Abstract
Free full text

‘Final common pathway’ of human cancer immunotherapy: targeting random somatic mutations
Abstract
Effective clinical cancer immunotherapies, such as administration of the cytokine IL-2, adoptive cell transfer (ACT) and the recent success of blockade of the checkpoint modulators CTLA-4 and PD-1, have been developed without clear identification of the immunogenic targets expressed by human cancers in vivo. Immunotherapy of patients with cancer through the use of ACT with autologous lymphocytes has provided an opportunity to directly investigate the antigen recognition of lymphocytes that mediate cancer regression in humans. High-throughput immunological testing of such lymphocytes in combination with improvements in deep sequencing of the autologous cancer have provided new insight into the molecular characterization and incidence of anti-tumor lymphocytes present in patients with cancer. Here we highlight evidence suggesting that T cells that target tumor neoantigens arising from cancer mutations are the main mediators of many effective cancer immunotherapies in humans.
Many approaches have been used over the past 25 years to identify antigens that are naturally processed and presented on human cancer cells. Most such studies have involved the immunological testing of tumor cDNA library pools introduced, along with genes encoding autologous major histocompatibility complex (MHC) molecules, into highly transfectable target cells such as COS7 monkey kidney cells and HEK293 human embryonic cancer cells. Rarely, antigens have been identified by evaluation of the responses of tumor-reactive T cells to target cells pulsed with purified tumor-cell proteins or with peptides eluted from MHC molecules isolated from the tumor-cell surface. Such techniques have identified individual protein antigens but have not been successful in enabling a wide survey of the antigens recognized by autologous human T cells.
A brief survey of non-mutant tumor antigens
Tumor antigens identified by the techniques noted above can be grouped into two broad categories: self antigens and non-self antigens. Self antigens, which represent products expressed by normal (non-cancerous) cells, have generally been further sub-divided into three general categories on the basis of their expression patterns in normal and tumor tissues. Cancer germline antigens represent natural proteins that are expressed during fetal development and are re-expressed in a variable proportion of many cancer histologies but often with limited expression in normal adult tissues. ACT with autologous lymphocytes genetically engineered to express a T cell antigen receptor (TCR) for the HLA*0201 epitope of the cancer germline antigen NY-ESO-1 led to durable tumor regression in a small number of patients with meta-static melanoma or synovial cell sarcoma1; however, broad applicability of this approach is limited by the low frequency (often 2–3%) of common cancers that homogenously express this epigenetically controlled antigen2. In addition, unanticipated toxicities have been observed in trials targeting more broadly expressed cancer germline antigens, such as MAGE-A3, due to the expression of similar proteins in vital normal tissues3,4.
Therapies have targeted differentiation antigens expressed in normal adult tissue as well as in tumors derived from that tissue; however, the normal tissues that express these products are then at risk of immunological attack. ACT with autologous T cells transduced with highly avid TCRs for epitopes of the melanocyte-melanoma differentiation antigens MART-1 and gp100 in patients with metastatic melanoma has led to transient tumor regression, but has simultaneously resulted in severe dose-limiting toxicity due to the recognition of normal melanocytes in the eyes and ears5. Targeting of the normal B cell signal-transduction receptor CD19 present on most B cell lymphomas through the use of autologous cells transduced with chimeric antigen receptors directed against CD19 has been effective in treating these tumors6–11, although the concomitant elimination of normal B cells and neurological toxicity require careful monitoring of patients. Unfortunately, there are few examples of solid cancers arising from non-essential organs that express shared intracellular or cell-surface differentiation proteins that can be targeted.
Therapies have also targeted overexpressed proteins, such as carcinoembryonic antigen, that represent cell products whose expression in normal tissues is lower than their expression in malignant cells. ACT with T cells genetically engineered to express a high-avidity TCR for carcinoembryonic antigen raised in mice with transgenic expression of HLA-A*02:01 was associated with limited cancer regression in humans but also mediated nearly fatal destruction of colonic mucosa; this demonstrates that targeting antigens with relatively low expression in normal cells can lead to severe toxicity12.
Cancer germline, differentiation and overexpressed proteins have been the predominant targets of many hundreds of human therapeutic vaccine trials. There is little to no evidence of their clinical effectiveness, which might be due in part to central tolerance that has eliminated cells bearing high-avidity TCRs for normal, non-mutant proteins13. The foregoing results highlight the limitations of immunologically targeting these classes of antigens and the need to more safely and effectively target other tumor antigens.
Non-self antigens include epitopes derived from viral gene products and neoepitopes encoded by non-synonymous mutations that arise during the process of tumorigenesis and are therefore not expressed by normal cells. The tumor-specific expression of these antigens suggests that immunotherapeutic attack of these antigens should be safe and potentially effective. Prophylactic vaccination against proteins and peptides expressed by viruses such as human papilloma virus (HPV) can be effective in preventing cancers caused by these viruses14. Moreover, vaccination with HPV peptides seems to prevent tumor progression in patients with premalignant disorders of the uterine cervix15. In the setting of metastatic cancer, durable tumor regression has been observed in two of nine patients with cervical cancer who received autologous transfer of tumor infiltrating lymphocyte (TIL) populations that included HPV-reactive T cells16; however, the role of the HPV-reactive T cells in mediating tumor regression in these patients is unclear, since the majority of the T cells infused did not target HPV antigens.
The development of technologies over the past 5–10 years that allow rapid and relatively inexpensive transcriptome and whole-exome sequencing (WES) of tumor DNA and matched normal DNA, in conjunction with the development of novel immunological screening methods, has facilitated the evaluation of T cell reactivity to cancer neoepitopes, which are mutant peptides encoded by random mutations expressed in the autologous cancer. As discussed below, mounting evidence suggests that the MHC-restricted recognition of these unique mutant epitopes by lymphocytes probably represents the ‘final common pathway’ that explains the efficacy of most cancer immunotherapies and provides clues to the extension of immuno-therapy to additional cancer types.
Mouse tumor neoepitopes as potent tumor-rejection antigens
Early studies of mouse tumor model systems indicated that neoepitopes represent potent tumor-rejection antigens17–22; however, the laborious and time-consuming techniques needed to identify neoepitope-reactive T cells have hampered efforts to broadly evaluate the role of these cells in anti-tumor responses. Advances in high-throughput sequencing methods have allowed more efficient investigation of the role of neoepitope-reactive T cells in antitumor immunity. In one of the first studies to use this approach, candidate neoepitopes were identified on the basis of an algorithm used to predict binding of neoepitopes to individual MHC class I molecules, combined with high-throughput sequencing of tumor-cell DNA and RNA obtained from mouse sarcomas generated in immunodeficient mice lacking the gene encoding the recombinase component RAG-2 (ref. 23). That approach led to the identification of a mutant spectrin-β2 neoepitope as a dominant tumor-rejection antigen in this mouse model. The results of additional mouse studies have indicated that vaccination against candidate tumor neoepitopes identified by WES can be used to treat mice with small tumor burdens24–26. Neoepitope-reactive T cells have also been identified by the analysis of tumor samples via WES and high-throughput RNA sequencing to identify candidate neoepitopes, coupled with mass spectrometry of peptides eluted from cell-surface MHC molecules. Analysis of the MC-38 and TRAMP-C1 mouse tumor-cell lines by this method has led to the identification of three neoepitopes expressed by MC-38 that elicit tumor-reactive T cells that seem to provide partial prevention and treatment of tumors in a mouse tumor-vaccine model system27.
Neoantigen burden and clinical benefit in metastatic cancer
In humans, several lines of correlative evidence suggest that T cells that target mutant neoantigens might serve an important role in mediating clinical responses to cancer immunotherapy. With the exception of renal-cell carcinoma28,29 and some virus-induced cancers30, immunological checkpoint inhibitors targeting the CTLA-4 and PD-1 pathways have shown the greatest clinical activity against cancer types with the greatest average number of somatic mutations, such as melanoma31–35, non-small-cell lung cancer36,37, bladder cancer38 and cancers with DNA-mismatch-repair deficiencies39. Even within the same cancer type, individual patients with melanoma33–35 or non-small-cell lung cancer37,40 whose tumors had a relatively high mutation burden were more likely to clinically benefit from checkpoint-blockade therapy than were those with a lower mutation load. Clinical benefit from immunological checkpoint blockade has also been associated with a relatively high burden of potential neoepitopes identified by algorithms used to predict the binding of peptides to MHC molecules33,34,37. Such findings have provided evidence of an association between mutation or potential neoantigen load and patient survival and are highly suggestive of, but do not directly demonstrate, the principle that specific targeting of cancer neoantigens can result in cancer regression.
TILs targeting neoepitopes and human melanoma
Clinical trials by the Surgery Branch of the National Cancer Institute studying ACT immunotherapy of 194 patients with metastatic melanoma receiving treatment with autologous TILs have provided reagents (cancer tissue and lymphocytes) that have allowed detailed evaluation of the role of neoantigen reactivity in mediating immunotherapy responses. Patients in these trials received a non-myeloablating lymphodepleting chemotherapy that eliminated circulating lymphocytes for about 8 days, at which time maximum lymphodepletion was achieved, followed by the adoptive transfer of autologous TIL populations expanded in vitro, plus IL-2. An objective response rate of 55%, as assessed by Response Evaluation Criteria In Solid Tumors, was seen; this included 44 patients (23%) with complete responses, 42 of whom have ongoing responses with a median potential follow-up of 65.4 months and are probably cured41,42. The great majority of those complete responses occurred in the absence of cell-induced off-tumor toxicity, which suggests that the transferred T cells targeted mainly molecules unique to the cancer23.
The observations noted above raised the possibility that T cells that target tumor-specific neoantigens might have a role in the clinical responses seen in some of those patients. Initial studies of a small number of patients from those trials used screening of human tumor cell cDNA libraries and have revealed the presence of neoepitope-reactive cells in the TILs administered to patients who experienced durable cancer regression43–46, consistent with the hypothesis that neoepitope-reactive T cells can mediate tumor regression. Further support for this hypothesis has been provided by studies using next-generation sequencing methods combined with high-throughput immunological screening approaches to identify immunogenic mutations. In one of the earliest studies to use this approach to identify neoepitopes expressed by human cancers, WES of cell lines from three patients with melanoma was coupled to the use of an algorithm to predict candidate minimal peptides able to bind to autologous MHC molecules. These peptides were then synthesized and pulsed onto HLA-matched antigen-negative target cells and evaluated for their ability to stimulate autologous TILs47. A total of seven different neoepitopes were retrospectively identified as targets of three autologous TILs administered to patients with melanoma, two of whom exhibited durable complete regression of multiple metastases following the transfer of TIL samples containing immunodominant populations of neoantigen-reactive T cells. In another patient with melanoma, the adoptive transfer of a single TIL product that was found to recognize ten distinct neoantigens was associated with durable complete tumor regression of all metastatic lesions48. The observation that T cells targeting five of the ten neoantigens composed nearly 30% of total peripheral T cells approximately 1 month after transfer provided further evidence that neoantigen-reactive T cells might have an important role in mediating the complete tumor regression seen in some patients receiving adoptive immunotherapy48. In an additional study, the objective clinical response observed in a patient with melanoma who received autologous TILs was associated with the persistence of CD4+ T cells present in the transferred T cells reactive with a neoepitope expressed by autologous tumor cells49.
Additional studies aimed at the characterization and isolation of neoepitope-reactive T cells have used algorithms to predict peptide-MHC binding, in conjunction with a high-throughput screening method and the use of an ultraviolet-irradiation-mediated peptide-exchange process to generate panels of tetrameric peptide-MHC complexes50. In one study, TILs generated from a tumor resected from a patient with melanoma who subsequently exhibited a partial response to ipilimumab (monoclonal antibody to CTLA-4) were screened for their ability to bind to a library of tetramers containing candidate neoepitopes in conjunction with an algorithm to identify neoepitopes peptides that were potentially able to bind to the patient’s HLA-A and HLA-B molecules32. Through this approach, T cells that recognized two neoepitopes were identified: one corresponded to approximately 3% of the cultured TILs, and a second corresponded to 0.003% of the cultured TILs. The frequency of cells targeting the dominant neoepitope underwent an increase of fivefold in the patient’s peripheral blood 1 month after treatment with ipilimumab, which suggested that they were involved in the tumor regression observed in that patient. Similarly, WES was carried out on eight patients with melanoma whose tumors expressed either one or two of the HLA-A class I molecules HLA-A*01:01, HLA-A*02:01 and HLA-A*11:01, followed by the use of an algorithm to predict the binding of peptides to MHC class I to identify candidate neoepitopes that were incorporated into tetramers through the use of ultaviolet-irradiation-mediated peptide exchange51. This approach led to the identification of T cells that recognized a total of nine distinct neoepitopes from five of the eight patients analyzed. Moreover, T cell populations reactive with eight of the nine epitopes identified as targets of TILs could be isolated and expanded from the patient’s peripheral blood before adoptive TIL transfer, at which time they represented between 0.002% and 0.4% of total peripheral blood T cells.
Through the use of an approach similar to that described above for the mouse tumor model system, coupling WES with mass spectrometry of peptides eluted from the tumor cell surface31, two neoepitopes were identified from a cultured melanoma cell line, one of which was strongly recognized by autologous cultured TILs52.
While the algorithms used for identifying candidate peptides able to bind to MHC class I molecules were helpful in the studies cited above, they were not robust enough to allow accurate identification of minimal epitopes that bound to infrequently expressed human MHC class I allelic products for which binding data is limited, or to human MHC class II molecules, which limits the comprehensive identification of cancer antigens. In an attempt to address this issue, an alternative approach was developed that simultaneously evaluated the reactivity of CD4+ and CD8+ T cells to any mutant peptide presented by any of the patient’s MHC class I and class II molecules without the need for any epitope predictions. In this approach (Fig. 1), next-generation sequencing of cancerous and normal tissue from the same patient was performed to identify all non-synonymous somatic mutations present in patient’s tumor samples. Minigene constructs were then designed and synthesized to encode each mutated codon plus the 12 additional upstream and downstream codons flanking the mutation, corresponding to a 25-amino-acid peptide containing the sequence of all possible 8- to 12-amino-acid peptides that included the mutant amino acid. In general, between 6 and 24 minigenes were then linked into tandem minigenes (TMGs) in a single open reading frame, and autologous antigen-presenting cells, such as dendritic cells or B cells, were transfected with in-vitro-transcribed RNA generated from the TMGs, which allowed the processing and presentation of all mutant peptides in the context of the patient’s own MHC class I and II molecules. In addition, pools of 25-amino-acid peptides, in which each peptide contained the mutant amino acid flanked on both sides by the 12 normal amino acids, were pulsed on autologous antigen-presenting cells. Transfected and peptide-pulsed antigen-presenting cells were then evaluated for their ability to stimulate patient T cells, and positive TMGs or peptide pools were then deconvoluted for the identification of the specific neoantigen recognized. Through this approach, the TMGs or the peptide pools representing all expressed cancer mutations served as an avatar of the tumor and obviated the need to use autologous tumor cell lines, which are difficult to generate from the majority of cancer types. As technologies improve, it might be possible to obtain robust WES data from circulating tumor DNA (liquid biopsy) or single circulating tumor cells from blood, which would allow a relatively non-invasive method with which to identify somatic mutations expressed by all tumor lesions from a patient.
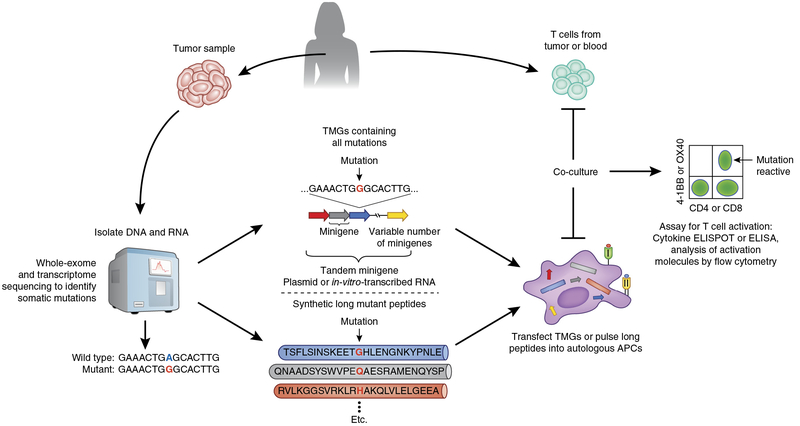
Identification of neoantigen-reactive T cells from patients with cancer. Next-generation sequencing (whole exome and whole transcriptome) is performed on tumor and matched normal cells to identify non-synonymous somatic mutations expressed by the cancer (left). Next, two approaches that do not rely on predictions of HLA–peptide binding can be used to investigate the reactivity of T cells to neoantigens encoded by the identified mutations. In the first approach (middle), minigenes encoding the mutation flanked by nucleotides encoding 12 amino acids from the wild-type gene can be synthesized in tandem to create TMG constructs, which are then cloned into an appropriate expression vector. Linking multiple minigenes in tandem allows a relatively large number of mutations to be evaluated at once. Plasmids encoding TMGs or TMG RNAs transcribed in vitro are then introduced into the appropriate antigen-presenting cells (APCs), such as autologous dendritic cells or B cells, through techniques such as electroporation or lipid-based transfection, to allow processing and presentation of the neoantigens in the context of the patient’s own HLA class I and II molecules. T cells derived from tumor (TILs) or from the blood (right) are then co-cultured with the antigen-presenting cells expressing the TMGs, and T cell reactivity is evaluated by immunological methods such as cytokine ELISPOT or ELISA or the analysis of T cell–activation molecules such as CD137 (4–1BB) or CD134 (OX40) by flow cytometry (far right). The second approach (bottom) is identical to the first approach, except that instead of genetic constructs encoding the mutations, long peptides containing the mutant amino acid flanked by 12 amino acids from the wild-type protein are synthesized and then pulsed onto antigen-presenting cells, which process and present the mutant peptides to T cells. Similar to the minigenes and TMG concept, in this approach, a variable number of individual long peptides can be combined to generate peptide pools, which increases the throughput of neoantigen screening.
Through the use of the techniques noted above, 75 neoantigens have been identified that are recognized by autologous TILs or peripheral lymphocytes from 29 of 31 patients with melanoma restricted by a wide variety of MHC class I and class II molecules44–47,51,53–55 (Table 1 and data not shown). None of the immunogenic mutant antigens were shared among patients; each neoantigen was unique to the autologous patient. The epitopes identified thus far were derived from a wide array of expressed genes with no clear association with a single recognized pathway, which indicates that almost any mutant intra-cellular protein can potentially serve as a cancer antigen in patients with melanoma.
Table 1
Neoantigens recognized by T cells from patients with melanoma
| Patient | Immunogenic neoantigens | HLA restriction element | Patient | Immunogenic neoantigens | HLA restriction element |
|---|---|---|---|---|---|
| 1 | 1 | DRB1*01:01 | 18 | 1 | B*51:01 |
| 2 | 1 | A*2402 | 18 | 1 | C*14:02 |
| 2 | 1 | DRB1*15:02 | 18 | 1 | B*44:02 |
| 3 | 1 | DRB1*04:01 | 18 | 1 | A*02:01 |
| 4 | 1 | A*01:01 | 18 | 1 | A*03:01 |
| 5 | 1 | DRB1*01:01 | 18 | 1 | Unknown class II |
| 6 | 1 | DRB1*01:01 | 19 | 1 | A*02:01 |
| 7 | 1 | A*02:01 | 20 | 3 | A*02:01 |
| 8 | 1 | HLA-A11 (mutant) | 20 | 6 | A*29:02 |
| 8 | 1 | A*11:01 | 20 | 2 | B*44:03 |
| 9 | 4 | A*02:01 | 21 | 2 | B*15:01 |
| 10 | 1 | A*02:01 | 22 | 4 | B*07:02 |
| 11 | 1 | A*02:05 | 23 | 2 | A*01:01 |
| 12 | 2 | A*01:01 | 24 | 1 | A*02:01 |
| 12 | 1 | A*26:01 | 25 | 2 | Unknown class I |
| 13 | 1 | A*01:01 | 26 | 3 | B*38:01 |
| 13 | 1 | A*02:01 | 27 | 1 | A*01:01 |
| 14 | 1 | C*07:01 | 27 | 1 | A*30:01 |
| 15 | 1 | Unknown class II | 27 | 1 | Unknown class II |
| 16 | 2 | A*11:01 | 28 | 1 | A*01:01 |
| 17 | 3 | A*02:01 | 28 | 1 | A*30:02 |
| 17 | 2 | B*39:01 | 28 | 1 | B*15:01 |
| 17 | 1 | B*44:03 | 28 | 1 | C*03:03 |
| 29 | 1 | A*02:01 | |||
| 29 | 4 | Unknown class I |
Total HLA class I neoantigens, 67; total HLA class II neoantigens, 8.
Human epithelial cancers and neoantigen-reactive T cells
Although cancer immunotherapies can mediate durable regression in some patients with metastatic melanomas, the large majority of patients with common epithelial cancers, which account for approximately 90% of cancer deaths in the USA, do not respond to immuno-therapies now in use. The correlation between mutation or neoepitope load and clinical benefit after immunotherapy suggests that the low response rate observed in many types of epithelial cancer might be due in part to the low frequency or absence of neoantigen-reactive T cells in these patients, due to the lower average number of mutations in these cancers56. Findings obtained from patients with melanoma, however, have raised the possibility that neoepitope reactivity, if observed in these cancers, might provide an opportunity for the development of effective immunotherapy for patients with additional cancer types.
Multiple studies carried out over the past two decades have demonstrated the presence of neoepitope-reactive T cells in patients with common epithelial malignancies, such as lung cancer37,57–60, bladder cancer61, head and neck cancer62,63, ovarian cancer64 or pancreatic cancer65, and in patients whose cancers have DNA-mismatch-repair deficiencies66. Most of the immunogenic neoepitopes identified were derived from individual case studies of patients with cancer and thus it was unclear whether or not most patients with common epithelial cancers harbor neoepitope-reactive T cells. In an attempt to address this issue, TILs cultured from a panel of metastatic cancers of the gastrointestinal tract that included esophageal, colon, pancreatic, gastric and bile-duct tumors were screened for their ability to recognize mutated TMGs and mutant peptides identified by WES of fresh autologous tumors (Fig. 1). This analysis led to the identification of CD4+ and/or CD8+ T cells that targeted 73 somatic neoantigens expressed by autologous tumors in 31 of 35 (89%) patients that have been evaluated so far67,68 (Table 2 and data not shown). The neoantigens identified in each patient were unique, with the exception of an identical neoepitope encoded by the KRASG12D hotspot-driver mutation that was recognized by CD8+ T cells from two patients with colorectal cancer67,68. Five of the five KRASG12D-specific CD8+ TCRs isolated from the two patients’ TILs were HLA-C*08:02 restricted and, intriguingly, the TCR α-chain variable sequence in one patient was identical to two of four of the KRASG12D-reactive TCRs in the second patient67,68. Together these observations indicate that most patients with common gastrointestinal cancers have T cells that target unique somatic neoantigens and suggest that immunotherapeutic strategies aimed at harnessing neoantigen-reactive T cells might represent a viable treatment option for many patients with common epithelial cancers.
Table 2
Neoantigens recognized by T cells from patients with epithelial gastrointestinal cancer
| Patient | Cancer type | Immunogenic neoantigens | HLA restriction element | Patient | Cancer type | Immunogenic neoantigens | HLA restriction element |
|---|---|---|---|---|---|---|---|
| 1 | Esophagus | 3 | Unknown class II | 16 | Colorectum | 2 | Unknown class I |
| 2 | Esophagus | 2 | Unknown class I | 17 | Colorectum | 2 | Unknown class I |
| 2 | Esophagus | 5 | Unknown class II | 18 | Colorectum | 1 | Unknown class I |
| 3 | Colorectum | 2 | Unknown class I | 18 | Colorectum | 1 | Unknown class II |
| 4 | Colorectum | 2 | Unknown class I | 19 | Colorectum | 1 | Unknown class I |
| 4 | Colorectum | 1 | Unknown class II | 20 | Colorectum | 2 | Unknown class I |
| 5 | Colorectum | 1 | C*08:02 | 20 | Colorectum | 1 | Unknown class II |
| 5 | Colorectum | 2 | Unknown class I | 21 | Pancreas | 1 | Unknown class I |
| 6 | Colorectum | 1 | A:03:01 | 22 | Pancreas | 2 | Unknown class II |
| 6 | Colorectum | 1 | Unknown class I | 23 | Pancreas | 1 | Unknown class II |
| 7 | Colorectum | 3 | Unknown class I | 24 | Pancreas | 2 | Unknown class II |
| 8 | Colorectum | 1 | Unknown class I | 25 | Pancreas | 1 | Unknown class I |
| 9 | Colorectum | 3 | Unknown class I | 26 | Bile duct | 1 | Unknown class I |
| 10 | Colorectum | 2 | Unknown class II | 27 | Bile duct | 1 | DQB1*06:01 |
| 11 | Colorectum | 2 | Unknown class I | 28 | Bile duct | 1 | Unknown class II |
| 12 | Colorectum | 2 | Unknown class I | 29 | Bile duct | 2 | A*02:01 |
| 12 | Colorectum | 1 | Unknown class I | 29 | Bile duct | 1 | Unknown class I |
| 13 | Colorectum | 1 | C*08:02 | 30 | Bile duct | 2 | Unknown class I |
| 13 | Colorectum | 2 | Unknown class I | 30 | Bile duct | 2 | Unknown class II |
| 14 | Colorectum | 2 | Unknown class II | 31 | Bile duct | 4 | Unknown class I |
| 15 | Colorectum | 1 | Unknown class I | 31 | Bile duct | 1 | Unknown class II |
| 15 | Colorectum | 1 | Unknown class II |
Total HLA class I neoantigens, 46; total HLA class II neoantigens, 27.
The polyclonal nature of the lymphocyte populations in the studies described above made it difficult to assign tumor regression to individual antigen reactivities. No objective responses were seen in 15 patients with a variety of gastrointestinal cancers treated with unselected autologous TILs (data not shown). However, 2 of 12 patients with metastatic gastrointestinal cancers treated with T cells that targeted mainly a single neoantigen expressed by the autologous tumor67,68 (data not shown) demonstrated objective clinical responses.
The first patient, whose metastatic bile-duct cancer contained 26 non-synonymous somatic mutations, exhibited disease stabilization for about 1 year following the transfer of 42 billion autologous TIL that were retrospectively determined to contain approximately 25% CD4+ T helper type 1 cells targeting a neoepitope derived from the putative tumor suppressor ERBB2IP. Upon disease progression, the patient was treated with a second infusion product that contained 126 billion T cells, approximately 95% of which recognized the ERBB2IP neoantigen. This patient then experienced a substantial regression of lung and liver metastases that lasted 35 months. This response was associated with greater in vivo persistence of the mutant-ERBB2IP-neoepitope-reactive T cells than such persistence after the first treatment69 (data not shown). These findings provided the first direct evidence that transfer of a highly enriched population of neoantigen-reactive T cells can mediate the regression of metastatic human cancer. In addition, although most cancer immunotherapies have emphasized the harnessing of tumor-reactive cytotoxic CD8+ T cells, these findings indicated that neoantigen-reactive, HLA-class-II-restricted CD4+ cells can mediate the regression of human tumors.
The second patient who responded to adoptive TIL therapy targeting a cancer neoantigen was one of the two patients with metastatic colorectal cancer whose T cells recognized the KRASG12D neoantigen in the context of HLA-C*08:02. The transfer of 148 billion T cells, approximately 75% of which targeted KRASG12D, was associated with the regression of all seven metastatic lung lesions and a relatively high persistence of T cells reactive to KRASG12D (ref. 68); however, a single lung lesion progressed 9 months after the transfer of TILs. This lesion was resected, and the patient remains clinically disease free 7 months later. Genomic analysis of the progressing lesion revealed the presence of tumor cells that had lost the copy of chromosome 6 encoding the HLAC*08:02 restriction element required for recognition by the KRASG12D- reactive T cells, which provided evidence for the immunoselection of a resistant tumor clonotype in response to TIL therapy. Nevertheless, the tumor regression observed following the administration of highly enriched populations of neoepitope-reactive T cells to these two patients provided evidence that both CD4+ T cells and CD8+ T cells targeting mutant antigens can mediate substantial clinical benefit to patients with metastatic cancer. Moreover, these responses occurred in the absence of any major toxicities, which highlights the idea that harnessing the T cell response to tumor-specific neoantigens will probably be safe.
Future approaches to targeting unique cancer neoantigens
All cancers contain mutant proteins that are potential targets of immunotherapy. Some cancer types, mainly those associated with known environmental carcinogens, such as melanoma and smoking-induced lung cancer, have higher median numbers of mutations than those of most common epithelial cancers. Although there would appear to be a greater chance of generating neoepitopes able to bind to autologous cell-surface MHC molecules in tumors with relatively high mutation rates, therapeutically relevant neoantigen-reactive T cells were generated in a patient with cholangiocarcinoma whose tumor, as noted above, harbored only 26 mutations. Nevertheless, novel approaches might be needed to enhance the low response rates to adoptive immunotherapy observed in patients bearing gastrointestinal tumors and to apply those therapies to patients bearing other common epithelial tumor types (Box 1). Optimal mutant antigens to target are those presumed to be vital to sustaining the malignant phenotype of the cells, such as driver mutations in KRAS, which are among the most common hotspot mutations involved in oncogenesis. Driver mutations are also more likely to be expressed by most, if not all, cancer cells, which further makes them attractive therapeutic targets; however, many epitopes encoded by driver mutations might not be naturally immunogenic (i.e., they might not be processed and/or bound to the patient’s autologous MHC molecules) and therefore might not give rise to neoantigen-reactive T cells. Moreover, tumor cells can escape immunological recognition by T cells reactive with an individual neoepitope through a variety of mechanisms that include loss of antigen expression and loss of heterozygosity at the HLA locus. Thus, successful immunotherapy might require the simultaneous harnessing of multiple T cell populations able to target multiple neoantigens expressed on different MHC molecules to counteract the extensive heterogeneity of tumor genomes and the consequent mechanisms by which human cancers escape the immune system.
The efficacy of adoptive transfer of TILs targeting neoepitopes might potentially be enhanced by combination with immunomodulators such as immunological checkpoint inhibitors, agonistic antibodies to T cell costimulatory molecules, neoepitope-targeted vaccines, or therapies that lead to the activation of other inflammatory immuno-logical effector cells in the tumor. An inherent limitation of the use of neoantigen-reactive TILs, however, is that they often represent highly differentiated effector cells with a limited proliferative capacity and ability to persist in vivo following adoptive transfer. Preclinical cancer models of ACT have clearly demonstrated that less-differentiated T cells engraft better than more-differentiated T cells and mediate superior anti-tumor responses than those of more-differentiated T cells70. If that observation holds true in humans, then the adoptive transfer of less-differentiated neoantigen-reactive T cells would be expected to improve therapeutic efficacy. This can be achieved through a personalized TCR-gene-therapy approach whereby genes encoding neoantigen-reactive TCRs are introduced into autologous, less-differentiated naive or central memory T cells, derived from blood followed by the adoptive transfer of these cells back into the patient. Alternative approaches for the generation of less-differentiated T cells include the culture of neoantigen-reactive TILs or peripheral blood T cells with small-molecule inhibitors such as an inhibitor of the kinase AKT71,72 or cytokines such as IL-21 (ref. 73) that can partially preserve the differentiation state of the T cells during population expansion. Alternatively, the introduction of DNA or RNA encoding transcription factors such as MYC, OCT3-OCT4, SOX2, and KLF4 to reprogram mature cells into pluripotent stem cells also holds promise for the generation of less-differentiated T cells from highly differentiated effector T cells74. The process of generating T cells for adoptive transfer also allows the unique opportunity to carry out additional manipulations of the patient’s T cells ex vivo before cell transfer. Gene-editing technologies such as ZFNs, TALENS and CRISPR-Cas9 could be used to specifically inactivate inhibitory genes and/or to introduce genes that enhance the effector function or survival of neoantigen-reactive T cells to potentially improve their in vivo efficacy. Studies have demonstrated that ZNF-mediated inactivation of the gene encoding PD-1 enhances in vitro the effector function of T cells from human melanoma TILs, which would theoretically render these cells resistant to inhibition mediated by the PD-1 ligands PD-L1 and PD-L2 in vivo75.
Cell-transfer immunotherapy approaches that target unique mutant neoantigens present challenges for the application of this treatment to large numbers of patients with cancer, since this therapy is highly personalized and cell manufacturing is a relatively complex process. However, multiple approaches for the commercialization of cell therapy are in progress that use a model in which tumor and/or lymphocytes are sent to a central facility that prepares the therapeutic cells for delivery to the primary site of care. Indeed, T-cell-transfer therapy targeting the shared antigen CD19 expressed by malignant B cells is on the verge of commercialization, which demonstrates that the generation of defined T cell products for the treatment of many patients with cancer is probably feasible.
Vaccine-based approaches that use patient-specific mutant peptides or minigenes encoding mutant epitopes are also being investigated in several ongoing clinical trials. In one report, three patients with metastatic melanoma were treated with a dendritic-cell vaccine loaded with mutant peptides specific for the patient’s cancer, and an increase in neoantigen-specific T cells was detected in these patients, although therapeutic effect could not be evaluated in this study76. Vaccination against mutant neoepitopes could also be used to potentiate the immune response of adoptively transferred T cells or cells activated through immunological checkpoint blockade.
Concluding remarks
The first report of T cell reactivity to a mutant epitope in human cancer was published over two decades ago77, but the role of neoantigen-reactive T cells in endogenous therapeutic anti-tumor responses has been appreciated only recently. High-throughput next-generation sequencing technologies have enabled the efficient investigation of T cell reactivity to the tumor ‘mutanome’ (all mutations in the tumor); this has revealed that most patients with melanoma and epithelial cancers mount immune responses to neoantigens. Correlative studies have demonstrated that patients with a higher mutation load are more likely to respond to immunological checkpoint inhibitors, while studies of the transfer of highly enriched populations of neoantigen-reactive T cells have provided direct evidence that these cells can indeed mediate tumor regression. Thus, it appears that the targeting of cancer neoantigens by T cells might represent the ‘final common pathway’ that results in cancer regression in response to a variety of cancer immunotherapies, which suggests that effectively harnessing this pathway holds promise for improving clinical outcomes in patients with metastatic cancers.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ni.3682
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6295671?pdf=render
Citations & impact
Impact metrics
Article citations
Consensus, debate, and prospective on pancreatic cancer treatments.
J Hematol Oncol, 17(1):92, 10 Oct 2024
Cited by: 0 articles | PMID: 39390609 | PMCID: PMC11468220
Review Free full text in Europe PMC
Natural TCRs targeting KRASG12V display fine specificity and sensitivity to human solid tumors.
J Clin Invest, 134(21):e175790, 17 Sep 2024
Cited by: 0 articles | PMID: 39287991 | PMCID: PMC11529987
Identification and validation of tumor-specific T cell receptors from tumor infiltrating lymphocytes using tumor organoid co-cultures.
Cancer Immunol Immunother, 73(9):164, 02 Jul 2024
Cited by: 0 articles | PMID: 38954022 | PMCID: PMC11219989
Downregulation of HNRNPA1 induced neoantigen generation via regulating alternative splicing.
Mol Med, 30(1):85, 12 Jun 2024
Cited by: 1 article | PMID: 38867190
Immunogenicity of Non-Mutated Ovarian Cancer-Specific Antigens.
Curr Oncol, 31(6):3099-3121, 30 May 2024
Cited by: 0 articles | PMID: 38920720 | PMCID: PMC11203340
Go to all (256) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The present status and future prospects of peptide-based cancer vaccines.
Int Immunol, 28(7):319-328, 28 May 2016
Cited by: 71 articles | PMID: 27235694
Review
Targeting neoantigens for cancer immunotherapy.
Int Immunol, 28(7):365-370, 19 May 2016
Cited by: 27 articles | PMID: 27208041 | PMCID: PMC5007611
Review Free full text in Europe PMC
Cancer treatment: The killer within.
Nature, 508(7494):24-26, 01 Apr 2014
Cited by: 29 articles | PMID: 24695297
Neoantigen heterogeneity: a key driver of immune response and sensitivity to immune checkpoint blockade?
Immunotherapy, 8(7):763-766, 01 Jun 2016
Cited by: 7 articles | PMID: 27349975
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: Z01 BC010984-01





