Abstract
Free full text

Single-cell transcriptome analysis reveals gene signatures associated with T-cell persistence following adoptive cell therapy
Abstract
Adoptive cell therapy (ACT) using tumor-infiltrating lymphocytes (TILs) can mediate responses in some patients with metastatic epithelial cancer. Identifying gene signatures associated with successful ACT might enable the development of improved therapeutic approaches. The persistence of transferred T cells in the peripheral blood is one indication of clinical effectiveness, but many T-cell and host factors may influence T-cell persistence. To limit these variables, we previously studied a patient with metastatic colorectal cancer treated with polyclonal TILs targeting the KRAS(G12D) hotspot mutation, who experienced a partial response for 9 months. Three dominant clonotypes specifically recognizing KRAS(G12D) epitopes were identified, but we found that only two clonotypes persisted 40 days after ACT. Because of these findings, in this study, we performed the single-cell transcriptome analysis of the infused TILs. The analysis revealed a total of 472 genes that were differentially expressed between clonotypes 9.1-NP and 9.2-P single cells, and 528 genes between 9.1-NP and 10-P. Following these clonotypes in the peripheral blood after ACT, the gene expression patterns changed, but IL7R, ITGB1, KLF2 and ZNF683 remained expressed in the persistent 9.2-P and 10-P cells, compared to the non-persistent 9.1-NP cells. Additionally, four autologous TILs, which were used for treatment but persisted poorly one month after ACT, did not express the gene profiles associated with persistence. These results suggest that certain TIL populations possess a unique gene expression profile that can lead to the persistence of T cells. Thus, this single-patient study provides an insight on how to improve ACT for solid cancer.
Introduction
Adoptive cell therapy (ACT) using tumor-infiltrating lymphocytes (TILs) has shown significant efficacy in patients with metastatic melanoma (1). Striking responses have been observed in individual patients with metastatic cholangiocarcinoma, cervical cancer, colorectal cancer, or breast cancer after ACT (2–5). However, clinical responses to ACT have been limited in the majority of patients with solid epithelial cancers (3,4,6). To increase the efficacy of the current treatments, multiple aspects of ACT are being explored, including the selection of effective targets and the avidity of antigen-reactive T cells (7,8). Our previous studies have shown that neoantigen recognition by T cells is a major factor associated with effective TIL-ACT (2,9–11). Other studies have also demonstrated that effective immune checkpoint blockade immunotherapies correlate with the abundance of neoantigens expressed by tumors (12–15). Despite efforts to expand and transfer selective TIL populations recognizing neoantigen targets, many of the TIL treatments have been ineffective (6,16).
The persistence of specific T-cell clonotypes in the peripheral blood after T-cell infusion has become an important parameter to monitor during the course of ACT (17). In addition to the antigen avidity of T cells, several factors can also influence T-cell persistence in the peripheral blood. Examples include the levels of homeostatic cytokines, endogenous T-cell reconstitution, tumor antigen expression, and the tumor microenvironment (17–20). Therefore, it is difficult to study persistence data because of many potential variables, unless the analysis is from a large number of patients treated with nearly the same cell product (17).
In a previous study, we observed that patient-4095 with metastatic colorectal cancer, who was treated with mutated KRAS(G12D)-reactive TILs, experienced a partial response for 9 months after ACT (4). A single non-responding pulmonary lesion was resected, and the patient has remained disease-free for three years after treatment. We found that the infused TILs comprised three dominant clonotypes (49.5%, 19.1%, 6.9%, respectively). The first clonotype, 9.1-NP, recognized a KRAS(G12D) 9-mer epitope (GADGVGKSA); the second clonotype, 10-P, recognized a KRAS(G12D) 10-mer epitope (GADGVGKSAL); and the third clonotype, 9.2-P, recognized the same 9-mer epitope as 9.1-NP. The most dominant clonotype, 9.1-NP, did not persist (NP) at all on day 40, but the 10-P and 9.2-P clonotypes continued to persist (P) up to day 266 after ACT. The 9.1-NP and 9.2-P clonotypes shared the same specificity and similar TCRα/β sequences, except one amino acid at the CDR3α region and six amino acids at the CDR3β region, and no significant difference in TCR avidity was observed (4).
Because these three clonotypes shared the same in vitro culture environment and in vivo host environment, we sought to investigate whether different gene expression profiles were associated with different durations of persistence after ACT. Because the majority of biological assays cannot distinguish the difference between 9.1-NP and 9.2-P T cells, we performed single-cell TCR and transcriptome analysis for this single-patient study. The results suggested that TILs that could persist in patient-4095 had distinguishable gene expression profiles, including key genes encoding surface markers and transcription factors.
Materials and Methods
Patients
Patients were enrolled in a clinical trial of TIL therapy (ClinicalTrial.gov ID: ). This trial was approved by the institutional-review board (IRB) of the National Cancer Institute (NCI), and the written informed consent was obtained from the patients, following NIH guidelines and Declaration of Helsinki.
The characteristics, treatment, and clinical response for patient-4095 with metastatic colorectal cancer have been published previously (4). We have also reported the summary of characteristics for patient-4007, −4071, and −4081 with metastatic colorectal cancer and patient-4069 with pancreatic cancer (6,16). Briefly, TILs were generated from the metastatic tumors of patients. TIL cultures were selected based on the reactivities against tumor-specific mutations, and selected TIL cultures were expanded for treatment. The patients were treated with a lymphodepleting chemotherapy regimen, a single infusion of TILs, followed by several doses of IL2. The peripheral blood lymphocyte (PBL) samples were obtained from the patients every 2–3 days during hospitalization and during follow-up visits. PBL samples were cryopreserved and stored in a liquid nitrogen container before use. The percentage of individual T-cell clonotypes in samples was obtained by an Immunoseq Assay service, provided by Adaptive Biotechnologies (Seattle, WA).
Generation of TILs
TILs used for this study were generated by methods described previously (21). Briefly, metastatic tumors were resected from patients, and tumor fragments were excised and cultured in RPMI medium supplemented with 10% in-house human AB serum, 2 mM L-glutamine, 25 mM HEPES, gentamicin (10 μg/mL), and IL2 (6000 IU/mL; Clinigen, Yardley, PA). TIL cultures were grown for 2–4 weeks and then screened for recognition of tumor-specific mutations (22). The screening results for patient-4095, −4007, −4071, −4081, and −4069 have been published (4,6,16). The mutation-reactive TIL cultures were selected and expanded using a rapid expansion protocol (REP) to large numbers for patient infusion (23). The REP culture contained 5×106 TILs and 5×108 irradiated PBMC feeder cells in RPMI/AIM V medium (50%/50% mixed), supplemented with 7.5% in-house human AB serum, 2 mM L-glutamine, IL2 (3000 IU/mL), and OKT3 antibody (30 ng/mL; Miltenyi Biotec, Bergisch Gladbach, Germany) in a G-Rex100 flask (Wilson Wolf, Saint Paul, MN). A small portion of TILs were cryopreserved and stored in a liquid nitrogen container for the experiments shown in this report.
Single-cell TCR/transcriptome sequencing
The samples of infused TILs were thawed and recovered overnight in RPMI/AIM V medium (50%/50%) supplemented with 5% human AB serum (Valley Biomedical, Winchester, VA). TILs were resuspended in PBS and then subjected to a 10X Chromium instrument (10X Genomics, Pleasanton, CA) for the single-cell analysis, as described below. 1X107 PBLs from patient-4095 were stained with KRAS-9mer tetramer or KRAS-10mer tetramer (NIH tetramer core facility, Atlanta, GA), together with CD8 antibody (clone RPA-T8, BD Biosciences, San Jose, CA) for 40 minutes. Stained cells were washed twice with PBS containing 5% fetal bovine serum (SAFC, St. Louis, MO). After washes, tetramer-positive cells were sorted by BD FACSAria II and subjected to a 10X Chromium instrument for the single-cell analysis.
We used the standard protocol and reagent kit for single-cell V(D)J analysis, provided by 10X Genomics. Up to 8 samples/reactions in 8 channels can be processed simultaneously by a 10X Chromium instrument. Briefly, 10,000 cells per reaction/channel were loaded, with the targeted cell recovery of 6,000 cells. For TIL4095, a total of 4 channels were loaded to obtain the desired number of cells. For patient-4095’s PBLs on day 12 after ACT, KRAS-9mer tetramer+ cells were loaded on 3 channels and KRAS-10mer tetramer+ cells were loaded on 1 channel. For patient-4095’s PBLs on day 40 after ACT, KRAS-9mer tetramer+ cells and KRAS-10mer tetramer+ cells were loaded on 2 channels each. Single cells were captured and lysed, and mRNA was reverse transcribed to cDNA using the provided reagents (10X Genomics). Single-cell cDNA samples were barcoded and amplified by 12 PCR cycles. The amplified cDNA samples were divided into two fractions for used in two analyses. The first fraction was target-enriched for TCRs (10X Genomics proprietary), and barcoded samples were pooled and sequenced by an Illumina NextSeq 550 sequencer (High Output v2 kit, Read1: 150 b.p., Read2: 150 b.p.). The second fraction was processed for 5’ gene expression libraries by following the manufacturer’s instruction (10X Genomics). Each channel was processed and sequenced separately by an Illumina NextSeq 550 sequencer (High Output v2 kit, Read1: 26 b.p., Read2: 98 b.p.).
Single-cell samples prepared by Fluidigm C1 systems
TIL4095 (TILs from patient-4095) single cells were processed by two identical Fluidigm C1 systems (Fluidigm, South San Francisco, CA), following the protocol published previously (24). Briefly, the infused TIL sample from patient-4095 was thawed and recovered overnight in RPMI/AIM V medium (50%/50%) supplemented with 5% human AB serum. TILs were stained with KRAS-9mer tetramer or KRAS-10mer tetramer, together with CD8 antibody for 40 minutes. After washes, tetramer-positive cells were sorted by BD FACSAria II and subjected to Fluidigm C1 systems for the single-cell analysis. 12,000 KRAS-9mer tetramer+ cells or KRAS-10mer tetramer+ cells were loaded separately on an IFC plate (Fluidigm). Because of the low-abundance of 9.2-P clonotype, KRAS-9mer tetramer+ cells were loaded on 6 additional IFC plates, and 9.2-P single cells were selected based on the CDR3 sequence information. Single-cell samples were processed by the Fludigm C1 system and barcoded by Nextera XT DNA Library Preparation Kit (Illumina). Single-cell samples were first sequenced by an Illumina MiSeq sequencer (High Output v3 kit, Read1: 250 b.p., Read2: 250 b.p.) to identify CDR3 sequences. Single-cell samples with correct CDR3 sequences (9.1-NP, 9.2-P or 10-P) were sequenced again by an Illumina HiSeq2500 sequencer (Read1: 100 b.p., Read2: 100 b.p.), in order to obtain sufficient sequence information for data analysis.
Single-cell bioinformatics analysis and statistical analysis
Single-cell sequencing data were first processed by Cell Ranger pipelines (v2.1.1)(10X Genomics), in order to obtain T-cell clonotypes and gene expression profiles associated with individual single-cell barcodes. T-cell clonotypes were defined based on TCR Vβ CDR3 nucleotide sequences. Single cells with two different TCR Vβ CDR3 nucleotide sequences were likely doublets and were excluded. Low-quality single cells, with less than 200 detectable genes, were also excluded. Genes with less than 10 read counts, as well as mitochondrial genes, were removed from the subsequent analysis. Non-coding RNA genes were not investigated in this study.
To construct the gene expression matrix for the datasets, we merged all uniquely aligned reads with the same unique molecular identifiers (UMI), cell barcode, and gene annotation in the HDF5 format. The raw single-cell data were then normalized with convolution methods (25). The normalized data were decomposed using randomized principal component analysis (26). The principal components were used to generate t-SNE projections, which were implemented with R package Seurat v.2.3 (https://github.com/satijalab/seurat). The differential expression comparisons were generated with edgeR package with selected genes (false discovery rate, FDR<0.05).
Gene Set Enrichment Analysis
The Persistence_Up and Persistence_Down gene sets were constructed based on the shared, differentially expressed genes (FC > ±1.5, FDR < 0.05), obtained from the comparisons of 9.1-NP vs 9.2-P and 9.1-NP vs 10-P (Supplementary Table S1). Gene Set Enrichment Analysis (GSEA (27)) software/scripts and instruction are publicly available from Broad Institute (http://software.broadinstitute.org/gsea/index.jsp). We made necessary in-house modifications for the scripts, in order to execute the GSEA scripts on the NIH Biowulf server ((http://hpc.nih.gov).
Single-cell bioinformatics analysis for single-cell data obtained by Fluidigm C1 systems
Single-cell whole-transcriptome sequences obtained by the Fluidigm C1 were first analyzed by an in-house bioinformatics pipeline to obtain TCR-CDR3 nucleotide sequences (24). The sequencing data of single cells with correct CDR3 were further analyzed by a bioinformatics pipeline, including the alignment using STAR (v2.4.2) and raw counts by subread (v1.4.6). Good-quality single cells were defined as cells in which more than 1,500 genes were expressed and detected. Individual genes expressed in more than two cells were selected for the subsequent analysis. The gene expression matrix was generated, and the p-values were calculated by the Wilcoxon signed-rank test.
Data availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (28) and are accessible through GEO Series accession number GSE136394 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136394).
Statistical analysis
Statistics for single-cell data and GSEA are described in the above sections.
Results
To study the gene expression profiles of persistent and non-persistent clonotypes, the infused TILs from patient 4095 were subjected to single-cell TCR/transcriptome sample preparation by a 10X Genomics single-cell instrument, followed by next-generation sequencing. Each single cell contained a unique barcode, which could be used to link the gene expression profiles to one of the three dominant clonotypes (9.1-NP, 9.2-P or 10-P), based on their unique TCR-CDR3β sequences. After filtering out single cells with low-quality data, we obtained 9707 single cells from the 9.1-NP clonotype, 1737 single cells from the 9.2-P clonotype and 2771 single cells from the 10-P clonotype for the downstream bioinformatics analysis (Fig. 1A).
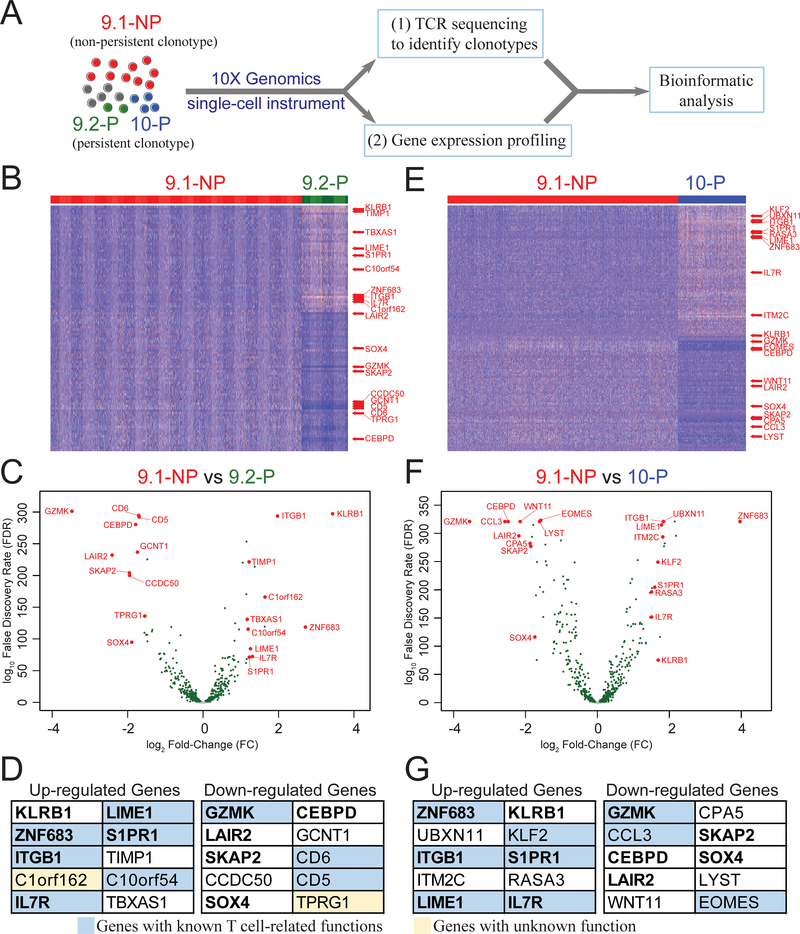
A, TIL4095, contained three dominant clonotypes 9.1-NP, 9.2-P and 10-P (4). TIL4095 T cells were subjected to a single-cell instrument (10X Genomics) to perform (i) single-cell TCR sequencing and (ii) single-cell gene expression profiling. Each single cell contained a unique barcode, which could be used to link the gene expression profiles to one of the three dominant clonotypes (9.1-NP, 9.2-P or 10-P), based on their unique TCR-CDR3β sequences. This set of information was used in the subsequent analysis. Next, single cells from 9.1-NP non-persistent clonotype were compared with single cells from 9.2-P persistent clonotype to generate (B) a heatmap and (C) a volcano plot. (D) Top 10 upregulated genes and top 10 downregulated genes based on the fold changes (FC) are listed. Genes with known T cell-related functions are shown in blue boxes. Genes with unknown function are shown in yellow boxes. Single cells from 9.1-NP non-persistent clonotype were then compared with single cells from 10-P persistent clonotype to generate (E) a heatmap and (F) a volcano plot. (G) Top 10 upregulated genes and top 10 downregulated genes based on the FC are listed. Genes with known T cell-related functions are shown in blue boxes. For the heatmaps, each vertical line represents a single cell. Only genes with FC>1.5 and false discovery rate (FDR)<0.05 are shown. For the volcano plots, genes with FDR<0.05 are shown in green. The eleven genes shared between (D) and (G) are in bold text.
Because 9.1-NP and 9.2-P clonotypes recognized exactly the same 9-mer epitope with similar avidity, we first compared the gene expression profiles of single cells from non-persistent 9.1-NP and persistent 9.2-P clonotypes. We identified a total of 472 differentially expressed genes with false discovery rate (FDR) less than 0.05 (Fig. 1B–C; Supplementary Table S2). By comparing the fold changes (FC) of gene expression between 9.1-NP and 9.2-P, we identified the top 10 upregulated genes and the top 10 downregulated genes. These genes are listed in Figure 1D, and these 20 genes are also highlighted in the heatmap and volcano plot (Fig. 1B–C). The brief descriptions of their known functions are listed in Supplementary Table S3, based on the summaries obtained from online databases Genecards (www.genecards.org) and OMIM (www.omim.org). Gene functions in 18 out of the 20 genes have been reported in previous studies, and nine of them have been shown to function significantly in T cells. However, the biological roles of C1orf162 and TPRG1 have been largely unknown (Fig. 1D).
We then compared the single-cell gene expression profiles from non-persistent 9.1-NP and persistent 10-P single cells, with the caveat that 10-P clonotype recognized a longer epitope with one additional leucine at the C-terminus. Similarly, a total of 528 differentially expressed genes (FDR<0.05) were identified in this comparison (Fig. 1E–F; Supplementary Table S2). The top 10 upregulated genes and the top 10 downregulated genes based on the FC of gene expression are listed in Figure 1G. These 20 genes are also highlighted in the heatmap and volcano plot (Fig. 1E–F). The brief descriptions of their known functions are listed in Supplementary Table S4. Gene functions in all 20 genes have been reported in previous studies, and nine of them have been shown to function significantly in T cells (Fig. 1G).
By assessing these two sets of differentially expressed genes, a total of 411 genes were shared between these two comparisons (Fig. 2A). A total of 11 genes in the top upregulated and downregulated gene lists were shared between both groups (bold text, Fig. 1D,,G).G). Because these gene signatures were also found in another persistent clonotype 10-P, these genes might be more significant in the biology of persistence than the 61 unique genes in the 9.2-P vs 9.1-NP comparisons. Using the shared 411 genes identified from these two comparisons, a t-distributed stochastic neighbor embedding (t-SNE) plot was generated. The majority of 9.2-P and 10-P single cells were separated from the 9.1-NP single cells (Fig. 2B).
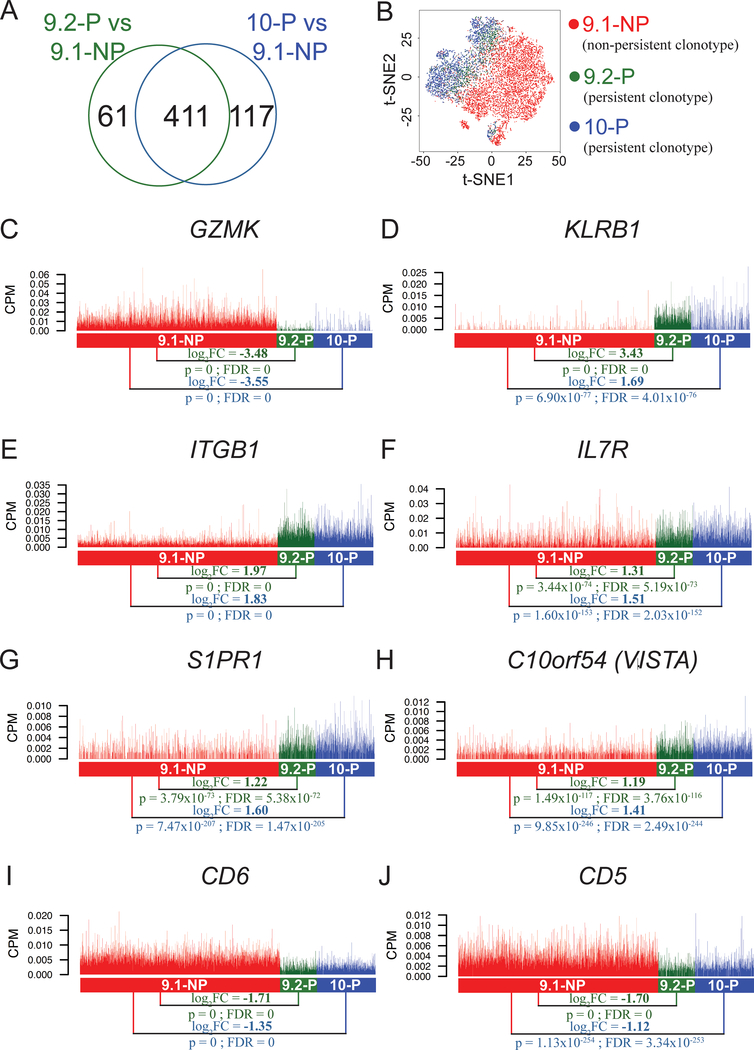
A, Venn diagram showing a total of 472 genes that were differentially expressed (FDR<0.05) in the comparison between 9.1-NP vs. 9.2-P single cells, and a total of 528 genes that were differentially expressed (FDR<0.05) in the comparison between 9.1-NP vs 10-P single cells. Among these genes, a total of 411 genes were shared in these two comparisons. B, The t-distributed stochastic neighbor embedding (t-SNE) plot of 9.1-NP, 9.2-P and 10-P single cells using the shared 411 genes obtained from the two comparisons. C-J, The expression of genes encoding cell surface markers are shown. Each vertical line represents a single cell. CPM: counts per million; GZMK: granzyme K; KLRB1: killer cell lectin like receptor B1; ITGB1: integrin subunit beta 1; IL7R: interleukin 7 receptor; S1PR1: sphingosine-1-phosphate receptor 1; C10orf54 (VISTA): chromosome 10 open reading frame 54 (the V domain‐containing immunoglobulin suppressor of T‐cell activation).
Among these shared gene signatures, we focused on cell surface markers and transcription factors in the top 10 upregulated and downregulated gene lists (Fig. 1D,,G).G). The cell surface markers could be potentially utilized to monitor or enrich T-cell populations associated with the persistence phenotype based on antibody staining, whereas the transcription factors could be the potential key factors defining the persistence phenotype. As shown in Figure 2C, the gene expression of granzyme K (GZMK) was much higher in the 9.1-NP non-persistent single cells, compared to the 9.2-P persistent single cells (log2FC = −3.48, FDR=0). Similarly, the GZMK expression was much higher in the 9.1-NP single cells, compared to the 10-P persistent single cells (log2FC = −3.55, FDR=0)(Fig. 2C). The expression of other granzymes, including GZMB, GZMH, and GZMM, were also significantly higher in the 9.1-NP, compared to the 9.2-P or 10-P single cells (Supplementary Table S2). These results suggested that 9.1-NP single cells showed a more effector-like phenotype, compared to 9.2-P or 10-P single cells. Conversely, NK cell marker KLRB1 (killer cell lectin-like receptor subfamily B, member 1, CD161) had high expression in 9.2-P single cells, compared to 9.1-NP single cells (log2FC = 3.43, FDR=0) (Fig. 2D). The 10-P single cells were also expressed higher KLRB1 than 9.1-NP single cells, but the difference was less than the 9.1-NP vs. 9.2-P comparison (log2FC: 3.43 vs. 1.69). The integrin beta 1 (ITGB1/CD29) gene also had high expression in both 9.2-P and 10-P single cells, compared to 9.1-NP single cells (log2FC = 1.97 and 1.83, respectively)(Fig. 2E). In these comparisons, we also found other differentially expressed genes related to T-cell mobility, including integrin ITGA1 (integrin alpha-1, CD49a), ITGAX (integrin alpha X, CD11c), chemokine CCL3, XCL2 and chemokine receptor CXCR4 and CCR5 (Supplementary Table S2).
Interleukin-7 (IL7) has been known as one of the master regulators for T-cell homeostasis (20). Higher expression of IL7 receptor (IL7R/CD127) was observed in 9.2-P single cells, compared to 9.1-NP single cells (log2FC = 1.31, FDR=5.19 × 10−73)(Fig. 2F). Similarly, higher IL7R expression was observed in 10-P single cells, compared to 9.1-NP single cells (log2FC = 1.51, FDR = 2.03 × 10−152). These results suggested that these 9.2-P T cells, as well as 10-P T cells, might be better equipped to compete for the limited resource of IL7, leading to better persistence (20). Sphingosine-1-phosphate receptor 1 (S1PR1) has been described as the predominant receptor for S1P, which is the central mediator of T-cell egress from the thymus or lymphoid organs into the blood (29). The expression of S1PR1 was higher in 9.2-P single cells, compared to 9.1-NP single cells (log2FC = 1.22, FDR = 5.38 × 10−72)(Fig. 2G). Similarly, higher S1PR1 expression was observed in 10-P single cells, compared to 9.1-NP single cells (log2FC = 1.60, FDR=1.47 × 10−205). C10orf54, also known as B7H5 or VISTA (the V domain‐containing immunoglobulin suppressor of T‐cell activation), is one of the B7 family members. Previous studies have suggested that VISTA can negatively regulate T-cell functions in antitumor responses, similar to other checkpoint inhibitors (30). The C10orf54/VISTA expression was lower in 9.1-NP single cells, compared to 9.2-P single cells or 10-P single cells (log2FC = 1.19 and 1.41, respectively)(Fig. 2H). The expression levels of other negative co-stimulatory molecules were low (Supplementary Fig. S1). Cell surface marker CD5 and CD6 have been suggested to negatively regulate TCR signaling, but the evidence for this is not conclusive. CD5 and CD6 were both downregulated in both 9.2-P and 10-P single cells, compared to 9.1-NP single cells (Fig. 2I–J).
Several transcription factors were differentially expressed in this analysis, including ZNF683, SOX4, CEBPD, KLF2 and EOMES (Fig. 3A–E; Supplementary Table S2). Zinc finger protein 683 (ZNF683), also known as HOBIT (homolog of BLIMP1 in T cells), has been suggested to serve as a key transcription factor expressed in tissue-resident memory T cells (31). The expression of ZNF683 was upregulated in 9.2-P single cells, compared to 9.1-NP single cells (log2FC = 2.71, FDR = 1.92 × 10−119)(Fig. 3A). Similarly, the ZNF683 expression was upregulated in 10-P single cells, compared to 9.1-NP single cells (log2FC = 3.97, FDR=0). These results suggested that ZNF683 might also play an important role in T-cell persistence. SOX4 (sex determining region Y-box 4) has been shown to play a key role for CXCL13 production in human CD4+ T cells under inflammatory conditions (32). CCAAT/enhancer binding protein δ (CEBPD) has been known to work synergistically with CEBPB to control the induction of proinflammatory cytokines IL6 and TNFα in macrophages (33). However, its role in T cells is largely unclear. Both SOX4 and CEBPD were downregulated in 9.2-P single cells, compared to 9.1-NP single cells (log2FC = −1.89 and −1.79, respectively)(Fig. 3B–C). Both SOX4 and CEBPD were also significantly downregulated in 10-P single cells, compared to 9.1-NP single cells (log2FC = −1.74 and −2.48, respectively).
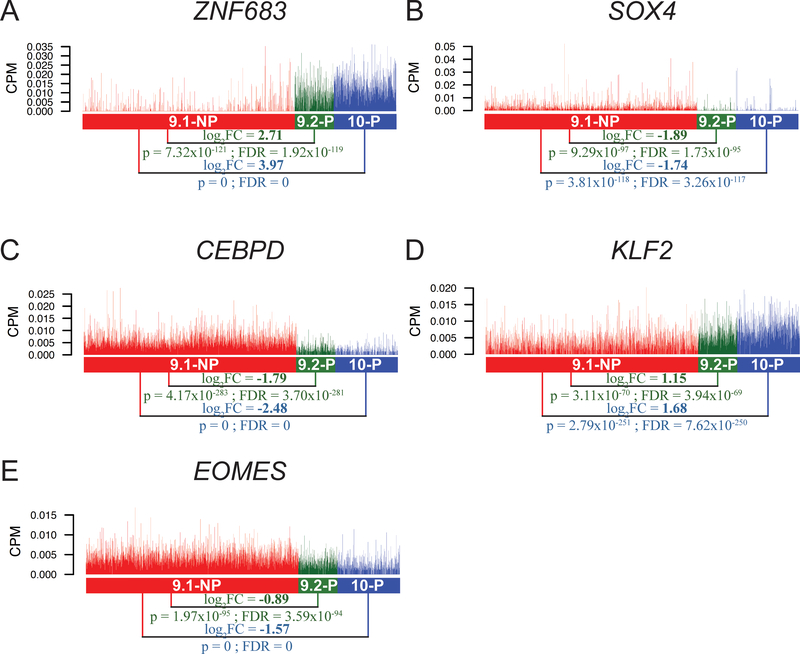
A-E, The expression of genes encoding transcription factors are shown. Each vertical line represents a single cell. ZNF683: zinc finger protein 683; SOX4: sex determining region Y-box 4; CEBPD: CCAAT/enhancer binding protein δ; KLF2: Kruppel like factor 2; EOMES: eomesodermin.
It has been reported that the transcription factor Klf2 (Kruppel like factor 2) can promote the expression of S1pr1 and restrain the differentiation of CD4+ T follicular helper cells in mice (34). The expression of KLF2 was higher in 9.2-P single cells, compared to 9.1-NP single cells (log2FC = 1.15, FDR = 3.94 × 10−69)(Fig. 3D). The KLF2 expression in 10-P single cells was also higher than 9.1-NP single cells (log2FC = 1.68, FDR = 7.62 × 10−250). Lastly, the transcription factor EOMES (eomesodermin) has been identified as one of the key markers among exhausted T-cell populations (35). Significantly less EOMES expression was observed in 9.2-P single cells, compared to the 9.1-NP single cells (log2FC = −0.89, FDR = 3.59 × 10−94)(Fig. 3E). The EOMES expression was also significantly downregulated in 10-P single cells, compared to the 9.1-NP single cells (log2FC = −1.57, FDR=0).
To test whether we could observe the same gene expression patterns using a different approach, we utilized another single-cell system developed by Fluidigm. This system uses a different molecular approach for the single-cell transcriptome analysis. Because Fluidigm C1 is a low-throughput system, TILs were first enriched using the binding of KRAS-9mer or KRAS-10mer tetramer and then subjected to the Fluidigm C1 single-cell system. The full-length, whole-transcriptome sequences from each single cell were obtained, including TCR-CDR3 information for clonotype identification (24). We obtained a total of 123 good-quality single cells from 9.1-NP, 42 single cells from 9.2-P and 72 single cells from 10-P. As illustrated in Figure 2–3, significant differences (p<0.05) were observed in 11 of the 13 genes when comparing 9.1-NP vs 9.2-P, except IL7R and KLF2, possibly due to the low number of cells obtained from 9.2-P single cells from this low-throughput approach (Supplementary Fig. S2). Significant differences were observed in all 13 genes comparing 9.1-NP vs 10-P (Supplementary Fig. S2).
To further observe the gene expression profiles of T cells after ACT, the patient’s peripheral blood lymphocytes (PBLs) were stained with KRAS-9mer tetramer or KRAS-10mer tetramer in the attempt to enrich the 9.1-NP, 9.2-P or 10-P populations. The non-persistent clonotype 9–1-NP was detectable on day 12, but not detectable on day 40 after ACT. Using the methods previously described (Fig. 1A), single-cell samples were prepared from the enriched populations by a 10X Genomics single-cell instrument (Fig. 4A). Using the peripheral blood sample from day 12 after ACT, we obtained a total of 7365, 525, and 3487 good-quality single cells associated with 9.1-NP, 9.2-P and 10-P clonotypes, respectively. From the peripheral blood sample on day 40 after ACT, we acquired a total of 7919 and 5806 good-quality single cells associated with 9.2-P and 10-P clonotypes, respectively. In this analysis, TILs were plotted together with PBLs to observe the changes of gene expression following ACT (Supplementary Table S5).
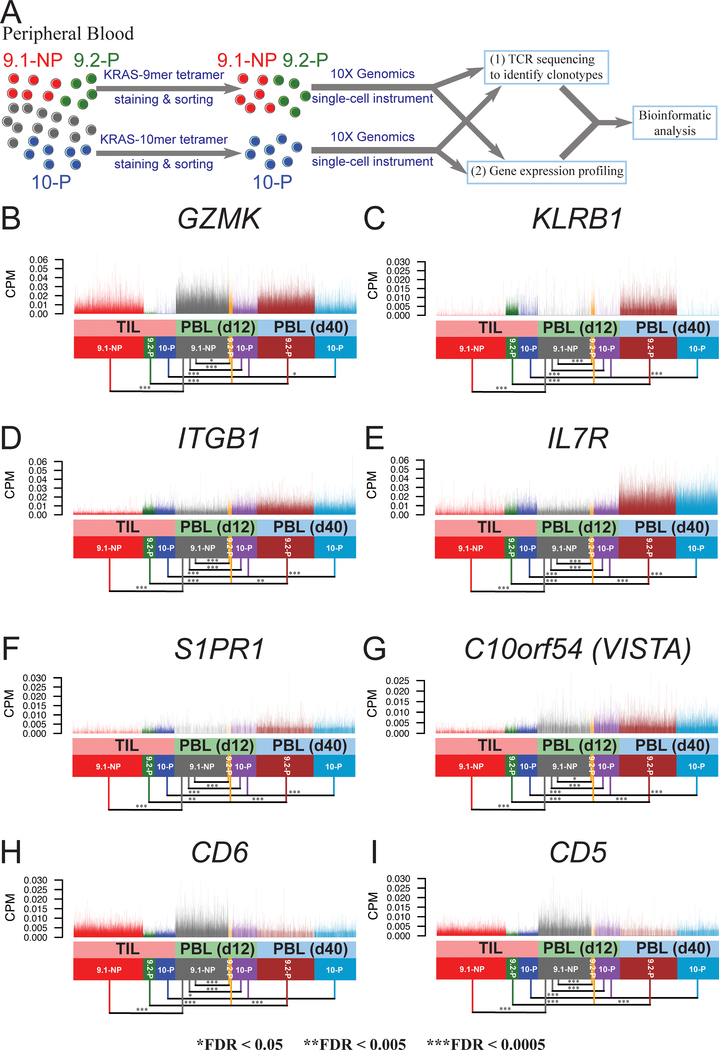
A, PBL samples on day 12 (d12) and day 40 (d40) were enriched by tetramer staining and sorting, followed by single-cell transcriptome analysis. B-I, The expression of genes encoding cell surface markers are shown. Single cells from TILs are plotted together with PBL samples on day 12 and day 40. Each vertical line represents a single cell.
As previously demonstrated, the 9.2-P and 10-P TILs expressed minimum amounts of GZMK, compared to 9.1-NP TILs. On day 12 after cell transfer, significant expression of GZMK was observed in the peripheral blood 9.2-P single cells, although the expression was slightly lower than 9.1-NP single cells in PBLs (log2FC = −0.16, FDR=9.69 × 10−3)(Fig. 4B). On day 40, 9.2-P single cells continued to express a high GZMK. Similar observations were found in 10-P single cells, compared to 9.1-NP single cells in the PBLs on day 12 (log2FC = −1.70, FDR=0). KLRB1 had high expression in 9.2-P TILs and 10-P TILs. After ACT, 9.2-P T cells in PBLs continued to express high KLRB1 on day 12 and day 40 (Fig. 4C). However, 10-P T cells lost the expression of KLRB1 on day 12 after ACT, and 10-P T cells continued to express low KLRB1 on day 40 (Fig. 4C). Similar to the observation in TILs, ITGB1 expression was higher in both 9.2-P and 10-P, compared to 9.1-NP on day 12 (log2FC = 1.12 and 0.96, respectively). For both 9.2-P and 10-P, the ITGB1 expression was slightly higher on day 40, compared to day 12 (log2FC = 0.21 and 0.31, respectively)(Fig. 4D). As one of the key regulators for T-cell homeostasis (20), IL7R expression remained higher in 9.2-P single cells, compared to 9.1-NP single cells on day 12 (log2FC = 0.62, FDR=2.62 × 10−12). The 9.2-P T cells significantly upregulated IL7R on day 40, compared to 9.2-P T cells on day 12 (log2FC = 1.66, FDR = 6.21 × 10−127) (Fig. 4E). Similar results were observed in peripheral blood 10-P T cells, compared to 9.1-NP T cells (log2FC = 0.86, FDR=6.4 × 10−122). These results suggested that 9.2P and 10-P were more competitive in terms of obtaining limited homeostatic cytokine IL7 in vivo. The S1PR1 expression was not significantly different between peripheral blood 9.1-NP and 9.2-P T cells on day 12, but the S1PR1 expression was significantly higher in 9.2-P T cells on day 40, compared to 9.2-P T cells on day 12 after ACT (log2FC = 0.95, FDR = 3.43 × 10−15)(Fig. 4F). On the other hand, the S1PR1 expression was significantly higher in peripheral blood 10-P T cells, compared to 9.1-NP T cells on day 12 (log2FC = 0.42, FDR = 8.06 × 10−23).The S1PR1 expression was significantly higher in 10-P T cells on day 40, compared to 10-P T cells on day 12 after ACT (log2FC = 0.83, FDR = 4.55 × 10−65). The expression of C10orf54 (VISTA) was slightly higher in 9.2-P T cells, compared to 9.1-NP T cells on day 12 after ACT (log2FC = 0.27, FDR = 1.48 × 10−2)(Fig. 4G). The expression of other negative co-stimulatory molecules were low (Supplementary Fig. S1). Both CD5 and CD6 expression was significantly lower in 9.2-P T cells, compared to 9.1-NP T cells on day 12 (log2FC = −1.23 and −1.24, respectively)(Fig. 4H–I).
Similar to the observation in TILs, the transcription factor ZNF683 also had high expression in 9.2-P T cells on day 12, compared to 9.1-NP T cells on day 12 (log2FC = 1.83, FDR = 2.14 × 10−72)(Fig. 5A). Similarly, 10-P T cells expressed higher ZNF683, compared to 9.1-NP T cells on day 12 (log2FC = 2.51, FDR=0). Both 9.2-P and 10-P T cells on day 40 expressed higher ZNF683, compared to 9.2-P and 10-P T cells on day 12 (log2FC = 0.54 and 0.61, respectively). The expression of SOX4 and CEBPD was minimum among the peripheral blood 9.1-NP, 9.2-P and 10-P T cells (Fig. 5B–C). The expression of transcription factor KLF2 was higher in 9.2-P T cells, compared to 9.1-NP T cells in the peripheral blood on day 12 (log2FC = 0.34, FDR=4.97 × 10−4)(Fig. 5D). Similarly, 10-P T cells expressed higher KLF2 than 9.1-NP T cells on day 12 (log2FC = 0.81, FDR=2.17 × 10−109). Lower levels of EOMES expression were observed in 9.2-P on day 12, compared to 9.1-NP (log2FC = −0.49, FDR = 7.38 × 10−6). However, the expression of EOMES in 9.2-P increased on day 40, compared to day 12 (log2FC = 0.26, FDR = 2.13 × 10−2)(Fig. 5E). Similarly, lower EOMES expression was observed in 10-P on day 12, compared to 9.1-NP (log2FC = –0.58, FDR = 3.05 × 10−43). However, the expression of EOMES in 10-P also increased on day 40, compared to day 12 (log2FC = 0.18, FDR = 1.3 × 10−5)(Fig. 5E). Taken together, we observed the continuing high expression of IL7R, ITGB1, ZNF683 and KLF2 in peripheral blood 9.2-P and 10-P T cells following ACT. On the other hand, 9.1-NP T cells had relatively lower expression of these genes in PBLs, compared to 9.2-P or 10-P T cells.
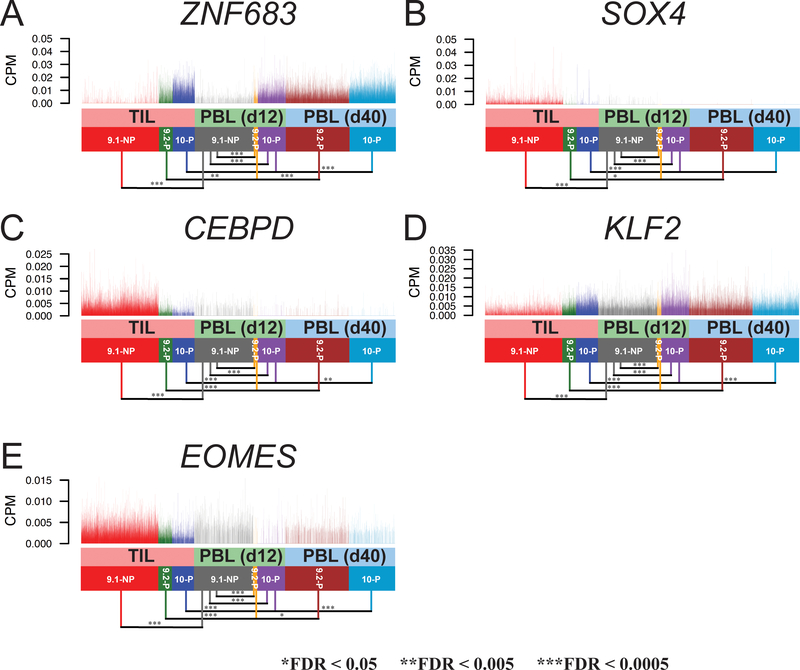
A-E, The expression of genes encoding transcription factors are shown. Single cells from TILs are plotted together with PBL samples on day 12 and day 40. Each vertical line represents a single cell.
The persistence of T cells can be influenced by the host condition, including the levels of homeostatic cytokines, endogenous T-cell reconstitution, the tumor microenvironment, as well as the binding affinity between TCR and antigen/HLA (17–20). With these caveats in mind, we attempted to test whether or not other infused TILs expressed a set of genes associated with persistence. From the comparisons of 9.1-NP vs 9.2-P and 9.1-NP vs 10-P, 38 shared genes were significantly upregulated, and 41 shared genes were significantly downregulated (FC > ±1.5, FDR<0.05), which were defined as the Persistence_Up gene set and Persistence_Down gene set, respectively (Supplementary Table S1.) To perform the Gene Set Enrichment Analysis (GSEA) analysis (27), we selected four autologous TILs, 4007, 4069, 4071, and 4081, obtained from patients with metastatic gastrointestinal cancers (6,16). These autologous TILs were used to treat these four patients within the five-month window prior to the treatment of patient 4095. The same as the majority of infused TILs in this clinical trial, these four TILs contained predominantly (32.3 ~ 91.9%) neoantigen-reactive CD8+ T cells but persisted poorly one month after ACT (Fig. 6A; Supplementary Table S6)(4,16). Following the same process as the previous experiments, the single-cell transcriptome analysis was performed to study gene expression profiles linked to neoantigen-reactive clonotypes. These single cells associated with neoantigen-reactive clonotypes were first compared to the persistent clonotypes (9.2-P and 10-P), and the differentially expressed genes were subjected to the GSEA analysis. As shown in Figure 6 ((BB–E), strong differences were observed in all eight GSEA tests for Persistence_Up and Persistence_Down gene sets. Therefore, these TILs obtained from four patients did not express the gene sets associated with the persistence, suggesting that these TILs might lack the “fitness” to persist in vivo.
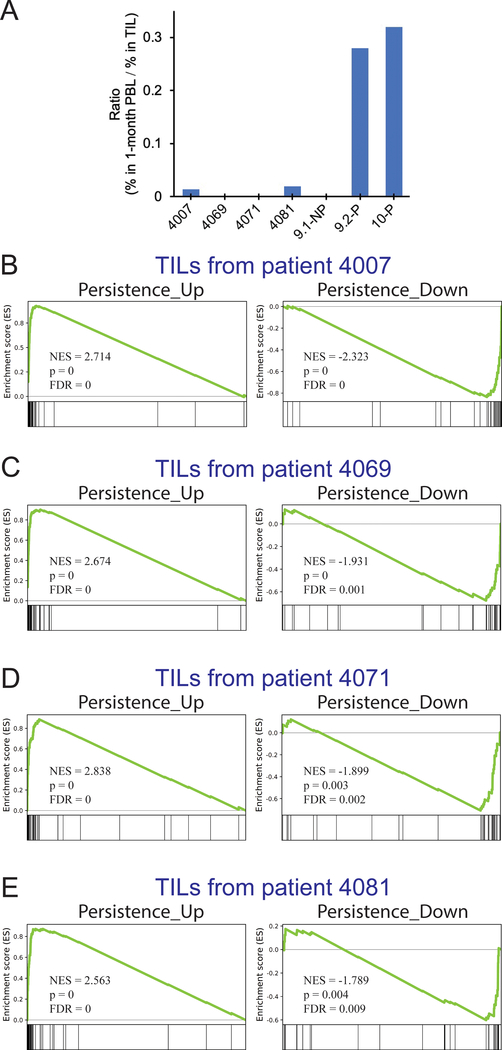
A, Four patients with metastatic gastrointestinal cancers, 4007, 4069, 4071 and 4081, were treated with autologous TILs within the five-month window prior to the treatment of patient 4095. The ratio of persistence was calculated based on the percentage of neoantigen-specific T cells in PBLs at approximately one month after ACT, divided by the percentage of neoantigen-specific T cells in the infused TILs (Supplementary Table S6). B-E, Gene set enrichment analysis (GSEA) was performed to compare 4095 persistent clonotypes (9.2-P and 10-P) with TIL products from the four patients. Two gene sets, Persistance_Up and Persistance_Down, were used in the GSEA analysis. The Persistence_Up and Persistence_Down gene sets were constructed based on the shared, differentially expressed genes (FC > ±1.5, FDR<0.05), obtained from the comparisons of 9.1-NP vs 9.2-P and 9.1-NP vs 10-P (Supplementary Table S1). NES: normalized enrichment score.
Discussion
In this study, we discovered a gene expression profile associated with T-cell persistence in a patient treated for KRAS(G12D)-expressing tumors. It has been known that IL7 is one of the key cytokines involved in T-cell homeostasis. (20). It is thus likely that T cells with high IL7R could have a survival advantage over the low IL7R cells. The 9.2-P and 10-P persistent T cells expressed high IL7R prior and after ACT, compared to the 9.1-NP non-persistent T cells. Similarly, a previous study suggested that in vitro expanded MART1- or gp100-specific T-cell clones that expressed the IL7R were more likely to persist longer after adoptive transfer to patients (36). Next, we studied S1PR1 and KLF2, the key regulators to promote T-cell egress from the thymus or lymphoid organs into the blood (29). A previous mouse study showed that Klf2 could induce the expression of S1pr1 and limit the differentiation of CD4+ T follicular helper cells (34). Another study suggests that CD8+ resident memory T cells express low Klf2 and S1pr1 (37). Consistent with these findings, we found that 9.2-P and 10-P persistent TILs expressed high KLF2 and S1PR1, whereas 9.1-NP non-persistent TILs expressed low KLF2 and S1PR1. After ACT, 9.2-P and 10-P T cells continued to express KLF2 and S1PR1. Because of the limitation in the human study, it is unclear whether or not T cells can downregulate KLF2 and S1PR1, and subsequently enter the tumor.
One of the most significant changes observed in this study was the expression of ZNF683 in T cells that were infused and persisted. Znf683 (Hobit) was identified as a homolog of Blimp1 and a key transcription factor for CD8+ tissue-resident memory T cells in mice (31). Zundler S et al demonstrates that Znf683 and Blimp-1 double-knockout mice are protected in CD4+ tissue-resident, memory T cell-mediated experimental colitis models (38). Conversely, a human study observes that ZNF683 (HOBIT) has high expression in viral-specific CD8+ effector T cells in peripheral blood, and it was proposed that ZNF683 might be a key transcription factor for long-lived effector-type T cells based on this observation (39,40). However, no further evidence has been published to support this hypothesis. In this study, we found that the ZNF683 expression in the infused CD8+ TILs was associated with their persistence in vivo. ZNF683 was highly expressed in persistent 9.2-P and 10-P T cells in peripheral blood 12 and 40 days after cell infusion. Therefore, ZNF683 might serve as a key transcription factor for T-cell persistence, potentially working together with other transcription factor(s), such as KLF2. Conversely, it is possible that not only a few key transcription factors, but the entire gene expression profile is responsible for the fitness of TILs.
It has been well-established that “younger” T cells, including naïve T cells, stem cell memory T cells, and central memory T cells, can persist better in vivo and induce stronger anti-tumor responses, compared to the “older”, terminally differentiated, effector memory T cells (41). However, it is puzzling that terminally differentiated T cells still can induce durable tumor regressions following TIL-ACT or checkpoint immunotherapy. To explain this, it has been proposed that mouse exhausted T cells can be divided into two subsets: (i) “Early” exhaustion subset, defined as EomesloPD-1intTcf1+ T cells; (ii) “Terminal” exhaustion subset, defined as EomeshiPD-1hiTbetloTcf1− T cells (42). The Eomeshi subset has poor proliferative potential but retains partial cytotoxicity, whereas Eomeslo subset retains moderate proliferative capacity but has limited cytotoxicity (35).The Eomeslo early exhaustion subset can convert to the Eomeshi terminal exhaustion subset after antigen stimulation and proliferation, and this process is not reversible (35). More importantly, PD-1 blockade can reverse and reinvigorate the exhausted phenotype of Eomeslo subset, but not the Eomeshi subset (43). However, the majority of evidence supporting this hypothesis was obtained from the LCMV chronic infection model, and there is little evidence in human cancer studies to support this hypothesis directly (42). In the present study, the persistent 9.2-P and 10-P TILs had lower EOMES expression than the non-persistent 9.1-NP TILs. Similarly, 9.2-P and 10-P TILs also expressed low GZMK, known to be associated with cytotoxicity. Following ACT, the persistent 9.2-P and 10-P T cells in the peripheral blood had lower EOMES than the non-persistent 9.1-NP T cells, and on day 40, 9.2-P and 10-P T cells upregulated EOMES, compared to 9.2-P and 10-P T cells on day 12 after ACT. Therefore, our observation in this study was in line with this hypothesis. However, further evidence is required, such as tracking the T-cell phenotypes in the peripheral blood and tumors through the course of immunotherapy, which can be difficult in human studies. It has also been hypothesized that Tcf1+ “memory-like” exhausted T cells are proliferation-competent and responsible for the anti-virus or likely antitumor responses after PD-1 blockade immunotherapy (44–46). However, the expression TCF1 (TCF7) were minimum in our single-cell analysis, and no significant difference of TCF1 (TCF7) expression was observed between 9.1-NP vs 9.2-P or 9.1-NP vs 10-P.
It remains an open question whether or not the persistence of T cells in the peripheral blood is required for tumor regression and clinical responses or is merely a byproduct of vigorous immune responses. Our previous ACT studies using melanoma TILs demonstrate a significant correlation between the persistence of TIL clonotypes and clinical responses (47). Conversely, in another ACT trial using peripheral blood-derived, MART-1-specific cytotoxic T cells, followed by a standard course of anti–CTLA-4, there was no relationship between the persistence of the infused T cells and the tumor responses (48). In ACT studies using CD19 CAR-modified T cells for patients with B-cell leukemias or lymphomas, several studies show the correlation between persistence and responses (17,18,49–52). However, no significant correlation was found in one of the studies (53). In one ACT study using NY-ESO-1 TCR-transduced T cells for patients with synovial sarcoma, a correlation between persistence and clinical responses was observed (54). However, the correlation was not observed in another study for patients with melanoma or synovial sarcoma (55). Taken together, the outcome of ACT is determined by many factors, which may or may not include the persistence of T cells. More direct evidence is needed to further understand these complicated observations.
In summary, we have identified two different TIL populations with distinctive gene expression profiles, which were associated with the persistence of TILs in a patient. Although we could obtain significant knowledge in a relatively defined environment in a single patient, the observation might not be generalized because of the diversity of patients. We plan to do this single-cell analysis for a large number of patients as more patients are accrued in this clinical trial. Given these caveats, the information obtained in this study has opened the possibility to improve TIL therapies against epithelial cancers by enriching TIL populations based on one or the combinations of the cell surface markers, such as KLRB1, ITGB1, IL7R, S1PR1 or C10orf54. Manipulating the expression of key transcription factors, such as ZNF683 and KLF2, may lead to better T-cell products for immunotherapy. These hypotheses need to be further tested in human studies.
Supplementary Material
1
2
3
Acknowledgements
The authors would like to thank Nicholas Restifo, Steven Shema, Valery Bliskovsky, Michael Kelly and Jinguo Chen for suggestions and technical support. The authors also thank Doug Joubert, NIH Library Writing Center, for manuscript editing assistance. Next-generation sequencing was conducted at the CCR Genomics Core at the National Cancer Institute. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Financial Support: This work was supported by the Intramural Research Program of National Cancer Institute.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/2326-6066.cir-19-0299
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6825592?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Distinct Immune Homeostasis Remodeling Patterns after HLA-Matched and Haploidentical Transplantation.
Adv Sci (Weinh), 11(39):e2400544, 03 Sep 2024
Cited by: 1 article | PMID: 39225336 | PMCID: PMC11497014
Research advances on TIL therapy for colorectal cancer.
Clin Transl Oncol, 26(12):2917-2923, 28 May 2024
Cited by: 0 articles | PMID: 38806995
Review
Neoantigen-specific CD4+ tumor-infiltrating lymphocytes are potent effectors identified within adoptive cell therapy products for metastatic melanoma patients.
J Immunother Cancer, 11(10):e007288, 01 Oct 2023
Cited by: 8 articles | PMID: 37802604 | PMCID: PMC10565316
Computational identification and clinical validation of a novel risk signature based on coagulation-related lncRNAs for predicting prognosis, immunotherapy response, and chemosensitivity in colorectal cancer patients.
Front Immunol, 14:1279789, 19 Oct 2023
Cited by: 0 articles | PMID: 37928532 | PMCID: PMC10620970
Combination of multiple omics techniques for a personalized therapy or treatment selection.
Front Immunol, 14:1258013, 27 Sep 2023
Cited by: 2 articles | PMID: 37828984 | PMCID: PMC10565668
Review Free full text in Europe PMC
Go to all (30) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE136394
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy.
J Immunother, 28(1):53-62, 01 Jan 2005
Cited by: 123 articles | PMID: 15614045 | PMCID: PMC2175172
Telomere length of transferred lymphocytes correlates with in vivo persistence and tumor regression in melanoma patients receiving cell transfer therapy.
J Immunol, 175(10):7046-7052, 01 Nov 2005
Cited by: 213 articles | PMID: 16272366 | PMCID: PMC1351312
Survival, persistence, and progressive differentiation of adoptively transferred tumor-reactive T cells associated with tumor regression.
J Immunother, 28(3):258-267, 01 May 2005
Cited by: 122 articles | PMID: 15838383 | PMCID: PMC2174599
Cell Therapy With TILs: Training and Taming T Cells to Fight Cancer.
Front Immunol, 12:690499, 01 Jun 2021
Cited by: 56 articles | PMID: 34140957 | PMCID: PMC8204054
Review Free full text in Europe PMC
Funding
Funders who supported this work.
HHS | National Institutes of Health (NIH) (1)
Grant ID: Z010984
Intramural NIH HHS (1)
Grant ID: Z99 CA999999





