhibition of rRNA transcription in fructose-1,6-diphosphate aldolase (fda) mutants. Bacteriol. 2003b;185:6192–6194. [Europe PMC free article] [Abstract] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A° resolution. Science. 2005;310:827–834. [Abstract] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. [Abstract] [Google Scholar]
- Shin D, Groisman EA. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in viv. J Biol Chem. 2005;280:4089–4094. [Abstract] [Google Scholar]
- Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: expression of cloned genes for three distinct Mg2+ transport systems. J Bacteriol. 1989a;171:4752–4760. [Europe PMC free article] [Abstract] [Google Scholar]
- Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J Bacteriol. 1989b;171:4761–4766. [Europe PMC free article] [Abstract] [Google Scholar]
- Soncini FC, Véscovi EG, Groisman EA. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. [Europe PMC free article] [Abstract] [Google Scholar]
- Soncini FC, Vescovi EG, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. [Europe PMC free article] [Abstract] [Google Scholar]
- Spinelli SV, Pontel LB, Véscovi EG, Soncini FC. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbio. Lett. 2008;280:226–234. [Abstract] [Google Scholar]
- Stouthamer AH. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek. 1973;39:545–565. [Abstract] [Google Scholar]
- Terasaki M, Rubin H. Evidence that intracellular magnesium is present in cells at a regulatory concentration for protein synthesis. Proc Natl Acad Sci USA. 1985;82:7324–7326. [Europe PMC free article] [Abstract] [Google Scholar]
- Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol. 2007 Chapter 1, Unit 1.17. [Abstract] [Google Scholar]
- Tosa T, Pizer LI. Biochemical bases for the antimetabolite action of L-serine hydroxamate. J Bacteriol. 1971;106:972–982. [Europe PMC free article] [Abstract] [Google Scholar]
- Véscovi EG, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulenc. Cell. 1996;84:165–174. [Abstract] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [Abstract] [Google Scholar]
- Wacker WE. The biochemistry of magnesium. Ann NY Acad Sci. 1969;162:717–726. [Abstract] [Google Scholar]
- Weiss RL, Morris DR. Cations and ribosome structure. I. Effects on the 30S subunit of substituting polyamines for magnesium ion. Biochemistry. 1973;12:435–441. [Abstract] [Google Scholar]
- Weiss RL, Kimes BW, Morris DR. Cations and ribosome structure. 3. Effects on the 30S and 50S subunits of replacing bound Mg2+ by inorganic cations. Biochemistry. 1973;12:450–456. [Abstract] [Google Scholar]
- Zhao G, Kong W, Weatherspoon-Griffin N, Clark-Curtiss J, Shi Y. Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J. 2011;30:1485–1496. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhi H, Wang X, Cabrera JE, Johnson RC, Jin DJ. Fis stabilizes the interaction between RNA polymerase and the ribosomal promoter rrnB P1, leading to transcriptional activation. J Biol Chem. 2003;278:47340–47349. [Abstract] [Google Scholar]
- Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. [Europe PMC free article] [Abstract] [Google Scholar]
- Zitomer RS, Flaks JG. Magnesium dependence and equilibrium of the Escherichia coli ribosomal subunit association. J Mol Biol. 1972;71:263–279. [Abstract] [Google Scholar]
- Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol. 2012;84:463–485. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2016.05.008
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276516301459/pdf
Citations & impact
Impact metrics
Article citations
Unlocking the electrochemical functions of biomolecular condensates.
Nat Chem Biol, 20(11):1420-1433, 26 Sep 2024
Cited by: 1 article | PMID: 39327453
Review
An intracellular phosphorus-starvation signal activates the PhoB/PhoR two-component system in <i>Salmonella enterica</i>.
mBio, 15(9):e0164224, 06 Aug 2024
Cited by: 1 article | PMID: 39152718 | PMCID: PMC11389368
Chaperone Hsp70 helps Salmonella survive infection-relevant stress by reducing protein synthesis.
PLoS Biol, 22(4):e3002560, 04 Apr 2024
Cited by: 0 articles | PMID: 38574172 | PMCID: PMC10994381
The Ribosome Hypothesis: Decoding Mood Disorder Complexity.
Int J Mol Sci, 25(5):2815, 29 Feb 2024
Cited by: 0 articles | PMID: 38474062 | PMCID: PMC10931790
Empowering Cartilage Restructuring with Biodegradable Magnesium Doped-Silicon Based-Nanoplatforms: Sustained Delivery and Enhanced Differentiation Potential.
Int J Nanomedicine, 19:491-506, 17 Jan 2024
Cited by: 0 articles | PMID: 38250188 | PMCID: PMC10800145
Go to all (49) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
When Too Much ATP Is Bad for Protein Synthesis.
J Mol Biol, 427(16):2586-2594, 04 Jul 2015
Cited by: 55 articles | PMID: 26150063 | PMCID: PMC4531837
Review Free full text in Europe PMC
RbfA and IF3 couple ribosome biogenesis and translation initiation to increase stress tolerance.
Nucleic Acids Res, 48(1):359-372, 01 Jan 2020
Cited by: 19 articles | PMID: 31728529 | PMCID: PMC7145577
The 100S ribosome: ribosomal hibernation induced by stress.
Wiley Interdiscip Rev RNA, 5(5):723-732, 18 Jun 2014
Cited by: 48 articles | PMID: 24944100
Protein synthesis controls phosphate homeostasis.
Genes Dev, 32(1):79-92, 01 Jan 2018
Cited by: 30 articles | PMID: 29437726 | PMCID: PMC5828397





