Abstract
Free full text

CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9orf72 dipeptide repeat protein toxicity
Abstract
Hexanucleotide repeat expansions in the C9orf72 gene are the most common cause of amyotrophic lateral sclerosis and frontotemporal dementia (c9FTD/ALS). The nucleotide repeat expansions are translated into dipeptide repeat (DPR) proteins, which are aggregation-prone and may contribute to neurodegeneration. We used the CRISPR-Cas9 system to perform genome-wide gene knockout screens for suppressors and enhancers of C9orf72 DPR toxicity in human cells. We validated hits by performing secondary CRISPR-Cas9 screens in primary mouse neurons. We uncovered potent modifiers of DPR toxicity whose gene products function in nucleocytoplasmic transport, the endoplasmic reticulum (ER), proteasome, RNA processing pathways, and in chromatin modification. One modifier, TMX2, modulated the ER-stress signature elicited by C9orf72 DPRs in neurons, and improved survival of human induced motor neurons from C9orf72 ALS patients. Together, this work demonstrates the promise of CRISPR-Cas9 screens to define mechanisms of neurodegenerative diseases.
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are devastating human neurodegenerative disorders. ALS is associated with progressive motor neuron loss from the brain and spinal cord, leading to muscle weakness, paralysis, and ultimately death, usually 2–5 years after symptom onset 1. FTD, the second most common cause of dementia in patients less than 65 years old, is caused by the degeneration of neurons from the frontal and temporal lobes of the brain and is associated with a range of cognitive and behavioral symptoms, including changes in personality. There is an emerging appreciation for clinical overlap between ALS and FTD, with evidence that FTD symptoms can be seen in ALS patients and motor neuron signs can be seen in FTD patients 2. The two disorders are also connected by pathology and genetics. Aggregates of the RNA-binding protein TDP-43 accumulate in neurons of nearly all ALS cases and almost half of FTD cases 3 and mutations in several genes can cause ALS, FTD, or even both (ALS/FTD) 4. Mutations in one such gene, C9orf72, are the most common cause of ALS and FTD 5,6.
The ALS and FTD causing mutation in the C9orf72 gene is a massively expanded hexanucleotide repeat (GGGGCC) 5,6, which produces sense and antisense RNA foci 5 and is translated into aggregation-prone dipeptide repeat (DPR) proteins through an unconventional form of AUG-independent translation (also called RAN translation) 7–10. Studies in flies and human cells suggest DPRs may be the main drivers of neuronal toxicity 11–13. The arginine-rich DPRs, Glycine-Arginine (GR) and Proline-Arginine (PR) are particularly toxic in experimental models 11,13–16. Synthetic PR or GR DPRs added exogenously to the culture media are rapidly transported to the nucleus, cause disruptions in RNA splicing – including in the canonical splicing and biogenesis of ribosomal RNA (rRNA) – and induce cell death in a dose-dependent manner 14. Subsequent studies have provided evidence through co-immunoprecipitation and mass-spectrometry that these DPRs preferentially bind proteins with low complexity domains, including RNA-binding proteins 17–19, ribosomal proteins, and translational elongation factors 20,21, as well as nuclear pore complex components 22.
Genetic screens in simple experimental model organisms like yeast, flies, and worms have empowered the discovery of fundamental biological processes including mechanisms of human disease 23. For example, we and others have used genetic screens in model systems to identify modifiers of toxicity elicited by aggregation-prone neurodegenerative disease proteins, such as TDP-43, FUS, Amyloid-β, alpha-synuclein, mutant huntingtin, and C9orf72 DPRs 15,16,24–34. Underscoring the impact of these simple model systems, some of the modifier genes from the genetic screens have been validated in mouse models and even connected to human disease through genetics and neuropathology 35–37.
While model systems have been powerful experimental tools for the study of human neurodegenerative disease mechanisms, it would be empowering to have access to the human genome to perform similar modifier screens in human cells. Recent technological advances in CRISPR-Cas9 genome editing have expanded the scope and reliability of genome-wide genetic deletion screens to the human genome using high complexity single-guide RNA (sgRNA) libraries 38–42. Here, we used the CRISPR-Cas9 system to perform comprehensive genome-wide knockout screens in human cells and mouse primary neurons to identify genetic modifiers of C9orf72 DPR toxicity.
Results
CRISPR-Cas9 screens for C9orf72 DPR toxicity modifiers
We engineered the human immortalized myelogenous leukemia cell line, K562, to stably express Cas9 43. We picked this cell line for the initial screen for several reasons. The cells grow in suspension and double rapidly, allowing us to expand the cultures to large numbers that are required to perform genome-wide screens with complex sgRNA libraries. Furthermore, this cell line has been used for several other genome-wide screens, allowing us to compare and contrast hits to test for specificity. Similar to previous reports in other human cell lines 14, synthetic polymers of PR and GR (PR20 and GR20) added to cell culture media were rapidly taken up by K562 cells, trafficked to the nucleus, and killed cells in a dose-dependent manner (Fig. 1a–d, Fig. S1a).
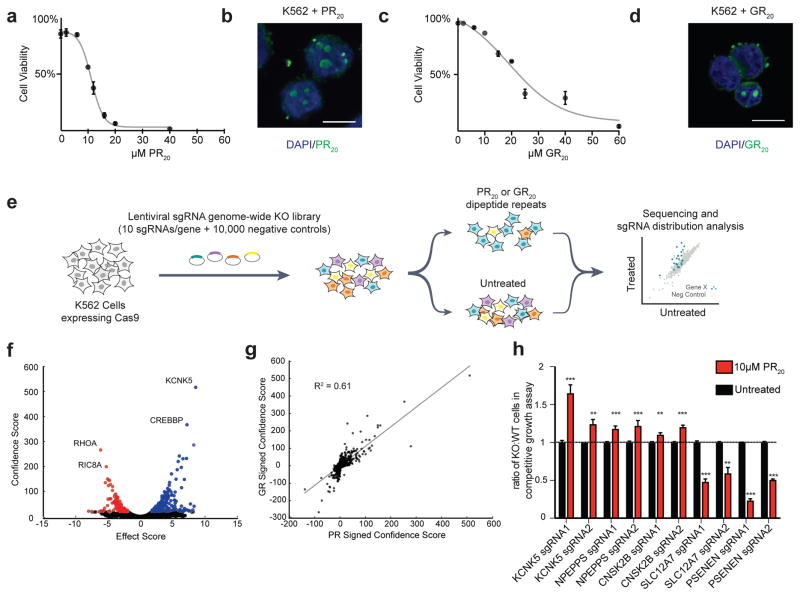
FLAG-tagged synthetic DPRs (PR20 or GR20) were added to the culture media of K562 cells, and dose-dependent cytotoxicity was measured using flow cytometry (a, c). The DPRs were internalized and localized to the nucleus of cells as visualized by immunocytochemistry (b, d, blue = DAPI, green = anti-FLAG, scale bar = 10μm). (e) Pooled CRISPR-Cas9 screening paradigm. K562 cells stably expressing Cas9 were infected with a lentiviral sgRNA library (10 sgRNAs/gene), the population was split and half the cells were treated with PR20 or GR20 for 4 pulses (at concentrations sufficient to kill 50% of cells each pulse), and then the resulting populations were subjected to deep sequencing and analysis. Screens were repeated 2 independent times for each DPR. (f) Volcano plot for all human genes in the GR20 screen. Colored in blue are all the gene conferring resistance to GR20 when knocked out (10% FDR) and colored in red are all the genes conferring sensitivity to GR20 when knocked out (10% FDR). (g) Correlation of signed confidence scores of significant hits between GR20 and PR20 screens (R2 = 0.61). (h) Validation of hits using independently generated single gene knockout K562 lines in co-culture competitive growth assays. Equal numbers of knockout cells (GFP+) and WT control cells (GFP−) were co-cultured in equal starting numbers and subsequently treated with PR20 (10 μM). The abundance of knockout cells (GFP+ cells) was quantified by flow-cytometry 48 hours after PR20 treatment and compared to the untreated population (n = 3, two-tailed t-test; *** p < 0.001, ** p < 0.01; error bars are ± S.D.) Each gene was validated using two distinct sgRNAs.
To identify genetic modifiers of both PR20 and GR20 mediated-toxicity, we conducted genome-wide CRISPR knockout screens. We used a lentiviral sgRNA library comprised of ten sgRNAs per gene, targeting ~20,500 human genes, along with ~10,000 negative control sgRNAs 44 (Fig. 1e). We used deep sequencing to track the effect (protective, sensitizing, neutral) of each sgRNA in a pooled population of knockout cells. That is, sgRNAs that protected cells from DPR toxicity were enriched and those that sensitized cells were depleted from the pooled population of cells. In order to identify both protective and sensitizing gene knockouts in the same screen, we treated the pooled population of cells with a dose of the DPR sufficient to kill 50% of cells and repeated this treatment four times over the course of the screen to amplify the selection. We performed separate screens for modifiers of PR20 and GR20 toxicity, each in duplicate, and repeated each screen two independent times.
To detect statistically significant suppressors and enhancers of DPR toxicity we used the casTLE algorithm as previously described 45. Using a false discovery rate (FDR) cut-off of 10%, the screens identified 215 genetic modifiers of PR20 toxicity and 387 genetic modifiers of GR20 toxicity (Fig. 1f, S2a–c, Supplementary Table 1). Results from the PR20 and the GR20 screen were well correlated (R2=0.61), suggesting similar mechanisms of toxicity (Fig. 1g). We validated individual knockout K562 lines by performing competitive growth assays with PR20 treatment. We tested five genes from the primary screen (two guides per gene) and all five genes validated in these assays (Fig. 1h). Furthermore, to assess the specificity of our screens, we compared the hits from our genome-wide screens to the hits from a similar screen performed for ricin toxicity, which used the same cell line, sgRNA library, and method of exogenous application of a toxic protein 44. We found zero correlation between hits in the two screens (R2 = 0), indicating that we are uncovering specific genetic modifiers, rather than non-specific genes involved in general peptide-mediated toxicity (Fig. S2d).
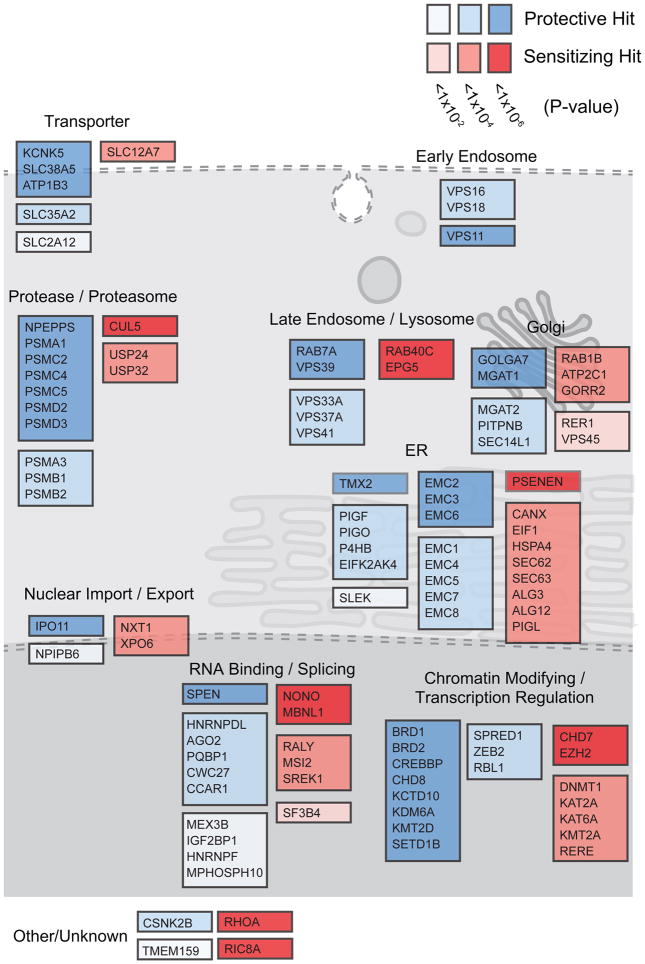
Among the hits from these knockout screens were genes encoding some of the same nuclear import and export factors discovered in DPR modifier screens in model organisms 15,16,46,47, such as XPO6 and IPO11 (Fig. 2), suggesting the same mechanisms are important in mammalian cells. In addition, we identified many novel genetic modifiers of PR and GR toxicity, including genes encoding RNA-binding proteins that have been shown to physically interact with PR and GR 17, such as NONO and HNRNPF. We also identified genes involved in RNA-splicing, such as SPEN and PQBP1, a process previously observed to be dysregulated in c9FTD/ALS patient brains 48 and dramatically disrupted in human cell lines treated with DPRs 14. In addition to these previously implicated genes, some of the strongest hits in the screen encoded endoplasmic reticulum (ER)-resident proteins, such as TMX2, CANX, and almost all members of the ER membrane protein complex (EMC), which is upregulated by ER stress and is thought to be involved in ER-associated degradation (ERAD) 49–51. Genes encoding proteasome subunits, as well as chromatin modifiers and transcriptional regulators, such as those involved in histone lysine methylation (KDM6A, KMT2D, KMT2A, and SETD1B) were also significant genetic modifiers of PR and GR toxicity (Fig. 2).
CRISPR-Cas9 screens in primary mouse neurons
To evaluate the role of the genetic modifiers identified in our human genome-wide screens in a more disease relevant context, we next designed a strategy to conduct pooled CRISPR-Cas9 screens in mouse primary cortical neurons. Similar to human cell lines, synthetic PR20 dipeptides were toxic to primary neurons in a dose-dependent manner (Fig. 3a). However, to better model the endogenous production of DPRs, like in patient brains, we also used a lentiviral construct expressing codon optimized PR50 driven by the neural-specific synapsin promoter 52. When expressed in primary neurons, PR50 localized to nuclear puncta and elicited neurotoxicity in a time-dependent manner (Fig. 3b, S3a). We constructed a custom lentiviral sgRNA library to test the top ~200 genes (5% FDR) that were modifiers of PR20 toxicity in the genome-wide CRISPR screens. The custom library contained ten sgRNAs targeting each mouse gene and ~1,000 negative control sgRNAs. Using primary neuron cultures isolated from mice constitutively expressing Cas9 53, we used immunoblotting to confirm efficient protein reduction of target genes after transduction with sgRNAs from this library following at least 10 days in culture (Fig. S4 a–c).
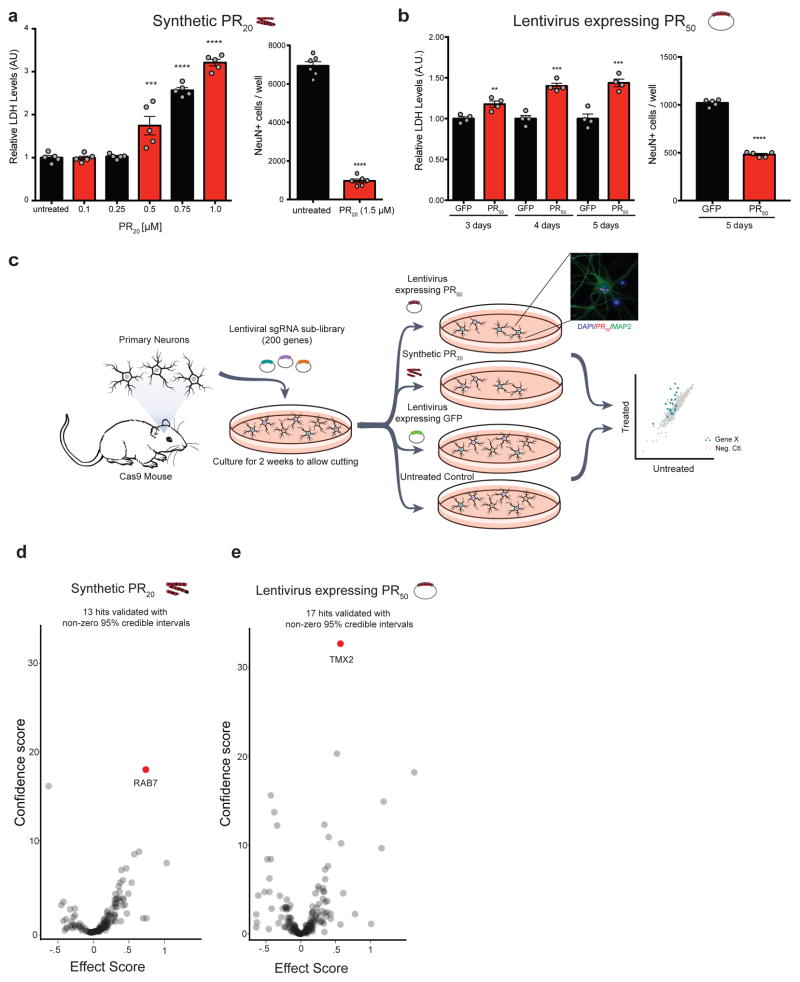
(a) Synthetic PR20 added to primary neuron cultures caused dose-dependent cytotoxicity within 24 hours measured by lactose dehydrogenase (LDH) release or counts of surviving NeuN+ neuronal nuclei by immunocytochemistry (two-tailed t-test; **** p < 0.0001, *** p < 0.001, ** p < 0.01; error bars are ± S.E.M). (b) Expression of PR50 using lentiviral transductions caused time dependent cytotoxicity measured by LDH release or counts of surviving NeuN+ neuronal nuclei by immunocytochemistry (two-tailed t-test; **** p < 0.0001, *** p < 0.001, ** p < 0.01; error bars are ± S.E.M). (c) CRISPR-Cas9 screening paradigm in primary mouse neuron cultures. Cortical neurons were plated and infected with sgRNA libraries (~200 genes passing a 5% FDR cutoff from the genome-wide PR screen with 10 sgRNAs/gene + ~1000 negative control sgRNAs). Four independent screening conditions were conducted, each in replicate: synthetic PR20 treated neurons (1.5μM, overnight), untreated neurons, lentiviral expression of PR50 (5 days), and lentiviral expression of GFP (5 days) as a control. The abundance of sgRNAs in the surviving cells was measured by sgRNA deep sequencing. PR50 localized to the nucleus of cultured neurons (blue = DAPI, red = anti-FLAG (PR50), green = anti-MAP2). (d, e) Volcano plots of effect scores and confidence scores of all genes for each screen. Effect and confidence scores are calculated using casTLE 45. The effect score is a gene-level summary of how protective or sensitizing the knockout is, while the confidence score is a log-likelihood ratio describing the significance of the effect. Genes were designated hits if their 95% credible intervals derived from casTLE did not contain zero; 13 genes in the synthetic PR20 screen and 17 genes in the lentiviral PR50 screen passed this criteria (Supplementary Table 2).
We performed the primary neuron CRISPR screens two ways: either expressing PR50 in neurons using lentivirus or treating neurons with exogenous synthetic PR20 (Fig. 3c). In this way, we could simultaneously validate hits from the genome-wide screens and disentangle the genes responsible for the uptake and spread of the DPRs throughout the cell from those responsible for mediating the toxic effects once inside the cell. We performed these targeted CRISPR KO screens such that there was a ~90% reduction in viability after one round of selection. Given the stringency of this screen, together with the fact that these neurons are non-dividing, we would only expect genetic modifiers with the strongest effects to become enriched. We harvested the surviving neurons and assessed sgRNA enrichment compared to the control populations by deep sequencing (Fig. 3c). We identified 13 modifier genes in the synthetic PR20 screen and 17 genes in the lentiviral PR50 expression screen (determined by having a non-zero estimated effect with a 95% credible interval) (Fig. 3d, e, S4d, Supplementary Table 2). The genes that validated in these neuronal cultures showed strong specificity to the method of PR delivery to the neurons. We further validated the two strongest modifier genes from these neuronal screens in an independent primary neuron culture model of DPR toxicity, using mouse dorsal root ganglion neurons. Knockout of either Rab7 or Tmx2 protected against PR toxicity in these primary neurons (Fig. 4a–d).
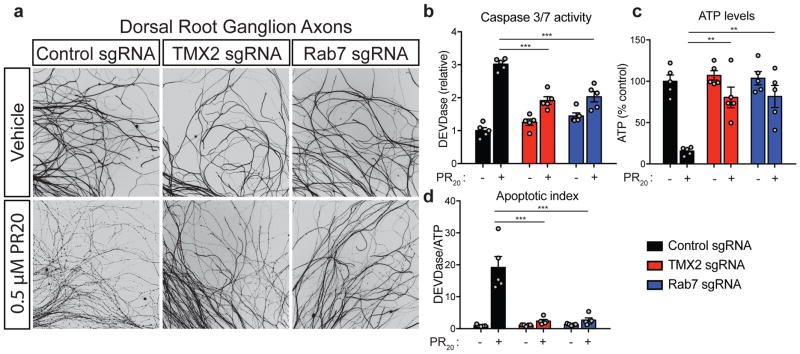
(a) Dorsal root ganglion (DRG) axon degeneration was evaluated by Tuj1 immunocytochemistry in the presence of 0.5 μM PR20 in cultures transduced with sgRNAs targeting a control genomic region, Tmx2, or Rab7. DRG axon degeneration was further quantified by caspase 3/7 activity (b), ATP levels (c), and an apoptotic index (d), which is the ratio of active caspase 3/7:ATP levels (one-way ANOVA, Dunnett’s multiple comparisons test, *** p < 0.001, ** p < 0.01; error bars are ± S.E.M).
RAB7A and intracellular transport of PR20
A top hit in the screen for modifiers of synthetic PR20-induced toxicity was the endo-lysosomal trafficking gene, RAB7A (Fig. 3d). We hypothesized that the endosomal trafficking pathway is necessary for the intracellular transport of exogenously applied PR20. We tested this hypothesis in WT and RAB7A knockdown HeLa cells by immunocytochemistry and found altered subcellular localization of PR20 (Fig. S3b), indicating that endo-lysosomal trafficking is important for the localization and subsequent toxicity of synthetic PR20. We used the CRISPR-i system to reduce levels of RAB7A in HeLa cells. This caused a stereotypical enlargement and mislocalization of LAMP1 positive lysosomal puncta. In control cells, at an early time point after PR20 addition (1hr, 1μM PR20), PR20 distributed diffusely throughout the cell, including the nucleus but was not yet enriched in the nucleus, as in Fig. 1b,d, S1. However, at this same early time point in RAB7A knockdown cells (Rab7-i.1), PR20 accumulated in numerous puncta speckled throughout the cytosol, indicative of an early endomembrane compartment (Fig. S3b). Since cell-to-cell spread of DPRs is possibly relevant to disease 54,55, understanding mechanisms of DPR cellular entry and trafficking could inform strategies to block this process.
C9orf72 DPRs induce an ER stress response
In contrast to the screen with synthetic PR20, none of the trafficking genes present in the mini-library were hits in the screen in which neuronal toxicity was induced by lentiviral mediated PR50 expression. In this screen, the strongest genetic modifiers were genes that encode proteins predominantly localized to the nucleus and ER; a top protective knockout was a poorly characterized ER-resident transmembrane thioredoxin protein, Tmx2 (Fig. 3e). Since this strongest hit was implicated in ER function, it suggested that DPR accumulation might induce an ER stress response. To test this hypothesis, we performed RNA sequencing (RNA-seq) on primary neuron cultures transduced with a lentivirus expressing either PR50 or GFP (Fig. 5a). Following four days of PR50 expression, 126 genes were significantly upregulated and 133 genes significantly downregulated (DESeq2, adjusted p-value < 0.05; Fig. 5b, S5, Supplementary Table 3). A significantly enriched Gene Ontology (GO) category among upregulated genes was ‘apoptotic signaling pathway in response to endoplasmic reticulum stress, GO:0070059’ (Fig. 5c). Genes in this enriched ontology category included Atf4, Bbc3 (Puma), Chac1, Bax, and the ER Ca2+ channel Itpr1, in which loss of function variants have been shown to cause spinocerebellar ataxia 56 (Fig. 5d). Other notable enriched GO categories among these differentially expressed genes included ‘neuronal apoptosis’, ‘ion homeostasis’, ‘ribonucleoprotein’, ‘translation’, and ‘DNA repair’.
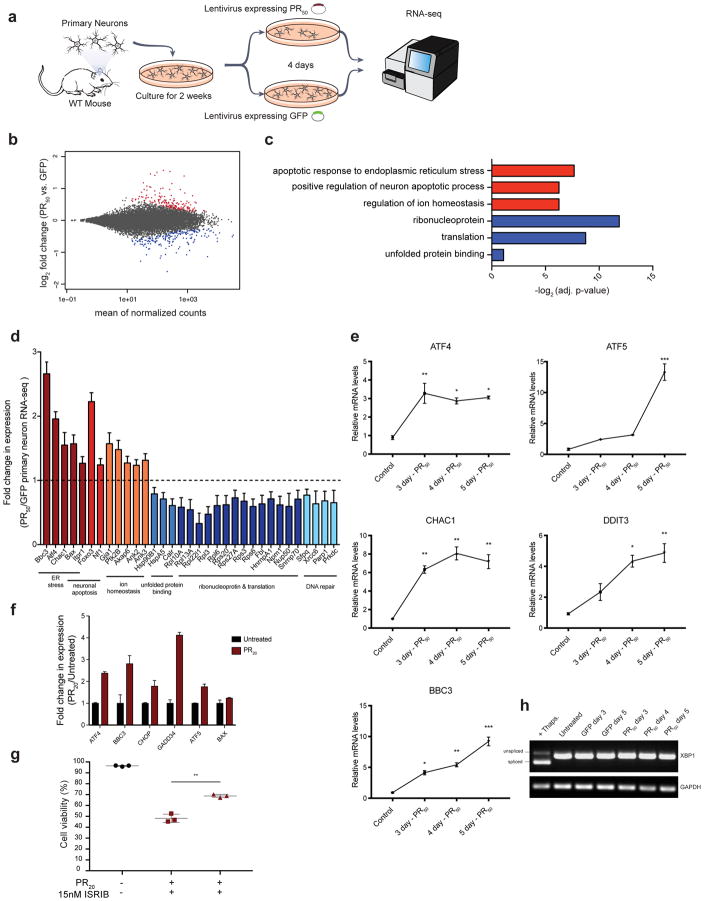
(a) RNA sequencing of primary mouse neurons treated with lentiviruses expressing either PR50 or GFP under control of the synapsin promoter for 4 days. Differentially expressed genes were determined using DESeq2. (b) MA plot of differential gene expression of PR50 expressing neurons compared to control GFP expressing neurons (red = significantly upregulated genes, adjusted p-value < 0.05; blue = significantly down regulated genes, adjusted p-value < 0.05). (c) Selected gene ontology (GO) terms enriched within significantly upregulated genes (red) or significantly downregulated genes (blue) determined using DAVID 6.8. (d) Fold change in expression of select genes that were found to be significantly differentially expressed by DESeq2 (n = 3, adjusted p-value < 0.001; error bars are ± S.E.M) grouped by GO category. (e) qRT-PCR validation of Atf4, Atf5, Chac1, Ddit3, and Bbc3 upregulation in neurons expressing PR50 for 3, 4, or 5 days compared to control neurons. (f) RNA sequencing in K562 cells treated for 24 hours with synthetic PR20 also show upregulation of ER stress related genes. Relative expression of significantly differentially expressed genes determined by DESeq2 (n = 3, adjusted p-value < 0.001; error bars are ± S.D.) plotted using FPKMs for each gene relative to the untreated population of cells. (g) K562 cells pre-incubated with 15nM ISRIB or equal amounts of DMSO were treated with 15 μM PR20 (red) or untreated (black) for 24 hours, then measured for viability by flow cytometry (FSC/SSC). Each measurement was performed in triplicate with mean and SD plotted by error bars (** p-value < 0.005, two-tailed t-test). (h) Reverse transcriptase PCR analysis to measure amounts of spliced XBP1 mRNA (relative to GAPDH levels) in cortical neurons transduced with PR50 for 3, 4, or 5 days (+ Thaps. = 200 nM thapsigargin treatment, 2hr.).
Based on these RNA-seq results, we confirmed a time dependent induction of ER stress related genes by PR50 expression using quantitative reverse transcription PCR (qRT-PCR) (Fig. 5e). The IRE1 stress sensor of the unfolded protein response was not activated by PR50 expression in neurons, as measured by levels of spliced XBP1 mRNA (Fig. 5h). These data are consistent with activation of the integrated stress response, which converges on eIF2 and the upregulation of transcription factor ATF4 57. To confirm that these ER stress responses are conserved in other models of DPR toxicity we performed RNA-seq on primary mouse neurons as well as human K562 cells, each treated with synthetic PR20. ATF4 and related ER stress response genes were also induced in each of these models (Fig. 5f, S6, Supplementary Table 3). Finally, in addition to observing genetic signatures of ER stress, we also used a pharmacological approach to test the importance of this pathway in response to PR20. The small molecule ISRIB is a potent inhibitor of the cellular response to ER stress 58, preventing ATF4 induction by inhibiting eIF2α phosphorylation during the integrated stress response 59. Pre-treatment of K562 cells with 15nM ISRIB mitigated PR20 toxicity (Fig. 5g), providing further evidence for a role of ER stress in DPR-mediated toxicity. Future experiments will be required to validate ISRIB’s efficacy in primary neurons and animal models of C9orf72 DPR toxicity.
TMX2 modifies the C9orf72 DPR-induced ER stress response
Several lines of evidence have suggested a potential role for ER stress in C9orf72 pathogenesis. Transcriptomic analysis of C9orf72 mutant ALS brain tissue reveals an upregulation of genes involved in the unfolded protein response (UPR) and ER stress 48. A specific PERK-ATF4 mediated ER stress response was reported in a separate study of c9ALS/FTD patient brain tissue when compared to sporadic ALS patient brain tissue 60. Additionally, ER stress and disruptions to ER Ca2+ homeostasis were observed in c9ALS/FTD patient iPSC-derived motor neurons 61. Given that a main transcriptional stress response to endogenous and exogenous DPR treatment was the ER stress response, we chose to further investigate TMX2, a gene encoding a poorly characterized ER-resident protein that was one of the strongest protective hits in both the PR and GR genome-wide CRISPR screens in K562 cells and in the targeted primary neuron screen expressing PR50. We first confirmed that the sgRNAs targeting TMX2 effectively knocked out the gene in K562 cells and primary neurons cultured from the Cas9-expressing mouse (Fig. 6a, S7a). Strikingly, reduction of TMX2 levels was sufficient to markedly suppress C9orf72 DPR toxicity both in K562 cells (Fig. S7b) and in primary mouse cortical neurons (Fig. 6b, c).
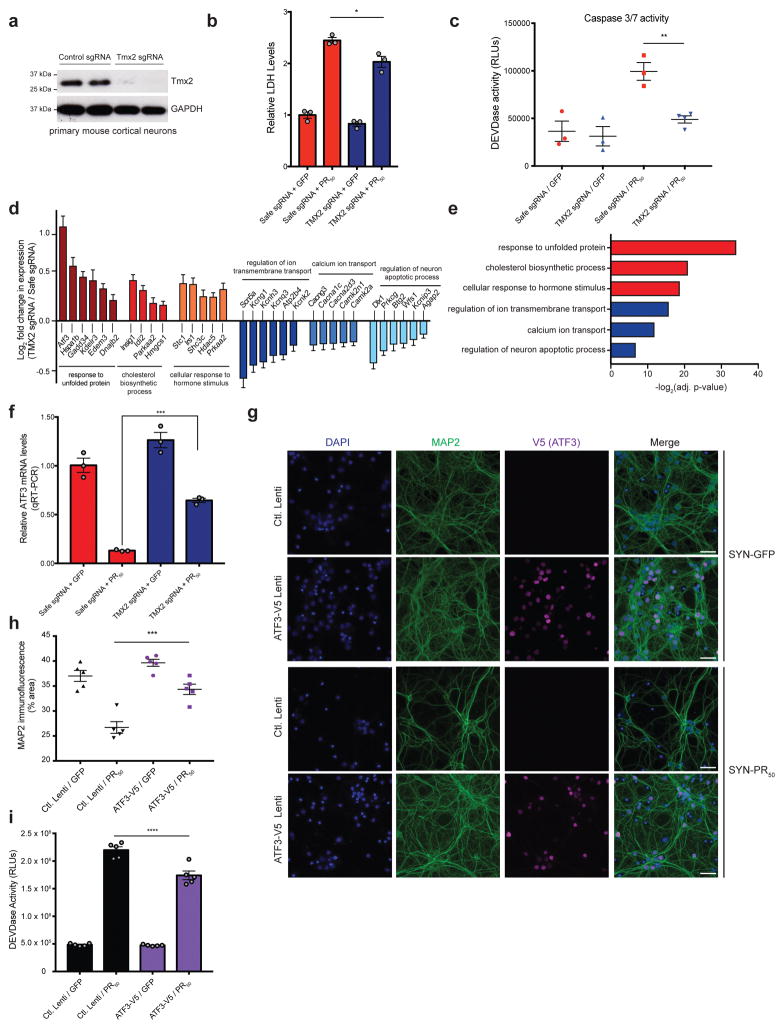
(a) Tmx2 protein levels measured in primary Cas9+ cortical neurons by immunoblot 10 days post sgRNA transduction (uncropped blot images are shown in Supplementary Data Set 1). (b) Quantification of lactose dehydrogenase levels (LDH) in primary cortical neurons 5 days post-transduction with PR50 or GFP lentiviruses as a measurement of cytoxicity. Tmx2 reduction by sgRNA lentivirus transduction on DIV1 (TMX2 sgRNA) protected primary neurons against PR50 induced cytotoxicity relative to control infected neurons (Safe sgRNA) (one-way ANOVA; n = 3; * p < 0.05; error bars are ± S.E.M). (c) Quantification of activated caspase 3/7 (DEVDase activity; RLU = relative light units) in primary neurons 4 days post-transduction with PR50 or GFP lentiviruses (n = 3–4, one-way ANOVA, Dunnett’s multiple comparisons test, ** p < 0.01). (d) RNA-seq analysis of neurons infected with Safe or TMX2 targeting sgRNAs 4 days post-transduction with PR50 or GFP lentiviruses. Log2 fold change (TMX2 sgRNA/Safe sgRNA in PR50 treated conditions) of differentially expressed genes determined using DESeq2 (n = 3; adjusted p-value < 0.001; error bars are ± S.E.M.) enriched in select GO categories (e) Select GO term enrichment determined using DAVID 6.8 (red = upregulated genes, blue = downregulated genes) of differentially expressed genes (adjusted p-value < 0.001) determined by comparing RNA-seq results between control (Safe sgRNA) and Tmx2 KO (TMX2 sgRNA) neuron cultures expressing PR50. (f) qRT-PCR validation of Atf3 expression changes after PR50 expression in control (Safe sgRNA) and Tmx2 KO (TMX2 sgRNA) neurons (n = 3; p < 0.0001; one-way ANOVA; error bars are ± S.E.M.). (g) V5-tagged ATF3 overexpression by lentiviral transduction of primary cortical neurons confers protection of PR50 induced dendrite degeneration, as measured by immunocytochemistry (blue = DAPI, green = anti-MAP2 (dendrite marker), magenta = anti-V5 (ATF3-V5 overexpression), SYN = lentiviral synapsin promoter, scale bar = 20μm). (h) Quantification of % area of MAP2 immunofluorescence (n=5, one-way ANOVA, Dunnett’s multiple comparisons test, *** p < 0.001). (i) Quantification of active caspase 3/7 (DEVDase) in PR50 or GFP expressing cortical neurons with or without ATF3 overexpression (n=5, one-way ANOVA, Dunnett’s multiple comparisons test, (**** p < 0.0001).
TMX2 is an ER resident transmembrane thioredoxin protein that may be enriched at the mitochondrial-associated membrane (MAM) of the ER 62. It is a member of the protein disulfide isomerase (PDI) family, but it is unclear whether it is catalytically active since it harbors an SXXC motif instead of the canonical CXXC motif necessary for the oxidoreductase activity used to catalyze the formation or rearrangement of disulfide bonds 63. To define the mechanism by which Tmx2 reduction mitigates DPR toxicity, we performed RNA-seq on primary Cas9+ neurons infected with control sgRNAs (Safe) or sgRNAs targeting Tmx2, with or without PR50 expression (Fig. 6d, e, S7c–h). Upon PR50 expression, Tmx2 KO neurons upregulated a distinct set of pro-survival unfolded protein response (UPR) pathway genes and downregulated calcium-binding and apoptotic genes, compared to primary neurons expressing control sgRNAs (Fig. 6d, e, Supplementary Table 3). We validated, by qRT-PCR, one of the upregulated ER stress genes that we observed in the Tmx2 KO neurons after PR50 expression, Atf3, which has been previously shown to be protective upon overexpression in mouse models of ALS 64 and neuronal models of excitotoxicity 65 (Fig. 6f). These results suggest that Tmx2 reduction may be protective by modulating the ER stress response elicited by the DPRs. Indeed, upregulation of Atf3 alone was sufficient to protect primary cortical neurons from PR toxicity (Fig. 6g–i).
Lowering TMX2 improves survival of C9-ALS motor neurons
Given the strong protective effect of Tmx2 knockdown on PR mediated toxicity in rodent primary neurons, we tested whether lowering TMX2 levels could modify survival phenotypes in neuronal cells from ALS patients harboring endogenous C9orf72 GGGGCC expansions. We generated induced motor neurons (iMNs) from iPSCs from C9orf72 ALS patients and healthy subjects using transcription factor-mediated lineage conversion as previously described 13,66. These C9-ALS patient derived iMNs show reduced survival after glutamate addition compared to control iMNs (Fig. 7, S8, and 13). Using a seven factor (7F) differentiation method to generate iMNs, we tested two independent shRNAs targeting TMX2, and found that both shRNAs significantly increased the proportion of surviving C9orf72 iMNs compared to control shRNAs in two independent C9-ALS lines (Fig. 7a, b), but did not increase the proportion of surviving control iMNs (Fig. S8). We repeated this experiment using a different differentiation method (Dox-NIL) to generate iMNs, and found a similar protective effect of TMX2 reduction in C9-ALS but not control iMNs, though there was a degree of variability of the extent of rescue between patient lines with this differentiation method (Fig. S9). Further studies will be required to assess the effect of TMX2 on C9orf72 related pathologies in additional patient cell lines and ALS models, but these results suggest our screening strategy may be useful to identify potent modifiers of ALS-related phenotypes.
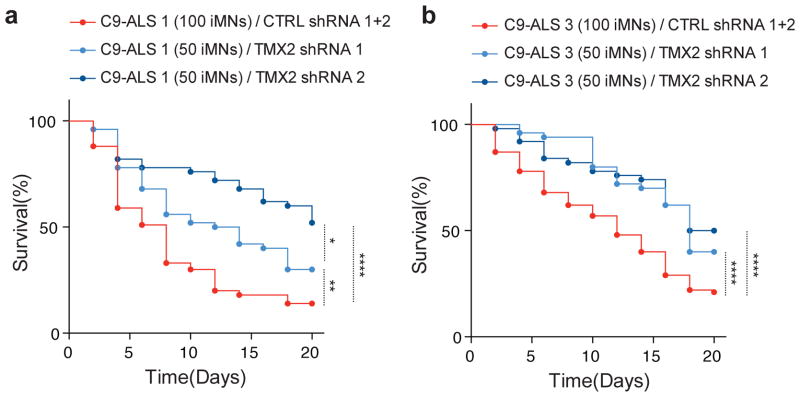
Quantification of surviving motor neurons with or without TMX2 reduction by lentiviral shRNA transduction in two independent C9-ALS iPSC lines (a = C9-ALS line 1, b = C9-ALS line 2). Induced motor neurons were generated from iPSCs using a seven factor (7F) differentiation system. The survival of HB9-RFP+/shRNA-GFP+ iMNs was tracked by imaging after the addition of 10 μM glutamate to the cultures. All iMN survival experiments were analyzed by log-rank test, and statistical significance was calculated using the entire survival time course. * p < 0.05, ** p < 0.01, **** p < 0.0001 (survival assays were performed in triplicate with the indicated number of iMNs analyzed for each group, Supplementary Table 4).
Discussion
Here, we used comprehensive CRISPR-Cas9 knockout screens in human cells with further validation screens in primary neurons to discover modifiers of C9orf72 DPR toxicity. We identified nucleocytoplasmic transport machinery, confirming previous studies in model organisms 15,16, and also identified new genes that point to ER function and ER stress as important for c9FTD/ALS pathogenesis. The gene knockouts that mitigated toxicity, such as TMX2, could represent therapeutic targets. It is notable that TMX2 reduction was able to confer strong protection from PR toxicity across multiple cellular assays, including primary rodent neurons.
There are five distinct DPRs produced from the C9orf72 GGGGCC repeat expansion: Glycine-Alanine (GA), Glycine-Arginine (GR), Glycine-Proline (GP), Proline-Alanine (PA), and Proline-Arginine (PR) (GP is produced from both the sense and antisense transcript). Our studies here have focused on two of these, GR and PR. The relative contribution of each DPR to neurodegeneration and the potential for synergistic effects between multiple DPRs remains to be defined. While the evidence from model systems for a role of DPRs in neurodegeneration is compelling, there are some caveats. These studies, including ours, use synthetic DPRs exogenously added to cells or high levels of transgenic expression of specific DPRs, which might not accurately reflect the physiological levels of DPRs. Moreover, studies of c9FTD/ALS neuropathology have also produced results that seem to not support a role of DPRs as the drivers of neurodegeneration. The abundance and localization of DPR pathology does not seem to correlate with neurodegeneration and clinical phenotypes 67–71. Finally, GA and GP DPRs seem to be the most abundant but these ones are the least toxic in model systems 11,15,16. On the other hand, the most toxic DPR, PR, is extremely rare 70. One possible explanation for this apparent disconnect is that the most toxic DPRs (e.g., GR and PR) do not accumulate to high enough levels (because they are so toxic) before causing neuron death whereas the other ones (e.g., GA and GP) can accumulate to higher levels because they are more benign 72.
Beyond the DPRs, RNA foci from the sense (GGGGCC) and antisense (GGCCCC) repeat transcripts, as well as loss of function of the C9orf72 gene could also contribute to disease. These different mechanisms could also interact with one another. For example, loss of C9orf72 function could make neurons or glia more vulnerable to the RNA foci or DPRs. Additionally, our work here provides evidence that DPRs can elicit an ER stress response and recent work shows that ER stress can increase RAN translation 73,74 – setting up the potential for a feedforward loop. It will be important to define the relative contributions of each of these facets of C9orf72 pathology (loss of function, RNA foci, DPRs) to disease so that effective therapeutic strategies can be developed. If the DPRs are indeed key contributors to disease, our work here identifies novel suppressors of DPR toxicity that represent potential therapeutic targets.
In addition to the types of CRISPR knockout screens we used here, the CRISPR-Cas9 system has been adapted to perform other types of genetic screens. Using Cas9 with a deactivated nuclease (dCas9), which is unable to generate double strand breaks, specific genomic regions can be targeted for transcriptional control 75,76. Fusing a KRAB effector domain to Cas9 results in transcriptional repression (CRISPRi, CRISPR interference) 77. Alternatively, CRISPRa (CRISPR activation) utilizes dCas9 fused with various transcriptional activation domains, empowering genome-wide transcriptional activation (i.e. overexpression) screens 76–79. Thus, the same logic of yeast, worm and fly genetic screens for up- or downregulation of genes can now be achieved in human cells on a genome-wide scale. Together, we anticipate that CRISPR screens in human cells and primary neurons will be a powerful addition to the experimental toolbox to study mechanisms of neurodegenerative disease.
Online Methods
Cell Culture
K562 cells (ATCC) were cultured in RPMI-1640 (Gibco) media with 10% fetal bovine serum (FBS) (Hyclone), penicillin (10,000 I.U./mL), streptomycin (10,000 μg/mL), and L-glutamine (2 mM). Cells were grown in log phase by maintaining the population at a concentration of 500,000 cells per mL. K562 cells were maintained in a controlled humidified incubator at 37°C, with 5% CO2.
Primary mouse cortical neurons were dissociated into single cell suspensions from E16.5 mouse cortices using a papain dissociation system (Worthington Biochemical Corporation). Neurons were seeded onto poly-L-lysine coated plates (0.1% w/v) and grown in Neurobasal media (Gibco) supplemented with B-27 serum-free supplement (Gibco), GlutaMAX, and Penicillin-Streptomycin (Gibco) in a humidified incubator at 37°C, with 5% CO2. Half media changes were performed every 4–5 days, or as required. For gene silencing experiments in Cas9+ cortical neurons, lentiviruses encoding sgRNAs expressed from a U6 promoter were transduced on DIV1. Cultures were allowed to incubate for at least 10 days with the sgRNAs in order to get adequate reduction of target protein levels before further experiments were performed.
Dorsal root ganglion cultures: 24 well plates were coated overnight with PDL (100 μg/ml) and following a water wash were coated with laminin (1.33 mg/ml) for at least 3 hours. DRGs were dissected from E12.5 mouse embryos and dissociated in 0.05% Trypsin-EDTA. Dissociated neurons were plated in Neurobasal supplemented with 2% B27, 0.45% D-glucose, 1% GlutaMAX, 100U/ml penicillin, and 100U/ml streptomycin. On DIV1, the cultures were fed with fresh Neurobasal/B-27 medium containing 5μM 5-fluoro-2′-deoxyuridine and 5μM uridine to inhibit the growth of non-neuronal cells. On DIV1, cells were transduced with lentivirus expressing sgRNAs targeting a safe genomic region (Safe) or TMX2 or Rab7. Cultures were treated with PR at 0.5 μM on DIV 10. For ATP measurement, neuronal cell bodies and proximal axon were removed, and axons were subjected to ATP measurement with Cell Titer Glo (Promega) or Caspase 3/7 Glo (Promega).
Generation of stable RAB7A knockdown cells: HeLa cells stably expressing dCas9 that is fused to a KRAB domain were infected with sgRNAs targeting RAB7A or a negative control sgRNA. Cells were selected with puromycin (1ug/mL) for stable sgRNA expression.
Lentivirus production
HEK293T (ATCC) cells were cultured under standard conditions (DMEM + 10% FBS + Penicillin-Streptomycin.) and used to package lentiviral particles following standard protocols with 3rd generation packaging plasmids. Lentiviral containing media was harvested after 48hrs., centrifuged at 300g for 5 min. to remove cellular debris, and concentrated 10-fold using Lenti-X concentrator (Clontech) before adding to cell cultures. GFP or PR50-FLAG constructs were cloned into a 3rd generation lentiviral vector that expresses the transgenes under the control of the neuron specific human synapsin 1 promoter (SYN).
Lentivirus production for iPSC derived motor neuron experiments: All shRNA and Hb9::RFP-encoding lentiviruses were produced as follows: HEK293T cells were transfected at 80–90% confluency with viral vectors containing the genes of interest and viral packaging plasmids (pPAX2 and VSVG for lentivirus) using polyethylenimine (PEI)(Sigma-Aldrich). The medium was changed 24h after transfection. Viruses were harvested at 48 and 72 hours after transfection. Viral supernatants were filtered with 0.45 μM filters and concentrated by incubating with Lenti-X concentrator (Clontech) for 24 hours at 4°C and centrifuging at 1,500 × g at 4°C for 45 minutes. The pellets were resuspended in 300 μl DMEM + 10% FBS and stored at −80°C.
Immunocytochemistry and antibodies
Cells were grown on poly-L-lysine coated glass coverslips (0.1% w/v) in standard multiwell cell culture plates and were stained using standard immunocytochemistry techniques. Briefly, cells were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, blocked with 5% normal goat serum, and stained with the following antibodies: mouse monoclonal anti-FLAG (1:1000, M2 Sigma cat.# F1804), mouse monoclonal anti-MAP2A (1:1000, Millipore cat.# MAB378), mouse monoclonal anti-V5 tag (1:500, Thermo Fisher Scientific cat.# R960-25), rabbit monoclonal anti-LAMP1 (1:200, Cell Signaling Technology, D2D11 cat.# 9091), or mouse monoclonal anti-NeuN (1:1000, Millipore cat.# MAB377). Coverslips were mounted using Prolong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Images were acquired using either a Leica DMI6000B inverted fluorescence microscope with a 40X oil immersion objective or a confocal Zeiss LSM710 microscope with a 63X oil immersion objective. Percent area of MAP2 immunofluorescence staining was quantified using ImageJ. Images in a stack were thresholded and the % area of the signal for each image was measured.
DRG immunocytochemistry: Neuronal cultures were fixed in 4% (w/v) paraformaldehyde/PBS at room temperature for 15 min, followed by permeabilization with 0.1% Triton X-100/PBS for 30 min. After blocking for 1 hour with 5% bovine serum albumin (BSA) / 0.1% Triton X-100 / PBS, axons were then stained with anti-Tuj1 (1:2000) in blocking buffer at 4ºC overnight. After washing two times with 0.1% Triton X-100 / PBS, axons were labeled with the appropriate Alexa-dye conjugated secondary antibodies (Life Technologies) for 2 hours at room temperature, washed three times with 0.1% Triton-X-100 / PBS, and mounted in Fluoromount-G.
Genome-wide CRISPR-Cas9 screens in K562 cells
The 10-sgRNA per gene CRISPR/Cas9 deletion library was synthesized, cloned, and infected into Cas9 expressing K562 cells as previously described 44. Briefly ~300 million K562 cells stably expressing SFFV-Cas9-BFP were infected with the 10 guide/gene genome-wide sgRNA library at an MOI < 1. Infected cells underwent puromycin selection (1ug/mL) for 5 days after which point puromycin was removed and cells were resuspended in normal growth media without puromycin. After selection, sgRNA infection was measured by flow cytometry confirming that > 90% of cells were mCherry+. Sufficient sgRNA library representation was confirmed by deep sequencing after selection. Cells were maintained for three weeks at 1000× coverage (~1000 cells containing each sgRNA) at a concentration of 500,000 cells/mL for the duration of each screen. Cells were split into two conditions: one control group that remained untreated, and one group that was treated with the respective synthetic DPR (PR20 at 8 μM or GR20 at 10 μM). This treatment occurred four times over three weeks, allowing for cells to recover to ~90% viability after each round of treatment. At the end of each screen genomic DNA was extracted for all screen populations separately according to the protocol included with QIAGEN Blood Maxi Kit and deep sequencing on an Illumina Nextseq was used to monitor library composition. Guide composition between PR or GR treated conditions was compared to the untreated condition using casTLE 45 version 1.0 (see list of URLs). Briefly, the enrichment of individual guides was calculated as log ratios between treated and untreated conditions, and gene-level effects were calculated from ten guides targeting each gene. A confidence score was then derived as a log-likelihood ratio describing the significance of the gene-level effect. P-values were then calculated by permutating the targeting guides as previously described 45.
K562 competitive growth assays
K562 cells stably expressing Cas9 were infected with sgRNAs targeting each gene of interest for validation from the genome wide screen. sgRNA plasmids also encoded for GFP expression. Cells were selected for stable sgRNA expression using puromycin (1 μg/mL) and were confirmed to be GFP positive before use in competitive growth assays. Equal numbers of GFP+/sgRNA expressing cells were co-cultured with wildtype/GFP- cells, and were subsequently treated with 8 μM PR20. The percentage of GFP+ cells were quantified by fluorescence activated cell sorting using a BD Accuri C6 flow cytometer after 48 hours.
Pre-treatment of ISRIB in PR20 exposed K562s
ISRIB (SIGMA SML0843) was dissolved in DMSO and added to the K562 cell line at a concentration of 15 nM 3 hours before treatment with PR20. An equal amount of DMSO was added to cells that did not get pre-treated with ISRIB. 15 μM of PR20 was then added to these cells and a third group of K562s were not treated with ISRIB or PR20 to serve as an untreated control. 24 hours after PR20 treatment cells were measured using the BD Accuri C6 flow cytometer for forward and side scatter analysis (FSC/SSC). All experiments were performed in triplicate.
CRISPR-Cas9 screen for DPR toxicity in primary mouse cortical neurons
Cortical tissue from mouse E.16.5 embryos [Gt(ROSA)26Sortm1.1(CAG-cas9*,-EGFP)Fezh/J; Jackson Laboratory Stock No: 024858] was dissected and dissociated into a single cell suspension using a papain dissociation system (Worthington Biochemical Corporation). 7 million cells were seeded onto 10cm dishes respectively in Neurobasal media (Gibco) supplemented with B-27 serum-free supplement (Gibco), GlutaMAX (Gibco), and Penicillin-Streptomycin (Gibco). Half the culture media was replenished every 4–5 days. One day after seeding, neurons were infected with a lentiviral library containing ~3000 sgRNA elements consisting of 178 genes (5% FDR for PR20 genome-wide screen hits), 10 sgRNAs/gene, and 1,000 negative control sgRNAs, synthesized and cloned as previously described 45. Transductions were performed using a titer resulting in a 50–70% infection rate. Cas9-mediated genome editing was allowed to occur for 2 weeks in culture, after which PR20 synthetic dipeptides or PR50 expressing lentivirus were applied. Synthetic DPR screen: half the cells were left untreated (control group), and the other half were treated with 1.5 μM synthetic PR20 for 24hr, which resulted in approximately 90% cell death. Lentiviral mediated DPR screen: half the cells were infected with a control lentivirus expressing eGFP from the synapsin promoter (control group), and the other half were infected with a lentivirus expressing codon optimized PR50 from the synapsin promoter. Lentiviral PR50 resulted in approximately 90% cell death between 4–6 days. To harvest DNA from the remaining live cells, cultures were washed 3 times in PBS, then treated with DNaseI (Worthington Biochemical Corporation) for 10 minutes at 37°C (to remove any extracellular gDNA from dead neurons), and then washed again 3 times in PBS to remove residual DNaseI. Genomic DNA was harvested using a DNeasy Blood and Tissue Kit (QIAGEN) including Proteinase K digestion to inactivate residual DNaseI. sgRNA amplification and library preparation for deep sequencing was performed as described above. Gene-level effects and confidence scores were calculated as in the primary screen. The significance of the gene-level effects was determined using the 95% credible intervals calculated using casTLE 45. A gene was considered a hit if its 95% credible interval did not contain zero.
RNA-sequencing
Total RNA quality control was performed using the Eukaryote Total RNA Nano assay on the Agilent 2100 Bioanalyzer System for all RNA samples prior to library preparation for RNA sequencing. mRNA libraries were prepared for Illumina paired-end sequencing using the Agilent SureSelect Strand Specific RNA-Seq Library Preparation kit on an Agilent Bravo Automated Liquid Handling Platform. Libraries were sequenced on an Illumina NextSeq sequencer. Alignment of RNA-sequencing reads to the transcriptome was performed using STAR with ENCODE standard options, read counts were generated using rsem, and differential expression analysis was performed in R using DESeq2 package 80. All bioinformatics analyses were performed on Sherlock, a Stanford HPC cluster.
qRT-PCR
cDNA was reverse transcribed from total RNA samples using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). Taqman assays (Applied Biosystems) for ATF3, ATF4, ATF5, CHAC1, DDIT3, BBC3, and BAX were used in qPCR reactions using inventoried Taqman Universal PCR Master Mix (ThermoFisher Scientific) and a StepOne or QuantStudio 3 real time PCR machine. In order to measure XBP1 splicing, cDNA was generated as described above and equal amounts of cDNA (2 μl) were used in a PCR reaction using KOD polymerase (EMD Millipore cat.# 71086). Primers used in these experiments are listed in Supplementary Table 5. The cycling conditions were as follows: 95°C for 5 min; 10 cycles of 94°C for 20 s, 65–55°C touchdown (−1 degree/cycle) for 20 s, and 72°C for 30 s; 35 cycles of 94°C for 20 s, 58°C for 20 s, and 72°C for 30 s, and a final extension at 72°C for 5 min. Products were analyzed using 2% agarose gel electrophoresis.
Western Blotting
Protein lysates were prepared using RIPA buffer supplemented with 1X Halt Protease Inhibitor Cocktail (Pierce). Crude lysates were centrifuged at 12,500g for 10 min. at 4°C to remove cellular debris. Clarified lysates were quantified using Pierce BCA protein assay. Equal amounts of protein were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted following standard protocols. The following antibodies were used: mouse anti-GAPDH (1:5000, clone GAPDH-71.1, Sigma G8795), rabbit polyclonal anti-TMX2 (1:1000, Novus NBP1-87305), and rabbit polyclonal anti-XPO5 (1:2000, Bethyl Laboratories A303-991A).
Cytotoxicity Assays
K562 cell viability after DPR treatment was measured by flow cytometry using a BD Accuri C6 flow cytometer by measuring number of events in forward scatter and side scatter gates. Cytotoxicity in primary neuron cultures was measured both by lactose dehydrogenase (LDH) release assays (Promega, CytoTox 96® Non-Radioactive Cytotoxicity Assay) and by NeuN+ quantification of surviving neurons using immunocytochemistry. To quantify numbers of NeuN+ neurons, cells were fixed and stained as described above, and quantified using ImageJ. Briefly, thresholds were applied to image stacks in order to detect NeuN positive neuronal nuclei, which were then automatically counted using built in ImageJ plugins. Additional cytotoxic assays were performed using the Caspase-Glo 3/7 Assay (Promega cat.# G8090), a luminescence based readout of caspase 3/7 activity (DEVDase activity) in neurons according to the manufacturer’s instructions.
Virus production for induced motor neuron experiments
Complementary DNAs (cDNAs) for the induced motor neuron (iMN) factors (Ngn2, Lhx3, Isl1, NeuroD1, Ascl1, Myt1l, and Brn2) were purchased from Addgene. Each cDNA was cloned into the pMXs retroviral expression vector using Gateway cloning technology (Invitrogen). The Hb9::RFP lentiviral vector was also purchased from Addgene (ID: 37081). Viruses were produced as follows: HEK293 cells were transfected at 80–90% confluency with viral vectors containing genes of interest and viral packaging plasmids (PIK-MLV-gp and pHDM for retrovirus; pPAX2 and VSVG for lentivirus) using polyethylenimine (PEI)(Sigma-Aldrich). The medium was changed 24h after transfection. Viruses were harvested at 48 hours and 72 hours after transfection. Viral supernatants were filtered with 0.45 μM filters, incubated with Lenti-X concentrator (Clontech) for 24 hours at 4 °C, and centrifuged at 1,500 × g at 4°C for 45 min. The pellets were resuspended in 300 μl DMEM + 10% FBS and stored at −80 °C.
Generation of Dox-NIL induced motor neurons
To generate Dox-NIL iMNs, the Dox-NIL construct was integrated into the AAVS1 safe harbor locus of the control and C9-ALS patient iPSC lines using CRISPR/Cas9 editing (gRNA sequence shown in Supplementary Table 4). The Conversion of NIL-iMNs was performed in 96-well plates (2 × 103 cells/well), which was sequentially coated with matrigel (1 hour) and laminin (2–4 hours) at room temperature. iPSCs were cultured in mTeSr and induced with 1 μg/ml doxycycline on day 1 after plating. On day 3 after plating, the culture was transduced with CTRL shRNA1, CTRL shRNA2, TMX2 shRNA1 and TMX2 shRNA2 with 5 μg/ml polybrene in N3 media containing DMEM/F12 (Life Technologies), 2% FBS, 1% penicillin/streptomycin, N2 and B27 supplements (Life Technologies), and 10 ng/ml each of GDNF, BDNF, and CNTF (R&D). For the detection of the motor neurons, the culture was transduced with Hb9::RFP reporter 48 hours after shRNA transduction. On day 5, primary mouse cortical glial cells from P1 ICR pups (male and female) were added to the transduced cultures in N3 media.
Generation of 7 transcription factors (7F) induced motor neurons
Reprogramming was performed in 96-well plates (8 × 103 cells/well) that were sequentially coated with gelatin (0.1%, 1 hour) and laminin (2–4 hours) at room temperature. To enable efficient expression of the transgenic reprogramming factors, iPSCs were cultured in fibroblast medium (DMEM + 10% FBS) for at least 48 hours and either used directly for retroviral transduction or passaged before transduction for each experiment. 7 iMN factors were added in 100–200 μl fibroblast medium per 96-well well with 5 μg/ml polybrene. For iMNs, cultures were transduced with lentivirus encoding the Hb9::RFP reporter 48 hours after transduction with transcription factor-encoding retroviruses. On day 5, primary mouse cortical glial cells from P1 ICR pups (male and female) were added to the transduced cultures in glia medium containing MEM (Life Technologies), 10% donor equine serum (HyClone), 20% glucose (Sigma-Aldrich), and 1% penicillin/streptomycin. On day 6, cultures were switched to N3 medium containing DMEM/F12 (Life Technologies), 2% FBS, 1% penicillin/streptomycin, N2 and B27 supplements (Life Technologies), 7.5 μM RepSox (Selleck), and 10 ng/ml each of GDNF, BDNF, and CNTF (R&D). The iMN neuron cultures were maintained in N3 medium, changed every other day, unless otherwise noted.
Induced motor neuron survival assay
Hb9::RFP+ neurons appeared between days 10–13 after initial seeding. The doxycycline was withdrawn at day 10 and the survival assay was initiated at day 20. For the glutamate treatment, 10 μM glutamate was added to the culture medium on day 20 and removed after 24 hours. Longitudinal tracking was performed by imaging neuronal cultures in a Nikon Biostation CT once every 24 hours starting at day 20. Tracking of neuronal survival was performed using SVcell 3.0 (DRVision Technologies). Neurons were scored as dead when their soma was no longer detectable by RFP and GFP fluorescence. All neuron survival assays were performed at least twice, with one of the trials being used for the quantification shown. All trials quantified were representative of other trials of the same experiment. When iMNs from multiple independent donors are combined into one survival trace in the Kaplan-Meier plots for clarity, the number of iMNs tracked from each line can be found in Supplementary Table 4.
Statistics
Statistical tests were performed using GraphPad Prism 7. Two-tailed t-tests, one-way ANOVAs, or log-rank tests for survival curves were used as indicated.
Data Availability
RNA sequencing data from this study have been deposited with NCBI GEO with the accession code: GSE109177
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
1
2
3
Acknowledgments
We thank Emily Crane and Billy Li for help with RNA-seq and helpful discussions, Kyuho Han for helpful discussions and image analysis, and the Stanford Neuroscience Microscopy Service, supported by NIH NS069375. This work was supported by NIH grants R35NS097263 (A.D.G.), DP2HD084069 (M.B.), and R01NS097850 (J.K.I.), the National Science Foundation Graduate Research Fellowship (N.J.K.), The National Human Genome Research Institute Training Grant (M.S.H.), the Robert Packard Center for ALS Research at Johns Hopkins (A.D.G.), Target ALS (M.B., A.D.G.), the Stanford Brain Rejuvenation Project of the Stanford Neurosciences Institute (M.B., A.D.G.), the Muscular Dystrophy Association (J.K.I.), and Department of Defense grant W81XWH-15-1-0187 (J.K.I.). J.K.I. is a New York Stem Cell Foundation-Robertson Investigator.
Footnotes
Author Contributions: N.J.K. and M.S.H. designed experiments, collected data, and wrote the manuscript. D.W.M. analyzed and performed statistics on screen data. A.J. helped design experiments for primary neuron screens. A.L and J.O. assisted with cloning. R.M. and G.B. contributed to primary neuron experiments. J.C. contributed to RNA-sequencing analyses. C.K.T. contributed to the RAB7A knockdown cell experiments. N.T.H. and M.T.-L. contributed to the dorsal root ganglia experiments. Y.S. and J.K.I. contributed to the ALS patient iMNs experiments. M.C.B. and A.D.G. supervised the study and wrote the manuscript.
Competing Financial Interests: J.K.I. is a co-founder of Acurastem, Inc. All other authors declare no competing interests.
URLs
casTLE 1.0 software: https://bitbucket.org/dmorgens/castle
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/s41588-018-0070-7
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5893388
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
CRISPR-Cas9 screens reveal regulators of ageing in neural stem cells.
Nature, 634(8036):1150-1159, 02 Oct 2024
Cited by: 1 article | PMID: 39358505 | PMCID: PMC11525198
An efficient low cost means of biophysical gene transfection in primary cells.
Sci Rep, 14(1):13179, 08 Jun 2024
Cited by: 0 articles | PMID: 38849388
Selenium Nanoparticles as Neuroprotective Agents: Insights into Molecular Mechanisms for Parkinson's Disease Treatment.
Mol Neurobiol, 05 Jun 2024
Cited by: 1 article | PMID: 38837103
Review
Amyloidogenic regions in beta-strands II and III modulate the aggregation and toxicity of SOD1 in living cells.
Open Biol, 14(6):230418, 05 Jun 2024
Cited by: 1 article | PMID: 38835240 | PMCID: PMC11285818
C9orf72 polyPR directly binds to various nuclear transport components.
Elife, 12:RP89694, 14 Mar 2024
Cited by: 1 article | PMID: 38483313
Go to all (123) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE109177
Quick GO
- (1 citation) GO - GO0070059
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.
Autophagy, 17(11):3306-3322, 26 Feb 2021
Cited by: 48 articles | PMID: 33632058 | PMCID: PMC8632097
Review Free full text in Europe PMC
CRISPR/Cas9-mediated excision of ALS/FTD-causing hexanucleotide repeat expansion in C9ORF72 rescues major disease mechanisms in vivo and in vitro.
Nat Commun, 13(1):6286, 21 Oct 2022
Cited by: 25 articles | PMID: 36271076 | PMCID: PMC9587249
Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS.
Nat Neurosci, 18(9):1226-1229, 01 Sep 2015
Cited by: 398 articles | PMID: 26308983 | PMCID: PMC4552077
CRISPR-Cas9 Screens Identify the RNA Helicase DDX3X as a Repressor of C9ORF72 (GGGGCC)n Repeat-Associated Non-AUG Translation.
Neuron, 104(5):885-898.e8, 03 Oct 2019
Cited by: 80 articles | PMID: 31587919 | PMCID: PMC6895427
Funding
Funders who supported this work.
NHGRI NIH HHS (1)
Grant ID: T32 HG000044
NICHD NIH HHS (1)
Grant ID: DP2 HD084069
NIGMS NIH HHS (1)
Grant ID: T32 GM007790
NINDS NIH HHS (3)
Grant ID: P30 NS069375
Grant ID: R01 NS097850
Grant ID: R35 NS097263