Abstract
Free full text

Alternative Splicing Coupled Nonsense-Mediated Decay Generates Neuronal Cell Type-Specific Expression of SLM Proteins
Abstract
The unique physiological and morphological properties of neuronal populations are crucial for the appropriate functioning of neuronal circuits. Alternative splicing represents an attractive mechanism for generating cell type-specific molecular repertoires that steer neuronal development and function. However, the mechanisms that link neuronal identity to alternative splicing programs are poorly understood. We report that cell type-specific, mutually exclusive expression of two alternative splicing regulators, SLM1 and SLM2, in the mouse hippocampus is achieved by a cross-repression mechanism. Deletion of SLM2 in vivo modifies alternative splicing of its paralog Slm1 and stabilizes its mRNA, resulting in expression of SLM1 in previously SLM2-expressing cells. Despite this ectopic upregulation of SLM1, loss of SLM2 severely disrupts the alternative splicing regulation of Nrxn1, Nrxn2, and Nrxn3, highlighting that the two SLM paralogs have partially divergent functions. Our study uncovers a hierarchical, SLM2-dependent mechanism for establishing cell type-specific expression of neuronal splicing regulators in vivo.
Introduction
The mammalian nervous system consists of a complex network of neuronal cell types, which are defined by unique molecular, physiological, and anatomical properties. Cell type-specific properties rely on gene expression programs that shape protein levels and isoform repertoires, including cell surface receptors, ion channels, signaling, and transport regulators. The transcriptional mechanisms that specify neuronal cell types and guide their growth during early development have been extensively studied (Dasen and Jessell, 2009; Hobert et al., 2010). By comparison, there is a lack of knowledge about cell type-specific gene expression programs associated with neuronal connectivity and function, in particular with respect to the complex recognition events between synaptic partners.
Significant diversification of protein function is achieved at the level of alternative pre-mRNA splicing (Norris and Calarco, 2012; Darnell, 2013; Zheng and Black, 2013). Temporally and spatially controlled alternative splicing programs can direct molecular and, thereby, functional diversity of gene products. More recently, alternative splicing-dependent nonsense-mediated mRNA decay (NMD) has emerged as a mechanism for controlling gene expression levels (McGlincy and Smith, 2008; Bicknell et al., 2012). To this end, alternative splicing directs the incorporation of exons, which contain premature translation termination codons, leading to the targeted degradation of the mRNA. Thus, alternative splicing not only contributes to the molecular diversification of gene products but also plays a major role in the regulation of levels and kinetics of protein expression.
A prerequisite for cell type-specific gene regulation through alternative splicing is the selective expression of alternative splicing regulators in defined cell populations. One interesting example are the KH-domain containing RNA-binding proteins Sam68, SLM1, and SLM2 (Sam68-like mammalian proteins), which belong to the STAR (signal transduction and activator of RNA) family. SAM68 (Src-associated substrate in mitosis of 68 kDa) is ubiquitously expressed in neuronal and non-neuronal cells, whereas SLM1 and SLM2 exhibit a remarkable, mutually exclusive expression in neuronal cell types (Di Fruscio et al., 1999; Venables et al., 1999; Stoss et al., 2004; Iijima et al., 2011, 2014). Notably, SLM1 and SLM2 are important regulators of neuron-specific alternative splicing, as both proteins repress incorporation of alternative exons at the alternatively spliced segment 4 (AS4) in Nrxn1, Nrxn2, and Nrxn3, a class of synaptic cell surface receptors (Iijima et al., 2011, 2014; Ehrmann et al., 2013). The resulting NRX AS4 protein variants differ in their interactions with cell surface ligands, and disruption of alternative splicing at AS4 results in defects in synapse formation and plasticity (Boucard et al., 2005; Chih et al., 2006; Graf et al., 2006; Aoto et al., 2013). Despite this critical importance of SLM1 and SLM2 function in generating cell type-specific repertoires of NRX isoforms, the mechanisms underlying the mutually exclusive expression of SLM1 and SLM2 proteins in the mammalian nervous system are unknown.
In this work, we uncover that deletion of Slm2 in mice results in an unexpected elevation of SLM1 expression, specifically in cells formerly expressing SLM2. In wild-type but not Slm2KO neurons, Slm1 mRNA is targeted for NMD by use of a previously uncharacterized splice donor site in the 3′UTR. Ectopic expression of Slm2 in dentate granule cells of the hippocampus is sufficient to suppress endogenous SLM1 expression. Surprisingly, deletion of Slm2 alone results in a profound disruption in alternative splicing regulation of Nrxn1, Nrxn2, and Nrxn3, despite the upregulation of Slm1. Our study highlights a key function for Slm2 in neuronal alternative splicing regulation by directing cell type-specific splice variant choices of Nrxns. Moreover, our results provide strong genetic evidence for alternative splicing coupled NMD in instructing cell type-specific gene expression patterns in vivo.
Materials and Methods
Knock-out mice.
Slm1KO mice were previously described (Iijima et al., 2014). An Slm2 conditional allele was generated by homologous recombination in mouse embryonic stem cells. Briefly, a genomic DNA fragment containing exon 2 (ENSMUSE00000328887) was flanked by a LoxP site and a FRT-PGK-neo-LoxP cassette encoding neomycin phosphotransferase under the control of the phosphoglycerate kinase 1 promoter. The targeting vector was electroporated into 129SvEvTac embryonic stem cells. Homologous recombination in G-418-resistant clones was confirmed, and selected cells were blastocyst injected. Chimeric animals were crossed with ROSA-26 Flpe mice (Farley et al., 2000) to remove PGK-neo sequences through Frt/Flp-mediated excision. Slm2flox/+ mice were crossed with CMVcre deleter mice (Schwenk et al., 1995) to create a germline deletion of exon 2. The Slm2flox allele was detected by PCR using primers 5′-GCTTTACAGTAGAGAAACTGG-3′ (Lox gtF), 5′-CATAGATGACCTGCTGTCAG-3′ (Lox gtR), 5′-CGGTTGGATGGTATACATGAC-3′ (Frt gtF), and 5′-CAATGCTGTTACTCATCAGG-3′ (Frt gtR). Slm1:Slm2DKO mice were generated by intercrossing of the individual mutant mice. The resulting homozygous mice were viable.
Antibodies.
Polyclonal antibodies for SAM68, SLM1, and SLM2 were previously described (Iijima et al., 2014). Commercially available antibodies were used: mouse anti-β-tubulin (E7, DSHB, 1:10000), mouse anti-GFP (Invitrogen, 1:1000), and mouse anti-NeuN (Millipore Bioscience Research Reagents, 1:2000).
Molecular biology procedures.
For immunoblotting, either HRP-conjugated secondary antibodies and Pierce ECL Western Blotting Substrate or fluorescent IRDye coupled secondaries were used. Signals were acquired using an image analyzer (Bio-Rad, ChemiDoc MP Imaging System and Li-Cor, Odyssey). Dissociated cultures of mouse hippocampal cells were prepared from P0 pups and maintained in Neurobasal medium (Invitrogen) containing 2% B27 (Invitrogen) supplement and 2 mm Glutamax (Invitrogen) at 37°C and 5% CO2. Cylcoheximide was applied at a final concentration of 50 μg/ml for 4 h before harvesting. Total RNA from cultured cells was extracted with Trizol and purified using QIAGEN RNeasy Mini Kit.
Alternative splicing analysis.
PCR primer sequences are as follows: Nrxn1 AS4, 5′-TGTTGGGACAGATGACATCGCC-3′ and 5′-GAGAGCTGGCCCTGGAAGGG-3′; Nrxn2 AS4, 5′GTGCGCTTTACTCGAAGTGGTG3′ and 5′CCCATTGTAGTAGAGGCCGGAC3′; Nrxn3 AS4, 5′TTGTGCGCTTCACCAGGAATG3′ and 5′AGAGCCCAGAGAGTTGACCTTG3′; Gapdh, 5′GCTTGTCATCAACGGGAAG3′ and 5′TTGTCATATTTCTCGTGGTTCA3′; and Slm1 Ex8-Ex9b, 5′CTACGGTCATGGAGTAAACGAG3′ and 5′ GGGAATACAAGAAATAGTGGGAGC3′; and Slm1 Ex8-Ex10, 5′CTACGGTCATGGAGTAAACGAG3 and 5′CCCTTTAGGCCATCATATCTGC3′.
Sequences of quantitative PCR primers.
Quantitative PCR was performed on a StepOnePlus qPCR system (Applied Biosystems). Gene expression assays were used with TaqMan Master Mix (Applied Biosystems) or Power SYBR Green Master Mix (Applied Biosystems) and comparative CT method. The cDNA levels were normalized to Gapdh cDNA.
Commercially available gene expression assays for Gapdh (Mm99999915g1), Nrxn1 (Mm00660298_m1), Nrxn2 (Mm01236851_m1), and Nrxn3 (Mm00553213_m1) were from Applied Biosystems; and assays for Slm1 (Mm.PT.58.41510930) and Slm2 (Mm.PT.58.10822025) were from Integrated DNA Technologies. Custom gene expression assays to interrogate Nrxn splicing at AS4 were previously described (Iijima et al., 2014).
Primer sequences used with Power SYBR Green PCR Mastermix were as follows: Slm1 Ex3-Ex4, 5′-GACCAAGAGGAAACTCCTTGAA-3′ and 5′-GGCATGACTCATCCGTGAATA-3; Slm1 Ex8-9a, 5′-GTGCCTGAATACTATGACTACGG-3′ and 5′-GAGACCACAGGCTATGAATTGT3′; Slm1 Ex9a-9b, 5′-CATCTGTGACCTCCTCAAAGAC-3′ and 5′-TGTCCACGTCACATTAACAGTAT-3′; and Slm1 Ex9a-10, 5′-CATCTGTGACCTCCTCAAAGAC-3′ and 5′-CCCTTTAGGCCATCATATCTGC-3′.
Immunohistochemistry, image acquisition, and statistical analysis.
Animals (male and female) were transcardially perfused with fixative (4% PFA/15% picric acid in 100 mm phosphate buffer, pH 7.2). Tissue was sectioned at 50 μm in PBS on a vibratome (VT1000S, Leica), and floating sections were immunostained following standard procedures. Images were acquired at room temperature on an inverted LSM700 confocal microscope (Zeiss) using 10× and 63× Apochromat objectives and controlled by Zen 2010 software. Images were assembled using Adobe Photoshop and Illustrator software. For quantitative assessment of protein upregulation or downregulation, ImageJ was used. Statistical analysis was done with Prism software (GraphPad software). All statistical data are mean ± SEM. Data were tested for normality using ANOVA and Bonferroni's or Dunnett's post hoc test for multiple comparisons.
All procedures related to animal experimentation were reviewed and approved by the Kantonales Veterinäramt Basel-Stadt.
Adeno-associated virus (AAV) production.
Viral supernatants were produced by cotransfection of HEK293T cells with a mixture of three plasmids. The plasmid mixture consisted of 70 μg of AAV helper plasmid (Rep/Cap, Serotype II), 200 μg of AAV pHGTI-adeno1 (Plasmid Factory), and 70 μg of vector plasmid Synapsin-SLM2-2A-Venus or Synapsin-EGFP (containing the human synapsin promoter). At 45–60 h after transfection, medium containing viral particles was harvested and purified using the Iodixanol purification method. The fraction samples were dialyzed and concentrated in Millipore Amicon 100K columns, washed 3× with PBS, and centrifuged at 3500 rpm at 4°C. Viruses were frozen in aliquots and stored at −80°C. Viruses were delivered by stereotaxic injection at postnatal day 8 into the dentate gyrus (DG) of the mouse hippocampus. Infected cells were marked by EGFP expression from the same vector. Animals were transcardially perfused at postnatal day 40, and 50-μm-thick vibratome slices were analyzed by immunohistochemistry.
Results
Ablation of Slm2 in vivo results in elevated expression of Slm1
To explore functions of the RNA-binding protein SLM2 in vivo, a novel mouse knock-out Slm2 allele was created in which exon2 of the mouse Khdrbs3 gene (which encodes for SLM2) was flanked by LoxP sites. Global Slm2KO mice were generated by cre-mediated recombination in the germline. Removal of exon 2 introduces multiple premature stop codons and disrupts the Slm2 open reading frame (Fig. 1A). SLM2 is mainly expressed in the brain and in testis (Stoss et al., 2004; Venables et al., 2004); we thus investigated loss of SLM2 in these body regions (Fig. 1B). Additionally, Western blotting analysis with two SLM2-specific antibodies directed against different epitopes further confirmed that Slm2KO mice lacked detectable SLM2 protein expression (Fig. 1C). Slm2KO mice were viable and fertile and did not exhibit obvious behavioral abnormalities, as previously reported for a different Slm2KO allele (Ehrmann et al., 2013).

A, Targeting strategy for generation of conditional Slm2KO mice. The Khdrbs3 gene encoding SLM2 protein was modified by insertion of LoxP sites (blue) flanking exon 2 (E2) and targeted ES cells isolated based on G418-resistance conferred by the PGKneo cassette (orange). PGKneo was subsequently excised by breeding to ROSA-26 Flpe mice. The Slm2 knock-out allele was generated by crossing Slm2 floxed animals with CMVcre delete mice. B, Successful ablation of SLM2 was verified by Western blot analysis of cortical and testis tissue. C, Western blot analysis of two sets of WT and Slm2KO littermates, tested with two different antibodies (A, B) against two different epitopes of the SLM2 protein. Slm2 encodes several protein variants detected as bands of different electrophoretic mobility.
Upregulation of SLM1 in previously SLM2-positive cells
Surprisingly, we detected a 2.5-fold upregulation of SLM1 protein levels in hippocampus and cortex of Slm2KO mice (Figure 2A; one-way ANOVA with Bonferroni post hoc test, p < 0.01). A significant increase was also detected for Slm1 mRNA in the hippocampus of Slm2KO mice (Fig. 2B; one-way ANOVA with Bonferroni post hoc test, p < 0.01). By contrast, no alterations were observed for the closely related RNA-binding protein SAM68 (data not shown). To explore whether SLM1 is globally elevated or whether its upregulation is restricted to specific cell populations, we performed detailed immunohistochemical analysis using SLM1- and SLM2-specific antibodies. Principal cells in the mature hippocampus exhibit a remarkable, cell type-specific expression pattern of SLM1 and SLM2: SLM1 is expressed in dentate granule cells, and a specific subgroup of inhibitory interneurons, however, is absent from the cornus ammonis-1 (CA1) and CA3 pyramidal cells. By contrast, SLM2 is highly expressed in the CA1 and CA3 pyramidal cells and a different set of interneurons, but not detectable in the DG (Stoss et al., 2004; Iijima et al., 2014) (Fig. 3A, top). In Slm2KO mice, SLM1 was significantly upregulated in CA1 and CA3, whereas expression in DG was unchanged (Fig. 3A,B; two-way ANOVA with Bonferroni post hoc test). Thus, in the absence of SLM2, formerly SLM2-positive cells selectively turn on SLM1 expression. This suggests that the mutually exclusive expression pattern of SLM1 and SLM2 in neuronal populations might be achieved through cross-repression of SLM1 by SLM2. Interestingly, we did not detect an analogous upregulation of SLM2 in the DG of Slm1KO mice (Fig. 3A,B). These findings suggest that SLM2 is required for spatially restricting SLM1 expression in a unidirectional manner.
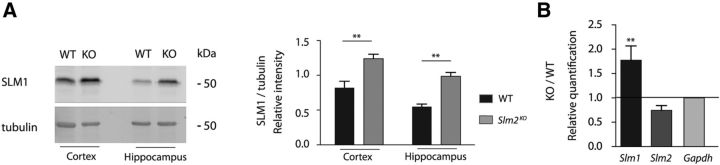
A, Upregulation of SLM1 in different brain regions in Slm2KO mice (left). Quantification of SLM1/tubulin ratios in WT and Slm2KO mice of the hippocampus and cortex (right) (n = 4 animals, 8 weeks old). **p < 0.01. B, Quantitative RT-PCR for total Slm1 and Slm2 mRNA of hippocampal tissue. WT mRNA/Gapdh values were set to 1.0 and compared with Slm2KO values (n = 4 animals, 8 weeks old).

A, Hippocampal sections of WT, Slm2KO, and Slm1KO mice were stained for the neuronal marker NeuN, as well as for SLM1 and SLM2. Scale bar, 200 μm. B, Quantification of mean intensities of SLM1 and SLM2 in DG, CA1, and CA3 areas for Slm1KO and Slm2KO animals (n = 4 animals, 3 months of age, 2 sections per animal were used). Relative values compared with NeuN are plotted. ns, Not significant (p > 0.05). *p < 0.5. **p < 0.01. ***p < 0.001. These values are the raw fluorescent values without background subtraction. The KO genotypes serve as an indication of respective background labeling.
Slm2 is required and sufficient to establish the mutually exclusive expression pattern of SLM proteins
Considering that SLM2 is an RNA-binding protein, we hypothesized that it may suppress Slm1 expression by targeting Slm1 mRNA for NMD in wild-type animals. To test this possibility, we treated cultured wild-type and Slm2KO hippocampal neurons with cycloheximide (CHX), an inhibitor of protein translation. As NMD depends on active translation, transcripts that are subject to NMD are stabilized upon translational arrest (Carter et al., 1995). Slm1 transcript levels increased threefold upon CHX treatment of wild-type neurons. (Fig. 4A; one-way ANOVA with Bonferroni post hoc test). Importantly, in Slm2KO cultures, Slm1 transcript levels were not further increased after CHX treatment (Fig. 4A; one-way ANOVA with Bonferroni post hoc test, not significant, p > 0.05).
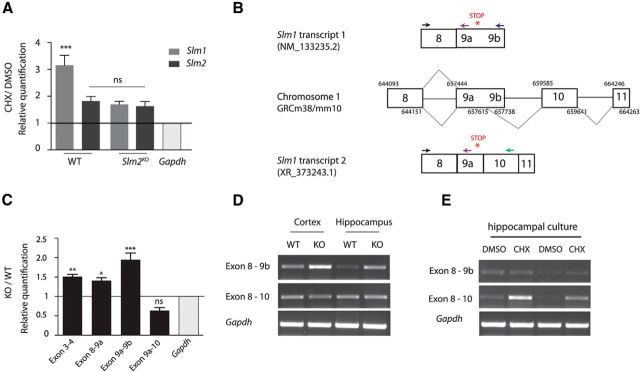
A, Quantitative RT-PCR for total Slm1 and Slm2 mRNA. Mouse hippocampal cultures were treated at day in vitro 7 for 4 h either with the translation blocker cycloheximide (CHX) or DMSO (n = 3 independently generated cultures). WT mRNA/Gapdh values in DMSO-treated control cells were set to 1.0 and compared with cells treated with CHX. There is a significant upregulation of Slm1 in the WT when treated with CHX; however, there is no upregulation of either Slm1 or Slm2 in the Slm2KO. ns, Not significant (p > 0.05). ***p < 0.001. B, Schematic representation of incorporation of exon 10 and exon 11 and primer binding sites for quantitative RT-PCR. Chromosome coordinates are taken from USCS Genome Browser (release date reference GRCm38/mm10). All chromosome coordinates start with 3217. C, Quantitative RT-PCR for increases in Slm1 transcripts at various exons comparing WT and Slm2KO animals (n = 4 animals, 8 weeks old). ns, Not significant (p > 0.05). *p < 0.5. **p < 0.01. ***p < 0.001. For the quantitative RT-PCR, primer sets flanking from exon 9a onwards instead of exon 8 were used because of the sizes of the exons. D, Exon spanning RT-PCR of WT and Slm2KO animals. E, The transcript targeted for NMD (Ex8–10) is stabilized in wild-type animals when treated with CHX.
In the same experiment, CHX treatment appears to slightly elevate Slm2 transcripts in wild-type and Slm2KO neurons (Fig. 4A), although this trend did not reach statistical significance.
These findings are consistent with NMD of Slm1 mRNA through a process that requires Slm2. One possible mechanism of how SLM2 could target Slm1 mRNA for NMD would be the incorporation of alternative exon sequences through regulated alternative splicing that attract exon junction complexes downstream of the stop codon in Slm1 transcripts. Through database searches, we identified a previously uncharacterized potential splice donor site in exon9 of Slm1, which would link two additional exons encoding the 3′UTR (exons 10 and 11; Fig. 4B). We hypothesized that an SLM2-driven alternative splicing event might promote incorporation of these exons. To test this possibility, we performed exon-spanning RT-PCRs for the presumptive exon 10 in the 3′ UTR. We used primer sets binding upstream and downstream of the potential internal splice donor site flanking various exons of the Slm1 transcript (Fig. 4B). We could confirm the novel transcript 2 variant linking exon 8 and exon 10 (PCR products exons 8–10), representing the Slm1 variant containing a 3′ UTR intron, predicted to undergo NMD (Fig. 4D). In Slm2KO mice, we observed an increase in the alternative transcript 1 variant containing exon 8–9b both with RT-PCR and quantitative RT-PCR, confirming that the presence of SLM2 suppresses formation of this (stable) form of Slm1 mRNA (Fig. 4C,D; Dunnett's multiple comparison test). Using primer pairs upstream of the internal splice donor site (exons 3 and 4, exons 8 and 9a), we further confirmed that the whole Slm1 transcript was stabilized (Fig. 4C). Products containing exons 8–10 showed a trend to decrease in Slm2KO cells (Fig. 4C). However, if these mRNAs are indeed targeted for translation-dependent decay, this is difficult to quantify. To further explore this possibility, we performed CHX treatment of wild-type hippocampal neurons. Exons 8–10-containing transcripts were significantly increased in CHX-treated cells, consistent with the hypothesis that they are targeted for NMD (Fig. 4E).
If Slm2 indeed targets Slm1 mRNA for degradation, then Slm2 should be sufficient to suppress SLM1 expression when introduced into SLM1-positive cell populations. AAV-mediated SLM2 mis-expression in the DG of the mouse hippocampus in vivo abolished SLM1 expression in dentate granule cells (Fig. 5A,B). SLM2 was introduced into the DG at postnatal day 8 (P8), and mis-expressing neurons were viable and developed apparently normal axonal and dendritic processes when analyzed at P40. However, their cell nuclei were devoid of SLM1 protein. Thus, SLM2 is required and sufficient to regulate the cell type-specific expression of SLM proteins in vivo.

A, Injection of AAV-viruses into the DG of WT animals at postnatal day 8 (P8). Mice were analyzed at P40. Scale bar, 100 μm. B, Immunohistochemistry analysis of WT animals injected either with pAAV-EGFP, as injection control, or pAAV-SLM2-Venus. Sections were stained for the nucleic acid stain Hoechst, and antibodies against EGFP, SLM1, and SLM2. Scale bar, 50 μm. Bottom, Higher-magnification views of fields from the overview panels. Color code for merge is GFP (green) and SLM1 (magenta).
Alternative splicing regulation in Slm1:Slm2DKO mice
SLM1 and SLM2 have partially overlapping targets; however, they are expressed in largely nonoverlapping cell populations (Stoss et al., 2004; Iijima et al., 2014). Thus, the upregulation of SLM1 in Slm2KO neurons needs to be considered when interpreting loss-of-function phenotypes. We set out to determine the impact of Slm1 and Slm2 loss-of-function in Slm1:Slm2DKO mice. Slm1:Slm2DKO mice were born at Mendelian frequencies and were viable. Successful ablation of SLM1 and SLM2 was verified by immunohistochemistry (Fig. 6A) and Western blotting analysis (Fig. 6B). Slm1KO as well as Slm2KO neurons fail to direct exon skipping of Nrxn1-3 at AS4, which correlates with their regional expression in the brain (Ehrmann et al., 2013, Iijima et al., 2014). To explore how simultaneous loss of both RNA binding proteins affects neuronal alternative splicing regulation, we examined the Nrxn AS4 regulation in the Slm1KO, Slm2KO, and Slm1:Slm2DKO mice. Analysis of cortical and hippocampal tissue revealed that splicing patterns were altered for all three Nrxn genes. Expression of AS4(−) forms was significantly reduced in Slm2KO and completely abolished in the Slm1:Slm2DKO. By contrast, the splicing patterns in the Slm1KO cortex and hippocampus remained unchanged (Fig. 6C,D). Quantitative RT-PCR revealed that overall Nrxn1, Nrxn2, and Nrxn3 levels were unaltered in all three mutants and confirmed a striking increase of AS4(+) in Nrxn1–3 for the Slm2KO and the Slm1:Slm2DKO (Fig. 6D; two-way ANOVA). Notably, there was a significant loss of Nrxn AS4- splice isoforms in Slm2KO hippocampus, despite the substantial upregulation of SLM1 in SLM2-deficient cells. Thus, in vivo, SLM1 upregulation cannot significantly compensate for loss of SLM2, at least with respect to alternative splicing regulation of Nrxns at AS4.
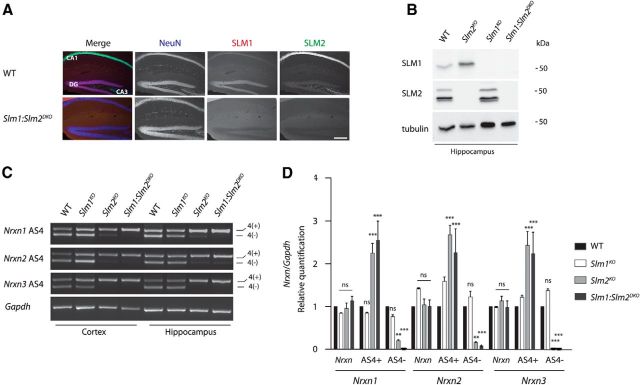
A, Loss of SLM1 and SLM2 expression in the Slm1:Slm2DKO was verified by SLM1 and SLM2-specific immunostaining. Scale bar, 200 μm. B, Loss of SLM1 and SLM2 expression verification in the Slm1KO, Slm2KO, and Slm1:Slm2DKO by Western blotting analysis. Two gels were loaded with equal amounts of sample for the individual genotypes; however, the tubulin control displayed derived from the gel also probed for SLM1. C, Representative images of semiquantitative RT-PCR with Nrxn AS4 in cortex and hippocampus from WT, Slm1KO, Slm2KO, and Slm1:Slm2DKO mice. D, Quantitative RT-PCR for Nrxn1, Nrxn2, and Nrxn3 AS4 variants in hippocampal tissue. Total Nrxn1, Nrxn2, and Nrxn3 levels are unchanged in the Slm1KO (n = 3 animals), Slm2KO (n = 4 animals), and Slm1:Slm2DKO (n = 3 animals). For Slm2KO and Slm1:Slm2DKO, significant upregulation could be detected for Nrxn1, Nrxn2, and Nrxn3 AS4(+) and corresponding downregulation of Nrxn1, Nrxn2, and Nrxn3 AS4(−), respectively. In hippocampus, the Slm1KO did not display any changes for all three Nrxn genes at AS4 compared with WT animals. ns, Not significant (p > 0.05). **p < 0.01. ***p < 0.001.
Discussion
In mammals, regulated alternative splicing events are widespread; however, the factors underlying the generation of neuronal cell type-specific alternative patterns are poorly understood. The ability of alternative splicing to generate mRNA isoforms that are targeted for NMD extends the functional impact of alternative splicing regulation from the generation of multiple isoforms derived from a single gene to the spatial and temporal control of transcript levels. In this study, we set out to uncover in vivo functions of SLM2 in neurons and identified an unexpected role in setting up the mutually exclusive expression of SLM1 and SLM2 paralogs in neuronal cell types.
We observed that SLM2 itself is required and sufficient for the mututally exclusive expression of SLM1 and SLM2 in genetically and anatomically defined populations of neurons in the mouse hippocampus. SLM2 promotes the incorporation of an alternative splice donor in the last coding exon of the Slm1 transcript. Notably, transcripts with 3′UTR introns have long been considered to be erroneous predictions as the resulting mRNAs would be predicted to be NMD substrates. However, recent studies have highlighted unexpected functions of some of these transcripts (Bicknell et al., 2012). We identified one such transcript containing a 3′UTR intron for Slm1. Our genetic gain- and loss-of-function analysis in mice demonstrates that this mRNA is indeed produced through SLM2-dependent alternative splicing and that the resulting destabilization of the alternative Slm1 transcript represents a fundamental mechanism underlying the neuronal cell type-specific expression of SLM1 protein in vivo.
Previous studies performed largely in cell lines demonstrated that splicing regulators are able to control their own expression by producing splice isoforms of their own mRNA that are targeted for NMD (Lareau et al., 2007; Ni et al., 2007). Further studies identified cross-regulation between paralogs of alternative splicing regulators, such as PTBP1 (polypyrimidine-tract-binding protein 1) and PTBP2 (Boutz et al., 2007; Spellman et al., 2007), to shift the alternative splicing patterns from precursors to postmitotic neurons during differentiation and maturation in development. Our results identify a similar cross-repression mechanism for SLM1 and SLM2 to generate a highly spatially restricted pattern of SLM paralogs in specific neuronal populations. SAM68, SLM1, and SLM2 bind similar RNA sequence motifs (Galarneau and Richard, 2009) and appear to regulate some common targets. However, these proteins also exhibit certain isoform-specific properties, including differential regulation by kinases and selective association with additional alternative splicing factors, which results in differential splicing activities (Stoss et al., 2004; Iijima et al., 2011, 2014). Thus, SAM68 requires activation through calcium/CaMK4 signaling to modify alternative splice isoform choices of Nrxns, whereas SLM1 and SLM2 seem to be constitutively active in a cell type-specific manner. In the midbrain, where SLM1 is highly expressed, Nrxn alternative splicing at AS4 is significantly altered in Slm1KO mice (Iijima et al., 2014). By contrast, in the hippocampus where SLM1 protein levels are lower, its deletion has little impact on Nrxn1, Nrxn2, and Nrxn3 AS4 regulation. Possible explanations of why neither SAM68 nor SLM1 rescued the splicing phenotype observed in Slm2KO cells include that these cells lack signaling-dependent activation or cofactors for SAM68 or SLM1 function, respectively.
Our work uncovers a mechanism underlying the mutually exclusive, cell type-specific expression pattern of STAR proteins in the mammalian brain. Loss of Slm2 function results in the production of Nrxn variants with altered affinity for their synaptic binding partners (Boucard et al., 2005; Chih et al., 2006; Baudouin and Scheiffele, 2010; Uemura et al., 2010). Further studies will be needed to determine the impact of Slm2-dependent alternative splicing with regards to changes in synaptic connectivity.
Footnotes
This work was supported by the Swiss National Science Foundation, EU-AIMS (which receives support from the Innovative Medicines Initiative Joint Undertaking of the EU FP7) and the Kanton Basel-Stadt to P.S., L.T. was supported by the Erasmus Student Mobility Program. We thank Oriane Mauger, Harald Witte, Le Xiao, and other members of the P.S. laboratory for advice and insightful comments on the manuscript.
The authors declare no competing financial interests.
References
- Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88. 10.1016/j.cell.2013.05.060. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Baudouin S, Scheiffele P. SnapShot: neuroligin-neurexin complexes. Cell. 2010;141:908. 10.1016/j.cell.2010.05.024. [Abstract] [CrossRef] [Google Scholar]
- Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ. Introns in UTRs: why we should stop ignoring them. Bioessays. 2012;34:1025–1034. 10.1002/bies.201200073. [Abstract] [CrossRef] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. 10.1016/j.neuron.2005.08.026. [Abstract] [CrossRef] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. 10.1101/gad.1558107. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. 10.1074/jbc.270.48.28995. [Abstract] [CrossRef] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. 10.1016/j.neuron.2006.06.005. [Abstract] [CrossRef] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. 10.1146/annurev-neuro-062912-114322. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. 10.1016/S0070-2153(09)88006-X. [Abstract] [CrossRef] [Google Scholar]
- Di Fruscio M, Chen T, Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci U S A. 1999;96:2710–2715. 10.1073/pnas.96.6.2710. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ehrmann I, Dalgliesh C, Liu Y, Danilenko M, Crosier M, Overman L, Arthur HM, Lindsay S, Clowry GJ, Venables JP, Fort P, Elliott DJ. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 2013;9:e1003474. 10.1371/journal.pgen.1003474. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. 10.1002/1526-968X(200011/12)28:3/4<106::AID-GENE30>3.3.CO;2-K. [Abstract] [CrossRef] [Google Scholar]
- Galarneau A, Richard S. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol Biol. 2009;10:47. 10.1186/1471-2199-10-47. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Graf ER, Kang Y, Hauner AM, Craig AM. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J Neurosci. 2006;26:4256–4265. 10.1523/JNEUROSCI.1253-05.2006. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33:435–445. 10.1016/j.tins.2010.05.006. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, Scheiffele P. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. 10.1016/j.cell.2011.11.028. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Iijima T, Iijima Y, Witte H, Scheiffele P. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. J Cell Biol. 2014;204:331–342. 10.1083/jcb.201310136. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. 10.1038/nature05676. [Abstract] [CrossRef] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. 10.1016/j.tibs.2008.06.001. [Abstract] [CrossRef] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. 10.1101/gad.1525507. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Norris AD, Calarco JA. Emerging roles of alternative pre-mRNA splicing regulation in neuronal development and function. Front Neurosci. 2012;6:122. 10.3389/fnins.2012.00122. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. 10.1093/nar/23.24.5080. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. 10.1016/j.molcel.2007.06.016. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Stoss O, Novoyatleva T, Gencheva M, Olbrich M, Benderska N, Stamm S. p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol Cell Neurosci. 2004;27:8–21. 10.1016/j.mcn.2004.04.011. [Abstract] [CrossRef] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRδ2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. 10.1016/j.cell.2010.04.035. [Abstract] [CrossRef] [Google Scholar]
- Venables JP, Vernet C, Chew SL, Elliott DJ, Cowmeadow RB, Wu J, Cooke HJ, Artzt K, Eperon IC. T-STAR/ETOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum Mol Genet. 1999;8:959–969. 10.1093/hmg/8.6.959. [Abstract] [CrossRef] [Google Scholar]
- Venables JP, Dalgliesh C, Paronetto MP, Skitt L, Thornton JK, Saunders PT, Sette C, Jones KT, Elliott DJ. SIAH1 targets the alternative splicing factor T-STAR for degradation by the proteasome. Hum Mol Genet. 2004;13:1525–1534. 10.1093/hmg/ddh165. [Abstract] [CrossRef] [Google Scholar]
- Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013;29:442–448. 10.1016/j.tig.2013.04.003. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from The Journal of Neuroscience are provided here courtesy of Society for Neuroscience
Full text links
Read article at publisher's site: https://doi.org/10.1523/jneurosci.3395-14.2014
Read article for free, from open access legal sources, via Unpaywall:
https://www.jneurosci.org/content/jneuro/34/50/16755.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
New perspective on sustained antidepressant effect: focus on neurexins regulating synaptic plasticity.
Cell Death Discov, 10(1):205, 01 May 2024
Cited by: 0 articles | PMID: 38693106 | PMCID: PMC11063156
Review Free full text in Europe PMC
Differential expression of paralog RNA binding proteins establishes a dynamic splicing program required for normal cerebral cortex development.
Nucleic Acids Res, 52(8):4167-4184, 01 May 2024
Cited by: 0 articles | PMID: 38324473 | PMCID: PMC11077083
A cell-type-specific alternative splicing regulator shapes synapse properties in a trans-synaptic manner.
Cell Rep, 42(3):112173, 01 Mar 2023
Cited by: 6 articles | PMID: 36862556 | PMCID: PMC10066595
mRNA isoform balance in neuronal development and disease.
Wiley Interdiscip Rev RNA, 14(3):e1762, 19 Sep 2022
Cited by: 7 articles | PMID: 36123820 | PMCID: PMC10024649
Review Free full text in Europe PMC
RNA and neuronal function: the importance of post-transcriptional regulation.
Oxf Open Neurosci, 1:kvac011, 07 Jul 2022
Cited by: 4 articles | PMID: 38596700 | PMCID: PMC10913846
Review Free full text in Europe PMC
Go to all (29) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1.
J Cell Biol, 204(3):331-342, 27 Jan 2014
Cited by: 59 articles | PMID: 24469635 | PMCID: PMC3912522
Neuroligin-induced presynaptic differentiation through SLM2-mediated splicing modifications of neurexin in cerebellar cultures.
Biochem Biophys Res Commun, 493(2):1030-1036, 20 Sep 2017
Cited by: 2 articles | PMID: 28939043
An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus.
Elife, 5:e22757, 13 Dec 2016
Cited by: 41 articles | PMID: 27960072 | PMCID: PMC5213383
Regulation of Neuronal Differentiation, Function, and Plasticity by Alternative Splicing.
Annu Rev Cell Dev Biol, 34:451-469, 20 Jul 2018
Cited by: 73 articles | PMID: 30028642 | PMCID: PMC6697533
Review Free full text in Europe PMC





