Abstract
Free full text

All-atom simulations disentangle the functional dynamics underlying gene maturation in the intron lariat spliceosome
Significance
Precursor messenger RNA (pre-mRNA) splicing is a crucial step of gene expression, enabling the maturation of pre-mRNA transcripts into protein-coding mRNAs. In humans, a majestic ribonucleoprotein machinery—the spliceosome—governs this fundamental process, the defects and misregulation of which lead to over 200 human diseases. A thorough understanding of splicing is pivotal for biology and medicine, holding the promise of harnessing it for genome modulation applications. Despite the recent breakthroughs gained by cryo-EM, an atomic-resolution picture of the spliceosome functional plasticity is still missing. Here, all-atom simulations elucidate the cooperative motions underlying the functional dynamics of the spliceosome at a late stage of the splicing cycle, suggesting the role of specific proteins involved in the spliceosome disassembly from an atomic-level perspective.
Abstract
The spliceosome (SPL) is a majestic macromolecular machinery composed of five small nuclear RNAs and hundreds of proteins. SPL removes noncoding introns from precursor messenger RNAs (pre-mRNAs) and ligates coding exons, giving rise to functional mRNAs. Building on the first SPL structure solved at near–atomic-level resolution, here we elucidate the functional dynamics of the intron lariat spliceosome (ILS) complex through multi-microsecond-long molecular-dynamics simulations of ~1,000,000 atoms models. The ILS essential dynamics unveils (i) the leading role of the Spp42 protein, which heads the gene maturation by tuning the motions of distinct SPL components, and (ii) the critical participation of the Cwf19 protein in displacing the intron lariat/U2 branch helix. These findings provide unprecedented details on the SPL functional dynamics, thus contributing to move a step forward toward a thorough understanding of eukaryotic pre-mRNA splicing.
Precursor messenger RNA (pre-mRNA) splicing is a process of paramount importance for living cells, spanning all domains of life. Splicing is a meticulous snip-and-stitch editing of primary RNA transcripts emerging from gene transcription, whereby the noncoding sections (introns) are removed, while the flanking coding regions (exons) are joined together to form functional mRNA filaments (1–4). Each splicing cycle entails two subsequent SN2 transesterification reactions facilitated by two Mg2+ ions, which enable a two-metal–aided catalysis (4). In eukaryotes, the tailor of pre-mRNA splicing is the spliceosome (SPL), a sophisticated multimegadalton ribonucleoprotein machinery composed of five small nuclear RNAs (snRNAs) (U1, U2, U4, U5, and U6) and hundreds of different proteins endowed with distinct functions (2). snRNAs associate with specific proteins to form the spliceosomal snRNPs subunits, which, together with other complexes like nineteen complex (NTC) and NTC-related (NTR), enzymes, and cofactors, tune the SPL catalytic, structural, and dynamical properties. In this entangled framework, U6 snRNA is directly in charge of catalysis (4), positioning the metals within a catalytic site strikingly similar to that of group II introns ribozymes (G2IRs) (4–7), the SPL evolutionary ancestors for which we have elucidated the catalytic mechanism (8, 9). SPL is assembled and disassembled at each splicing cycle and undergoes a continuous conformational and compositional remodeling aimed at processing the large and diverse RNA transcripts with single-nucleotide precision. Since genome fidelity is a key prerequisite for healthy cells, it is not surprising that more than 200 human diseases, among which are cancer and neurodegeneration, are associated with splicing defects and misregulations (10, 11). This highlights the urgent need of an in-depth knowledge of the SPL structural and functional cycle (11). High-resolution cryo-electron microscopy (cryo-EM) has prompted a transformative era in the SPL structural biology, providing groundbreaking information on its assembly, composition, and reactivity (3, 5, 6, 12–24). However, the mechanistic understanding of this majestic machinery at atomic level of detail is far from being complete. In particular, the SPL conformational plasticity, its impact on splicing activity, and the role of many proteins engaged at different stages of splicing are still poorly understood (10, 12, 25). Among the recent SPL cryo-EM models (5, 6, 11–23), here we focus on a structure solved at near-atomic resolution from the yeast Schizosaccharomyces pombe, representing the SPL at the latest stage of splicing, that is, the intron lariat spliceosome (ILS) complex (5, 6). In this study, we provide relevant insights on the SPL functional dynamics via multi-microsecond-long molecular-dynamics (MD) simulations of two large models built on this cryo-EM structure (5, 6) (Fig. 1). Our work unveils the central role of Spp42 (Prp8 in Saccharomyces cerevisiae), which appears to act as an orchestra conductor of the “gene maturation symphony” by tuning and governing the motions of different and specific SPL components. Remarkably, we show that, cooperatively with Spp42, the NTR Cwf19 protein marks the ILS disassembly by displacing the intron lariat (IL)/U2 snRNA double helix (also named branch helix). Intriguingly, our findings perfectly match with the stage of the splicing cycle explored here.
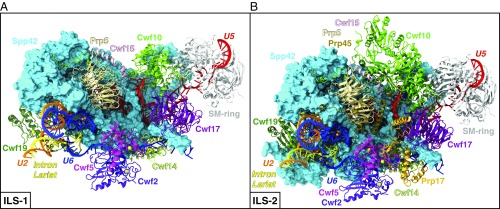
Spliceosomal (A) ILS-1 and (B) ILS-2 models built on the yeast S. pombe intron lariat spliceosome (ILS) cryo-EM structure (PDB entry 3JB9) (5, 6). Spp42 is shown as surface (cyan), while the other proteins and RNAs (snRNAs and IL) are depicted as cartoons using different colors. Mg2+ (orange) and Zn2+ (yellow) are shown as spheres, while GDP molecule is represented with atom-colored van der Waals spheres.
Results
SPL Dynamics.
Our MD simulations are based on the ILS complex solved from S. pombe at 3.6-Å average resolution and exceeding 3.2 Å in the core region (5, 6). Although this high-resolution cryo-EM map offers an exquisite structural reconstruction of the SPL, it must be noted that it is still at the limit of what can be used for atomic-level simulations. Two model systems were built, taking into account the central macromolecular constituents (i.e., proteins and RNAs) and ions of the Protein Data Bank (PDB) structure, and simulated in explicit water (SI Appendix, Fig. S1). These models, hereafter referred to as ILS-1 (Fig. 1A) and ILS-2 (Fig. 1B), count 721,089 and 914,099 atoms, respectively (for further details, see SI Appendix, Tables S1 and S2). Overall, three MD replicas of 750 ns for ILS-1 and one replica of 1 μs for ILS-2 resulted in a cumulative simulation time of 3.25 μs. Despite adopting the same simulation protocol, a longer sampling was allowed for the ILS-2 model. This was mostly a precautionary choice related to the larger size of this system (~200,000 atoms).
To date, these are the first SPL models investigated from an atomic-resolution perspective with force field (FF)-based MD simulations (26). From the inspection of root-mean-square deviation (rmsd) and radius of gyration (Rg), both models exhibit converged and comparable structural and dynamical profiles, which assess the reliability of the computational approach adopted to build the models (SI Appendix, Fig. S2). Moreover, our simulations reveal a stable four-Mg2+–ion catalytic site, showing the hallmark architecture also displayed by G2IRs in our previous studies (SI Appendix, Fig. S3) (8, 9). Notably, not only the U6 ligands coordinating the catalytic Mg2+ but also the characteristic triple-helix formed by U6 and U2 are conserved throughout the MD simulations (SI Appendix, Fig. S4). Correlation analyses, principal-component analysis (PCA), and electrostatic calculations were then applied to disentangle the cooperative motions at the basis of the SPL functional dynamics and to unravel their possible link with the electrostatic properties. In the following, we mostly discuss the results obtained out of three MD replicas of ILS-1. The MD simulation of ILS-2 is used as a further confirmation of the functional dynamics observed in ILS-1 and to check the impact of the proteins additionally included in this larger model.
The SPL: A Protein-Directed Ribozyme.
To characterize the SPL functional dynamics and the leading role of Spp42, the largest and the most important protein of the SPL, we have computed the cross-correlation scores (CSs) for each pair of SPL components (i.e., RNAs and proteins, explicitly considering the Spp42 domains and N-terminal motifs), which enable to qualitatively measure their interdependent dynamical coupling (27–29). The CSs histogram for ILS-1 model (Fig. 2B) is generated from the per-residue Pearson’s coefficients (CCs) cross-correlation matrix (Fig. 2A) and provides a coarse and prompt picture of the most relevant linearly coupled motions occurring across the system during the MD simulations. Among others, the following crucial connections disclose important results: (i) all of the snRNAs (U2, U5, U6) and the IL are overall positively correlated with Spp42 (especially U5) and most of the other proteins (blue squares highlighted with colored boxes in Fig. 2B), thus indicating that the SPL acts as a protein-directed ribozyme; (ii) Spp42 (Fig. 2C) is highly correlated with specific proteins, therefore governing and tuning the SPL plasticity through its unique multibranched architecture. In particular, the closest dynamical ties are established with two proteins of U5 snRNP (Cwf10 and Cwf17) and with the NTC DExD/H-box Prp5 protein. Indeed, Cwf10, a GTPase involved in SPL remodeling (6), moves in lock-step with the “lasso,” a peculiar N-terminal extended loop of Spp42, whereas Cwf17 and Prp5 are strongly coupled with two protruding “arms” of Spp42, the N-terminal α-helices binding to Cwf17 (B-Cwf17), and to reverse-transcriptase (B-RT), respectively; (iii) finally, U2 and IL positively correlate with Cwf19 and Spp42, pointing to a cooperative functional role involving these two proteins and the branch helix.
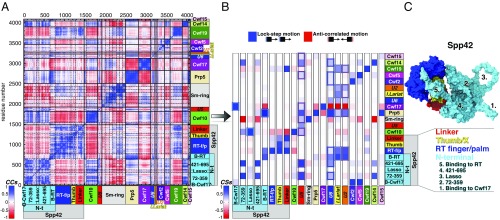
Cooperative motions underlying the functional dynamics in the spliceosomal ILS-1 model system. (A) Per-residue Pearson’s coefficients (CCs) cross-correlation matrix derived from the mass-weighted covariance matrix constructed over the last 670 ns of MD simulations of ILS-1 (replica #1) for Cα and P atoms. CCs are comprised between −1 (anticorrelation, red) and +1 (correlation, blue). The domains of Cwf10 highlighted in the matrix are (from Left to Right): N-t and G; proteins of Sm-ring are (from Left to Right): D3, B, D1, D2, E, F, and G. (B) Histogram reporting the (per-column) normalized correlation scores (CSs) calculated for each pair of spliceosomal components. Each column reports the CSs calculated between a specific component and all of the others (listed on rows). Spp42 domains and N-terminal motifs are distinctly considered. CS value ranges from −1 (anticorrelated motions) to +1 (lock-step motions). The most relevant correlations are highlighted using colored boxes. (C) Close-up of Spp42 (surface representation) in ILS-1, with the four domains shown/listed with different colors. The five N-terminal motifs are numbered and listed. Abbreviations: B-Cwf17/RT, binding to Cwf17/RT; I. Lariat, intron lariat; RT-f/p, reverse-transcriptase finger/palm; Thumb, Thumb/X.
The IL/U2 Helix Displacement.
Strikingly, behind the intriguing coupling discussed above in iii, the essential dynamics extracted from the PCA unveiled a displacement of the IL/U2 helix, whose unrolling is copromoted by Cwf19 and Spp42 (Fig. 3 and Movie S1). This original finding sheds lights on the so-far-unclear functional annotation of Cwf19 and is fully consistent with the late stage of the splicing cycle under study. In the ILS complex, in fact, the IL/U2 branch helix, formed before the catalysis to bulge out the branch point (BP) adenosine, must be displaced and unwound before IL linearization (i.e., debranching) and degradation (30–33).
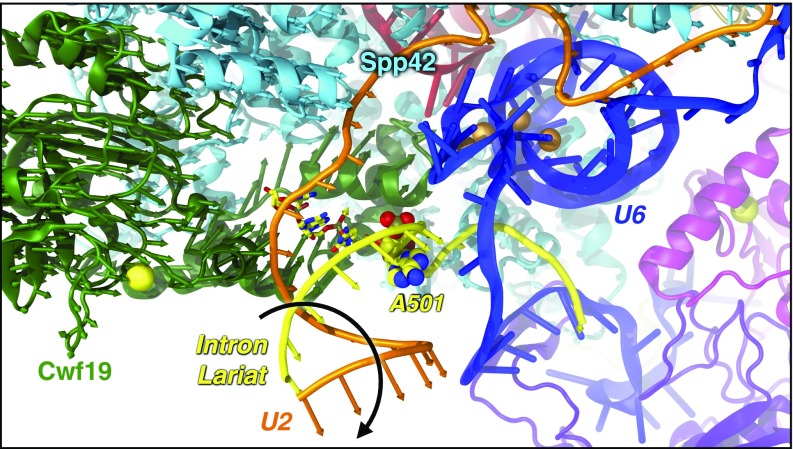
Displacement (highlighted by a black arrow) of the intron lariat (IL)/U2 helix, which unrolls away from the SPL, as revealed by the PCA of MD trajectories. Arrows, colored by protein/RNA, show the motion of Cα/P atoms along the first eigenvector. Cwf19 (green) and Spp42 (cyan) are depicted as cartoon; U2 (orange) and IL (yellow) are shown as tube; U6 (blue) and the other proteins are represented as cartoons. Mg2+ (orange) and Zn2+ (yellow) are shown as spheres.
Interestingly, electrostatic calculations performed on the cryo-EM structure and on representative MD snapshots reveal the presence of a positively charged cavity, rich in arginines and lysines, exposed by the RT domain of Spp42 and by Cwf19 (SI Appendix, Fig. S5). This pocket acts as an electrostatic trap for the negatively charged nucleotides of U2 and IL. The gradual IL/U2 displacement (Fig. 4) appears to be triggered by two key events: (i) the electrostatic recruitment of IL/U2 in the vicinity of the positive pocket; (ii) the electrostatic lock of the BP region, that is, the BP adenosine (A501), and the flanking G100 and C502 nucleotides. This lock is modulated by K364, K366, and R388 of Cwf19, which form a polar tweezers stably anchoring the BP region (Fig. 5). The 3′-terminus of the IL is firmly embedded in the positive pocket, thus favoring the 5′-downstream unrolling of the branch helix. The positive clamp surrounding the BP region constitutes the pivot of the IL/U2 unrolling (Fig. 5), which, assisted by the concomitant action of Cwf19 and Spp42, promotes the IL/U2 displacement. Remarkably, the IL/U2 helix is truncated in the cryo-EM structure (SI Appendix, Fig. S1) (5, 6). This, consistently with our results, may be related to a significant mobility of IL/U2, which most likely hampered its complete reconstruction.
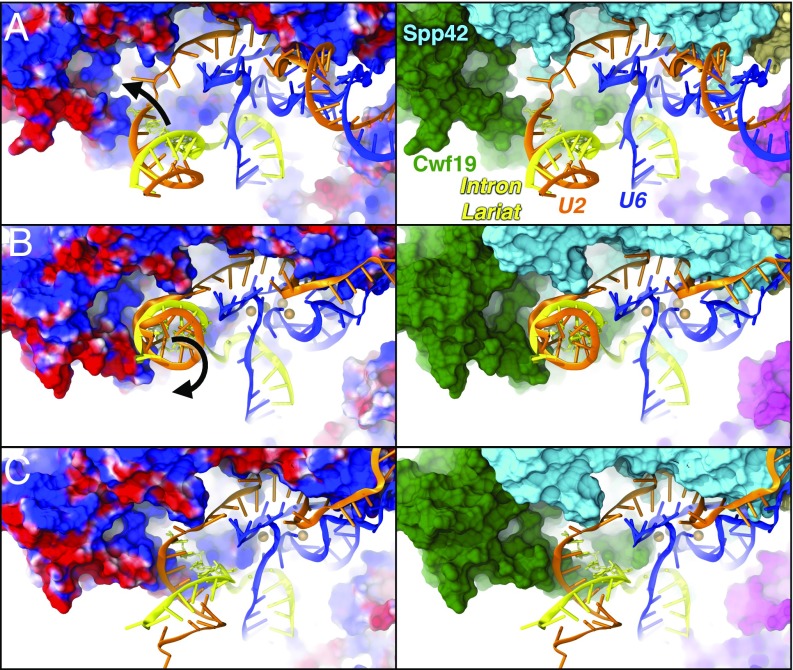
The electrostatic origin of the branch helix displacement. Snapshots from MD simulations showing the gradual displacement of the intron lariat (IL)/U2 helix. (Left) Proteins are shown with electrostatic surface (blue/red colors for positive/negative charges, respectively). (Right) Cwf19 (green) and Spp42 (cyan) are represented with surface; U2 (orange), IL (yellow), and U6 (blue) are shown as cartoon. (A) The positively charged pocket, formed by Cwf19 and Spp42 recruits IL/U2. (B) IL/U2 interacts with this pocket and starts unrolling. (C) IL/U2 displacement due to the action of Cwf19 and Spp42.
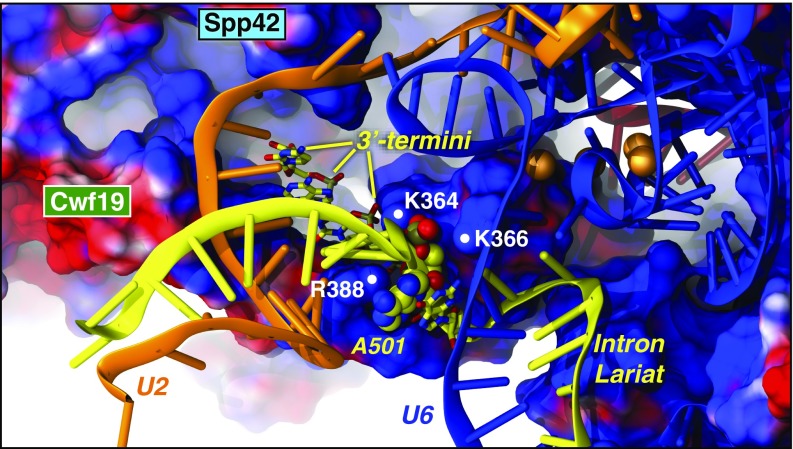
The polar tweezers formed by K364, K366, and R388 of Cwf19. Cwf19 and Spp42 are shown with the electrostatic surface (blue/red colors for positive/negative charges, respectively). U2 (orange), U6 (blue), and IL (yellow) are depicted as cartoons. IL A501 (spheres) and 3′-termini (licorice) are colored by atoms.
The Enlarged (ILS-2) Model.
Here, results from the MD simulation of the enlarged ILS-2 model (Fig. 1B) are compared with those of ILS-1. The PCA scatterplot shows that the conformational space explored along the first two PCs is similar in the two systems (SI Appendix, Fig. S6). A narrower distribution of points is observed for ILS-2, with the contribution of the first three PCs to the overall SPL motion being roughly equivalent to that of PC1-only in ILS-1 (SI Appendix, Fig. S7). The contraction of the conformational space explored by ILS-2 can be ascribed to the proteins (Prp45 and Prp17) and domains (of Spp42 and Cwf10) additionally included in this model. Prp45 in particular appears to stabilize the overall SPL structure, damping and softening the functional motions of ILS-2 (at least within the timescale investigated). Despite this, the coarse CSs histogram (Fig. 6B) obtained from the per-residue cross-correlation matrix (Fig. 6A) confirms the central role of Spp42 (Fig. 6C) in directing the motions of distinct SPL components as observed in ILS-1. Interestingly, in ILS-2, the IL/U2 displacement is shown by PC2 and PC3 (Fig. 7), which display an increased relative contribution to the total variance with respect to that observed in the ILS-1 model (SI Appendix, Fig. S7). Finally, the MD simulation of ILS-2 further assesses the critical role exerted by the polar tweezers, which forms the hinge of the IL/U2 unrolling by tightening the BP region (SI Appendix, Fig. S8). Here, in addition to K364, K366, and R388, also the R385 residue exposed by Cwf19 contributes to lock the BP region.
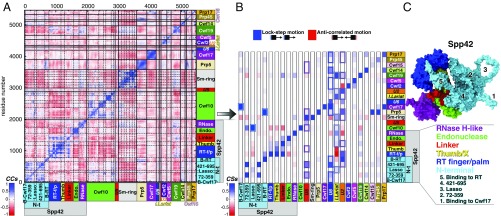
Cooperative motions underlying the functional dynamics in the spliceosomal ILS-2 model system. (A) Per-residue Pearson’s coefficients (CCs) cross-correlation matrix derived from the mass-weighted covariance matrix constructed over the last 920 ns of MD simulations of ILS-2 (replica #1) for Cα and P atoms. CCs are comprised between −1 (anticorrelation, red) and +1 (correlation, blue). The domains of Cwf10 highlighted in the matrix are (from Left to Right): N-t, G, D-II, D-III, D-IV, D-V, and C-t; proteins of Sm-ring are (from Left to Right): D3, B, D1, D2, E, F, and G. (B) Histogram reporting the (per-column) normalized correlation scores (CSs) calculated for each pair of spliceosomal components. Each column reports the CSs calculated between a specific component and all of the others (listed on rows). Spp42 domains and N-terminal motifs are distinctly considered. CS value ranges from −1 (anti-correlated motions) to +1 (lock-step motions). The most relevant correlations are highlighted using colored boxes. (C) Close-up of Spp42 (surface representation) in ILS-2, with the six domains shown/listed with different colors. The five N-terminal motifs are numbered and listed. Abbreviations: B-Cwf17/RT, binding to Cwf17/RT; Endo., endonuclease; I. Lariat, intron lariat; RNase, RNase H-like; RT-f/p, reverse-transcriptase finger/palm; Thumb, Thumb/X.

Displacement (highlighted with a black arrow) of the intron lariat (IL)/U2 branch helix, which unrolls away from the SPL, as revealed by the PCA of MD trajectory of ILS-2 model. Arrows, colored according to the protein/RNA filament to which they belong, show the motion of Cα/P atoms along the second eigenvector (PC2, thicker arrows) and third eigenvector (PC3, thinner arrows). Cwf19 (green) and Spp42 (cyan) are depicted as cartoon; U2 (orange) and IL (yellow) are shown as tube; U6 (blue) and the other proteins are represented as cartoons. Mg2+ (orange) and Zn2+ (yellow) are shown as spheres.
Discussion
The SPL is an extremely dynamic machinery, undergoing continuous conformational and compositional changes. Although recent cryo-EM experiments have provided significant insights on this sophisticated macromolecule, the SPL structural/functional plasticity still calls for an in-depth understanding, aimed at better dissecting the specific role of the different proteins recruited along the splicing cycle. In the present study, we disentangle the most important cooperative motions underlying the functional dynamics of the ILS complex from an unprecedented atomic-resolution perspective. As major results, our work elucidates the leading role of the Spp42 protein and supplies key hints for a functional annotation of the NTR Cwf19 protein.
As highlighted by correlation and PCA analyses, our MD simulations corroborate that the SPL acts as a protein-directed ribozyme. Indeed, the three snRNAs of the ILS complex, all included in our models, are extremely tied with the proteins (Spp42 in particular) in their dynamical evolution, suggesting that their motions are directed by the protein scaffold (Figs. 2B and and6B).6B). Significantly, this unique trait of the SPL, so far highlighted by structural considerations based on the cryo-EM models solved in the last years (12, 25), is now sustained also by our simulations. In this scenario, the large Spp42 (Prp8 in S. cerevisiae) protein (Figs. 2C and and6C),6C), which exhibits a peculiar structural organization made of seven distinct domains (i.e., N-terminal, RT finger/palm, thumb/X, linker, endonuclease-like, RNase H-like, Jab1/MPN), is the central hub of the SPL (1–4, 25). Despite the dramatic compositional and conformational remodeling observed along the splicing cycle, the cryo-EM models of the SPL (mostly from S. cerevisiae) have pictured that Prp8 (Spp42) constitutes a rigid scaffold from the Bact to the ILS complex, with the exception of its highly mobile RNase H-like domain and some peculiar structural motifs and loops (25). Apparently, small conformational changes involving these local structural elements and RNase H-like domain of Prp8 (Spp42) may contribute to finely control the progression along the SPL cycle (25). Remarkably, these experimental observations are fully consistent with the functional dynamics of Spp42 captured from our simulations, showing that Spp42 heads the gene maturation in the ILS by modulating the motions of specific proteins through its sprawling multibranched architecture. The leading role of Spp42 is particularly emphasized by the plasticity of its N-terminal domain, which bears peculiar structural motifs such as the lasso loop or α-helices acting as protruding arms. Indeed, the CSs histograms calculated for ILS-1 (Fig. 2B) and ILS-2 model (Fig. 6B) reveal that, through these N-terminal motifs/loops, Spp42 establishes a close dynamical connection with specific proteins, among which are Cwf10, Cwf17, and Prp5. These couplings are remarkably evident and conserved in both ILS-1 and ILS-2 models. Since it has been hypothesized that the stage-specific splicing factors and enzymes are recruited onto a generally similar SPL scaffold (12, 25), it is tempting to suggest that the functional dynamics of Spp42 (Prp8) elucidated here may be shared also by other SPL complexes assembled along the cycle.
Intriguingly, the fine modulation of ILS cooperative motions occurs while preserving a stable catalytic site placed at the core of the SPL and composed of four Mg2+ ions coordinated by U6 snRNA phosphate ligands (1–4). This intricate RNA-based active site has been extraordinarily conserved across evolution from bacteria to humans for catalyzing the pre-mRNA splicing reactions (1–4). However, at variance with G2IRs, where the active site is stabilized by a network of RNA/RNA interactions, in the SPL the catalytic center is embedded in a protein cavity formed by Spp42 (Prp8) (25). In line with these experimental observations, our simulations confirm the stability and the integrity of the catalytic site even at the late stage of splicing studied here (ILS), further assessing the reliability of the adopted simulation protocol.
Of utmost importance is that the cryo-EM structure investigated here provides structural information of Cwf19 (5, 6), whose function has remained so far obscure (32). The S. pombe Cwf19 (CWF19L2 in humans) (19) is the paralog of the S. cerevisiae Drn1, as they show only a conserved C-t domain (32). In S. cerevisiae, Drn1 enhances the activity of its homolog Dbr1 (32), the enzyme that linearizes the lariats by cleaving the 2′,5′-phosphodiester linkage (33). Garrey et al. (32) hypothesized that Cwf19 (as well as CWF19L2 in humans), lacking a complete evolutionary homology with Drn1, may have developed supplemental functions that go beyond the regulation of Dbr1 debranching activity. In this respect, our results appear to foster this theory, suggesting that Cwf19 is effectively involved in the displacement of the branch helix (Figs. 3, ,4,4, and and7),7), a critical event preceding (and necessary for) IL debranching. As a final remark, the electrostatics, which plays a fundamental role in the SPL for the recruitment and the placement of the snRNAs, as well as for the formation of the catalytic cavity (5, 6, 25), is here suggested to be also at the origin of this functional motion affecting the ILS disassembly (Fig. 5 and SI Appendix, Figs. S5 and S8).
In summary, multi-microsecond-long all-atom MD simulations of the heart of the ILS complex provide unprecedented insights on the SPL functional plasticity. This work repaints the key role of Spp42 as that of an orchestra conductor of a gene maturation symphony by strikingly revealing its fine modulation of the functional motions of many distinct SPL components. Among those, the essential dynamics analysis has unveiled an electrostatically driven unrolling and displacement of the branch helix copromoted by Cwf19 and Spp42. This finding provides evidences for a functional involvement of Cwf19 in the ILS disassembly. A detailed comprehension of the eukaryotic splicing has potential breakthrough implications for revolutionary gene modulation therapies to fight complex diseases. In this scenario, our work represents an unprecedented attempt to characterize the SPL functional dynamics with MD simulations and dispenses a noteworthy piece of knowledge for a thorough mechanistic understanding of this fundamental step of gene expression.
Materials and Methods
MD simulations were based on the S. pombe SPL reconstructed with cryo-EM at the average resolution of 3.6 Å (PDB entry 3JB9) (5, 6). The ILS-1 and ILS-2 models were built on the core region of this structure, representing the most important and conserved part of the SPL throughout the splicing cycle. De novo model building, as implemented in Modeler 9, version 16 (34), was used to reconstruct short missing loops where necessary. MD simulations were performed with Gromacs 5 (35) software package. The AMBER-ff12SB FF was adopted for proteins (36), while the ff99+bsc0+χOL3 FF was used for RNAs (37, 38), since these are the most validated and recommended FFs for protein/RNA systems (26). Mg2+ ions were described with the nonbonded fixed point charge FF due to Åqvist (39) as it was shown to properly describe binuclear sites (9). Na+ ions parameters were taken from Joung and Cheatham (40), while Zn2+ ions were modeled with the cationic dummy atoms approach developed by Pang (41). A slow and careful equilibration protocol was adopted before the productive phase as suggested by Šponer et al. (26). The trajectories were inspected and visualized with VMD software (42). All of the analyses, including rmsd, Rg, PCA, and the calculation of the cross-correlation matrices, were done with cpptraj module of Ambertools 16 (43) and with Gromacs 5 (35) suite. The correlation scores (CSs) were derived from the cross-correlation matrices (Figs. 2A and and6A6A and SI Appendix, Figs. S9 and S10), following the approach adopted in previous studies (27–29). Electrostatic calculations were performed on the proteins included in ILS-1 and ILS-2 with the adaptive Poisson-Boltzmann solver (APBS 1.4) software (44). A detailed description of the structural models (ILS-1 and ILS-2), systems setup, MD simulations, PCA, correlation analysis, and electrostatic calculations are provided in SI Appendix.
Supplementary Material
Supplementary File
Supplementary File
Acknowledgments
We are grateful for Italian SuperComputing Resource Allocation Grant HP10B1YXX3 for computational resources. A.M. thanks Italian Association for Cancer Research for financial support (MFAG 17134).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1802963115/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1802963115
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/115/26/6584.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.1802963115
Article citations
Metastasis-Associated Lung Adenocarcinoma Transcript 1 (<i>MALAT1</i>) lncRNA Conformational Dynamics in Complex with RNA-Binding Protein with Serine-Rich Domain 1 (RNPS1) in the Pan-cancer Splicing and Gene Expression.
ACS Omega, 9(41):42212-42226, 03 Oct 2024
Cited by: 0 articles | PMID: 39431102 | PMCID: PMC11483381
CWF19L2 is Essential for Male Fertility and Spermatogenesis by Regulating Alternative Splicing.
Adv Sci (Weinh), 11(31):e2403866, 18 Jun 2024
Cited by: 1 article | PMID: 38889293 | PMCID: PMC11336944
Assessing the mechanism of fast-cycling cancer-associated mutations of Rac1 small Rho GTPase.
Protein Sci, 33(4):e4939, 01 Apr 2024
Cited by: 0 articles | PMID: 38501467
Monovalent metal ion binding promotes the first transesterification reaction in the spliceosome.
Nat Commun, 14(1):8482, 20 Dec 2023
Cited by: 5 articles | PMID: 38123540 | PMCID: PMC10733407
RNA-protein complexes and force field polarizability.
Front Chem, 11:1217506, 22 Jun 2023
Cited by: 1 article | PMID: 37426330 | PMCID: PMC10323139
Go to all (33) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(2 citations)
PDBe - 3JB9View structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structure of a yeast spliceosome at 3.6-angstrom resolution.
Science, 349(6253):1182-1191, 20 Aug 2015
Cited by: 214 articles | PMID: 26292707
Endogenous U2·U5·U6 snRNA complexes in S. pombe are intron lariat spliceosomes.
RNA, 20(3):308-320, 17 Jan 2014
Cited by: 29 articles | PMID: 24442611 | PMCID: PMC3923126
Structural basis of pre-mRNA splicing.
Science, 349(6253):1191-1198, 20 Aug 2015
Cited by: 114 articles | PMID: 26292705
Group II intron lariat: Structural insights into the spliceosome.
RNA Biol, 12(9):913-917, 01 Jan 2015
Cited by: 11 articles | PMID: 26121424 | PMCID: PMC4615233
Review Free full text in Europe PMC





