Abstract
Free full text

Opinion: Genetic Conflict With Mobile Elements Drives Eukaryotic Genome Evolution, and Perhaps Also Eukaryogenesis
Abstract
Through analyses of diverse microeukaryotes, we have previously argued that eukaryotic genomes are dynamic systems that rely on epigenetic mechanisms to distinguish germline (i.e., DNA to be inherited) from soma (i.e., DNA that undergoes polyploidization, genome rearrangement, etc.), even in the context of a single nucleus. Here, we extend these arguments by including two well-documented observations: (1) eukaryotic genomes interact frequently with mobile genetic elements (MGEs) like viruses and transposable elements (TEs), creating genetic conflict, and (2) epigenetic mechanisms regulate MGEs. Synthesis of these ideas leads to the hypothesis that genetic conflict with MGEs contributed to the evolution of a dynamic eukaryotic genome in the last eukaryotic common ancestor (LECA), and may have contributed to eukaryogenesis (i.e., may have been a driver in the evolution of FECA, the first eukaryotic common ancestor). Sex (i.e., meiosis) may have evolved within the context of the development of germline–soma distinctions in LECA, as this process resets the germline genome by regulating/eliminating somatic (i.e., polyploid, rearranged) genetic material. Our synthesis of these ideas expands on hypotheses of the origin of eukaryotes by integrating the roles of MGEs and epigenetics.
Overview
Based on observations of dynamic genomes (i.e., cyclical polyploidy, genome rearrangements) in diverse eukaryotic lineages, we have previously argued that last eukaryotic common ancestor (LECA) used epigenetic mechanisms to distinguish germline from somatic DNA, even in the context of a single nucleus (McGrath and Katz 2004; Zufall et al. 2005; Parfrey et al. 2008; Parfrey and Katz 2010; Maurer-Alcala and Katz 2015; Weiner et al. 2020). As discussed in this series of papers from our lab, examples of such germline/soma distinctions include: sequestered germline nuclei in animals, ciliates, and some foraminifera; cyclical polyploidization throughout life cycles of apicomplexans such as Plasmodium (the causative agent of malaria); generation of extrachromosomal DNA, including amplification of ribosomal RNA loci in many eukaryotes; developmentally regulated genome rearrangements, for example, in trypanosomes and immune cells of vertebrates (i.e., V(D)J recombination); and even the mis-regulation of DNA through polyploidization in cancer cells (Erenpreisa et al. 2017). Despite this long list, examples of genome dynamics in diverse lineages of eukaryotic microbes are still limited as the bulk of their life cycle data come from a small number of model lineages (e.g., Tetrahymena, Plasmodium). However, promising recent evidence of chromatin extrusion and depolyploidization in Amoeba proteus (Goodkov et al. 2020) suggests that more examples of such dynamics are on the horizon.
We have also argued that germline/soma distinctions in eukaryotes are regulated by epigenetic tools including histone modification, DNA methylation, and scanning by small nonprotein-coding RNAs (Zufall et al. 2005; Parfrey et al. 2008; Parfrey and Katz 2010; Maurer-Alcala and Katz 2015; Weiner et al. 2020). Here, we extend this hypothesis by combining it with two observations: (1) the widespread occurrence of mobile genetic elements (MGEs) (e.g., transposable elements [TEs], viruses) and (2) data on the epigenetic regulation of MGEs within eukaryotes. Synthesis of these observations leads to the hypothesis that genetic conflict has shaped the evolution of eukaryotic genomes and, as others have also argued (e.g., Aravind et al. 2012; Koonin 2017; Massey and Mishra 2018; Havird et al. 2019), perhaps the evolution of eukaryotes themselves.
MGEs are Widespread
The function and abundance of MGEs such as viruses and TEs has been extensively reviewed, and we provide only a few highlights here. TEs are present in genomes across the tree of life (e.g., Kidwell and Lisch 2001; Suzuki and Bird 2008; Kejnovsky et al. 2012; Campbell et al. 2017) and can constitute more than half the genome of many eukaryotic lineages (e.g., Kazazian 2004; Fedoroff 2012; Song and Schaack 2018). Viruses are the most abundant biological entities on Earth (e.g., Edwards and Rohwer 2005; Koonin 2017), and, like TEs, they are able to integrate into eukaryotic genomes (Chalker and Yao 2011; Koonin 2017; Song and Schaack 2018).
Though early studies characterized MGEs as “parasitic” and/or “selfish” because of the harm they can cause to host genomes, it is now clear that MGEs also generate novel genetic variation that can be the source of adaptation (e.g., Fedoroff 2012; Koonin and Krupovic 2018). Some of the damage TEs can cause includes mutations, DNA breaks, and rearrangement of chromosomes as they move through host genomes (e.g., Kazazian 2004; Fedoroff 2012; Parhad and Theurkauf 2019). Similarly, rapid evolution and replication of viruses create an “arms race” with the host genomes that have evolved to eliminate them (e.g., Bruscella et al. 2017; Koonin and Krupovic 2018). Consequently, replication and mobilization of MGEs is a substantial source of genetic variation in eukaryotes, and these abilities allow MGEs to both resist elimination and create an immediate and lasting impact on host evolution (e.g., Kidwell and Lisch 2001; Schaack et al. 2010; Campbell et al. 2017; Koonin and Krupovic 2018).
MGEs are Regulated by Epigenetics
Epigenetic mechanisms are key to eukaryotic responses to MGEs (e.g., Levine et al. 2016; Campbell et al. 2017; Song and Schaack 2018; Parhad and Theurkauf 2019). In many cases, epigenetic responses protect the host’s germline by limiting TE mobilization (Chung et al. 2008; Suzuki and Bird 2008; Parhad and Theurkauf 2019). Drosophila exemplify this through expansion of the HP1D gene family, which silences TEs in the female germline (Levine et al. 2016). While under epigenetic regulation, TEs display a spectrum of fitness effects within host genomes from parasitism to mutualism (Kidwell and Lisch 2001; Vogt et al. 2013; Cosby et al. 2019). This relationship can also change over time as the epigenetic systems that regulate them may evolve such that transposons ultimately become domesticated (e.g., neutral or used for host function, Kidwell and Lisch 2001; Vogt et al. 2013; Piegu et al. 2015; Cosby et al. 2019; Doyle and Coate 2019).
Epigenetic mechanisms can also regulate viruses within eukaryotic genomes. Endogenous retroviruses, like TEs, occur at various levels of mobility and can be epigenetically regulated via processes like histone methylation (Manghera and Douville 2013; Collins et al. 2015; Meyer et al. 2017). Viruses have also been observed to regulate their replication cycles through epigenetic mechanisms of their own (Woellmer and Hammerschmidt 2013; Balakrishnan and Milavetz 2017; Bruscella et al. 2017). The human Epstein–Barr herpesvirus represents one such intimate relationship, as the latent virus is restrained by Polycomb proteins, but in the lytic replication stage, when Polycomb repression is erased, the virus escapes from the methylation network of the host (Woellmer and Hammerschmidt 2013). This type of multilayered epigenetic relationship reflects the complexity of interactions between viral replication systems and eukaryotic hosts.
Genetic Conflict is Foundational to Eukaryotic Genome Evolution, and Perhaps Eukaryogenesis
The widespread occurrence and epigenetic regulation of MGEs engender the hypothesis that genetic conflict between host and MGEs led to the evolution of a dynamic eukaryotic genome that distinguishes germline and soma (Figure 1). Genetic conflict, the competitive relationship between MGEs and host genomes, has been well described as a driving force of evolutionary change (e.g., Hurst et al. 1996; Werren 2011; McLaughlin and Malik 2017; Massey and Mishra 2018; Song and Schaack 2018). Hurst et al. (1996) argued for a “gene’s-eye view” of such conflict to describe the strategies that MGEs and hosts deploy in the struggle over inheritance and proliferation. Nearly two decades later, Song and Schaack (2018) provide an extensive review on the nature of genetic conflict between hosts and MGEs and the possible mechanisms of resolution. In light of this conflict, we and others (e.g., Aravind et al. 2012; Fedoroff 2012; Koonin 2017) propose that epigenetic mechanisms resulting from interactions with MGEs were likely fundamental to eukaryotic evolution. Indeed, the genetic mechanisms that underlie epigenetic regulation (i.e., the epigenetic toolkit) clearly predate the evolution of eukaryotes (e.g., Oliverio and Katz 2014; Weiner et al. 2020), though the specific machinery may have been replaced and/or elaborated on over time (Maurer-Alcala and Katz 2015). Here, we expand on these ideas by linking them explicitly to the origin of germline–soma distinctions during eukaryogenesis.
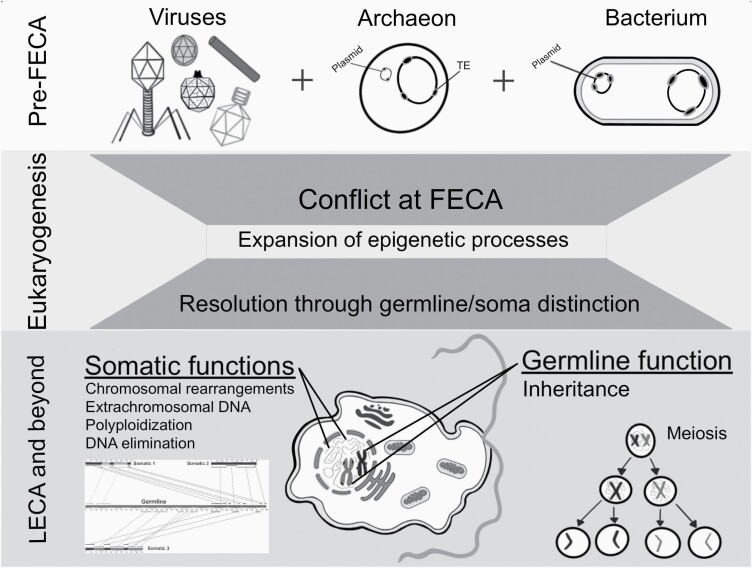
Genetic conflict during eukaryogenesis resulted in epigenetically regulated germline–soma distinctions in eukaryotes. This figure depicts the players at the origin of eukaryotes, namely the diversity of viruses and the presence of TEs integrated within both bacteria, including the ancestor of mitochondria, and archaea, including the likely host cell of FECA (top panel). Conflict among these genomes and mobile genetic elements (MGEs; middle panel) resulted in eukaryotes that distinguish germline (i.e., marked for inheritance, capable of meiosis to reset genome, represented by the condensed chromosomes in LECA) and somatic (e.g., cyclical polyploidy, extrachromosomal DNA, developmentally regulated genome rearrangements, DNA elimination, represented by the thinner lines within the nucleus of LECA) material (bottom panel). The inset under the somatic functions in LECA represents three somatic chromosomes generated from a single germline region in the ciliate Chilodonella uncinata (redrawn from Gao et al. 2015). Additional details and references can be found in the text.
Consistent with the idea that genetic conflict between host and MGEs specifically led to distinction of germline and somatic genome material are observations on the differential epigenetic regulation of MGEs in extant eukaryotic lineages. For example, flowering plant pollen possesses the ability to epigenetically regulate and de-regulate transcription of TEs in a cyclical manner (Slotkin et al. 2009). In animals like Drosophila, TEs are silenced in the germline through female-specific RNA silencing mechanisms (Levine et al. 2016), while a different set of small interfering RNAs regulate TEs in the soma (Chung et al. 2008). In the nematode Caenorhabditis elegans, piRNA epigenetic silencing networks suppress TE mobility in the germline, and this silencing can be inherited across more than 20 generations (Ashe et al. 2012). In ciliates, epigenetic mechanisms including small RNAs and transposases co-opted from transposons are used to shape somatic genomes following conjugation (e.g., Chalker and Yao 2011; Bracht et al. 2013; Maurer-Alcala and Nowacki 2019). The observation of differential epigenetic regulation of MGEs between germline and somatic nuclei in diverse extant eukaryotes raises the possibility that such a mechanism was present in LECA and perhaps even FECA (the first eukaryotic common ancestor).
A special case of conflict at the origin of eukaryotes stems from the acquisition of mitochondria, an event extensively reviewed in the literature (though there remain debates on the timing and physiology of the events; e.g., Pittis and Gabaldon 2016; Lopez-Garcia et al. 2017; Martin 2017; Gabaldon 2018; Lopez-Garcia and Moreira 2019; Wein et al. 2019). At the time of the acquisition of mitochondria, the chimeric cell had to navigate two distinct genomes in a shared cytoplasm. Certainly, there is evidence of conflict between mitochondria and nuclei of extant organisms; for example, in humans, nucleocytoplasmic conflict can lead to disease (e.g., Cummins 2001; Havird et al. 2019) and there are data indicating epigenetic interactions between mitochondria and nuclei (Harvey 2019). Hence, it is possible that conflict from a single but significant “mobile” event, the acquisition of an alphaproteobacterial symbiont in FECA, contributed to the invasion/expansion of MGEs (Krupovic and Koonin 2015) and ultimately the evolution of eukaryotic genome structures.
We suggest that eukaryogenesis resulted in the evolution of a genome that distinguishes germline from soma, which was fueled by genetic conflict between MGEs and hosts (Figure 1). Our hypothesis does not specify the timing of events between FECA and LECA, nor do we address the origin of the eukaryotic cytoskeleton, the synapomorphy of eukaryotes that allowed for the evolution of diverse morphologies and life histories. Instead, we suggest that germline–soma distinctions evolved as a response to genetic conflict with MGEs and contributed to the second major epoch of evolution, the origin of eukaryotes with meiotic sex, as described in Bonner (2019). Under such a scenario, the nucleus may have evolved to “protect” the genome from viruses (e.g., Bell 2009; Aravind et al. 2012; Forterre and Gaia 2016; Poole and Hendrickson 2019) or may have resulted from selection to separate transcription from translation, allowing for the excision of mobile elements (Martin and Koonin 2006; Brunk and Martin 2019). It may also be the case that the nuclear envelope is just a byproduct of events at the time (i.e., resulting from the chaos of the acquisition of mitochondria with its genome [including its own MGEs], or some other autogenous event).
Sex (i.e., meiosis and syngamy) is argued to be ancestral in eukaryotes based on the widespread distribution of meiotic genes coupled with other evidence (i.e., cell fusion, cryptic sexual cycles) in lineages previously thought to be asexual (Lahr et al. 2011; Tekle et al. 2017; Hofstatter et al. 2018) but see Maciver (2019). Kondrashov (1994, 1997) argued that meiosis evolved as a means to regulate cycles of polyploidy, which are part of what we refer to as somatic genome content (i.e., cyclical polyploidization, along with the generation of extrachromosomal DNA and developmental regulated rearrangements, all represented by the thin lines within the nucleus of LECA in Figure 1). In fact, Kondrashov (1994) suggested that sex may have evolved as a means for “orderly genetic reduction,” which would be required in novel eukaryotic lineages with complex genome dynamics (e.g., McGrath and Katz 2004; Zufall et al. 2005; Parfrey et al. 2008; Parfrey and Katz 2010; Maurer-Alcala and Katz 2015; Goodkov et al. 2020; Weiner et al. 2020). Despite open questions (e.g., on the timing of events, the origin of the nuclear envelope and cytoskeleton), we believe consideration of our hypothesis—that genetic conflict between host and MGEs at the time of the origin of eukaryotes led to dynamic genomes in which germline–soma distinctions are regulated by epigenetics and reset through meiosis—provides an important expansion on models of eukaryogenesis.
Acknowledgments
We thank Maria Orive (University of Kansas) for organizing the 2019 American Genetics Association symposium that inspired this manuscript and several members of the Katzlab for comments on earlier versions. We are grateful to Jailene C. Gonzalez for creating the illustration (Figure 1).
Funding
L.A.K. is supported by grants a National Institute of Health award (R15HG010409) and three awards from the National Science Foundation (OCE-1924570, DEB-1651908, and DEB-1541511).
References
- Aravind L, Anantharaman V, Zhang DP, De Souza RF, Iyer LM. 2012. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front Cell Infect Microbiol. 2:21. [Europe PMC free article] [Abstract] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. . 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 150:88–99. [Europe PMC free article] [Abstract] [Google Scholar]
- Balakrishnan L, Milavetz B. 2017. Epigenetic regulation of viral biological processes. Viruses. 9:346. [Europe PMC free article] [Abstract] [Google Scholar]
- Bell PJL. 2009. The viral eukaryogenesis hypothesis: a key role for viruses in the emergence of eukaryotes from a prokaryotic world environment. In: Witzany G, editor. Natural genetic engineering and natural genome editing (Annals of the New York Academy of Sciences). Vol. 1178. Oxford, England: Blackwell Publishing. p. 91–105. [Abstract] [Google Scholar]
- Bonner JT. 2019. The evolution of evolution. J Exp Zool B Mol Dev Evol. 332:301–306. [Abstract] [Google Scholar]
- Bracht JR, Fang WW, Goldman AD, Dolzhenko E, Stein EM, Landweber LF. 2013. Genomes on the edge: programmed genome instability in ciliates. Cell. 152:406–416. [Europe PMC free article] [Abstract] [Google Scholar]
- Brunk CF, Martin WF. 2019. Archaeal histone contributions to the origin of eukaryotes. Trends Microbiol. 27:703–714. [Abstract] [Google Scholar]
- Bruscella P, Bottini S, Baudesson C, Pawlotsky JM, Feray C, Trabucchi M. 2017. Viruses and miRNAs: more friends than foes. Front Microbiol. 8:824. [Europe PMC free article] [Abstract] [Google Scholar]
- Campbell S, Aswadl A, Katzourakis A. 2017. Disentangling the origins of virophages and polintons. Curr Opin Virol. 25:59–65. [Abstract] [Google Scholar]
- Chalker DL, Yao MC. 2011. DNA elimination in ciliates: transposon domestication and genome surveillance. In: Bassler BL, Lichten M, Schupbach G, editors. Annual review of genetics. Vol. 45. Palo Alto, CA: Annual Reviews. p. 227–246. [Abstract] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 18:795–802. [Europe PMC free article] [Abstract] [Google Scholar]
- Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. 2015. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc Natl Acad Sci USA. 112:8367–8372. [Europe PMC free article] [Abstract] [Google Scholar]
- Cosby RL, Chang NC, Feschotte C. 2019. Host-transposon interactions: conflict, cooperation, and cooption. Genes Dev. 33:1098–1116. [Europe PMC free article] [Abstract] [Google Scholar]
- Cummins JM. 2001. Mitochondria: potential roles in embryogenesis and nucleocytoplasmic transfer. Hum Reprod Update. 7:217–228. [Abstract] [Google Scholar]
- Doyle JJ, Coate JE. 2019. Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. Int J Plant Sci. 180:1–52. [Google Scholar]
- Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat Rev Microbiol. 3:504–510. [Abstract] [Google Scholar]
- Erenpreisa J, Salmina K, Belyayev A, Inashkina I, Cragg MS. 2017. Survival at the brink. In: Hayat MA, editor. Autophagy: cancer, other pathologies, inflammation, immunity, infection, and aging. Elsevier. p. 275–294. [Google Scholar]
- Fedoroff NV. 2012. Presidential address transposable elements, epigenetics, and genome evolution. Science. 338:758–767. [Abstract] [Google Scholar]
- Forterre P, Gaia M. 2016. Giant viruses and the origin of modern eukaryotes. Curr Opin Microbiol. 31:44–49. [Abstract] [Google Scholar]
- Gabaldon T. 2018. Relative timing of mitochondrial endosymbiosis and the “pre-mitochondrial symbioses” hypothesis. IUBMB Life. 70:1188–1196. [Europe PMC free article] [Abstract] [Google Scholar]
- Gao F, Roy SW, Katz LA. 2015. Analyses of alternatively processed genes in ciliates provide insights into the origins of scrambled genomes and may provide a mechanism for speciation. mBio. 6:e01998-14. [Europe PMC free article] [Abstract] [Google Scholar]
- Goodkov AV, Berdieva MA, Podlipaeva YI, Demin SY. 2020. The chromatin extrusion phenomenon in Amoeba proteus cell cycle. J Eukaryot Microbiol. 67:203–208. [Abstract] [Google Scholar]
- Harvey AJ. 2019. Mitochondria in early development: linking the microenvironment, metabolism and the epigenome. Reproduction. 157:R159–R179. [Abstract] [Google Scholar]
- Havird JC, Forsythe ES, Williams AM, Werren JH, Dowling DK, Sloan DB. 2019. Selfish mitonuclear conflict. Curr Biol. 29:R496–R511. [Europe PMC free article] [Abstract] [Google Scholar]
- Hofstatter PG, Brown M, Lahr DJG. 2018. Comparative genomics supports sex and meiosis in diverse amoebozoa. Genome Biol Evol. 10:3118–3128. [Europe PMC free article] [Abstract] [Google Scholar]
- Hurst LD, Atlan A, Bengtsson BO. 1996. Genetic conflicts. Q Rev Biol. 71:317–364. [Abstract] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science. 303:1626–1632. [Abstract] [Google Scholar]
- Kejnovsky E, Hawkins JS, Feschotte C. 2012. Plant transposable elements: biology and evolution. In: Wendel J, Greilhuber J, Dolezel J, Leitch IJ, editors. Plant genome diversity. Vol. 1. Wien: Springer-Verlag. p. 17–34. [Google Scholar]
- Kidwell MG, Lisch DR. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution. 55:1–24. [Abstract] [Google Scholar]
- Kondrashov AS. 1994. The asexual ploidy cycle and the origin of sex. Nature. 370:213–216. [Abstract] [Google Scholar]
- Kondrashov AS. 1997. Evolutionary genetics of life cycles. Ann Rev Ecol Syst. 28:391–435. [Google Scholar]
- Koonin EV. 2017. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol Direct. 12:5. [Europe PMC free article] [Abstract] [Google Scholar]
- Koonin EV, Krupovic M. 2018. The depths of virus exaptation. Curr Opin Virol. 31:1–8. [Abstract] [Google Scholar]
- Krupovic M, Koonin EV. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. 13:105–115. [Europe PMC free article] [Abstract] [Google Scholar]
- Lahr DJG, Parfrey LW, Mitchell EA, Katz LA, Lara E. 2011. The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proc Biol Sci. 278:2081–2090. [Europe PMC free article] [Abstract] [Google Scholar]
- Levine MT, Vander Wende HM, Hsieh E, Baker ECP, Malik HS. 2016. Recurrent gene duplication diversifies genome defense repertoire in Drosophila. Mol Biol Evol. 33:1641–1653. [Europe PMC free article] [Abstract] [Google Scholar]
- Lopez-Garcia P, Eme L, Moreira D. 2017. Symbiosis in eukaryotic evolution. J Theor Biol. 434:20–33. [Europe PMC free article] [Abstract] [Google Scholar]
- Lopez-Garcia P, Moreira D. 2019. Eukaryogenesis, a syntrophy affair. Nat Microbiol. 4:1068–1070. [Europe PMC free article] [Abstract] [Google Scholar]
- Maciver SK. 2019. Ancestral eukaryotes reproduced asexually, facilitated by polyploidy: a hypothesis. BioEssays. 41:1900152. 10.1002/bies.201900152. [Abstract] [CrossRef] [Google Scholar]
- Manghera M, Douville RN. 2013. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology. 10:16. [Europe PMC free article] [Abstract] [Google Scholar]
- Martin WF. 2017. Symbiogenesis, gradualism, and mitochondrial energy in eukaryote evolution. Period Biol. 119:141–158. [Google Scholar]
- Martin W, Koonin EV. 2006. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 440:41–45. [Abstract] [Google Scholar]
- Massey SE, Mishra B. 2018. Origin of biomolecular games: deception and molecular evolution. J R Soc Interface. 15:20180429. 10.1098/rsif.2018.0429. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Maurer-Alcala XX, Katz LA. 2015. An epigenetic toolkit allows for diverse genome architectures in eukaryotes. Curr Opin Genet Dev. 35:93–99. [Europe PMC free article] [Abstract] [Google Scholar]
- Maurer-Alcala XX, Nowacki M. 2019. Evolutionary origins and impacts of genome architecture in ciliates. Ann N Y Acad Sci. 1447:110–118. [Europe PMC free article] [Abstract] [Google Scholar]
- McGrath CL, Katz LA. 2004. Genome diversity in microbial eukaryotes. Trends Ecol Evol. 19:32–38. [Abstract] [Google Scholar]
- McLaughlin RN, Malik HS. 2017. Genetic conflicts: the usual suspects and beyond. J Exp Biol. 220:6–17. [Europe PMC free article] [Abstract] [Google Scholar]
- Meyer TJ, Rosenkrantz JL, Carbone L, Chavez SL. 2017. Endogenous retroviruses: with us and against us. Front Chem. 5:23. [Europe PMC free article] [Abstract] [Google Scholar]
- Oliverio AM, Katz LA. 2014. The dynamic nature of genomes across the tree of life. Genome Biol Evol. 6:482–488. [Europe PMC free article] [Abstract] [Google Scholar]
- Parfrey LW, Katz LA. 2010. Dynamic genomes of eukaryotes and the maintenance of genomic integrity. Microbe. 5:156–164. [Google Scholar]
- Parfrey LW, Lahr DJG, Katz LA. 2008. The dynamic nature of eukaryotic genomes. Mol Biol Evol. 25:787–794. [Europe PMC free article] [Abstract] [Google Scholar]
- Parhad SS, Theurkauf WE. 2019. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 9:180181. 10.1098/rsob.18.0181. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Piegu B, Bire S, Arensburger P, Bigot Y. 2015. A survey of transposable element classification systems - a call for a fundamental update to meet the challenge of their diversity and complexity. Mol Phyl Evol. 86:90–109. [Abstract] [Google Scholar]
- Pittis AA, Gabaldon T. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature. 531:101–104. [Europe PMC free article] [Abstract] [Google Scholar]
- Poole AM, Hendrickson HL. 2019. Response: commentary: manifold routes to a nucleus. Front Microbiol. 10:2585.10.3389/fmicb.2019.02585. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25:537–546. [Europe PMC free article] [Abstract] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 136:461–472. [Europe PMC free article] [Abstract] [Google Scholar]
- Song MJ, Schaack S. 2018. Evolutionary conflict between mobile DNA and host genomes. Am Nat. 192:263–273. [Abstract] [Google Scholar]
- Suzuki MM, Bird A. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 9:465–476. [Abstract] [Google Scholar]
- Tekle YI, Wood FC, Katz LA, Ceron-Romero MA, Gorfu LA. 2017. Amoebozoans are secretly but ancestrally sexual: evidence for sex genes and potential novel crossover pathways in diverse groups of amoebae. Genome Biol Evol. 9:375–387. 10.1093/gbe/evx002. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vogt A, Goldman AD, Mochizuki K, Landweber LF. 2013. Transposon domestication versus mutualism in ciliate genome rearrangements. PLoS Genet 9:e1003659. 10.1093/gbe/evx002. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wein T, Picazo DR, Blow F, Woehle C, Jami E, Reusch TBH, Martin WF, Dagan T. 2019. Currency, exchange, and inheritance in the evolution of symbiosis. Trends Microbiol. 27:836–849. [Abstract] [Google Scholar]
- Weiner AKM, Yan Y, Ceron Romero M, Katz LA. 2020. Phylogenomics of the eukaryotic epigenetic toolkit reveals punctate retention of genes across lineages and functional categories. Gen Biol Evol. 12:evaa198. [Europe PMC free article] [Abstract] [Google Scholar]
- Werren JH. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci USA. 108:10863–10870. [Europe PMC free article] [Abstract] [Google Scholar]
- Woellmer A, Hammerschmidt W. 2013. Epstein-Barr virus and host cell methylation: regulation of latency, replication and virus reactivation. Curr Opin Virol. 3:260–265. [Europe PMC free article] [Abstract] [Google Scholar]
- Zufall RA, Robinson T, Katz LA. 2005. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool B Mol Dev Evol. 304B:448–455. [Abstract] [Google Scholar]
Articles from Journal of Heredity are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/jhered/esaa060
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jhered/article-pdf/112/1/140/36584171/esaa060.pdf
Citations & impact
Impact metrics
Article citations
A decade of dinoflagellate genomics illuminating an enigmatic eukaryote cell.
BMC Genomics, 25(1):932, 04 Oct 2024
Cited by: 0 articles | PMID: 39367346 | PMCID: PMC11453091
Review Free full text in Europe PMC
Foraminifera as a model of eukaryotic genome dynamism.
mBio, 15(3):e0337923, 08 Feb 2024
Cited by: 0 articles | PMID: 38329358
Salmonidae Genome: Features, Evolutionary and Phylogenetic Characteristics.
Genes (Basel), 13(12):2221, 27 Nov 2022
Cited by: 3 articles | PMID: 36553488 | PMCID: PMC9778375
Review Free full text in Europe PMC
Evolving Perspective on the Origin and Diversification of Cellular Life and the Virosphere.
Genome Biol Evol, 14(6):evac034, 01 May 2022
Cited by: 7 articles | PMID: 35218347 | PMCID: PMC9169541
Review Free full text in Europe PMC
Epigenetic influences of mobile genetic elements on ciliate genome architecture and evolution.
J Eukaryot Microbiol, 69(5):e12891, 19 Feb 2022
Cited by: 2 articles | PMID: 35100457 | PMCID: PMC9421606
Review Free full text in Europe PMC
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An epigenetic toolkit allows for diverse genome architectures in eukaryotes.
Curr Opin Genet Dev, 35:93-99, 30 Nov 2015
Cited by: 9 articles | PMID: 26649755 | PMCID: PMC4864960
Review Free full text in Europe PMC
The eukaryotic ancestor had a complex ubiquitin signaling system of archaeal origin.
Mol Biol Evol, 32(3):726-739, 17 Dec 2014
Cited by: 42 articles | PMID: 25525215 | PMCID: PMC4327156
A Field Guide to Eukaryotic Transposable Elements.
Annu Rev Genet, 54:539-561, 21 Sep 2020
Cited by: 194 articles | PMID: 32955944 | PMCID: PMC8293684
Review Free full text in Europe PMC
Diverse genome structures among eukaryotes may have arisen in response to genetic conflict.
Genome Biol Evol, evae239, 07 Nov 2024
Cited by: 0 articles | PMID: 39506510
Funding
Funders who supported this work.
NHGRI NIH HHS (1)
Grant ID: R15 HG010409
National Institutes of Health (1)
Grant ID: R15HG010409
National Science Foundation (3)
Grant ID: DEB-1651908
Grant ID: DEB-1541511
Grant ID: OCE-1924570
 1,2
1,2




