Abstract
Free full text

Migrant memory B cells secrete luminal antibody in the vagina
Abstract
Antibodies secreted into the mucosal barriers serve to protect the host from a variety of pathogens, and are the basis for successful vaccines1. In type I mucosa such as the intestinal tract, dimeric IgA secreted by local plasma cells is transported through polymeric Ig receptors (pIgR)2, and mediates robust protection against viruses in the vaccinees3,4. However, due to the paucity of pIgR and plasma cells, how and whether antibodies are delivered to the type II mucosa represented by the lower female reproductive tract (FRT) lumen remains unclear. Here, using genital herpes infection in mice, we show that primary infection does not establish plasma cells in the lamina propria of FRT. Instead, upon secondary challenge with herpes simplex virus 2 (HSV-2), circulating memory B cells that enter the FRT serve as the source of rapid and robust antibody secretion into the FRT lumen. CD4 tissue-resident memory T cells (TRM) secrete interferon gamma (IFN-γ), which induces expression of chemokines including CXCL9 and CXCL10. Circulating memory B cells are recruited to the vaginal mucosa in CXCR3-dependent manner, and secrete virus-specific IgG2b, IgG2c and IgA into the FRT lumen. These results reveal circulating memory B cells as a rapidly inducible source of mucosal antibodies for the FRT.
Antibodies delivered inside the lumen of type II mucosa are capable of blocking infections. In the FRT, IgG but not IgA are the most protective isotypes against HSV-2 (Ref. 5). HSV-2 specific IgG, when inoculated inside the vaginal cavity, confers protection against intravaginal (ivag) HSV-2 challenge6,7 (Extended Data Fig. 1a). However, the same antibodies injected intravenously had no protective effects6,7 (Extended Data Fig. 1a), due to the lack of access of circulating antibodies to the FRT lumen6,7. We examined the ability of circulating antibodies (FITC-conjugated IgG) to enter different tissues including the FRT lumen. FITC-IgG was detected in the spleen and lung 2 hours after intravenous injection, while it was barely detectable in the vaginal parenchyma or mucosa, even after 24 h post injection (Extended Data Fig. 1b). This is consistent with the levels of antigen-specific IgG in the cervicovaginal secretion of women immunized with HPV vaccine8, influenza vaccine9 and tetanus toxoid10 being less than 1–0.1% of those found in circulation.
For certain viruses like the human papillomavirus that requires breach of the epithelial barrier and minor abrasion for infection, serum antibodies can access the site of infection to confer protection11. We tested whether serum antibodies enter the vaginal lumen in response to a minor breach in the barrier. In intact mice, virus-specific Ab was not detected in the vaginal lumen of mice immunized subcutaneously with an attenuated thymidine kinase mutant (TK−) HSV-2 (Extended Data Fig. 1c), despite the presence of serum antibodies (Extended Data Fig. 1d). However, virus-specific Ab was detected in the vaginal lumen (Extended Data Fig. 1c) after epithelial barrier breach with a cervical brush (Extended Data Fig. 1e). Thus, at steady state, circulating antibodies do not access the vaginal lumen.
Systemic inoculation of live attenuated SIV establishes plasma cells in the FRT12. Whether vaginal IgG secretion can be enhanced by other means of immunization remains unclear, and is a key question in the field of vaccines against sexually transmitted infections. To address this question, we first examined the presence of B cells within the FRT following ivag immunization with TK− HSV-2. No increase in the percentage or the number of plasmablasts (CD138+CD19+), plasma cells (CD138+CD19−), or CD138− CD19+ B cells was detected in the vagina five weeks after immunization (Fig. 1a). B cell number remained low in the vagina even after inducing local inflammation with intravaginal CpG inoculation 5 days after priming with TK− HSV-2 (Extended Data Fig. 2a). Analysis of vaginal tissue section showed very few B220+ cells in naïve or immunized mice. We detected rare CD138+B220− plasmablast/plasma cells scattered throughout the vaginal lamina propria after immunization (Fig. 1b), consistent with a previous study13. Nevertheless, the presence of B cells or plasma cells paled in comparison to the robust formation of the CD4 T cells within the memory lymphocyte cluster (MLCs)14 (Fig. 1b). We also examined the presence of B cells within the upper FRT. Similar to the vaginal mucosa, no increase in the number of plasmablasts, plasma cells, or CD138− CD19+ B cells was observed in the cervix and uterus five weeks after immunization (Extended Data Fig. 3; immune group).
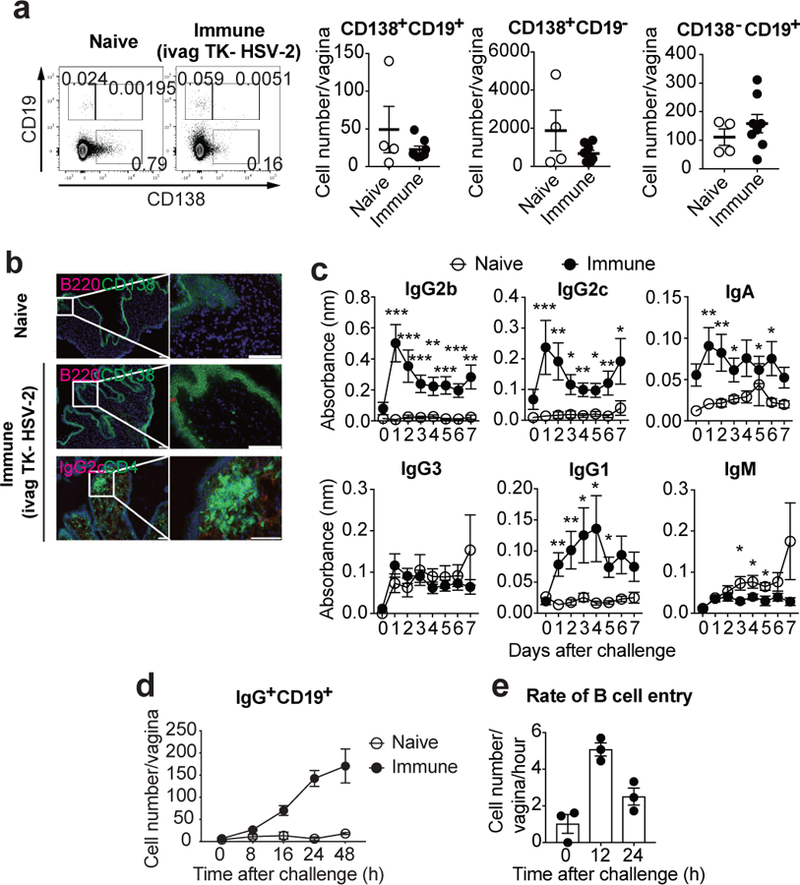
a-b, C57BL/6 mice were immunized intravaginally with TK− HSV-2. a, Five weeks later, CD138+CD19+, CD138+CD19−, and CD138−CD19+ cells in vaginal tissues were analyzed in both naïve (n=4) and immunized (n=8) mice by flow cytometry. b, Six weeks later, frozen sections of vagina were stained with antibodies against CD138 and CD4 (green), B220 and IgG2c (red), and DAPI (blue). Scale bars indicate 100 μm. c–d, C57BL/6 mice with or without immunization with TK− HSV-2 five weeks prior were challenged with WT HSV-2 intravaginally. c, Following challenge, HSV-2-specific antibodies in vaginal wash were measured by ELISA (naïve; n=9, immune; n=13). Sample dilution was 1:7. d, After challenge, the number of B cells in vagina was analyzed by flow cytometry (naïve; n=3, immune; n=4). e, B cells isolated from DsRed mice immunized with TK− HSV-2 five weeks prior were adoptively transferred into mice immunized with TK− HSV-2 five weeks prior at the indicated hours after secondary challenge with WT HSV-2. Vaginal tissues were collected 2 h after adoptive transfer, and the number of DsRed+IgG+ B cells in vaginal tissues was analyzed by flow cytometry (n=3). Data are mean ± SEM. Data are pooled from two (a) and four (c) independent experiments, or are representative of three (b,d) and two (e) independent experiments. *, p<0.05; **, p<0.01; ***, p<0.001 (two-tailed Mann-Whitney U test).
Next, we examined the levels of secreted HSV-2-specific antibodies in the vaginal lumen and in circulation five weeks after immunization (Extended Data Fig. 1f and andg).g). Intranasal (i.n.) immunization with influenza virus A/PR8 was included as a reference. In bronchoalveolar lavage (BAL) fluid, high levels of flu-specific antibodies were detected even after 5 weeks of immunization (Extended Data Fig. 1f). In contrast, vaginal secretion contained low levels of HSV-2-specific antibodies, despite the high levels of circulating antibodies (Extended Data Fig. 1f and andg).g). Antibody levels in vaginal lumen were not significantly enhanced by local inflammation induced by CpG ivag inoculation 5 days post immunization (Extended Data Fig. 2b). These results indicated that unlike the respiratory mucosa, very limited amount of circulating antibodies enter the vaginal lumen following immunization.
Next, we investigated luminal Ab secretion following secondary challenge with vaginal wild-type (WT) HSV-2 in mice previously vaginally immunized with TK− HSV-2 (Fig. 1c). Notably, virus-specific IgG2b, IgG2c, IgG1, and IgA were rapidly secreted into the vaginal lumen, as early as one-day post challenge. Naïve mice challenged for the first time with HSV-2 ivag had no detectable virus-specific antibodies in the time frame examined (Fig. 1c). Following secondary viral challenge, a rapid and robust recruitment of IgG+ B cells to the vagina was observed (Fig. 1d). Increased number of plasma cells in cervix and uterus was also observed early after secondary challenge (Extended Data Fig. 3). These results are consistent with the recruitment of B cells observed after restimulation of CD4 TRM or CD8 TRM in the FRT15,16. Primary infection of naïve mice with HSV-2 resulted in minimal increase in B cell migration within the first 48 hours (Fig. 1d), indicating that the immune status of the host was required for this rapid B cell migration into the FRT. Memory B cells can be categorized into CD80+PD-L2+ cells, which differentiate rapidly into antibody forming cells, and CD80−PD-L2− memory B cells that generate few early antibody forming cells but robustly seed germinal centers17. Notably, about half of IgD−IgG+ B cells (memory B cells) in the FRT of secondary challenged mice expressed both CD80 and PD-L2 (Extended Data Fig. 4a), and accumulated in the FRT (Extended Data Fig. 4b). To measure the rate of B cell entry into the vagina following secondary challenge, B cells from DsRed transgenic immune animals were adoptively transferred to host mice previously immunized with TK− HSV-2. The entry of transferred B cells peaked at 12 h post re-challenge and still remained high at 24 h (Fig. 1e). An increasing proportion of IgG+ B cells that are recruited to the vaginal tissue in response to secondary HSV-2 challenge expressed costimulatory molecule, CD86, as well as activation marker, CD69 (Extended Data Fig. 5a). Further analysis demonstrated that VCAM-1 expression on endothelial cells in lamina propria rapidly increased following HSV-2 challenge (Extended Data Fig. 5b), consistent with a previous study18, possibly providing an adhesion molecule for the recruitment of B cells to the vaginal mucosa. Long-term tracking of B cells in the vagina showed peak accumulation around 5 days post re-challenge, followed by their decline over several weeks (Extended Data Fig. 5c). In contrast to mice immunized vaginally, those immunized through subcutaneous (SQ) injection with TK− HSV-2 did not show B cell recruitment into the FRT upon vaginal challenge with WT HSV-2 (Extended Data Fig. 6a), despite the two routes of immunization (SQ and IVAG) leading to similar levels of circulating Ab (Extended Data Fig. 6b). Thus, in mice immunized locally with an attenuated HSV-2, IgG+ B cells are recruited rapidly to the vagina shortly after secondary challenge and become activated in the tissue. These cells remain within the vaginal tissue for several weeks thereafter.
We next investigated whether antigen-specific, active virus challenge is necessary for the recruitment of B cells to the FRT. Intravaginal challenge with a heterologous virus, influenza A/PR8, or stimulation with CpG1826 to induce inflammation in the FRT19 failed to recruit B cells into the vagina of immunized mice (Extended Data Fig. 7a–c). In contrast, vaginal inoculation of heat-inactivated WT HSV-2 induced significant recruitment of B cells into the tissue, with an accompanying increase in luminal secretion of HSV-2-specific antibodies (Extended Data Fig. 7d and ande). Thesee). These data indicated that a local antigen-specific recall response is required for the recruitment of IgG+ B cells and secretion of antigen-specific IgG.
To determine whether B cell migration is required for local Ab secretion, we examined the secretion of virus-specific immunoglobulin in the vaginal wash following treatment with FTY720 (S1P1R agonist)20. This treatment resulted in a vast reduction of B cells in the FRT (Fig. 2a), while having no effects on resident DC population (Fig. 2a) or TRM population14. Notably, HSV-2 ivag challenge of FTY720-treated immune mice resulted in severe reduction in the levels of IgG2b, IgG2c and IgA secreted into the vaginal lumen (Fig. 2b). FTY720 treatment did not affect serum antiviral Ig levels (Extended Data Fig. 8), as most of the serum Abs are secreted by stationary plasma cells in the bone marrow21. These data indicated that migration of B cells to the FRT is a prerequisite for local secretion of IgG2b, IgG2c, and IgA.
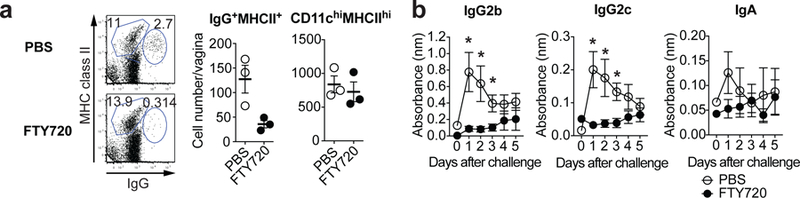
a-b, C57BL/6 mice were infected vaginally with TK− HSV-2 five weeks prior. Mice were given drinking water containing 4 μg/ml of FTY720 for two weeks. a, One day after secondary challenge, the number of IgG+ MHC class II+ cells and CD11chi MHC class IIhi cells were analyzed by flow cytometry (n=3). b, After challenge, HSV-2-specific antibodies in vaginal wash were measured by ELISA (n=5). Sample dilution for ELISA was 1:7. Data are mean ± SEM. Data are representative of two (f) independent experiments, or are pooled from two (g) independent experiments. *, p<0.05 (two-tailed Mann-Whitney U test).
To probe the nature of IgG+ B cells recruited into the FRT following secondary HSV-2 challenge, we collected vagina infiltrating lymphocytes for single cell RNA-sequencing. We also included a control group receiving primary vaginal infection with WT HSV-2 one day earlier. Compared to the primary infected mice, the FRT of previously immunized mice after secondary challenge contained a larger population of CD4 and CD8 T cells with tissue-resident phenotype (Extended Data Fig. 9a), consistent with previous findings14. A focused clustering of CD19 B cells were performed to identify B cell subtypes (Extended Data Fig. 9b). B cells infiltrating the FRT after immunization belonged to a greater diversity of clusters than in the primary infected FRT (seven populations vs. three) (Extended Data Fig. 9c). These cells included cells that resembled follicle associated B cells, plasma cells and memory B cells, with expression of genes such as Sdc1 (Cd138), Ighg2b and Il9r22,23 (Extended Data Fig. 9c and andd).d). Expression of chemokine receptors Cxcr3 and Ccr6 localized to populations that resembled memory, light zone and dark zone B cell populations and showed high expression of activation markers Cd86, Cd80 and Cd69 (Extended Data Fig. 9d). To further elucidate differences between B cells infiltrating the FRT after primary or secondary infection, we focused on FRT associated B cells and identified five distinct populations including one that was unique to immunized mice (cluster 4) (Extended Data Fig. 9e). This population expressed Ighg2b, Cd69, Lax1 (negative regulator of BCR expressed by activated B cells24) and Cxcr3 (Extended Data Fig. 9f). Furthermore, FRT of mice with the secondary challenge lost cluster 3 and instead gained three other populations (clusters 0–2), with enrichment of genes such as Nr4a1 (Nur77; a marker of surface BCR engagement25), Il21r, Cd69, as well as IFNγ-stimulated genes (Extended Data Fig. 9f).
To test whether B cell clones in the FRT after re-challenge are clonally related to the spleen, B cells were sorted from the spleen and clonal assignments were made using Change-O26 (See Methods). We identified a significant difference in the number of clones that spanned both compartments during secondary and primary infection (Extended Data Fig. 9g). One clone, clone IGHV5–17 IGHJ4/45, represented the largest clone present in the spleen (Extended Data Fig. 9h). Moreover, the splenic relative and vaginal descendants had gene expression profiles consistent with plasma cells and IgG production (Extended Data Fig. 9h).
To determine the migration cues responsible for recruiting circulating memory B cells following secondary viral challenge, we examined chemokine receptor expressed on memory B cells in the blood. Five weeks after immunization, a subset of IgG+CD19+IgD− memory B cells in circulation expressed CCR6, CXCR3, and CXCR5 chemokine receptors (Fig. 3a), consistent with our scRNAseq results (Extended Fig. 9d–f), but not others (Extended Data Fig. 10a). In contrast, naïve B cells (IgG−CD19+IgD+) did not express CXCR3 (Fig. 3a). Analysis of vaginal chemokine expression 8 hours following secondary challenge showed that CXCL9, CXCL10 and CXCL11 (CXCR3 ligand) were highly expressed, whereas CCL20 (CCR6 ligand) and CXCL13 (CXCR5 ligand) were not (Fig. 3b). The proportion of CXCR3 expressing memory B cells in circulation in secondary challenged mice was high compared to those in primary infection (Extended Data Fig. 10b). Primary infection also showed delayed expression of CXCR3 ligand, CXCL9, in the vagina compared to secondary challenged immunized mice (Extended Data Fig. 10c), providing possible explanation as to why the primary infection failed to elicit B cell recruitment in the FRT lumen (Fig. 1d). To examine whether CXCR3 expression on B cells are required for FRT recruitment, we generated mixed bone marrow chimeric mice lacking CXCR3 only on B lymphocytes (Extended Data Fig. 10d). We confirmed that CXCR3 was deficient only on B cell but not T cells in CXCR3-B cell knock out chimeric mice (Extended Data Fig. 10e). CXCR3 ablation on B cells did not affect the number of circulating memory B cells and T cells in the blood (Extended Data Fig. 10f). However, CXCR3-B cell KO chimeric mice showed reduction in the number of memory B cells entry into the vagina, while comparable number of naïve B cells were detected in the vagina following secondary ivag HSV-2 challenge (Fig. 3c). Moreover, secretion of virus-specific antibodies IgG2b and IgG2c was also impaired in CXCR3-B cell KO chimeric mice after secondary HSV-2 challenge (Fig. 3d). Taken together, these results indicated that CXCR3 is required for recruitment of circulating memory B cells and their subsequent secretion of antibodies into the vaginal cavity following secondary challenge.
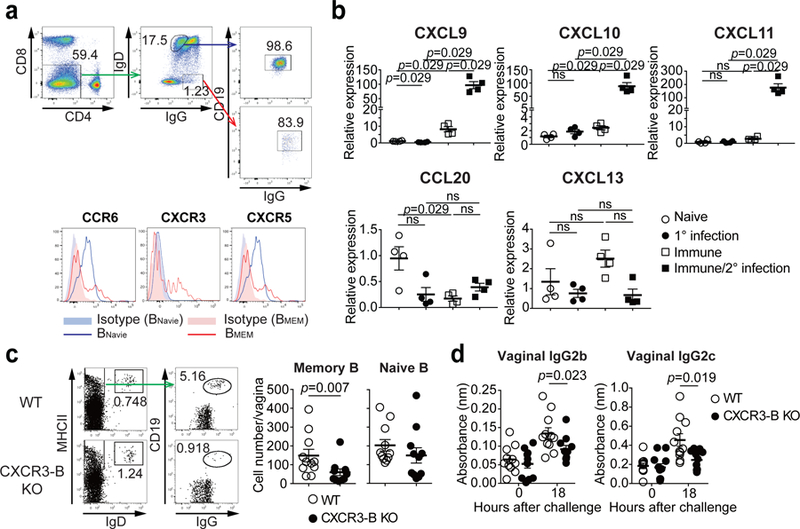
a, C57BL/6 mice were immunized ivag with TK− HSV-2. Five weeks later, expression of various chemokine receptors on circulating IgD−IgG+ memory B cells or IgD+IgG− naive B cells in blood was analyzed by flow cytometry. b, Eight hours after challenge in naïve or TK− HSV-2 immunized mice, mRNA expression of various chemokines (relative to naïve mice) was measured in vaginal tissues by real-time qPCR (n=4 mice). c-d, Mixed BM chimeric mice were immunized intravaginally with TK− HSV-2. Five weeks later, mice were challenged with WT HSV-2. c, Eighteen hours after challenge, the number of IgD+ naive B cells and IgG+ memory B cells in vaginal tissues was analyzed by flow cytometry (n=11). d, Eighteen hours after challenge, HSV-2-specific antibodies in vaginal wash were measured by ELISA (n=11). Sample dilution for ELISA was 1:10. Data are mean ± SEM. Data are representative of four (a) and two (b) independent experiments or are pooled from two independent experiments (c,d). ns, non-significant (two-tailed Mann-Whitney U test).
Our data showed that only immune but not naïve mice are competent for memory B cell recruitment into the FRT (Fig. 1d). Thus, we next focused on the role of MLC and CD4 TRM in recruitment of memory B cells in the vagina. CXCR3 ligands are interferon (IFN) inducible chemokines and CD4 T cells in the vagina are important source of IFN-γ production27 (Fig. 4a). CD4 TRM in the vagina of immunized mice are maintained in MLCs by a local network of macrophages, and depletion of CD11b+ cells in mice previously immunized with TK− HSV-2 ivag results in loss of MLCs14. Depletion of CD11b+ cells resulted in block of memory B cell, but not naïve B cell, recruitment after secondary challenge (Fig. 4b). In addition, CXCL9 expression in the vagina following secondary challenge in immunized mice was defective after depletion of CD11b+ cells (Fig. 4c). Depletion of CD4 T cells in immunized mice showed a reduction of the number of memory B cells, but not naïve B cells, in the vagina after secondary challenge (Fig. 4d), without affecting the number of circulating memory B cells (Fig. 4e). Moreover, the production of CXCR3 ligands in the vagina was impaired in CD4 T cell depleted mice upon secondary challenge (Fig. 4f). In addition, neutralization of IFN-γ in immunized mice impaired the recruitment of B cells into the vagina following secondary HSV-2 challenge (Fig. 4g). These results indicated that CD4 TRM cells maintained by the macrophages in MLCs play a critical role in recruiting circulating memory B cells into the vagina following secondary challenge by secreting IFN-γ which induces CXCL9 and CXCL10.
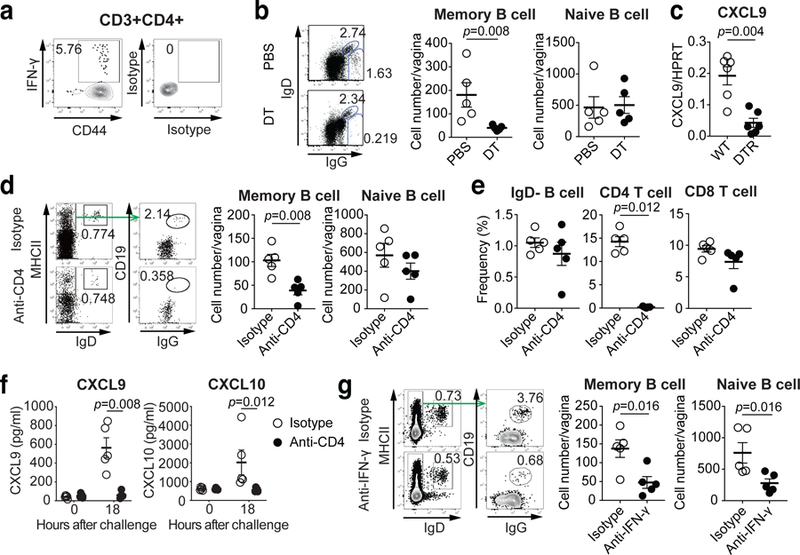
a, C57BL/6 mice were immunized intravaginally with TK− HSV-2. Five weeks later, HSV-2-specific IFN-γ+ CD44+CD4+ T cells in the vagina were analyzed by flow cytometry. b-c, CD11bDTR BM chimeric mice were immunized intravaginally with TK− HSV-2. b, Eight weeks later, these mice were treated with DT to deplete CD11b+ macrophages and challenged with WT HSV-2. One day later, the number of IgD+ naive B cells and IgG+ memory B cells in vaginal tissues was analyzed by flow cytometry (n=5). c, Five weeks after immunization, DT-treated chimeric mice were challenged with WT HSV-2. Eight hours later, mRNA expression of CXCL9 in vaginal tissue were measured by real-time qPCR (n=6). d-f, C57BL/6 mice were immunized intravaginally with TK− HSV-2. Five weeks later, mice were treated with anti-CD4 antibody to deplete CD4 T cells. d, One day after challenge with WT HSV-2, the number of IgD+ naive B cells and IgG+ memory B cells in vaginal tissues was analyzed by flow cytometry (n=5). e, The numbers of circulating memory B cells, CD4 T cells and CD8 T cells in blood after CD4 depletion were analyzed by flow cytometry (n=5). f, Eighteen hours after challenge, the level of CXCL9 and CXCL10 chemokines in vaginal wash was measured by ELISA (n=5). g, C57BL/6 mice immunized intravaginally with TK− HSV-2 were treated with anti-IFN-γ antibody. One day after challenge with WT HSV-2, the number of IgD+ naive B cells and IgG+ memory B cells in vaginal tissues was analyzed by flow cytometry (n=5). Data are mean ± SEM (two-tailed Mann-Whitney U test). Data are representative of two (a–c,g) and three (d–f) independent experiments.
Antibody secretion into the FRT is an inefficient process at the steady state, in part due to the lack of tissue-resident plasma cells. Our results show that local immunization with an attenuated HSV-2 strain induces a microenvironment conducive to rapid and robust recruitment of circulating memory B cells upon secondary challenge, which secrete high levels of virus-specific antibodies into the vaginal lumen.
Antibody-based vaccines against HSV-2 have failed to confer protection in humans28. None has achieved the supraphysiological levels of antibodies needed within the vaginal lumen to protect the host from HSV-2 (Ref.11). Our study showed that high level of anti-viral antibodies in blood does not translate into antibody presence on the vaginal lumen. Thus, despite the presence of neonatal Fc receptor, circulating antibodies fail to enter the vaginal lumen and thus has little protective effect in the vagina. For pathogens including HSV-1 or HSV-2 that do not require barrier compromise for infection, high existing titers of luminal antiviral antibodies is key to conferring protection. Our current results reveal an inducible means to provide robust antibody secretion into the FRT. Such understanding may be leveraged to create robust Ab-mediated vaccines against sexually transmitted infections.
Methods
Mice
Six to eight-week old female C57BL/6 (CD45.2+), congenic C57BL/6 B6.SJL-PtprcaPep3b/BoyJ (B6.Ly5.1) (CD45.1+), B6.129S2-IghtmICgn/J (µMT), B6.FVB-Tg [ITGAM-DTR/EGFP] 34Lan/J (CD11bDTR), B6.Cg-Tg(CAG-DsRed*MST)1Nagy/J (DsRed), and B6.129P2-Cxcr3tm1Dgen/J (CXCR3−/−) mice were purchased from the National Cancer Institute and Jackson Laboratory. All procedures used in this study complied with federal guidelines and institutional policies by the Yale animal care and use committee.
Viruses
HSV-2 strains 186syn− TK− and 186syn+ were generous gifts from Dr. David Knipe (Harvard Medical School, Boston, MA). These viruses were propagated and titrated on Vero cells (ATCC CCL-81) as previously described29. Influenza virus strain A/Puerto Rico/8/34 (H1N1) was a gift from Dr. Peter Staeheli (University Medical Center Freiburg, Germany). Influenza virus strain A/PR/8/34 (H1N1) was propagated as previously described30.
Virus infection
Six- to eight-week-old female mice were injected subcutaneoulsy with Depo Provera (Pharmacia Upjohn) at 2 mg per mouse in 100 μL at 5 – 7 d before infection, swabbed with calcium alginate, and inoculated intravaginally with 105 pfu of HSV-2 (186syn− TK−) or 2500 pfu of WT HSV-2 (186syn+) in 10 μL using a blunt-ended micropipette tip as previously described31. In some experiments, mice were inoculated subcutaneously with 106 pfu of TK− HSV-2 in 100 μL. For secondary challenge, immunized mice were challenged vaginally with 104–105 pfu of WT HSV-2 (186syn+) (100% lethal dose for naïve mice). The severity of disease was scored as; 0, no sign; 1, slight genital erythema and edema; 2, moderate genital inflammation; 3, purulent genital lesions; 4, hind-limb paralysis; 5, pre-moribund. Due to humane concerns, the animals were euthanized prior to reaching moribund state29. For boosting with inactive virus, immunized mice were injected intravaginally with 105 pfu of heat-inactivated WT HSV-2. For influenza A virus infection, mice were anesthetized by intraperitoneal injection of ketamine and xylazine, and inoculated intranasally with 10 – 20 pfu of highly virulent variant of A/PR8/34 (H1N1) in 20 μL PBS as previously described30.
Vaginal viral titers
Vaginal fluids were collected on days 1 – 5 after infection using calcium alginate swabs and PBS. To measure virus titer, vaginal washes were collected in ABC buffer (0.5 mM MgCl26H2O, 0.9 mM CaCl22H2O, 1% glucose, 5% HI FBS and penicillin-streptomycin). Viral titers were obtained by titration of vaginal wash samples on Vero cell monolayer as described previously29.
Antibodies
Anti-CD45.2 (104), anti-CD45.1 (A20), anti-CD45 (30-F11), anti-CD3ε (145–2C11), anti-CD4 (GK1.5 and RM4–5), anti-CD8α (53–6.7), anti-CD19 (6D5 and 1D3), anti-CD45R/B220 (RA3–6B2), anti-CD138 (281–2), anti-CD38 (90), anti-GL7 (GL7), anti-MHC class II (I-A/I-E, M5/114.15.2), anti-CD11c (N418), anti-IgD (11–26c.2a), anti-CD44 (IM7), anti-PD-L2 (TY25), anti-CD80 (16–10A1), anti-CD69 (H1.2F3), anti-CD86 (GL1) were purchased from BD Biosciences, e-Bioscience, or BioLegend. Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) was purchased from Invitrogen (ThermoFischer Scientific). Alexa Fluor 647-conjugated donkey anti-mouse IgG (H+L) Fab fragment was purchased from Jackson ImmunoResearch Laboratories Inc. For chemokine receptor staining, anti-CCR1 (643854), anti-CCR2 (475301), anti-CCR3 (83103), anti-CCR4 (2G12), anti-CCR5 (HM-CCR5), anti-CCR6 (29–2L17), anti-CCR7 (4B12), anti-CCR8 (SA214G2), anti-CCR9 (CW-1.2), anti-CCR10 (248918), anti-CXCR1 (1122A), anti-CXCR2 (TG11), anti-CXCR3 (CXCR3–173), anti-CXCR4 (2B11/CXCR4), anti-CXCR5 (L138D7), anti-CXCR6 (SA051D1) were purchased from BioLegend, BD Biosciences, or R&D systems. For intracellular staining, anti-IFN-γ (XMG1.2) were purchased from BioLegend.
Isolation of leukocytes from vaginal tissues and cervix/uterus
The genital tracts of vaginal tissues and upper reproductive tracts including cervix and uterus treated with Depo-Provera were dissected. Tissues were then incubated with 0.5 mg/mL Dispase II (Roche) for 15 min at 37 °C. Thereafter, Tissues were digested with 1 mg/mL collagenase D (Roche) and 30 μg/mL DNase I (Sigma-Aldrich) at 37 °C for 25 min. The resulting cells were filtered through a 70-μm filter and single cells were isolated as previously described32.
Flow cytometry
Preparation of single cell suspensions from vagina and cervix/uterus were described previously. Blood collected in PBS including 2mM EDTA was treated with ACK lysis buffer three times for single cell preparation. Single cells were pretreated with anti-CD16/32 antibody (2.4G2) to block Fc receptors, and stained with the antibodies. Live/dead cells were stained with LIVE/DEAD™ fixable Aqua dead cell stain kit (Invitrogen). Multiparameter analyses were carried out on the LSR II flow cytometer (Becton Dickinson) and were analyzed using the FlowJo software (Tree Star, Ashland, OR). To stain intracellular IFN-γ, single cell suspensions from vaginal tissues of TK− HSV-2 immunized mice were stimulated in the presence of 5 μg/mL Brefeldin A with naïve splenocytes (CD45.1+) loaded with heat-inactivated HSV-2 antigen (0.5 pfu equivalent/cells) for 10–12 hr14. Cells were surface stained with anti-CD3ε (145–2C11), anti-CD4 (GK1.5 and RM4–5), and anti-CD44 (IM7), and then fixed and permeabilized using a Cytofix/Cytoperm™ Fixation/Permeabilization kit (BD Biosciences) according to the manufacturer’s instructions. These cells were intracellular stained with anti-IFN-γ (XMG1.2).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Vaginal tissues were lysed in RLT lysis buffer and RNA was isolated using RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Contaminating DNA was removed using recombinant DNaseI (Roche), and cDNA was generated with iScript™ cDNA synthesis kit (Bio-Rad), each according to the manufacturer’s instructions. Quantitative PCR was performed using SYBR green-based quantification (Qiagen). Expression of mRNA of various chemokines in vaginal tissues was quantitated by RT-qPCR using the following set of primers. CXCL9 (F, cgagaccatttactgcaacag; R, taattgggccaacagtagcc), CXCL10 (F, gctgcaactgcatccatatc; R, gtggcaatgatctcaacacg), CXCL11 (F, cgggatgaaagccgtcaa; R, tatgaggcgagcttgcttgg), CCL20 (F, atggcctgcggtggcaagcgtctg; R, taggctgaggaggttcacagccct), and CXCL13 (F, ctctccaggccacggtatt; R, taaccatttggcacgaggat). Expression of target genes was normalized against housekeeping genes HPRT.
Bone marrow chimera
BM chimeric mice using C57BL/6, CD11bDTR, µMT, and CXCR3−/− mice were constructed by a standard method29. Briefly, the recipient mice were irradiated with two doses of 475 rad or 600 rad each, 3 h apart. Irradiated recipient mice were reconstituted with 5× 106 – 1 × 107 cells of the appropriate cell suspension by means of i.v. injection. The reconstituted mice were maintained in a clean facility for at least 8 weeks before use.
Adoptive transfer of B cells
B cells were isolated from spleen of DsRed mice immunized intravaginally with 105 pfu of TK− HSV-2 five weeks prior using EasySep™ Mouse B cell Isolation Kit (STEMCELL Technologies) according to the manufacturer’s instructions. At the indicated hours post challenge, 5 × 106 isolated B cells were transferred into C57BL/6 mice immunized intravaginally with 105 pfu of TK− HSV-2 five weeks prior. Animals were sacrificed 2 hours later and vaginal tissue were analyzed for the presence of donor B cells.
In vivo treatment with toxins, antibodies, FTY720, and CpG1826
To deplete CD11b+ cells, CD11bDTR chimeric mice are treated with 10 ng/g body weight intravenously on days −4, −1 and 50 ng diphtheria toxin (Sigma) intravaginally on days −4, −3, −2, −1. To deplete CD4 T cells, mice were treated with 300 µg intravenously on day −4, −1 and 10 µg anti-mouse CD4 Ab (GK1.5, BioXCell) intravaginally for four consecutive days before virus challenge. For neutralization of IFN-γ, mice were treated with 500 µg intravenously on day −4, −1 and 10 µg anti-mouse IFN-γ Ab (R4–6A2, BioXCell) intravaginally for four consecutive days before virus challenge. For fluorescent antibody injection, mice were injected intravenously with 200 µg of FITC-conjugated mouse IgG antibody (Jackson ImmunoResearch). For HSV-2 monoclonal Ab (MAb) treatment, mice were injected intravenously or intravaginally with 1 µg of mouse IgG1 MAb specific for glycoprotein D of HSV (E317, Absolute Antibody). Thirty seconds after ivag injection or 3 hours after i.v. injection of MAb, mice were infected intravaginally with 2500 pfu of WT HSV-2. For FTY720 treatment, immunized mice were given drinking water containing 4 µg/mL of FTY720 (Cayman Chemical) for 2 weeks. For CpG1826 treatment, mice were injected intravaginally with 100 µg of CpG1826 (TriLink BioTechnologies).
ELISA
Vaginal fluids were collected from the indicated days after infection by using calcium alginate swabs and pipetting PBS into and out of the vagina 20 times. BAL were collected by washing the trachea with 1 mL of PBS containing 0.1% BSA. Ninety six-well EIA/RIA plate was coated with 100 µl of heat-inactivated or PFA-inactivated purified HSV-2 (104 to 105 pfu equivalent per 100 µl) or PFA-inactivated A/PR8 virion (105 pfu equivalent per 100 µl) for virus-specific Ig measurement or goat anti-mouse Ig (1:1000; SouthernBiotech, 1010–01) for total Ig measurement in carbonate buffer (pH 9.5) and then incubated overnight at 4°C. On the following day, these plates were washed with PBS-Tween 20 and blocked 2hr with 5% FBS in PBS. Wash and serum samples were then plated in the wells and incubate for at least four hours at ambient temperature. After washing in PBS-Tween 20, HRP-conjugated anti-mouse IgG1, IgG3, IgM, IgA, IgG2a, IgG2b or IgG2c (SouthernBiotech) was added in the wells for 1 h, followed by washing and adding TMB solution (eBioscience). Reactions were stopped with 1N H2SO4 and absorbance was measured at 450 nm. The total Ab titers were defined by using Ig standard (C57BL/6 Mouse Immunoglobulin Panel; SouthernBiotech). The levels of CXCL9 and CXCL10 in vaginal fluids were measured using DuoSet ELISA kit (R&D Systems) according to manufacturer’s instructions.
Immunofluorescence staining
For tissue staining, frozen sections of vagina, spleen and lung collected from various experimental groups were stained with the Ab (anti-CD4 [H129.19]) in a previously described procedure29. Likewise, FITC, CD138, B220, CD31, VCAM-1, IgG2b, IgG2c, total IgG and HSV-2 antigens were detected using Alexa Fluor 647-conjugated anti-FITC Ab, biotin-conjugated anti-mouse CD138 Ab (281–2), PE-conjugated anti-mouse B220 Ab (RA3–6B2), biotin-conjugated anti-mouse CD31 Ab (390), biotin-conjugated anti-VCAM-1 Ab (429/MVCAM.A), HRP-conjugated IgG2b&IgG2c (SouthernBiotech), Alexa Fluor 568-conjugated anti-mouse IgG (H+L) (Invitrogen) and FITC-conjugated pAbs against HSV 1 and 2 (ViroStat). Stained slides were washed and incubated with DAPI and mounted with Fluoromount-G (SouthernBiotech). Images were captured using a 10x or 40x objective lens from fluorescence microscopy (BX51; Olympus).
Induction of breach of the epithelial layer and H&E stain
Vaginal tract of mice immunized subcutaneously with 106 pfu of TK− HSV-2 five weeks prior was brushed with interdental brush. Vaginal tract was washed with PBS after brushing for Ig ELISA and vaginal tissues were embedded in paraffin blocks and sectioned by the Yale Pathology Tissue Services. Hematoxylin and eosin (H&E) staining was performed by the Yale Pathology Tissue Services. H&E images were captured using light microscopy (BX51; Olympus).
Single-cell RNA-seq sample preparation
Naïve mice or mice immunized with 105 pfu of TK− HSV-2 5 weeks prior were infected with 105 pfu of WT HSV-2. One day later, single cells from vaginal tissue and spleen were isolated and surface stained with anti-CD45 (30-F11), anti-CD3ε (145–2C11), anti-CD4 (GK1.5), anti-CD8α (53–6.7), and anti-CD19 (1D3) for vagina and anti-CD45 (30-F11), anti-CD19 (6D5 and 1D3), anti-CD38 (90), anti-GL7 (GL7), anti-MHC class II (I-A/I-E, M5/114.15.2), and anti-IgD (11–26c.2a) for spleen. Stained cells were sorted by FACS Aria (BD Biosciences, Mountain View, CA). CD45+CD3ε+CD4+, CD45+CD3ε+CD8α+, CD45+CD3ε−CD19+ cells (vagina) and CD45+CD19+IgD−IgG+MHCII+GL7−CD38+ cells (spleen) were collected for further analysis. For B cell-sorted spleen samples, cell numbers were counted and 6,000 cells were prepared. Cells from primary and secondary infected FRT samples were prepared at a cell count proportional to the immune cell infiltration observed by flow cytometry. Single cell suspensions were loaded onto the Chromium Controller (10x Genomics) for droplet formation. Single-cell RNA-seq libraries were prepared using the Chromium Single Cell 5’ Reagent Kit (10x Genomics) and Single Cell V(D)J Kit according to manufacturer’s protocol. Samples were sequenced on the HiSeq4000 with 28 bp Read 1, 8 bp i7 index and 98 bp Read 2 for the gene expression library, and on the NovaSeq with 150 paired reads for BCR library.
Single-cell RNA-seq analysis
R v3.4.2 (R Core Team 2013) and python 3.5.4 (2017) was used for all statistical analysis. Sequencing results were demultiplexed into Fastq files using the Cell Ranger (10x Genomics, 2.2.0) mkfastq function. Samples were aligned to mm10–2.2.0 10x genome. The count matrix was generated using the count function with default settings. Matrices were loaded into Seurat v233 for downstream analysis. Cells with less than 500 UMIs, or high mitochondrial content were discarded. For principle component analysis, variable genes were analyzed using the Jackstraw method. An estimate of 13,763 cells were sequenced (2 conditions, 2 organs) with 45,838 mean reads per cell. PC values that were statistically significant were identified and a cutoff point was determined using the inflection point after using the PCElbowPlot function. Clusters were determined using the FindClusters function in Seurat v2 which implements a shared nearest neighbor (SNN) clustering algorithm. For analysis, Wilcoxon rank sum test was used for all differentially expressed genes. After this initial analysis, clusters expressing CD19 or CD3 were separated, and CD19 expressing clusters were re-clustered using the approach described above. Several iterations of the clustering was performed with different resolution and each version’s cluster was validated by gene expression and relative population size compared to FACS data to finally determine the resolution. Cell types were determined using previously published data sets, GSEA B cell subtypes (GSE11386, Broad Institute) and ImmGen (Immunological Genome project, Harvard medical school) as references. An additional clustering was also performed on FRT B cells using the method described above.
B cell repertoire analysis
V(D)J germline segments were assigned with IgBLAST v1.7.034 using the March 30, 2017 version of the IMGT gene database. Following V(D)J annotation, IGH V(D)J sequences were assigned into clonal groups using Change-O v0.4.226. IGH V(D)J sequences were partitioned by common IGHV, IGHJ gene annotations, and nucleotide junction region length. In addition, IGH V(D)J sequences were also grouped based on whether associated cells shared common combinations of IGKV with IGKJ or IGLV with IGLJ gene annotations. Within these larger groups, sequences differing from one another by a length normalized Hamming distance of less than 0.1 were defined as clones by single-linkage clustering. This threshold was identified by the local minima between the two modes of the within-sample bimodal distance-to-nearest histogram. Germline sequences were reconstructed for each clonal cluster (IGHV) with masked D segment and N/P regions (Ns replaced); ambiguous gene assignments within clonal clusters were assigned to majority gene. To calculate confidence intervals for clonal overlap and account for the effect of different sampling depth from each repertoire, clonal overlap of IGH V(D)J sequences was bootstrapped (2,000 replicates) using stratified bootstrapping implemented in the boot v1.3–20 R package. Significance from bootstrap analysis was computed from the empirical cumulative distribution function using a one-tailed test for clonal overlap analysis with a null hypothesis of non-zero overlap. Lineage trees were constructed for clones with greater than one sequence using PHYLIP and Alakazam v0.2.11 in R. All R codes and python codes used in this analysis will be available upon request.
Statistical Analysis
Survival curves were analyzed using the log-rank test. Disease score and virus titer were analyzed using two-way ANOVA. Others were analyzed using two-tailed Mann-Whitney U test. Significance from bootstrap analysis was computed from the empirical cumulative distribution function using a one-tailed test for clonal overlap analysis with a null hypothesis of non-zero overlap, and a two-tailed test for clonal diversity analysis.
Extended Data
Extended Data Fig.1
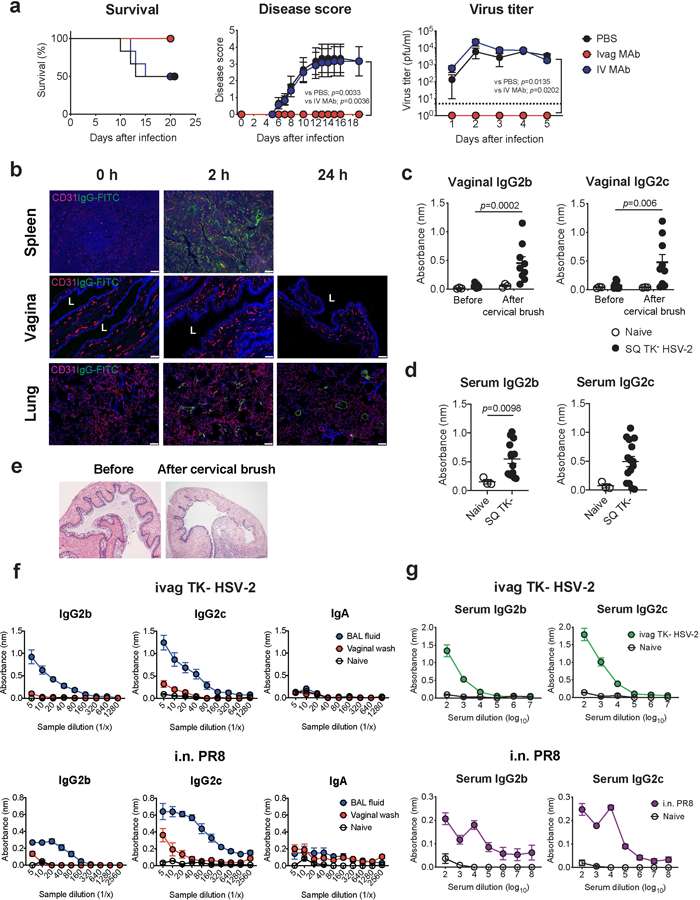
a, C57BL/6 mice treated intravaginally or intravenously with HSV gD-specific MAb were infected with intravaginal WT HSV-2. Survival, disease severity, and virus titer in vaginal wash were analyzed (n=6). Dashed line indicates limit of detection. b, C57BL/6 mice treated with depo-provera were injected intravenously with FITC-conjugated mouse IgG antibody. Frozen sections of vagina, spleen and lung were stained with anti-CD31 (red), anti-FITC (green) antibodies, and DAPI (blue). L indicates vaginal lumen. Scale bars indicate 100 μm. c-e, C57BL/6 mice were immunized subcutaneously with TK− HSV-2. Five weeks later, the vaginal tract of immunized mice with depo treatment was brushed with cervical brush to make a breach or minor abrasions of the epithelial barrier of the vaginal mucosa. HSV-specific antibodies in vaginal wash before and after cervical brush (c, naïve; n=3, SQ TK− HSV-2; n=9) and in serum (d, naïve; n=3, SQ TK− HSV-2; n=15) were measured by ELISA. Sample dilution for ELISA was 1:2 (vaginal wash) or 1:103 (serum). e, Hematoxylin and eosin (H&E) stain of the vagina before and after breach was performed in mice with depo treatment. f-g, C57BL/6 mice were immunized intravaginally with TK− HSV-2 or intranasally with influenza A/PR8 (H1N1) virus. Five weeks later, virus-specific antibodies in bronchoalveolar lavage (BAL) fluid, vaginal wash (f, naïve; n=4, ivag TK− HSV-2; n=6, i.n. PR8; n=4), and blood (g, naïve; n=4, ivag TK− HSV-2; n=13, i.n. PR8; n=8) were measured by ELISA. Data are mean ± SEM. Data are representative of two independent experiments (a,b,e) or are pooled from two independent experiments (c-d,f-g). Statistical significance was analyzed by two-way ANOVA (a; disease score, virus titer), log- rank (Mantel-Cox) test (a; survival) or two-tailed Mann-Whitney U test (c,d).
Extended Data Fig. 2
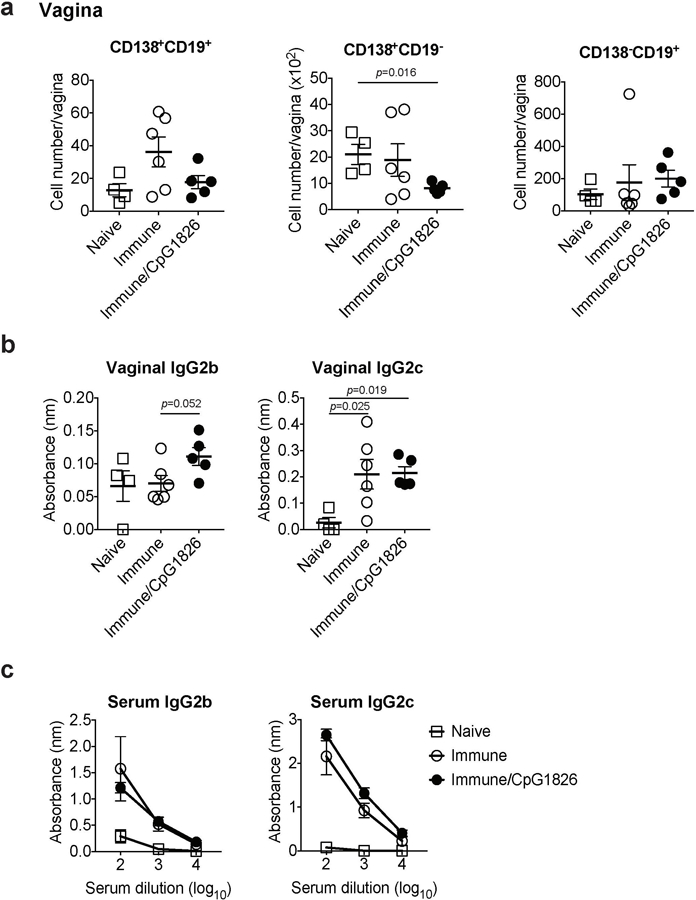
C57BL/6 mice were immunized intravaginally with TK− HSV-2. On day 5 following immunization, CpG1826 was injected into the vagina to boost local inflammation of the vaginal mucosa. a, Five weeks later, the number of CD138+CD19+, CD138+CD19−, and CD138−CD19+ cells in vaginal tissues was analyzed by flow cytometry (naïve; n=4, immune; n=6, immune/CpG1826; n=5). b-c, HSV-2-specific antibodies in vaginal wash (b, naïve; n=4, immune; n=6, immune/CpG1826; n=5) and in serum (c, naïve; n=3, immune; n=3, immune/CpG1826; n=5) were measured by ELISA. Sample dilution for vaginal wash ELISA was 1:5. Data are mean ± SEM. Data are representative of two independent experiments. Statistical significance was analyzed by two-tailed Mann-Whitney U test.
Extended Data Fig. 3
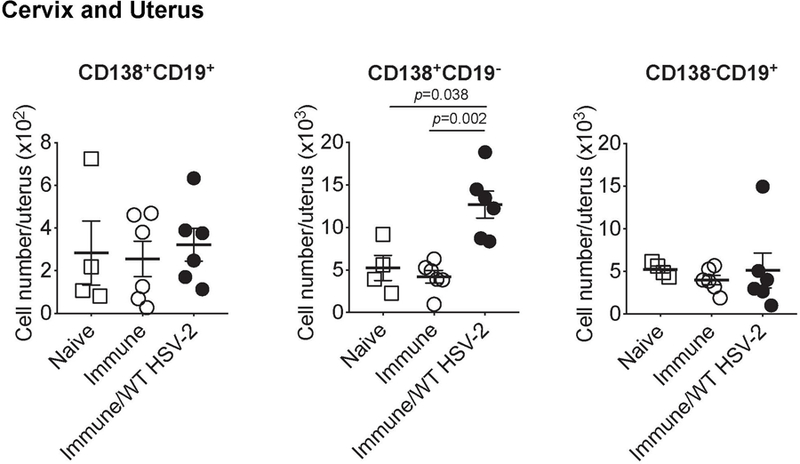
C57BL/6 mice were immunized intravaginally with TK− HSV-2. Five weeks later, mice were challenged with ivag WT HSV-2. One day after, the number of CD138+CD19+, CD138+CD19−, and CD138−CD19+ cells in cervix and uterus was analyzed by flow cytometry (naïve; n=4, immune; n=6, immune/WT HSV-2; n=6). Data are mean ± SEM. Data are pooled from two independent experiments. Statistical significance was analyzed by two-tailed Mann-Whitney U test.
Extended Data Fig. 4
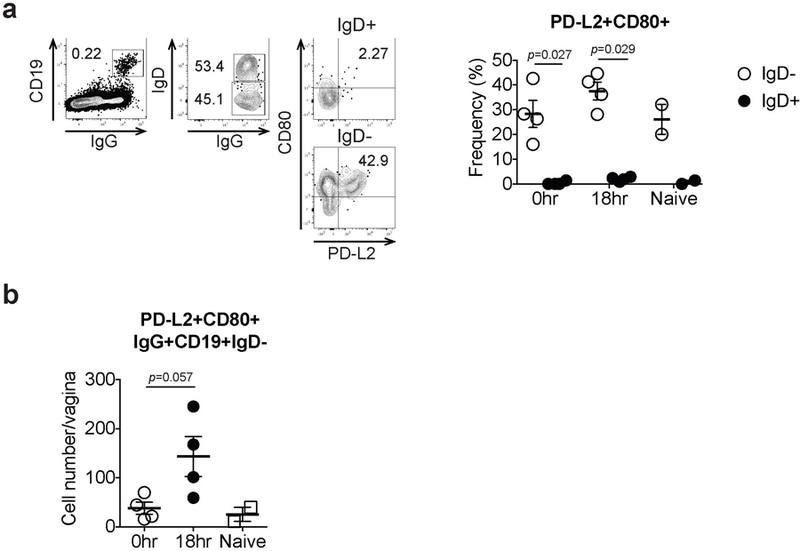
C57BL/6 mice immunized with TK− HSV-2 5 wk prior were challenged with WT HSV-2. a, Eighteen hours later, CD80 and PD-L2 expression on both IgD+ and IgD− B cells in vaginal tissues was analyzed by flow cytometry. b, The number of CD80+PD-L2+ memory B cells in the vagina on 18 hours after secondary challenge was analyzed by flow cytometry (naïve; n=2, immune; n=4). Data are mean± SEM. Data are representative of two independent experiments. Statistical significance was analyzed by two-tailed Mann-Whitney U test.
Extended Data Fig. 5
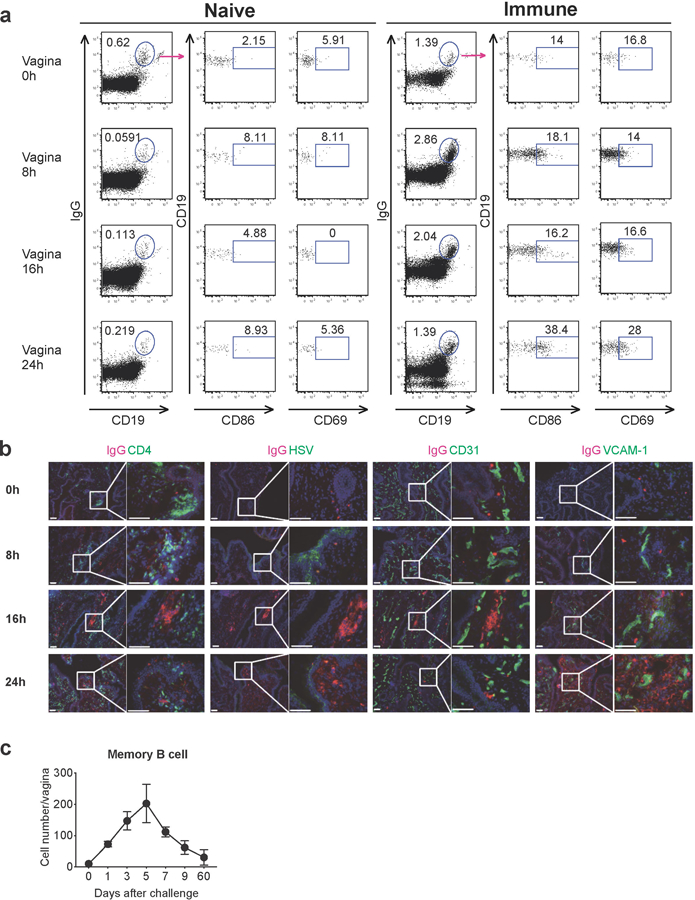
a, Naive C57BL/6 mice or C57BL/6 mice immunized with TK− HSV-2 5 wk prior were challenged with WT HSV-2. Eight, sixteen and twenty-four hours after challenge, CD86 and CD69 expression on IgD−IgG+ memory B cells in vaginal tissues was analyzed by flow cytometry. b, C57BL/6 mice were immunized vaginally with TK− HSV-2 five weeks prior. After challenge, frozen sections of vagina were stained with antibodies against IgG (red), CD4, HSV, CD31 and VCAM-1 (green), and DAPI (blue). Scale bars indicate 100 μm. c, C57BL/6 mice immunized with TK− HSV-2 5 wk prior were challenged with WT HSV-2. At the indicated days after challenge, the number of memory B cells in vaginal tissues was analyzed by flow cytometry (n=2–4 mice in each time point). Data are mean ± SEM. Data are representative of two independent experiments.
Extended Data Fig. 6
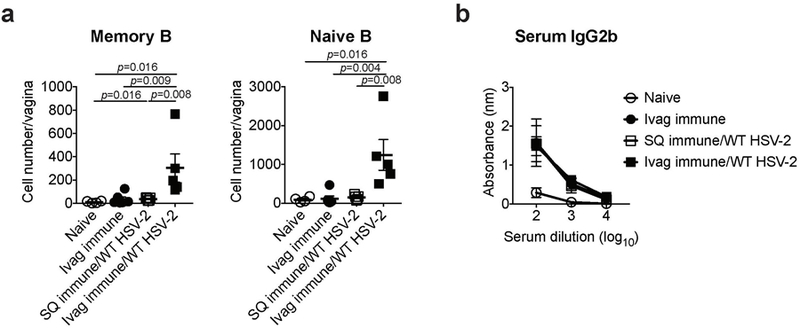
C57BL/6 mice were immunized intravaginally or subcutaneously with TK− HSV-2. Five weeks later, mice were challenged with WT HSV-2. a, One day after challenge, the number of memory and naïve B cells in vaginal tissues was analyzed by flow cytometry (naïve; n=4, ivag immune; n=6, SQ immune/WT HSV-2; n=5, ivag immune/WT HSV-2; n=5). b, HSV-2-specific IgG2b antibody in serum was measured by ELISA (naïve; n=3, ivag immune; n=3, SQ immune/WT HSV-2; n=5, ivag immune/WT HSV-2; n=3). Data are mean ± SEM. Data are representative of two independent experiments. Statistical significance was analyzed by two-tailed Mann-Whitney U test.
Extended Data Fig. 7
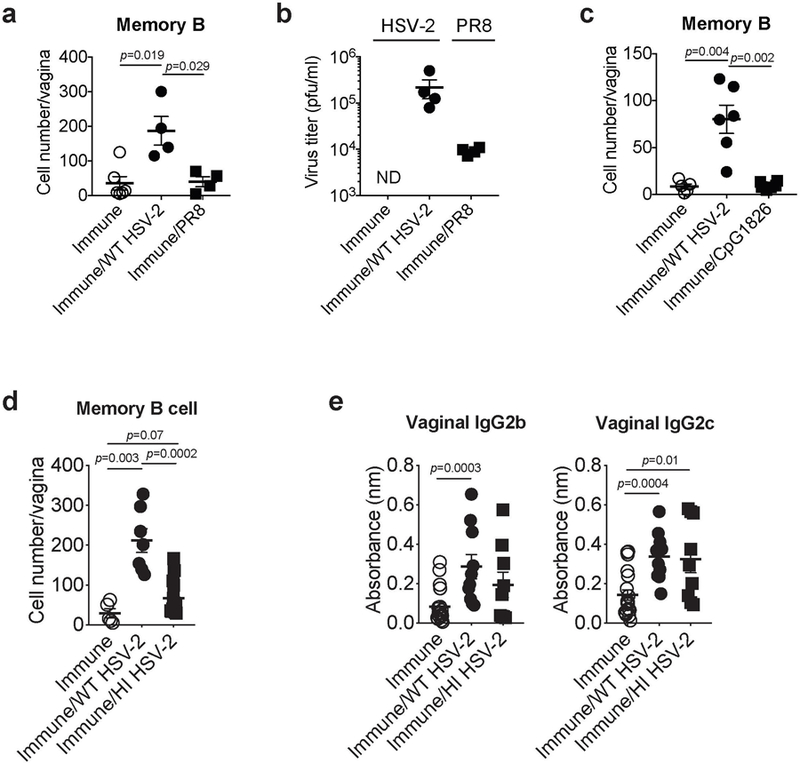
a-b, C57BL/6 mice immunized ivag with TK− HSV-2 five weeks prior were challenged with WT HSV-2 or influenza A/PR8 virus intravaginally (immune; n=6, immune/WT HSV-2; n=4, immune/PR8; n=4). a, One day after secondary challenge, the number of memory B cells in vaginal tissues was analyzed by flow cytometry. b, Indicated virus titer in vaginal wash was measured one day after secondary challenge. c, C57BL/6 mice immunized ivag with TK− HSV-2 five weeks prior were challenged with WT HSV-2 or CpG1826 intravaginally. One day after secondary challenge, the number of memory B cells in vaginal tissues was analyzed by flow cytometry (immune; n=5, immune/WT HSV-2; n=6, immune/CpG1826; n=6). d-e, C57BL/6 mice immunized ivag with TK− HSV-2 five weeks prior were challenged with active or heat-inactivated (HI) WT HSV-2 intravaginally. d, One day after secondary challenge, the number of memory B cells in vaginal tissues was analyzed by flow cytometry (immune; n=5, immune/WT HSV-2; n=7, immune/HI HSV-2; n=14). e, One day after challenge, HSV-2-specific antibodies in vaginal wash were measured by ELISA (immune; n=19, immune/WT HSV-2; n=10, immune/HI HSV-2; n=9). Sample dilution for ELISA was 1:5. Data are mean ± SEM. Data are representative of two independent experiments (a-c) or are pooled from three independent experiments (d,e). Statistical significance was analyzed by two-tailed Mann-Whitney U test.
Extended Data Fig. 8
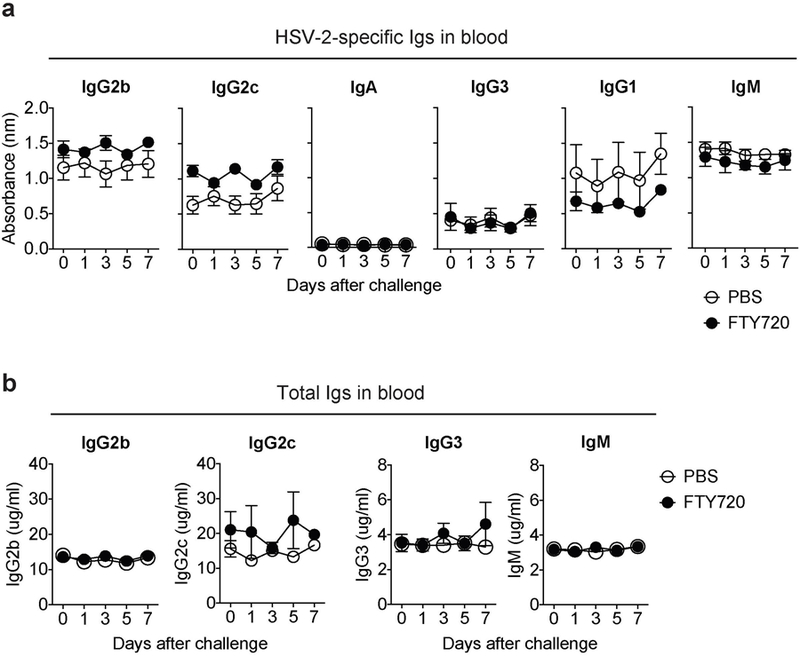
C57BL6 mice were infected vaginally with TK− HSV-2. Six weeks later, immunized mice were given drinking water containing 4 μg/ml of FTY720. Two weeks later, FTY720-treated mice were challenged with WT HSV-2 (104 pfu) intravaginally. At the indicated days after challenge, HSV-2-specific antibodies (a) and total antibodies (b) in the blood were measured by ELISA. Sample dilution for virus-specific ELISA was 1:140. Data are mean ± SEM. Data are representative of two independent experiments with 3 mice per group.
Extended Data Fig. 9
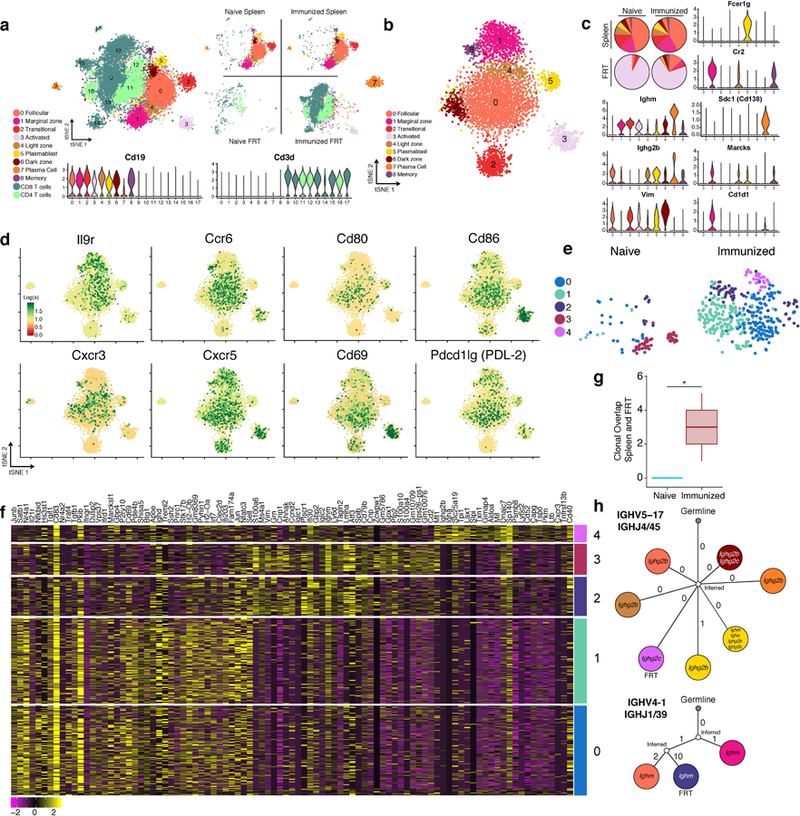
Spleens and FRT of C57BL/6 mice were collected for 10x single cell RNA-Seq one day after (re-)challenge with HSV-2; spleens were sorted for B cells and FRT were sorted for B and T cells (n= 8 pooled mice). a, tSNE plot associated with 5’ gene expression profiling of 13,763 cells captured on the 10x Chromium platform. B cell identities were determined by re-clustering CD19+ populations after initial clustering and reassigning them according to analysis in b, refer to methods for more detail. Violin plots depict kernel density estimates to show the distribution of expression values and was generated using the default VlnPlot function in Seurat (n = 13,763 cells). b, tSNE plot associated with CD19+ clusters identified from a after re-clustering (n = 5,390 cells). c, Relative cell population frequency in the different compartments and conditions are shown in pie charts and gene expression of each cluster is shown in violin plots. Violin plots depict kernel density estimates to show the distribution of expression values and was generated using the default VlnPlot function in Seurat (n = 5,390 cells). d, Heatmaps of genes in B cells associated with activation and chemokine/cytokine receptors (n = 5,390 cells). e, tSNE plots of B cells found in the FRT of naïve infected and immunized re-challenged mice after re-clustering only FRT B cells (n = 517 cells). f, Heatmap of top genes differentially expressed in each cluster in e. g. Clonal overlap between spleen and FRT of mice after primary or secondary challenge was measured using single cell BCR sequencing data, confidence intervals for clonal overlap were computed by bootstrap analysis and significance was assessed for non-zero overlap (p=0.003 by clone, p=0.0005 by sequence). Clonal overlap was computed from all productive BCR sequences from the 5,390 cells. Box plots depict number of clonal overlap calculated from bootstrap analysis with whiskers depicting upper percentile of 95th and lower percentile of 5th and box depicting 80th and 20th percentile. h. Lineage tree of clones that were shared in the spleen and FRT compartment, colors of cell types for splenic B cells and FRT B cells correspond to legends in c and e respectively.
Extended Data Fig. 10
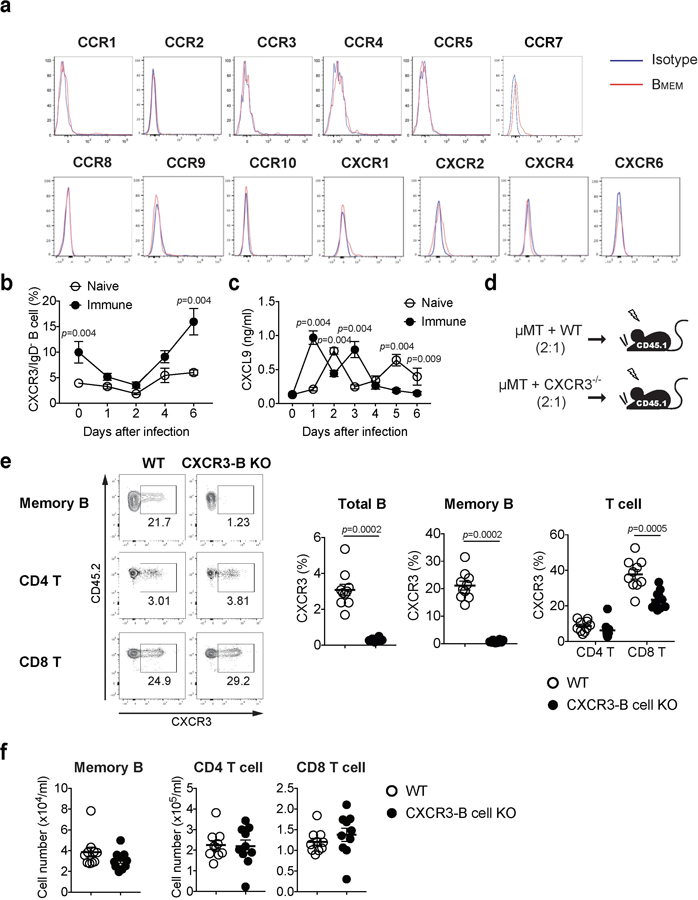
a, C57BL/6 mice were immunized ivag with TK− HSV-2. Five weeks later, expression of various chemokine receptors on circulating IgD−IgG+ memory B cells in blood was analyzed by flow cytometry. b-c, C57BL/6 mice with (n=6) or without (n=5) immunization with TK− HSV-2 five weeks prior were challenged with WT HSV-2 intravaginally. At the indicated days following challenge, CXCR3 expression on IgD−IgG+ memory B cells was analyzed by flow cytometry (b) and CXCL9 secretion in vaginal wash was measured by ELISA (c). d, Strategy for generating mixed BM chimeric mice. e-f, WT and CXCR3-B KO chimeric mice were immunized ivag with TK− HSV-2 (n=10). e, Five weeks later, CXCR3 expression on IgD−IgG+ memory B cells, CD4 T cells, and CD8 T cells were analyzed by flow cytometry. f, Five weeks later, the number of circulating memory B cells, CD4 T cells and CD8 T cells was analyzed by flow cytometry. Data are mean ± SEM. Data are representative of four (a) and two (b,c) independent experiments or are pooled from two independent experiments (e,f). Statistical significance was analyzed by two-tailed Mann-Whitney U test.
Acknowledgements
We thank Melissa Linehan and Huiping Dong for technical assistance, Yale Flow Cytometry Facility, especially Geoffrey Lyon, for cell sorting, Yale Center for Genome Analysis (YCGA), especially Guilin Wang and Christopher Castaldi, for 10x Chromium library preparations and sequencing help, Patrick Wong and Tianyang Mao for helpful discussions regarding the single cell RNA-seq data, Yong Kong for helpful advice in statistical analysis, and Kenneth B Hoehn for advice on BCR lineage tree drawing. This study was supported by awards from NIH AI054359, AI062428, AI064705, AI102625 (to A.I.), Basic Science Research Program through the National Research Foundation of Korea (NRF-2016R1A6A3A03010349) funded by the Ministry of Education (to J.E.O.), the Japan Agency for Medical Research and Development (AMED) JP18fm0208011h0002, the Japan Society for the Promotion of Science (JSPS) JP18H02857, Takeda Science Foundation (to N.I.), and NIH T32GM007205 (MSTP training Grant) (to E.S. and R.J.). A.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interests.
Data Availability
RNA sequencing data are available at BioProject under accession number PRJNA524497. All datasets generated and/or analyzed during the current study are presented in this published article, the accompanying Source Data or Supplementary Information, or are available from the corresponding author upon reasonable request.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/s41586-019-1285-1
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6609483
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/s41586-019-1285-1
Article citations
The emerging role of effector functions exerted by tissue-resident memory T cells.
Oxf Open Immunol, 5(1):iqae006, 14 Jun 2024
Cited by: 0 articles | PMID: 39193473 | PMCID: PMC11213632
Review Free full text in Europe PMC
The vaginal immunoproteome for the prediction of spontaneous preterm birth: A retrospective longitudinal study.
Elife, 13:e90943, 24 Jun 2024
Cited by: 1 article | PMID: 38913421 | PMCID: PMC11196114
Regulation of pulmonary plasma cell responses during secondary infection with influenza virus.
J Exp Med, 221(7):e20232014, 25 Apr 2024
Cited by: 1 article | PMID: 38661717 | PMCID: PMC11044945
HPV-specific antibodies in female genital tract secretions captured via first-void urine retain their neutralizing capacity.
Hum Vaccin Immunother, 20(1):2330168, 03 Apr 2024
Cited by: 2 articles | PMID: 38567541 | PMCID: PMC10993920
From neglect to spotlight: the underappreciated role of B cells in cutaneous inflammatory diseases.
Front Immunol, 15:1328785, 15 Feb 2024
Cited by: 1 article | PMID: 38426103 | PMCID: PMC10902158
Review Free full text in Europe PMC
Go to all (60) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (1 citation) BioProject - PRJNA524497
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Dendritic cells and B cells maximize mucosal Th1 memory response to herpes simplex virus.
J Exp Med, 205(13):3041-3052, 01 Dec 2008
Cited by: 110 articles | PMID: 19047439 | PMCID: PMC2605233
Novel Role for Interleukin-17 in Enhancing Type 1 Helper T Cell Immunity in the Female Genital Tract following Mucosal Herpes Simplex Virus 2 Vaccination.
J Virol, 91(23):e01234-17, 14 Nov 2017
Cited by: 30 articles | PMID: 28956763 | PMCID: PMC5686749
Estradiol Enhances Antiviral CD4+ Tissue-Resident Memory T Cell Responses following Mucosal Herpes Simplex Virus 2 Vaccination through an IL-17-Mediated Pathway.
J Virol, 95(1):e01206-20, 09 Dec 2020
Cited by: 10 articles | PMID: 33028712 | PMCID: PMC7737739
Vaginal immunity in the HSV-2 mouse model.
Int Rev Immunol, 22(1):43-63, 01 Jan 2003
Cited by: 37 articles | PMID: 12710503
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: F30 CA239444
NIAID NIH HHS (7)
Grant ID: R01 AI104739
Grant ID: R56 AI062428
Grant ID: R01 AI054359
Grant ID: R01 AI064705
Grant ID: R01 AI062428
Grant ID: R01 AI102625
Grant ID: R01 AI127429
NIBIB NIH HHS (1)
Grant ID: R01 EB000487
NIGMS NIH HHS (1)
Grant ID: T32 GM007205
NIH HHS (1)
Grant ID: S10 OD018521





