Abstract
Free full text

A decade of dinoflagellate genomics illuminating an enigmatic eukaryote cell
Associated Data
Abstract
Dinoflagellates are a remarkable group of protists, not only for their association with harmful algal blooms and coral reefs but also for their numerous characteristics deviating from the rules of eukaryotic biology. Genome research on dinoflagellates has lagged due to their immense genome sizes in most species (~ 1-250 Gbp). Nevertheless, the last decade marked a fruitful era of dinoflagellate genomics, with 27 genomes sequenced and many insights attained. This review aims to synthesize information from these genomes, along with other omic data, to reflect on where we are now in understanding dinoflagellates and where we are heading in the future. The most notable insights from the decade-long genomics work include: (1) dinoflagellate genomes have been expanded in multiple times independently, probably by a combination of rampant retroposition, accumulation of repetitive DNA, and genome duplication; (2) Symbiodiniacean genomes are highly divergent, but share about 3,445 core unigenes concentrated in 219 KEGG pathways; (3) Most dinoflagellate genes are encoded unidirectionally and are not intron-poor; (4) The dinoflagellate nucleus has undergone extreme evolutionary changes, including complete or nearly complete loss of nucleosome and histone H1, and acquisition of dinoflagellate viral nuclear protein (DVNP); (5) Major basic nuclear protein (MBNP), histone-like protein (HLP), and bacterial HU-like protein (HCc) belong to the same protein family, and MBNP can be the unifying name; (6) Dinoflagellate gene expression is regulated by poorly understood mechanisms, but microRNA and other epigenetic mechanisms are likely important; (7) Over 50% of dinoflagellate genes are “dark” and their functions remain to be deciphered using functional genetics; (8) Initial insights into the genomic basis of parasitism and mutualism have emerged. The review then highlights functionally unique and interesting genes. Future research needs to obtain a finished genome, tackle large genomes, characterize the unknown genes, and develop a quantitative molecular ecological model for addressing ecological questions.
1-250 Gbp). Nevertheless, the last decade marked a fruitful era of dinoflagellate genomics, with 27 genomes sequenced and many insights attained. This review aims to synthesize information from these genomes, along with other omic data, to reflect on where we are now in understanding dinoflagellates and where we are heading in the future. The most notable insights from the decade-long genomics work include: (1) dinoflagellate genomes have been expanded in multiple times independently, probably by a combination of rampant retroposition, accumulation of repetitive DNA, and genome duplication; (2) Symbiodiniacean genomes are highly divergent, but share about 3,445 core unigenes concentrated in 219 KEGG pathways; (3) Most dinoflagellate genes are encoded unidirectionally and are not intron-poor; (4) The dinoflagellate nucleus has undergone extreme evolutionary changes, including complete or nearly complete loss of nucleosome and histone H1, and acquisition of dinoflagellate viral nuclear protein (DVNP); (5) Major basic nuclear protein (MBNP), histone-like protein (HLP), and bacterial HU-like protein (HCc) belong to the same protein family, and MBNP can be the unifying name; (6) Dinoflagellate gene expression is regulated by poorly understood mechanisms, but microRNA and other epigenetic mechanisms are likely important; (7) Over 50% of dinoflagellate genes are “dark” and their functions remain to be deciphered using functional genetics; (8) Initial insights into the genomic basis of parasitism and mutualism have emerged. The review then highlights functionally unique and interesting genes. Future research needs to obtain a finished genome, tackle large genomes, characterize the unknown genes, and develop a quantitative molecular ecological model for addressing ecological questions.
Background
Dinoflagellates are one of the most important and unique groups of protists, notable for their ecological success and many peculiar characteristics that are a constant source of fascination [1, 2]. Generally believed to have emerged in the Silurian period (~ 400 MYA), the phylum of Dinoflagellata has over 2,300 extant species and roughly 2,000 fossil species [3, 4]. Morphologically, dinoflagellates are highly diverse (Fig. (Fig.1),1), but generally grouped into two major morphotypes: the thecate group in which cells contain cellulose-filled vesicles (amphiesma) underneath the cell membrane and the athecate (i.e. naked) group in which cells have weak or no discernible cellulose-filled amphiesma.
400 MYA), the phylum of Dinoflagellata has over 2,300 extant species and roughly 2,000 fossil species [3, 4]. Morphologically, dinoflagellates are highly diverse (Fig. (Fig.1),1), but generally grouped into two major morphotypes: the thecate group in which cells contain cellulose-filled vesicles (amphiesma) underneath the cell membrane and the athecate (i.e. naked) group in which cells have weak or no discernible cellulose-filled amphiesma.
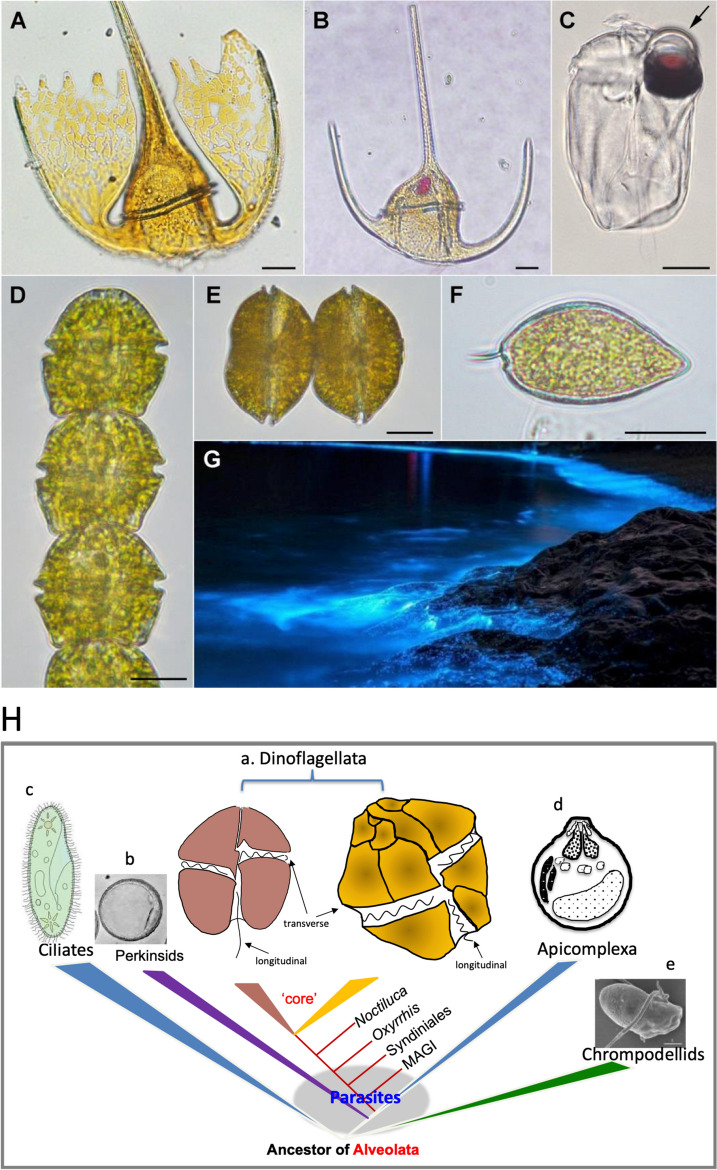
Light micrographs of dinoflagellates and a bioluminescence scene (A-G) and phylogenetic relationship of dinoflagellates, apicomplexa, and other alveolates (H). A. Tripos platycorne. B. Tripos muelleri. C. Erythropsidinium agile. The arrow points at the ocelloid. D. Gymnodinium catenatum. E. Gessnerium (=Alexandrium) monilatum. F. Prorocentrum micans. Scale bars = 20 µm. G. Bioluminescence by Noctiluca scintillans at Zhejiang, China. Images A-F courtesy of Dr. Fernando Gómez; all in apex facing up view except E and F being lateral view. Image G courtesy of Dr. Hao Luo. In H), core dinoflagellates (a) include naked dinoflagellate with two flagella (longitudinal and transverse) and thecate dinoflagellate with two flagella (longitudinal and transverse) and the conspicuous thecal plates. Non-core dinoflagellates include Marine Alveolate Group I (MAGI, euduboscquellids), Syndiniales (also known as MAGII), and the basal lineages Oxyrrhis and Noctiluca. Modified from Lin 2011 with up-to-date information
Ecologically, dinoflagellates are highly adaptable to vastly different habitats, from small ponds, rivers, lakes, and estuaries, to the ocean, and from the Antarctic to the Arctic, with lineages evolutionarily migrating between freshwater and marine environments [5, 6]. Many species in the Symbiodiniaceae are indispensable endosymbionts of reef-building corals. Many species from various lineages cause harmful algal blooms and produce toxins, devastating marine ecosystems and coastal economies as well as posing public health risks. Additionally, dinoflagellates live in diverse trophic modes (from autotrophic, heterotrophic, to mixotrophic) and lifestyles (from parasitic, symbiotic, to free-living), with photoautotrophic lineages (~ 50% of species) contributing substantially to photosynthetic carbon fixation and oxygen production and heterotrophic/mixotrophic species being important trophic links in the food chain. Dinoflagellates are the only group of protists that produce bioluminescence, which occurs in both photosynthetic (e.g. Alexandrium spp., Lingulodinium polyedrum) and heterotrophic or mixotrophic (e.g. red and green Noctiluca scintillans) species.
50% of species) contributing substantially to photosynthetic carbon fixation and oxygen production and heterotrophic/mixotrophic species being important trophic links in the food chain. Dinoflagellates are the only group of protists that produce bioluminescence, which occurs in both photosynthetic (e.g. Alexandrium spp., Lingulodinium polyedrum) and heterotrophic or mixotrophic (e.g. red and green Noctiluca scintillans) species.
From an evolutionary perspective, dinoflagellates encompass parasitic (Syndiniales) lineages at the base followed by the typical dinoflagellate lineages composed of athecate and thecate lineages (Fig. (Fig.1),1), which have acquired various types of plastids originating from secondary, serial secondary, or tertiary endosymbiosis. At the cellular and genomic levels, dinoflagellates possess numerous peculiarities such as permanently (or nearly so) condensed chromosomes with cholesteric liquid crystalline state of DNA, lack of nucleosomes, closed mitosis, and extranuclear mitotic spindle [7]. Due to these characteristics, how dinoflagellates regulate DNA duplication in the cell division cycle and regulate gene expression in response to growth conditions remain to be demystified.
Due to the enormous genome sizes (up to 250 Gbp), genome sequencing for dinoflagellates has lagged behind many other algal phyla. However, thanks to the precipitous drop in sequencing costs and significant advances in bioinformatic methodology, dinoflagellate genome sequencing has finally taken off and reached a significant milestone over the past decade. Along with exponentially growing transcriptome and metatranscriptome data, the dinoflagellate -omic (Dinomic) data has grown to an appreciable database (Table (Table1).1). Although the database is strongly biased towards small dinoflagellate genomes, it contains rich information that illuminates various aspects of dinoflagellate biology. This wealth of data warrants a synthesis to bring the last reviews up to date [2, 8]. Some of the findings have challenged long-standing notions regarding dinoflagellate nuclear biology and molecular genetics, while others have revealed unsuspected physiologies. This review focuses on the most exciting discoveries from the last decade and anticipates what lies ahead for the next decade of Dinomic research.
Table 1
Published genomes of dinoflagellates
| Species | Assembled genome size (Mbp) | Assembled rate (%) | Gene No. | Scaffolds N50 (kbp) | GC (%) | Repetitive DNA (%) | Reference |
|---|---|---|---|---|---|---|---|
| Amoebophrya sp. ex Karlodinium veneficum | ~ 130 130 | 94.62 | 41,936 | 21 | > 58 58 | Bachvaroff 2019 [9] | |
| Amoebophrya ceratii AT5.2 | 87.7 | 75.86 | 19,925 | 83.9 | 55.9 | 23.8 | John et al. 2019 [10] |
| Amoebophrya ceratii A120 | 115.5 | 92.21 | 26,441 | 9,243 | 51.2 | 13.1 | Farhat et al. 2021 [11] |
| Amoebophrya ceratii A25 | 116 | 97.8 | 28,091 | 1,082 | 47.8 | Farhat et al. 2021 [11] | |
| Amphidinium gibbosum | 6,300 | 98.44 | 85,139 | 166.4 | 47.1 | 29.9 | Beedessee et al. 2020 [12] |
| Breviolum minutum | 616 | 41.07 | 41,925 | 126.2 | 43.6 | Shoguchi et al. 2013 [13] | |
| Cladocopium goreaui (original) | 1,030 | 85.55 | 35,913 | 98 | 44.8 | Liu et al. 2018 [14] | |
| Cladocopium goreaui (improved) | 1,200 | 45,322 | 354 | 44.4 | 36.5 | Chen et al. 2022 [15] | |
| Cladocopium spp. | 704.8 | 65,832 | 43 | Shoguchi et al. 2021 [16] | |||
| Durusdinium trenchii | 670 | 30,054 | 97.5 | 47.4 | Shoguchi et al. 2021 [16] | ||
| Effrenium voratum RCC1521 | 1,200 | 85.71 | 32,108 | 720 | 50.8 | Shah et al. 2024 [17] | |
| Effrenium voratum CCMP3420 | 1,300 | 39,878 | 252 | 50.6 | Shah et al. 2024 [17] | ||
| Effrenium voratum CCMP421 | 1,100 | 57.89 | 32,615 | 304 | 50.9 | Shah et al. 2024 [17] | |
| Fugacium kawagutii CCMP2468 | 935 | 79.24 | 36,850 | 381 | Lin et al. 2015 [18] | ||
| Fugacium kawagutii CCMP2468 | 937 | 79.41 | 45,192 | 13,533.5 | Li et al. 2020 [19] | ||
| Fugacium kawagutii CCMP2468 | 1,050 | 88.98 | 26,609 | 268.8 | 16 | Liu et al. 2018 [14] | |
| Hematodinium sp.a | 4,769 | 29,510 | 17,235 | Gornik et al. 2015 [20] | |||
| Polarella glacialis CCMP1383 | 2,980 | 58,232 | 170.3 | 45.91 | 68 | Stephens et al. 2020 [21] | |
| Polarella glacialis CCMP2088 | 2,760 | 51,713 | 129.2 | 46.15 | 68 | Stephens et al. 2020 [21] | |
| Prorocentrum cordatum | 4,750 | 87.37 | 85,849 | 349.2 | 59.7 | Dougan et al. 2023 [22] | |
| Symbiodinium linucheae CCMP2456 | 695 | 75.96 | 58.1 | 50.36 | González-Pech et al. 2021 [23] | ||
| Symbiodinium microadriacticum CCMP2467 | 808 | 73.45 | 49,109 | 573.5 | 50.5 | 34.8 | Aranda et al. 2016 [24] |
| Symbiodinium microadriacticum CassKB8 | 813 | 72.65 | 43 | 51.91 | González-Pech et al. 2021 [23] | ||
| Symbiodinium microadriaticum 04-503SCI.03 | 775 | 73.62 | 50 | 50.46 | 27.9 | González-Pech et al. 2021 [23] | |
| Symbiodinium natans CCMP2548 | 762 | 100 | 610.5 | 51.79 | González-Pech et al. 2021 [23] | ||
| Symbiodinium necroappetens CCMP2469 | 768 | 76.26 | 11.4 | 50.85 | González-Pech et al. 2021 [23] | ||
| Symbiodinium pilosum CCMP2461 | 1,089 | 54.64 | 62.4 | 48.21 | González-Pech et al. 2021 [23] | ||
| Symbiodinium spp. clade C | 767 | 69,018 | 50 | Shoguchi et al., 2018 [25] | |||
| Symbiodinium spp. clade A | 705 | 65,832 | 43 | Shoguchi et al., 2018 [25] | |||
| Symbiodinium tridacnidorum CCMP2592 | 1,103 | 85.71 | 651.3 | 51.01 | González-Pech et al. 2021 [23] |
aInformation came from transcriptome and incomplete draft genome
Dinoflagellate genomics landscape and incredible inter-species dissimilarities
To date, 28 dinoflagellate genomes have been sequenced (with two sequenced more than once), mostly from the family Symbiodiniaceae (Table (Table1).1). Sequenced taxa other than Symbiodiniaceae included Amoebophrya sp. ex Karlodinium veneficum [9], Amoebophrya ceratii [10, 11], Amphidinium gibbosum [12], Hematodinium [20], Polarella glacialis [21], and Prorocentrum cordatum (formerly P. minimum) [22]. These taxa cover a wide range of environmental conditions and different lifestyles, despite the obvious bias toward small end of the dinoflagellate genome size spectrum. Species from the Symbiodiniaceae are endosymbionts of tropical corals or their free-living counterparts. Amoebophrya species are intracellular parasites, infecting other dinoflagellates. Amphidinium gibbosum and P. cordatum form harmful algal blooms and A. gibbosum is toxigenic. P. glacialis thrives in the Antarctic and Arctic, can survive temperature shock to 15ºC [26] and DNA barcoding suggests that this and closely related species occur in the temperate estuary Long Island Sound [27].
Most of these genome assemblies were based on Illumina Hi-Seq technology and hence are largely fragmentary. However, using PacBio to generate long sequence reads and Hi-C to establish linkages between scaffolds, chromosome level assemblies have been achieved for part of the F. kawagutii genome (> 120Mbp; [19]) and the genomes of Symbiodinium microadriaticum [28] and Breviolum minutum [29].
120Mbp; [19]) and the genomes of Symbiodinium microadriaticum [28] and Breviolum minutum [29].
The number of chromosomes varies vastly between species, ranging from several to more than 200 [30]. By microscopic count of DAPI-stained chromosomes and telomere end count (divided by 2), F. kawagutii has between 7 [31] and 11 chromosomes [18]. In sharp contrast, there are 94 chromosomes in S. microadriaticum [28] and 91 in B. minutum [29]. While the number for S. microadriaticum is comparable to a previous microscopic estimate (100; [32]), that for B. minutum exceeds five-fold the microscopic count of ~ 17 (Fig. (Fig.11 in [13]). This inter-species disparity is remarkable given that the genome size among these species differs by no more than 2-fold. Equally striking are their compensating differences in chromosome length. The chromosome length is markedly shorter in S. microadriaticum (16Kb to 16.6 Mb, median 6.6 Mb; [28]) and in B. minutum (median 6.7 Mb, longest
17 (Fig. (Fig.11 in [13]). This inter-species disparity is remarkable given that the genome size among these species differs by no more than 2-fold. Equally striking are their compensating differences in chromosome length. The chromosome length is markedly shorter in S. microadriaticum (16Kb to 16.6 Mb, median 6.6 Mb; [28]) and in B. minutum (median 6.7 Mb, longest ~
~ 11 Mb; [29]) than in F. kawagutii (maximum
11 Mb; [29]) than in F. kawagutii (maximum >
> 120 Mb; [19]). These data suggest an interesting evolutionary trend of chromosome number decrease and chromosome length increase from the basal S. microadriaticum to later-diverging F. kawagutii [33]. Based on the existing data, the small non-parasitic dinoflagellate genomes such as those of Symbiodiniaceae contain approximately 1–3 Gbp DNA distributed in 10–100 chromosomes of 100
120 Mb; [19]). These data suggest an interesting evolutionary trend of chromosome number decrease and chromosome length increase from the basal S. microadriaticum to later-diverging F. kawagutii [33]. Based on the existing data, the small non-parasitic dinoflagellate genomes such as those of Symbiodiniaceae contain approximately 1–3 Gbp DNA distributed in 10–100 chromosomes of 100 −
− 10 Mbp each; the largest dinoflagellate genome contains approximately 250 Gbp DNA likely distributed in 200–1000 chromosomes of 1.25 Gbp-250 Mbp each.
10 Mbp each; the largest dinoflagellate genome contains approximately 250 Gbp DNA likely distributed in 200–1000 chromosomes of 1.25 Gbp-250 Mbp each.
Genome sequencing combined with fluorescent in situ hybridization (FISH; Fig. Fig.2A,2A, B) revealed that the telomere sequence in F. kawagutii consists of repeats of TTTAGGG, depicted as (TTTAGGG)n, which is identical to the typical plant telomere (Richards and Ausubel 1988), as is also true for other dinoflagellates [34, 35]. The length of the telomere tract was estimated at 25–80 Kbp in Karenia papilionacea and Crypthecodinium cohnii [35] but is unclear in other species. Using non-denaturing fluorescent in situ hybridization, Cuadrado et al. (2022) [36] revealed bipolar distribution of clustered multiple foci of telomeric DNA: one in the nucleolus organization area and the other peripherally along the convex side of the nucleus, suggesting that the telomeres change their positions during cell division.
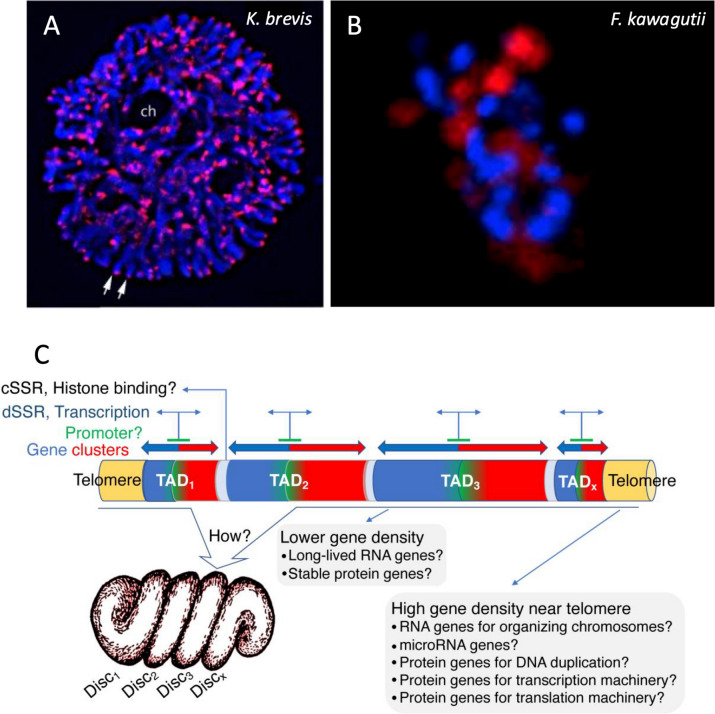
Distribution pattern of telomeres in a tertiary endosymbiosis dinoflagellate and schematic of TADs in dinoflagellate chromosomes and associated gene distribution and expression machinery. A Chromosomes of Karenia brevis stained with fluorescent in situ hybridization of telomere probe. B DNA DAPI stain (Blue) and telomere FISH (red) in F. kawagutii. C TAD organization highlighting gene distribution and gene expression machinery. Reproduced with permission from Cuadrado et al. 2019 [37] (A), Lin et al. 2015 [18] (B), and Lin et al. 2021 [33] (C)
Dinoflagellate genomes are highly divergent. This is reflected in the genomes of Symbiodiniaceae, among which a >
> 98% dissimilarity is consistently found by mapping reads from one genome to the assembly of the other genome [18, 23]. Whether this extreme divergence reflects rapid genome evolution in dinoflagellates in general or is due to severe isolation between Symbiodiniaceae species by their facultative symbiotic lifestyle remains to be investigated.
98% dissimilarity is consistently found by mapping reads from one genome to the assembly of the other genome [18, 23]. Whether this extreme divergence reflects rapid genome evolution in dinoflagellates in general or is due to severe isolation between Symbiodiniaceae species by their facultative symbiotic lifestyle remains to be investigated.
Topologically associated domains (TAD)
Dinoflagellate genomes form topologically associated domains (TADs; Fig. Fig.2C),2C), a feature widely found in eukaryotic genomes [28, 29]. TAD is a genomic region where DNA sequences within the same TAD interact with each other more frequently than with sequences outside of it [38]. Between the two species that have been examined, S. microadriaticum has about 450 TADs with a median size of 1.8 Mb, whereas B. minutum contains 580 TADs with a median size of 5.7 Mb. These are markedly larger than mammalian TADs (~ 0.18 Mb) and bacterial CIDs (~
0.18 Mb) and bacterial CIDs (~ 0.17 Mb), and do not show interactions with more distant regions of the chromosome in the Hi-C maps.
0.17 Mb), and do not show interactions with more distant regions of the chromosome in the Hi-C maps.
In these two genome assemblies, genes are distributed along the chromosomes as an alternating series of unidirectional clusters [28, 29]. TADs show two types of strand switch regions (SSR) (Fig. (Fig.2C).2C). One, where values of the DNA contact probability plot are closest to the diagonal axis in Hi-C interaction maps, corresponds to the end of transcription (a convergent strand switch region, cSSR). The other, where regions of the TADs show DNA interactions furthest from the diagonal corresponds to the initiation of transcription (a divergent SSR, dSSR). Roughly half the genes in the genome are found in groups of at least nine, all sharing a common direction. TAD boundaries are also associated with decreased GC content.
The dinoTAD structure suggests that transcriptional promoters might be found in the central region of the TADs where genes are oriented outward in both directions (Fig. (Fig.2C).2C). These sites could represent histone attachment points, reconciling the conservation of histone sequences with their paucity [39].
The average interaction frequency between two regions of the chromosome decreases when the distance between them increases. The trend in S. microadriaticum [28] suggests that DNA in this species is organized as linear rods. Comparison to similar analyses with other genome organizations shows that although dinoflagellate nuclei presumably use viral nuclear protein (DVNP) and bacterium-originated histone-like proteins more than histones (due to their higher expression levels), the chromosome organization appears more like eukaryotic chromosomes, with the highest similarity to mitotic chromosomes in metaphase when the chromosomes are highly condensed [28, 33].
Marinov et al. (2021) [29] and Nand et al. (2021) [28] noted that when the transcription was blocked, the TADs disappeared, revealing transcription’s critical role in maintaining the TADs in dinoflagellate chromosomes. This aligns with earlier findings on RNA’s importance in chromosomal DNA structure [40], though recent data suggest transcription activity may be more crucial than RNA in stabilizing domain structure.
Mechanisms of genome expansion: Duplication, (retro)transposition, and repetitive DNA
Dinoflagellate genome sizes vary widely among species, from < 0.1 Gbp in the parasitic taxon Amoebophrya, <
0.1 Gbp in the parasitic taxon Amoebophrya, < 3 Gbp for most coral symbionts in the family Symbiodiniaceae, to ~
3 Gbp for most coral symbionts in the family Symbiodiniaceae, to ~ 250 Gbp for free-living taxa [10, 41, 42]. There is no phylogenetic trend in the vast genome size variation, however (Fig. (Fig.3A).3A). For example, the coral symbiotic lineage Symbiodiniaceae has the smallest genomes among documented dinoflagellates despite its later-diverging position in the phylogenetic tree [43]. The large genome sizes and wide range among dinoflagellates suggest multiple independent events of genome expansion in evolution. Alternatively, genome contraction might have occurred occasionally, independent of symbiosis lifestyle [17].
250 Gbp for free-living taxa [10, 41, 42]. There is no phylogenetic trend in the vast genome size variation, however (Fig. (Fig.3A).3A). For example, the coral symbiotic lineage Symbiodiniaceae has the smallest genomes among documented dinoflagellates despite its later-diverging position in the phylogenetic tree [43]. The large genome sizes and wide range among dinoflagellates suggest multiple independent events of genome expansion in evolution. Alternatively, genome contraction might have occurred occasionally, independent of symbiosis lifestyle [17].
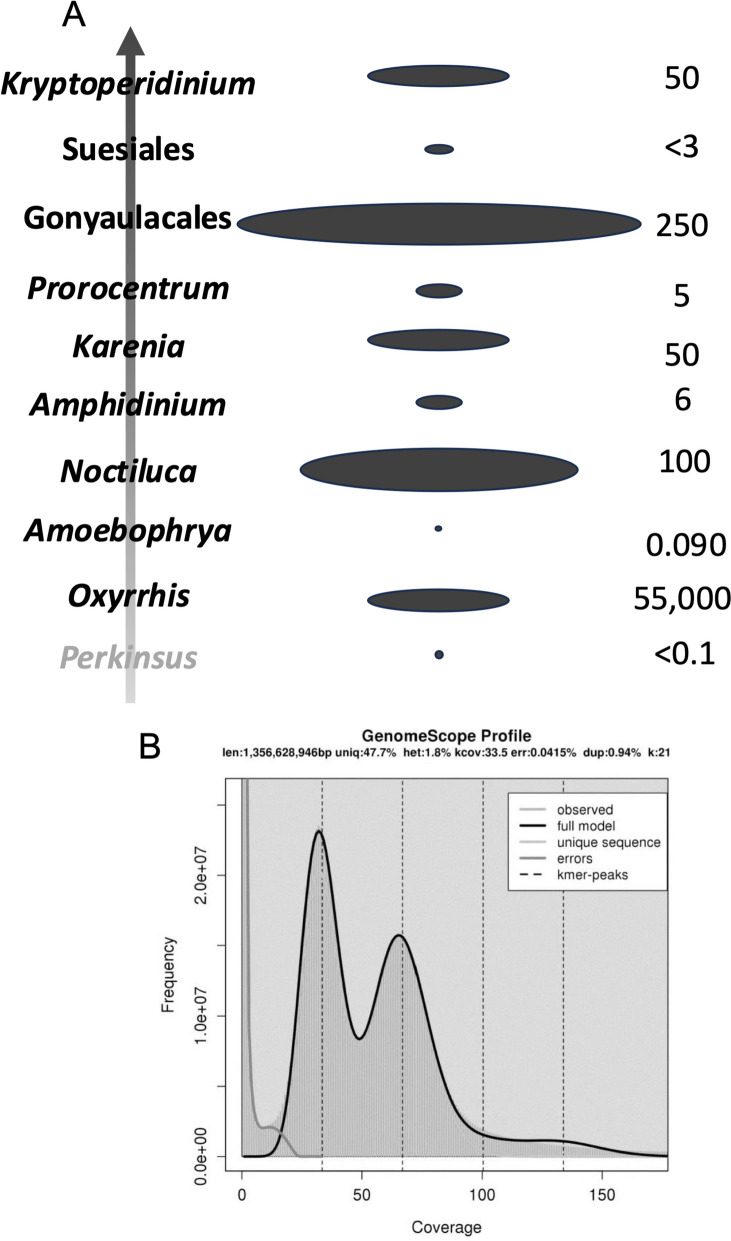
Evolutionary dynamics of dinoflagellate genome size showing no phylogenetic trend (A) and double peak in 21-mer profile of Polarella glacialis showing evidence of genome duplication (B). A Vertical arrow indicates phylogenetic trend from the dinoflagellate sister group perkinsids, basal dinoflagellate Oxyrrhis, to later diverging taxa. Size of oval in the middle illustrates genome size (not to scale) as depicted by numbers shown on the right. B Reproduced from Stephans et al. 2020 [21] under the terms of the Creative Commons CC BY license
Whole genome duplication (WGD) is the most efficient way to achieve chromosome multiplication and genome expansion [44, 45]. The immense dinoflagellate genomes might have resulted from WGD. However, K-mer profile and scaffold synteny (proxy of chromosome synteny) analyses for F. kawagutii did not show any signs of WGD [18]. Nevertheless, the genome of Polarella glacialis (~ 3 Gbp) is about double the size of that in Symbiodiniaceae [21]. Its k-mer profile, based on sequencing reads, shows double peaks characteristic of diploid genomes [21], suggesting a genome duplication (Fig. (Fig.3B).3B). “Diploid”-like chromosome pairs have been reported for Pyrocystis lunula as well [46]. There is a good chance that WGD is widespread in large dinoflagellate genomes. However, the original interpretation of these observations as diploid is not consistent with the traditional view that dinoflagellates are haploids.
3 Gbp) is about double the size of that in Symbiodiniaceae [21]. Its k-mer profile, based on sequencing reads, shows double peaks characteristic of diploid genomes [21], suggesting a genome duplication (Fig. (Fig.3B).3B). “Diploid”-like chromosome pairs have been reported for Pyrocystis lunula as well [46]. There is a good chance that WGD is widespread in large dinoflagellate genomes. However, the original interpretation of these observations as diploid is not consistent with the traditional view that dinoflagellates are haploids.
Besides WGD, genome expansion can result from processes like the proliferation of mobile DNA elements, including transposons and retrotransposons. The role of transposable elements in remodeling genomes has been demonstrated in plants [47]. Indeed, there is evidence that transposons [10] and retrotransposons [48] play roles in dinoflagellate genome expansion. The role of retrotransposition is corroborated by the prevalent gene paralogs and DinoSL (complete or truncated) at the upstream region of genes, which account for > 20% of total genes [49], made possible by the large families of reverse transcriptase and integrase genes in the genomes [18, 28]. In contrast, transposition is rare in the small genome (<
20% of total genes [49], made possible by the large families of reverse transcriptase and integrase genes in the genomes [18, 28]. In contrast, transposition is rare in the small genome (< 0.1 Gbp) of the parasitic species Amoebophrya ceratii [10]. González-Pech et al. (2021) [23] compared genome features of S. tridacnidorum and S. natans and postulated that retrotransposition might be a major driving force for the two genomes to diverge.
0.1 Gbp) of the parasitic species Amoebophrya ceratii [10]. González-Pech et al. (2021) [23] compared genome features of S. tridacnidorum and S. natans and postulated that retrotransposition might be a major driving force for the two genomes to diverge.
Stephens et al. (2020) detected a substantial proportion of long terminal repeat (LTR) elements (~ 12%) in P. glacialis genomes [21]. Transposable elements (such as LTRs) can comprise up to 80% of plant genomes and are induced by genome shock and polyploidization, leading to genome restructuring [50]. The abundance of LTRs in P. glacialis and their role in genome restructuring may explain the difference in genome size between populations. In S. microadriaticum, heat stress-activated Ty1-copia-type LTR retrotransposons were detected, and recent expansion was documented [51], suggesting proliferation of retrotransposable elements for adaptation to hot habitats. The survival of retroposed genes may be promoted by their methylation [52], as DNA methylation is thought to silence and promote transposition [53]. Similar TE domestication by epigenetic regulation seems common in eukaryotes [54], and methylation of transposable elements is presumably a major driving force of genome expansion in all eukaryotes [55].
12%) in P. glacialis genomes [21]. Transposable elements (such as LTRs) can comprise up to 80% of plant genomes and are induced by genome shock and polyploidization, leading to genome restructuring [50]. The abundance of LTRs in P. glacialis and their role in genome restructuring may explain the difference in genome size between populations. In S. microadriaticum, heat stress-activated Ty1-copia-type LTR retrotransposons were detected, and recent expansion was documented [51], suggesting proliferation of retrotransposable elements for adaptation to hot habitats. The survival of retroposed genes may be promoted by their methylation [52], as DNA methylation is thought to silence and promote transposition [53]. Similar TE domestication by epigenetic regulation seems common in eukaryotes [54], and methylation of transposable elements is presumably a major driving force of genome expansion in all eukaryotes [55].
The abundance of repetitive DNA also contributes to genome size variation. Using FISH, Cuadrado et al. (2022) [36] revealed clusters of dense trinucleotide repeats in the bean-shaped nuclei of Karenia selliformis and K. mikimotoi. Larger dinoflagellate genomes tend to contain more repetitive elements [21]. For instance, repetitive elements account for 23.8% of the 0.1156-Gbp Amoebophrya A120 genome, 13.1% of the 0.1160-Gbp Amoebophrya A25 genome, 27.9% of the ~ 1.10-Gbp Symbiodinium microadriaticum genome, and 16% of the ~
1.10-Gbp Symbiodinium microadriaticum genome, and 16% of the ~ 1.2-Gbp Fugacium kawagutii genome [14]. For P. glacialis, with genome sizes of ~
1.2-Gbp Fugacium kawagutii genome [14]. For P. glacialis, with genome sizes of ~ 3 Gbp, the level of repetitive elements increases to ~
3 Gbp, the level of repetitive elements increases to ~ 68% [21]. Crypthecodinium cohnii has a
68% [21]. Crypthecodinium cohnii has a ~
~ 7 Gbp genome [56], with 55–60% composed of repetitive DNA [57]. A survey of the ~
7 Gbp genome [56], with 55–60% composed of repetitive DNA [57]. A survey of the ~ 115 Gbp Alexandrium ostenfeldii genome indicated that repetitive DNA accounted for ~
115 Gbp Alexandrium ostenfeldii genome indicated that repetitive DNA accounted for ~ 58% of the genome [58]. Overall, these results suggest that repetitive elements significantly contribute to genome size evolution in dinoflagellates.
58% of the genome [58]. Overall, these results suggest that repetitive elements significantly contribute to genome size evolution in dinoflagellates.
Genomic innovation by rampant horizontal gene transfer (HGT)
Horizontal gene transfer (HGT) might be a significant driver of gene innovation in dinoflagellates [59]. HGT is widespread in eukaryotes, accounting for 0.1-6.4% of total genes, depending on the species [60]. While the full extent of HGT in dinoflagellates awaits the sequencing of large-genome species, current data provide ample evidence. Using EST data, Norsenko and Bhattacharya (2007) [61] identified 16 proteins acquired through ancient HGTs in the common ancestor of the genera Karenia and Karlodinium, and one protein from a more recent HGT. Eight of these proteins appear to have resulted from independent HGTs in several eukaryotic lineages, with many involved in energy metabolism. Most of these genes were transferred directly from bacteria, but some originated from another chromalveolate that had acquired them from bacteria. Some HGT-derived genes serve essential functions in dinoflagellates and warrant further discussion.
Genome sequencing for both the Antarctic (CCMP1383) and the Arctic (CCMP2088) strains of Polarella glacialis, compared with other dinoflagellate genomes, revealed putative bacteria-to-dinoflagellate HGT for ice-binding domain-containing proteins [21]. Several bacterium-originated genes were identified in the plastid genomes of Pyrocystis lunula and Ceratium horridum, including FtsY, Rpl28, Rpl33, Yef16, and Ycf24 [62]. F. kawagutii genome analysis also revealed 56 putative bacterium-originated HGT genes (0.12% of total gene models), of which 22 are flanked by eukaryotic genes on at least one side [18]. These included the erythromycin esterase superfamily gene, radical SAM protein, and Phosphoadenosine phosphosulphate (PAPS) reductase. Erythromycin esterases (IPR007815) disrupt erythromycin by hydrolyzing the macrolactone ring, thereby conferring antibiotic resistance (InterPro). Radical SAM proteins (IPR007197) catalyze diverse reactions, such as unusual methylations, isomerisation, sulfur insertion, ring formation, anaerobic oxidation, and protein radical formation in biosynthesis and biodegradation pathways of DNA precursors, vitamins, cofactors, antibiotics, and herbicides (InterPro). PAPS reductase (IPR002500) is part of the adenine nucleotide alpha hydrolases superfamily, which includes N-type ATP PPases and ATP sulfurylases; this enzyme uses thioredoxin as an electron donor for the reduction of PAPS to phosphor-adenosine-phosphate (PAP) (InterPro).
Form II Rubisco
One of the most notable bacterium-originated HGT genes in dinoflagellates is Form II Rubisco (Rubisco II). Rubisco II exists only in peridinin-containing dinoflagellates and anaerobic bacteria [63–65]. Possibly because of its anaerobic bacterial origin, Rubisco II is more sensitive to O2 and less efficient at CO2 fixation than Form I Rubisco, which is prevalent in oxygenic photosynthetic organisms.
Proton pump rhodopsin (PPR)
PPR (also known as proteorhodopsin) is another remarkable bacterium-originated HGT genes in dinoflagellates. Initially found in prokaryotes from a metagenomic library [66], PPR is now known to be widespread in bacteria [67] and various protists lineages, including dinoflagellates [68] and diatoms [69]. This transmembrane protein, carrying an all-trans retinal chromophore, functions as a photoreceptor harvesting light energy.
Dinoflagellate PPR expression increases in response to dim light, starvation, and phosphorus (P) deficiency [70–72]. In Prorocentrum shikokuense, PPR was upregulated 40-fold during a P-depleted bloom outbreak (Zhang et al. 2019). Studies consistently indicate PPR’s potential role in providing energy during nutrient stress.
PPRs in dinoflagellates localize on plasma or endosomal membranes. GFP-fused PPR genes from P. shikokuense and Alexandrium catenella (Group I) showed cell membrane localization in HEK293 cells [73]. Oxyrrhis marina possesses both proton pump and sensory rhodopsin [70, 74], with unique distribution on endosomal, plasma, and vacuolar membranes [74–76].
Some dinoflagellate genomes encode variants of microbial rhodopsin genes, including a giant ion channel formed by a rhodopsin gene fused with a bestrophin gene [77].
Dinoflagellate viral nuclear protein (DVNP)
Frequent viral infection [78–80] and colonization in a non-lytic fashion [81] provides copious opportunities for invasion into dinoflagellate genomes. The recent detection of a CRISPR-Cas enzyme in Symbiodinium pilosom [82] suggests that dinoflagellates might have evolved an anti-viral defense mechanism similar to that in bacteria. Frequent viral infection creates opportunities for HGT. The acquisition of DVNP [83], which will be further discussed in the next section, is hitherto the most prominent case of virus-to-dinoflagellate HGTs. The probable functional replacement of DVNP for histones in structuring chromatin [84] testifies for the remarkable impact of HGT on the biology of the host.
Loss of nucleosome and gains of nuclear proteins: A nuclear drama like no other
Chromosome structure is influenced by DNA binding proteins, cations, and RNA. Mg2+ and Ca2+ are important in DNA configuration [85–87]. RNA may also be involved in DNA packaging [40, 87]. Abundant RNA binding proteins were detected in the extracts of purified chromatin by high throughput proteomic analysis in Lingulodinium polyedra [88].
Dinoflagellate nuclei have a 10-fold lower nuclear protein to DNA ratio (1:10) than typical eukaryotes (1:1) and lack nucleosome (Rizzo 1991) despite presence of core histones [39, 68]. Recent studies suggest low amounts of nucleosomes may occur [89, 90]. Dinoflagellates have acquired alternative proteins through HGT. One of these proteins is the major basic nuclear protein (MBNP), the coding gene of which is one of the most highly expressed genes in dinoflagellates [2]. Two related proteins have abundantly reported: histone-like proteins (HLP) and bacterial DNA binding protein (HU)-like protein (Hcc) [91, 92]. Phylogenetic analysis (Fig. (Fig.4)4) suggests that MBNP, HLP, and HCc belong to the same protein family, thus warranting a unifying name. MBNP appears to be the most suitable to represent this gene family.
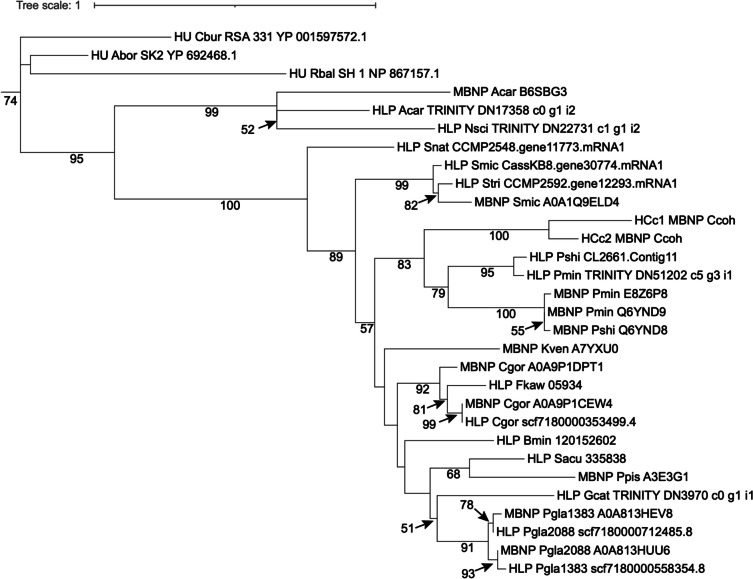
Phylogenetic tree of major basic nuclear protein (MBNP), histone-like protein (HLP), and bacterial DNA binding protein (HU)-like protein (Hcc). The tree is rooted with bacterial DNA binding protein (HU). The lack of monophyletic separation indicates that these three proteins belong to the same protein family under different names. Species name abbreviations: Abor, Alcanivorax borkumensis; Acar, Amphidinium carterae; Bmin, Breviolum minutum; Cbur, Coxiella burnetiid; Ccoh, Crypthecodinium cohnii; Cgor, Cladocopium goreaui; Fkaw, Fugacium kawagutii; Gcat, Gymnodinium catenatum; Kven, Karlodinium veneficum; Nsci, Noctiluca scintillans; Pgla, Polarella glacialis; Pmin, Prorocentrum minimum; Ppis, Pfiesteria piscicida; Pshi, Prorocentrum shikokuence; Rbal, Rhodopirellula baltica; Sacu, Scrippsiella acuminata; Smic, Symbiodinium microadriacticum; Snat, Symbiodinium natans; Stri, Symbiodinium tridacnidorum
Another acquired protein is dinoflagellate/Viral nucleoproteins (DVNP). DVNP acquisition coincided with nucleosome disappearance early in dinoflagellate evolution [83]. Heterologous expression of a DVNP in Saccharomyces cerevisiae showed that DVNP expression inhibited growth, diminished histones, disrupted nucleosomes, and impaired transcription, detrimental effects that were relieved by histone depletion [93]. These results suggest that histone abandonment might be due to the ‘colonization’ of the ‘invading’ viral protein.
Dinoflagellates appear to have lost histone H1 [94]. HCc exhibits about 10% similarity to H1, implying that HCc might be a functional replacement of H1 [95]. The similarity could cause misidentification of HCs as H1, which can explain reports of H1 presence [89]. Stringent in silico studies suggest that H1 is absent in dinoflagellates [94]. Experimental evidence is required to nail the issue.
Gene structure: Unidirectionality and intron dynamics
Unlike typical eukaryotes, dinoflagellate genes are dominantly unidirectionally arranged in the genome, forming co-oriented clusters [13, 18, 22]. This characteristic is widespread in dinoflagellates, including basal Amoebophrya spp [11]. Such organization might facilitate transcription in unison and in foci on certain areas of the TADs [28, 29, 33].
Contrary to previous belief, dinoflagellate genes are not intron-depleted. Even in streamlined parasitic Amoebophrya ceratii genomes, 67-71% of genes contain introns, with 64–99% in Symbiodiniaceae species (Table (Table1).1). The ~ 6.4 Gbp (A) gibbosum genome has fewer introns per gene but longer total intronic regions compared to smaller dinoflagellate genomes. Average intron lengths vary: 335–345 bp in Amoebophrya spp., 505, 517, and 893 bp for S. microadriaticum, (B) minutum, and F. kawagutii, respectively [11]. Notably, a 26,372 bp intron was detected in (C) cohnii [96].
6.4 Gbp (A) gibbosum genome has fewer introns per gene but longer total intronic regions compared to smaller dinoflagellate genomes. Average intron lengths vary: 335–345 bp in Amoebophrya spp., 505, 517, and 893 bp for S. microadriaticum, (B) minutum, and F. kawagutii, respectively [11]. Notably, a 26,372 bp intron was detected in (C) cohnii [96].
Intron-exon boundaries in dinoflagellates also differ from typical eukaryotes [11]. Besides the canonical GT-AG boundary, dinoflagellates use GCIGA-AG (25–74% in Symbiodiniaceae; <3% in Amoebophrya), and other boundaries (up to 67% in Amoebophrya). Interestingly, 99.98% of introns in Amoebophrya strain AT5 have canonical GT-AG splice sites, while over 60% in strains A25 and A120 have non-canonical splice sites.
Intron creation may be driven by introners (specialized transposable elements). Analysis of five dinoflagellate genomes revealed: (1) evidence of historical intron creation, (2) recently active introners in four of five species, (3) dynamic intron gain and loss, with higher gain rates than most eukaryotes, and (4) In P. glacialis, 12,253 introns from recent introner insertion, with 15 distinct families accounting for at least 100 introns each [97].
Data to date show that dinoflagellates use standard codons. However, exceptions have been reported, mostly in the stop codon. In Amoebophrya sp. ex Karlodinium veneficum, as demonstrated in dynein heavy chain genes, all three stop codons are inferred to code for amino acids [9]. UAA and UAG appear to encode glutamate whereas UGA either serves as the stop codon or encode tryptophan.
High gene content, complex organization, and abundant “dark” genes
High gene content
A predictive correlation between protein-coding genes and genome size was developed in Hou and Lin (2009) [98]: log (gene number) =
= LN (-46.200
LN (-46.200 +
+ 22.217*log[genome size]), where genome size is in Kbp. This empirical model predicts 38,188 to 87,688 protein-coding genes for dinoflagellate genomes of ~
22.217*log[genome size]), where genome size is in Kbp. This empirical model predicts 38,188 to 87,688 protein-coding genes for dinoflagellate genomes of ~ 2 to 250 Gbp. However, recent genome data show higher gene numbers than predicted in most cases (Table (Table2),2), suggesting dinoflagellate genome expansion disproportionately favors gene increase compared to other eukaryotes.
2 to 250 Gbp. However, recent genome data show higher gene numbers than predicted in most cases (Table (Table2),2), suggesting dinoflagellate genome expansion disproportionately favors gene increase compared to other eukaryotes.
Table 2
Comparison of identified gene number with predicted gene number based on Hou and Lin (2009) [98]
| Species | Genome size (Mbp) | Detected gene no. | Model-predicted gene no. | % deviationa |
|---|---|---|---|---|
| Parasitic | ||||
 Amoebophyra ceratii AT5.2 Amoebophyra ceratii AT5.2 | 120 | 19,925 | 15,834 | 26% |
 Amoebophyra sp. A25 Amoebophyra sp. A25 | 116 | 28,091 | 15,656 | 79% |
 Amoebophyra sp. A120 Amoebophyra sp. A120 | 115.5 | 26,441 | 15,633 | 69% |
 Hematodinium sp.a Hematodinium sp.a | 4800 | 29,510 | 42,412 | -30% |
| Symbiotic | ||||
 Breviolum minutum Breviolum minutum | 616 | 41,925 | 25,830 | 62% |
 Cladocopium goreaui Cladocopium goreaui | 1,200 | 45,322 | 30,710 | 48% |
 Durusdinium trenchii Durusdinium trenchii | 670 | 30,000 | 26,419 | 14% |
 Fugacium kawagutii Fugacium kawagutii | 935 | 45,192 | 28,828 | 57% |
 Symbiodinium microadriacticum CCMP2467 Symbiodinium microadriacticum CCMP2467 | 808 | 49,109 | 27,758 | 77% |
| Free-living | ||||
 Effrenium voratum RCC1521 Effrenium voratum RCC1521 | 1,200 | 32,108 | 30,710 | 5% |
 Effrenium voratum CCMP3420 Effrenium voratum CCMP3420 | 1,300 | 39,878 | 31,328 | 27% |
 Effrenium voratum CCMP421 Effrenium voratum CCMP421 | 1,100 | 32,615 | 30,046 | 9% |
 Polarella glacialis CCMP1383 Polarella glacialis CCMP1383 | 2,980 | 58,232 | 38,147 | 53% |
 Prorocentrum cordatum Prorocentrum cordatum | 4,750 | 85,849 | 42,316 | 103% |
a(detected-modeled)/modeled x 100
Shared and unique genes and gene families
Current genome sequencing efforts for dinoflagellates have biased toward species with smaller genomes, precluding identification of phylum-wide common genes. Nevertheless, comparative genomic analyses still provide interesting insights into differential dynamics and diversification of dinoflagellate genomes. Analysis of 15 Suessiales genomes found 555,682 predicted protein sequences, with 42,539 forming gene families [23]. Only 2,500 (5.88%) have homologs in all 15 isolates, indicating most gene families are lineage-specific. Within Symbiodiniaceae, 3,445 unigenes are shared by eight species, representing core genes enriched in 219 KEGG pathways (Fig. (Fig.55).
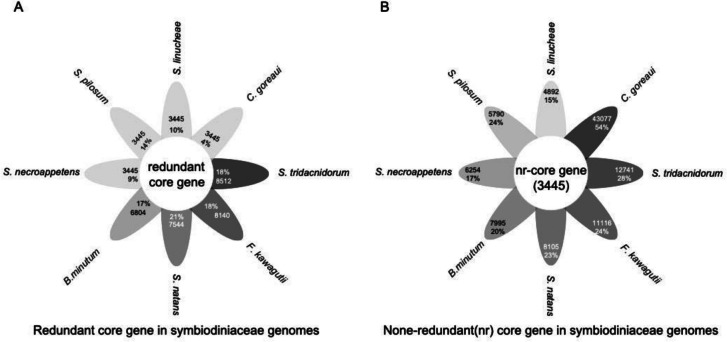
Shared and unique genes in Symbiodiniaceae genomes. A Core genes (3,445 after de-redundancy) in proportion of total genes in each species. B Unique genes in each species. Darker colors indicate a larger number of genes
Highly expressed and most abundant genes
Previous transcriptomics data showed highly expressed genes including MBNP, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), peridinin-Chl a-binding protein (PCP), heat shock protein (HSP) 90, HSP70, HSP40, elongation factor EF-1α, S-adenosyl-L-methionine (SAM) synthetase, A-adenosyl-homocysteine hydrolase (SAHase), and calmodulin [2]. Now with more data, the highly expressed genes exhibit more inter-specific difference (Table (Table33).
Table 3
Ten most highly expressed genes in dinofalgellates (shading indicates grouping by gene name)
| Species | Ranking | Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| Non-Symbiodiniaceae | MBNP | GAPDH | PCP | HSP90 | HSP70 | HSP40 | EF-1α | SAMS | SAHase | CaM | Lin et al. 2011 [2] | ||
| Cladocopium goreaui | PCP | atpE | unk1,2 | CaM | MBNP | FAM135A | unk3 | PSMG1 | psbD | unk4 | unk5 | RPL28e | Huang and Lin unpubl |
| Durusdinium trenchii | PEX16 | EFG | GAPDH | UBE | USP7 | PCP | unk8 | psbJ | EF-1α | petJ | unk9 | TUBB | Huang and Lin unpubl |
| Effrenium voratum | PCP | psbA | petJ | unk10 | petF | unk11–12 | LHCP | unk13 | RPL | unk14–16 | HSP40 | Ub | Li T et al. 2021 [99] |
| Fugacium kawagutti | psbA | petF | NRT2.5 | PCP | unk18–19 | psbV | rbcL | PGK | psbC | psaA | LHCP | Li et al. 2020 [19] | |
| Karenia mikimotoi | HLP | MBNP | LHCP9 | LHCP12 | Ub | unk20 | LHCP4 | LHCP | LHCP8 | unk21 | actin | unk22 | Lin et al. 2022 [100] |
| Karenia mikimotoi | unk23 | psbA | unk24 | SAG-L | MBNP | HLP | psbD | psaB | atpH | petB | rbcL | psbV | Lin et al. 2022 [100] |
| Prorocentrum shikokuense | SAG-L | unk25–26 | LSU-rRNA | unk27–28 | SKP1 | MBNP2 | psbA | psbE | psbD | PCP | unk29–30 | petB | Zhang et al. 2019 [101] |
| Prorocentrum shikokuense | unk25 | SRm300 | psbA | unk31–32 | psbD | unk33–34 | MBNP2 | RPS15A | RPS30 | unk35–37 | RPL36 | RHO | Yu et al. 2020 [102] |
| Prorocentrum shikokuense | MBNP2 | SRm300 | PCP | unk41 | FCP | unk42 | atpE | unk34 | RHO | MBNP | HSP40 | petB | Li H et al. 2021 [103] |
Pfam analysis for A. gibbosum genome revealed Leucine-rich repeat (LRR), Ankyrin, Tetratricopeptide (TPR), and Pentatricopeptide repeat (PPR) domains as the most abundant domains in this species [12].
Tandem gene arrays and polyprotein expression
Many dinoflagellate genes occur in tandem arrays, likely a result of unequal crossing over in meiosis, replication slippage during DNA replication, retroposition, DNA transposons, and gene conversion [104]. Some of these tandem-arrayed genes are transcribed multi-gene transcripts, which is then trans-spliced, with the addition of a 22-nt highly conserved spliced leader (DinoSL; [105]), to generate individual mature mRNA with DinoSL at the 5’ end and poly A tail at the 3’ end. Others are expressed as polyprotein: each array is transcribed as a poly-gene transcript and translated into a poly-protein peptide, which is subsequently cleaved into individual proteins. Genes expressed as polyproteins documented to date in dinoflagellates include the chlorophyll a-c binding protein [106], luciferase [107], as well as Rubisco II [104, 108]. The Rubisco II polyprotein contains a signal peptide at the N-terminus of the first protein unit guiding the polyprotein’s transport to the chloroplast. At the chloroplast, the polyprotein is cleaved into individual Rubisco with a transit peptide at the N terminus to facilitate entrance into the plastid, where the transit peptide is cleaved [2, 104]. The polyprotein gene arrays were also found from the recent genome sequencing [22]. While tandem repeats are common in dinoflagellates, polycistronic expression appears rare, as manifesting in L. polyedrum [109].
Polyprotein genes such as Rubisco II in dinoflagellates likely have resulted from retroposition-based gene duplication, because the gene cluster contains only one regulatory system, without stop codons between individual proteins in the tandem array, characteristic of retroposed genes [104]. Polyprotein expression is generally less common in eukaryotes than viruses, but TY1 retrotransposon in yeast and certain plant storage proteins such as in seeds, and prohormones in mammals are expressed as polyproteins. Interestingly, viruses express both polycistronic and polyprotein genes; some viruses use polycistronic mRNAs, while others use polyproteins. Further research is required to address the mechanism underlying the formation of tandem repeats and polyprotein genes in dinoflagellates.
“Dark” genes
About 50% or more of predicted genes in dinoflagellate genomes lack functional annotation. This applies to both small (Amoebophrya) and large (P. cordatum, A. gibbosum) genomes [11, 12, 22]. Some dark genes are common across taxa, but most are species-specific [23, 110]. These may have crucial functions, as shown in a bloom metatranscriptomic study [101].
Gene expression and regulation: Roles of cold shock proteins, microrna, and alternative splicing
Dinoflagellates are generally known to have limited transcriptional regulation (5–27% of genes; for review see Lin 2011 [2]). However, significant transcriptional responses to nutrient changes have been reported [111, 112]. For example, alkaline phosphatase gene expression and enzyme activity increase under phosphate limitation [112–115]. Nitrogen deficiency elicits significant upregulation of acquisition genes and downregulation of nitrogen associated protein gene (NAP50) [116]. In addition, Rubisco, antenna protein, and some cell cycle protein genes show strong diel dynamics of transcript abundance [72, 108]. Earlier studies indicated that several percentage of genes exhibited circadian rhythms of transcript abundance at 2- or more fold variations [117]. These findings suggest that dinoflagellates may have more dynamic transcriptional regulation than previously thought, particularly in response to environmental changes and daily cycles.
General gene expression
General transcriptional machinery in eukaryotes includes RNA polymerase II transcription initiation factors (TFII), TFIIA, TFIIB, TFIID, TFIIE, and TFIIH. These factors assemble on promoter DNA alongside polymerase II to form a multiprotein-DNA complex crucial for precise initiation of basal-level transcription [118]. Additionally, transcriptional activators may bind to the enhancer region and coactivators may mediate interactions between enhancer-bound activators and promoter-bound gene-specific transcription factors (TFs), regulating transcription responses to environmental cues [119].
Of the five TFII genes, only one TFIID and three TFIIH genes were identified in the F. kawagutii genome [18, 118]. Dinoflagellates lack the typical eukaryotic TATA box (consensus sequence TATAAA) that interacts with TATA-binding protein (TBP), but instead use TTTT interacting with a TBP-like protein (TBL), as previously proposed and confirmed by genome data [18, 120].
Cold shock domain (CSD)
CSD-containing proteins (CSPs) were initially identified in Escherichia coli during cold shock stress and are now known to occur in major groups of organisms. In prokaryotes, CSPs act mainly as RNA chaperones and can regulate transcription by binding to the gyrA promoter. In eukaryotes, CSPs usually contain additional domains and play roles in cold stress response, nutrient limitation, and growth. In plants, CSPs are essential for freezing tolerance and regulate translation during cold stress, seed germination, and flowering [121].
CSPs are ubiquitous in dinoflagellates. In L. polyedrum, CSP genes were the only gene family with higher transcriptional levels than in other protists [109], suggesting their significant role. Some LpCSPs contain only the CSD domain while others have additional domains, but neither showed cold induction [122]. In F. kawagutii, at least eight of 121 identified transcription factors (TFs) contain CSD (Skav204535, Skav204536, Skav217190, Skav218283, Skv218284, Skv223430, Fkav224338, Skv231215), suggesting that CSPs may be the functional domains of TFs in dinoflagellates.
CSPs were identified in Scrippsiella acuminata, showing increased expression in resting cysts [123]. Prorocentrum cordatum has two CSD genes that respond significantly to low temperatures [124]. Dinoflagellate CSPs contain a ZF domain and a glycine-rich motif besides the conserved CSD, with varying ZF and GR order between species. This organization resembles bacterial and plant CSPs, suggesting possible bacterial origin through HGT.
DNA and RNA binding assays showed dinoflagellate CSD proteins prefer binding RNA over DNA, and single-stranded over double-stranded DNA, without sequence specificity [118]. This suggests they are unlikely to act as transcription factors but may be involved in DNA unwinding. Some CSPs contain calcium-regulated mRNA-binding or helicase ATP-binding domains in certain species (Fig. (Fig.66).
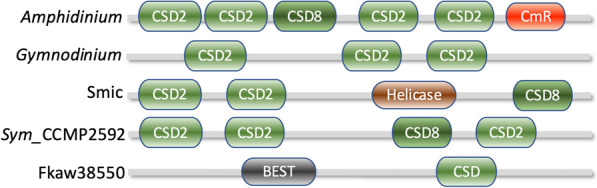
Organization of cold shock domain (CSD) in various proteins in dinoflagellates. Amphidinium, A. massarti, data source MMETSP0689; CSD2, domain PS51857; CSD8, domain SM00357; CmR, Calcium regulated mRNA-binding domain (IPR052069); Gymnodinium, G. catenatum, data source MMETSP0784-20121206; Smic, Symbiodinium microadriaticum, data source CAE7767176.1 dbp2, Helicase = helicase superfamily ATP-binding domain; Sym_CCMP2592, Symbiodinium strain CCMP2592; Fkaw38550, Fugacium kawagutii, BEST = bestrophin
MicroRNA (miRNA)
The scarce and weak transcriptional regulation in dinoflagellates suggests other mechanisms must exist. MicroRNA repertoires have been documented in the transcriptomes of Symbiodinium microadriaticum [125] and Prorocentrum shikokuence [102, 114] and the genome of F. kawagutii [18].
Functional microRNA machinery is evidenced by effective RNA silencing (Zhang and Lin 2019). A. gibbosum genome analysis suggests microRNA involvement in regulating responses to nitrogen and phosphate starvation [12].
In silico analyses of F. kawagutii miRNAs indicated potential influence on coral host gene expression [18]. The genome contains a putative RNA transporter homologous to Systemic RNA Interference Deficiency–1 (SID-1) in nematodes and mammals. Close relationship between Symbiodiniaceae and cnidarian homologs suggests possible HGT (Fig. (Fig.7A).7A). Potential target genes of symbiodiniacean miRNA were identified in coral hosts, showing broad interactive protein network (Fig. (Fig.7B)7B) and GO terms (Fig. (Fig.77C).
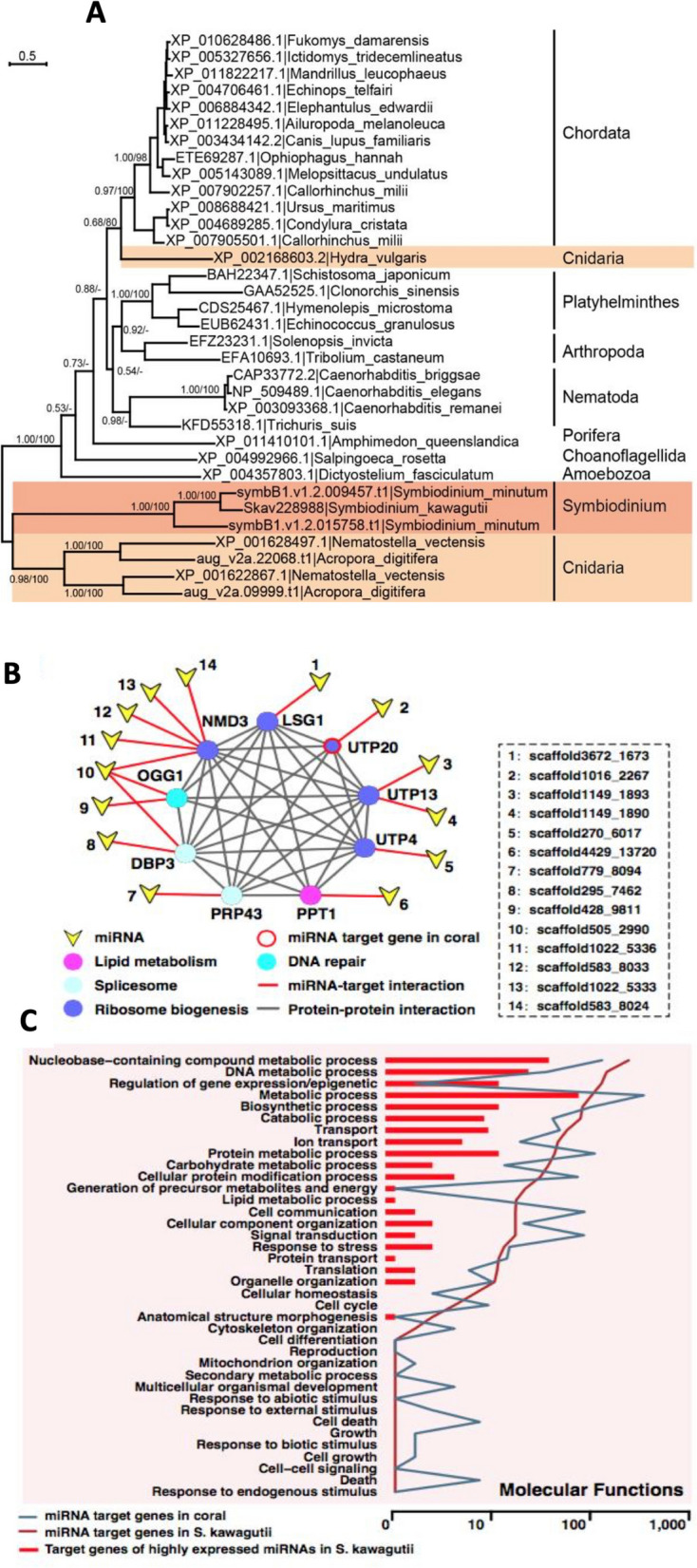
Evidence that Symbiodiniaceae miRNA may interact with coral host to modulate host gene expression. A Phylogenetic tree of double-stranded RNA channel (SID-1). The close relationship between the homologs in Symbiodinium and the cnidarians suggests horizontal gene transfer between the two lineages. B Protein interaction network and metabolic pathways predicted to be impacted by miRNA. C GO categorization of predicted miRNA target genes in both F. kawagutii and coral
These findings suggest Symbiodiniaceae microRNA may cross over into the invertebrate host and modulate gene expression, potentially impacting symbiosis mechanisms like nutrient uptake and photosynthate translocation. While not experimentally demonstrated, this is consistent with findings in other symbiotic and pathogenic systems. Vibrio fischeri produces noncoding small RNA that controls host gene expression in squid light organs [126, 127]. Pathogen-to-host miRNA transfer silences host immunity genes in plants [128].
Alternative splicing
Genome analyses have detected alternative splicing in dinoflagellates. In A. gibbosum, a high number of alternative splicing events were identified, potentially responsible for gene diversification, particularly in secondary metabolite biosynthesis pathways [12]. The majority (77%) of the alternative splicing events occur by skipping exons, and the rest uses alternative 3′splice sites (A3SS) or alternative 5′splice sites (A5SS) (6.8% and 11.3%, respectively). These events mainly impact ion transport, nucleic acid metabolism, and RNA metabolic process. Notably, polyketide synthase (PKS) genes have alternatively spliced isoforms and the genes are expressed as polycistronic. Alternative splicing is also suspected in F. kawagutii and other dinoflagellates [19, 110]. However, the extent and function of alternative splicing in dinoflagellates remain largely unknown.
Post-transcriptional regulation
Ribosome-protected mRNA profiling revealed pulses of synchronous translation of gene clusters throughout the diel cycle [129], indicating circadian or light/dark modulation of gene expression. Pearson correlation analysis of rRNA count versus mRNA count shows that on average, translation of an mRNA requires 3.6 ribosomes (Fig. (Fig.8).8). Deviations from this may indicate above or below average translational efficiency, with exceptionally high translation activity occurring at LD20 (four hours before light-on).
A study of 11 core dinoflagellate transcriptomes revealed diversity in the eIF4E family [130]. Each species contained 8–15 (average 11) different eIF4E family members, phylogenetically segregated into three clades. Dinoflagellate eIF4E-1 likely contains at least one translational initiation factor. eIF4E-2 lacks mRNA cap-binding residues and may have a non-essential regulatory role. eIF4E-3 genes share attributes with metazoan Class I eIF4E. The three clades were further divided into nine subclades, six containing members from all eleven species, suggesting ancestral duplication.
Powerhouse of RNA editing
RNA editing is a gene modification process where single or multiple nucleotides in a gene are altered during or after transcription. This can be identified by comparing the nucleotide sequence of a gene with its corresponding mRNA. RNA editing occurs in mRNA, tRNA, and rRNA, involving mitochondrial, plastid, and nuclear genes [131].
In dinoflagellates, RNA editing was first documented in mitochondrial protein-coding gene transcripts, showing extensive and mechanistic diversity [132]. Mitochondrial mRNA editing has increased in the percentage of impacted nucleotides throughout dinoflagellate evolution [27]. Most nucleotide changes are synonymous and do not affect the encoded proteins but tend to increase the GC content of the impacted genes. This may enhance translation efficiency since dinoflagellate mitochondria import cytoplasmic tRNA for translation, which may not function well with AT-rich mitochondrial genes [27]. This explanation, however, does not apply to rRNA editing.
The field of RNA editing in dinoflagellates has advanced significantly in the last decade. mRNA editing of mitochondrial protein-coding genes has been extended to basal, parasitic dinoflagellates such as Amoebophrya [9]. Furthermore, editing was also found in chloroplast-encoded transcripts, including mRNA and rRNA [133, 134]. For coral symbiotic Symbiodiniaceae, RNA editing was detected in in-hospite populations [135]. Interestingly, mitochondrial gene transcripts are more frequently edited than plastid genes [135]. RNA editing occurs not only in typical peridinin-containing dinoflagellate plastids but also in Chl b-containing plastids in Lepidodinium spp. and fucoxanthin-containing tertiary plastids of haptophyte origin in Karlodinium veneficum [136–138]. Whether plastids in the Dinotom group, such as Durinskia kwazulunatalensis with diatom-origin plastids, also undergo RNA editing remains unclear. This, along with the accelerated evolution of plastid protein-encoding genes in peridinin- and fucoxanthin-containing species, indicates a significant host effect [139].
Furthermore, mRNA editing of nucleus-encoded genes has also been detected. For instance, one of the saxitoxin biosynthesis genes, sxt4, was found to undergo editing in Alexandrium pacificum [140]. Additionally, using high-throughput sequencing, mRNA editing has been detected in nuclear genes. Up to 10,486 and 69,953 putative RNA editing sites were identified in the nuclear genomes of the coral symbiont D. trenchii CCMP2556 and the free-living bloom-forming species P. cordatum CCMP1329, respectively [22]. These events included all 12 possible types of RNA edits, with more transitions than transversions and a dominance of A-to-T transversion in noncoding regions, many of which were condition-specific. Furthermore, A-to-T editing within untranslated regions appears to be associated with the upregulation of edited genes under heat stress [15]. If experimentally verified, the high number of RNA editing events suggests that RNA editing may be a more influential molecular mechanism regulating gene expression in dinoflagellates than previously thought.
New insights on dinoflagellate chloroplasts
Nucleomorphs, relic nuclei of eukaryotic endosymbionts, have been found in some dinoflagellate chloroplasts. Previously known only in the phylum Cryptophyta [141], a recent study discovered nucleomorphs in two newly described dinoflagellate species with green chloroplasts [142]. These chloroplasts, like those in Lepidodinium spp., originate from prasinophytes, although Lepidodinium spp. do not contain nucleomorphs.
Typical dinoflagellate chloroplast genomes are composed of minicircles, each containing one or two genes (occasionally none), totaling 16–17 genes per plastid genome [143]. In contrast, plastid genomes in Durinskia baltica and Kryptoperidinium foliaceum are large macrocircles, similar to typical algal plastids, and more similar to those of diatoms [144]. Chloroplast proteins in dinoflagellates evolve faster than in other algal lineages, indicating a strong host effect on plastid evolution [139].
Dinoflagellates have conserved photosystem cores (PSI and PSII) but varied light-harvesting complexes (LHCI and LHCII). Dinoflagellates have 12 subunits in PSI, ten of which are shared by all photosynthetic organisms, and 24 in PSII, 20 of which are shared by all photosynthetic organisms (Fig. (Fig.9).9). Dinoflagellate LHCs consist of a thylakoid intrinsic Chl a-Chl c2-peridinin-protein complex (acpPC) and a water-soluble peridinin-Chl a-protein (PCP) complex [145, 146]. Peridinin is the major accessory pigment (carotenoid) unique to dinoflagellates, which is used to define typical dinoflagellates. PCP exists in two forms, differing in absorption and fluorescence properties [147, 148].
Comparison of dinoflagellate photosystem (PS) components with counterparts in other algal lineages. A PSI. Reproduced from Lin et al. 2024 [149]. B PSII. Information source: Fromme et al. 2001 [150], Pi et al. 2018 [151], Qin et al. 2019 [152], Gisriel et al. 2022 [153], Su et al. 2019, 2022 [154, 155], Zhao et al. 2023 [156], Li et al. 2024 [82]
Single-particle electron microscopy of PSI-LHCI supercomplexes in Breviolum minutum revealed 25 LHCs, including LHCF and LHCR families, and indicated a role in photoprotection via nonphotochemical quenching [157]. Recent cryo-EM studies identified novel PSI subunits (PsaT and PsaU) and suggested enhanced electron transport and energy quenching efficiency [82, 158].
Photochemical experiments have shown high quenching efficiency in dinoflagellates, such as symbiodiniacean species. Under stress conditions, Cladopodium cells activate a “super-quenching” mechanism, transferring excess excitation energy to PSI and converting it into heat, potentially triggering symbiont expulsion or bleaching [159].
Adaptation to symbiotic lifestyle
Comparative genomic analyses reveal that genes involved in nutrient uptake, photosynthate transport, stress response, and infection are present and expanded in symbiodiniacean dinoflagellates [18, 24]. In the genomes of B. minutum and F. kawagutii, genes important for mutualism, such as those related to sugar and fatty acid metabolism, oxidative stress, transport, photosynthesis, and adhesion, have undergone duplication during two major retroposition events (Fig. (Fig.10;10; [49]). Comparison of symbiodiniacean genomes with those of other algae, the land plant Arabidopsis thaliana, and the parasite malaria Plasmodium falciparum identified genes unique to Plasmodium and Symbiodiniaceae, such as those involved in cell recognition (e.g. merozoite surface protein 1) and genes with unknown functions (e.g. uncharacterized protein PF11_0213, protein dpy-19 homolog 1) [18]. In Plasmodium, merozoite surface proteins are crucial for red blood cell invasion (Beeson et al. 2016).

Two episodes of retroposition discovered in two Symbiodiniacea genomes coincident with two major periods of symbiosis evolution. A) Fugacium kawagutii. B) Breviolum minutum. a, first episode led to expansion of sugar and fatty acid metabolism, oxidative stress response, and transport. b, second episode that promoted photosynthesis and adhesion. Based on Song et al. 2017 [49]
The F. kawagutii genome encodes 16 putative circumsporozoite protein (CSP)-coding genes and a CSP-like gene with a forkhead domain, potentially regulating vesicle-mediated transport and CSP secretion. In the chlorophyte symbiont Symbiochlorun hainanensis, a CSP-like protein is upregulated by acidification and heat stress [160]. Protein secretion via the T2S system, encoded by at least 12 genes, is a major virulence mechanism in bacterial infections and may participate in bacteria-eukaryote mutualism [161–163].
UV protection is crucial in coral-Symbiodiniaceae mutualism. Mycosporine-like amino acids (MAAs) prevent UV damage, initially believed to be synthesized only by the basal lineage (clade A) of Symbiodiniaceae [164]. Genome analyses confirmed the presence of MAA biosynthesis genes in S. tridacnidorum and absence in later-diverging lineages like F. kawagutii and Cladocopium [18, 25], although an improvement of the genomes later revealed partial MAA biosynthesis genes in F. kawagutii [19]. Furthermore, the MAA gene cluster was found to be present in D. trenchii, in addition to Symbiodinium, but absent in B. minutum, consistent with the original notion of ancestral synthesis ability with secondary loss [16].
Insights into mutualism were gained from green Noctiluca scintillans, which harbors prasinophyte Protoeuglena noctilucae as endosymbionts. This mixotrophic dinoflagellate forms massive blooms in the Arabian Sea, feeding on phytoplankton and producing ammonium for symbiont photosynthesis [165, 166]. During blooms, symbiont photosynthesis is active, but cell division is suppressed, suggesting the host promotes symbiont photosynthesis while inhibiting symbiont proliferation to maximize photosynthate translocation [166].
Adaptation to parasitic lifestyle
Amoebophrya species are intracellular parasites of marine dinoflagellates, radiolarians, ciliates, and other Amoebophrya strains. The genomes of Amoebophrya ceratii strain AT5.2 and Amoebophrya sp. strains A25 and A120 are significantly streamlined, with sizes (~ 120 Mbp) much smaller than other dinoflagellates (~
120 Mbp) much smaller than other dinoflagellates (~ 1-250 Gbp). They have few transposable elements, short introns and intergenic regions, and a limited number of gene families, encoding 19,925, 28,091, and 26,441 genes, respectively [10, 11]. These genomes are gene-rich, with 26%, 27%, and 80% more genes than predicted for AT5.2, A25, and A120, respectively, comparable to some other dinoflagellates (7-89% above model prediction; Table Table22).
1-250 Gbp). They have few transposable elements, short introns and intergenic regions, and a limited number of gene families, encoding 19,925, 28,091, and 26,441 genes, respectively [10, 11]. These genomes are gene-rich, with 26%, 27%, and 80% more genes than predicted for AT5.2, A25, and A120, respectively, comparable to some other dinoflagellates (7-89% above model prediction; Table Table22).
Most genes in all three species (56%, 63.7%, and 59%) lack functional annotation in public databases, suggesting some may be specific to parasitism. The intron density (about 1.3, 1.47, and 1.42 introns per kb of coding sequence in AT5.2, A25, and A120, respectively) is typical of dinoflagellates, but the introns are mostly non-canonical, including repeated introns with highly variable splicing motifs. The canonical GT-AG splice sites account for only 34.02% and 30.41% in strains A25 and A120, respectively, with the majority (> 65%) being various other boundaries. In these strains, 11-30% of the non-canonical introns contain 8–20 nt inverted repeat motifs and 3–5 nt direct repeat motifs, spreading similarly to transposable elements (TEs).
65%) being various other boundaries. In these strains, 11-30% of the non-canonical introns contain 8–20 nt inverted repeat motifs and 3–5 nt direct repeat motifs, spreading similarly to transposable elements (TEs).
High gene block synteny (> 49%) but low gene sequence similarity (<
49%) but low gene sequence similarity (< 50%) was observed among the Amoebophrya strains, suggesting strong selection pressure for protein changes due to parasitic interaction with different hosts. All three strains exhibit organelle reduction, including loss of the plastid and potential loss of mitochondrial genome and functions, indicating adaptive evolution. This is consistent with findings in Hematodinium, another parasitic dinoflagellate, which infects shellfish and other organisms [18].
50%) was observed among the Amoebophrya strains, suggesting strong selection pressure for protein changes due to parasitic interaction with different hosts. All three strains exhibit organelle reduction, including loss of the plastid and potential loss of mitochondrial genome and functions, indicating adaptive evolution. This is consistent with findings in Hematodinium, another parasitic dinoflagellate, which infects shellfish and other organisms [18].
Transcriptome sequencing of Hematodinium revealed 29,510 unique ORFs with no evidence of nucleus-encoded plastid-targeted protein genes or plastid gene homologs. The same BLAST workflow revealed 94 putative mitochondrial proteins. Genes involved in fatty acid, lysine, and tetrapyrrole synthesis lack organelle-targeting signal peptides, suggesting these pathways occur in the cytosol. Interestingly, the type I fatty acid synthase (FAS) responsible for cytosolic FA synthesis appears to have taken the form of plastid type II FAS by fusing multiple protein genes [20].
Highlight of newly found genes and physiologies
The rapidly growing dinoflagellate genomic data constitute a rich resource for exploring unique physiologies or metabolic features of this unique group of protists. The many functional unknown genes predicted from their genomes will also serve as a target of future studies. A few of these genes, which are highly interesting but have not received adequate attention, deserve some highlight here. They are involved in acquisition and storage of nutrients, defense (toxin production), response to stress, and life cycle.
Use of Cyanate as a Nitrogen Nutrient
Alexandrium catenella ( formerly A. fundyense) actively expresses cyanate lyase (cyanase) during toxic bloom outbreaks. Zhuang et al. (2015) observed cyanase expression in a Northport Harbor, Long Island Sound bloom, indicating nitrogen stress due to rapid population growth [116]. Metatranscriptomic data showed downregulation of nitrogen-associated proteins (NAPs) and active expression of urea and nickel transporters (nickel is a cofactor of urease) and urease. This suggests A. catenella utilizes all nutrient acquisition mechanisms, including cyanate metabolism. Metagenomics data further revealed that cyanase is globally prevalent in plankton, underscoring the role of cyanate metabolism in the nitrogen biogeochemical cycle [167].
formerly A. fundyense) actively expresses cyanate lyase (cyanase) during toxic bloom outbreaks. Zhuang et al. (2015) observed cyanase expression in a Northport Harbor, Long Island Sound bloom, indicating nitrogen stress due to rapid population growth [116]. Metatranscriptomic data showed downregulation of nitrogen-associated proteins (NAPs) and active expression of urea and nickel transporters (nickel is a cofactor of urease) and urease. This suggests A. catenella utilizes all nutrient acquisition mechanisms, including cyanate metabolism. Metagenomics data further revealed that cyanase is globally prevalent in plankton, underscoring the role of cyanate metabolism in the nitrogen biogeochemical cycle [167].
Storage of Nitrogen as Uric Acid or Polyguanine
Dinoflagellates have long been thought to store nitrogen in uric acid crystals. NanoSIMS and transmission electron microscopy studies confirmed that Symbiodiniaceae species store nitrogen as uric acid crystals following sudden environmental nitrogen increases [168]. Pulses of ammonium, nitrate, or aspartic acid promoted uric acid crystal accumulation, which formed quickly and were remobilized within 24 h. Genome analysis of Fugacium kawagutii found genes promoting uric acid biosynthesis, such as xanthin dehydrogenase/oxidase [18].
Recent studies, however, showed that dinoflagellates also synthesize guanine polymers. Jantschke et al. (2019) [169] and Mojzes et al. (2020) [170] observed guanine crystals in Calciodinellum operosum and Amphidinium carterae, suggesting their function in light scattering into chloroplasts. In A. carterae, guanine supported growth as the sole nitrogen source for several generations, suggesting guanine crystals as nitrogen storage may be functionally equivalent to cyanophycin in cyanobacteria.
Phagotrophy Gene Repertoire
Phagotrophy, a mode of nutrition involving particle ingestion through endocytosis, is common among dinoflagellates. This process includes four major steps: endocytosis, phagosome formation, fusion of phagosomes with lysosomes, and autophagy [171, 172]. Phagotrophy is an ancient feature found in various organisms, including heterotrophic and photosynthetic protists [173–175].
Dinoflagellates, comprising 50% heterotrophic species, exhibit mixotrophy with phagotrophic capabilities [176, 177]. The molecular mechanisms of phagotrophy in dinoflagellates are still being uncovered. Genes involved in autophagy and endocytosis, such as ATG3, ATG8, ATG12, PIP5K, PLD, AP2 adaptor complex, dynamin, epsin, Eps15, EH domain-containing protein 1, VPS4, VPS35, VPS45, Rab8, and Rab11, have been documented in laboratory cultures and natural assemblages [101–103, 178]. In a natural bloom of Prorocentrum shikokuense, genes involved in endocytosis, phagosome, peroxisome, and lysosome were actively expressed, particularly at night, suggesting increased feeding activity during nighttime [102].
These findings highlight the complexity and diversity of nutrient acquisition and storage mechanisms in dinoflagellates, emphasizing their adaptability and ecological significance.
Polyketide Synthase (PKS)/Non-ribosomal Peptide Synthetase (NRPS) Enzyme System and Secondary Metabolites
PKS/NRPS enzymes are crucial for producing specialized metabolites, including toxins, in dinoflagellates [179]. PKSs consist of an acyl-transferase (AT) domain, an acyl-carrier protein (ACP), and a ketosynthase (KS) domain and are classified into Type I, II, and III based on their organization [180]. Type I PKS has all catalytic domains on a single polypeptide, which is used in a processive fashion for chain elongation, as in animals and fungi. Type II PKS is a multiprotein complexe where each catalytic domain is found on a separate polypeptide. Type III PKSs, functioning iteratively similarly to Type II, are known from the plant chalcone/stilbene synthases (CHS/STS) producing compounds such as naringenin chalcone. NRPSs synthesize non-ribosomal peptides and include adenylation (A-domain), thiolation (T-domain), and condensation (C-domain) domains [181]. These enzymes can work together to produce hybrid natural products [179]. PKS and NRPS enzymes are found in toxin-producing dinoflagellates like Karenia brevis, Gambierdiscus spp., and Amphidinium gibbosum [12, 182–185] and in the symbiotic lineage Symbiodiniaceae [24, 186]. Dinoflagellates are unique in possessing both canonical Type I PKS and an unusual Type I sequence single-domain PKS (e.g. KS) [187, 188] and significantly expanded the PKS family relative to other protists [189]. A study scanning 47 dinoflagellate transcriptomes for modular synthase domains and their co-occurrence with thiolation domains revealed widespread presence, but in particularly high abundance in Gymnodiniales, of the thiolation domains [190]. Some of these occur alongside tetratricopeptide repeats (especially hexa- and hepta-repeats), which are unique to dinoflagellates.
Cyclophilins
Cyclophilins are a family of proteins known for binding cyclosporin A (CsA), an immunosuppressant drug in humans [191]. These proteins possess peptidyl-prolyl cis-trans isomerase activity, essential for correct protein folding. Cyclophilins have diverse functions, including protein folding, signaling, transcriptional regulation, cell cycle control, and stress response [192]. In algae, cyclophilins have been reported in Chlamydomonas [193] and Ulva [194], where they are implicated in immunity and stress response. For dinoflagellates, cyclophilin genes were detected over two decades ago (Lin and Zhang 2010), but their functions remain unclear. Recent studies indicate cyclophilins in P. cordatum are induced by copper and polychlorinated biphenyl exposure, suggesting a role in stress response [195]. Cyclophilin B in Margalefidinium polykrikoides is implicated in environmental stress responses [196].
Macrophage Migration Inhibitory Factors (MIF)
MIFs are cytokines and chemokines essential for the vertebrate immune system, mediating innate immune responses [197]. These proteins are conserved across the tree of life, serving various cellular functions [198]. In free-living organisms, MIFs activate immune responses, promote immune cell proliferation, and inhibit p53-mediated apoptosis [199]. Parasitic species produce multiple MIF proteins to modulate host immune responses, facilitating infection [200–203]. A homolog of MIF (PmMIF) was identified in the cyanobacterium Prochlorococcus marinus, showing high structural homology with mammalian MIFs but lacking the Cys-X-X-Cys motif necessary for oxidoreductase activity [204].
A MIF-like protein was detected in L. polyedrum (LpMIF), but instead of a soluble single-domain protein as a typical MIF is, LpMIF is a transmembrane protein with a cytosolic domain [205]. The MIF domain is localized in cell wall-associated membranes, vesicular bodies, and membranes of extracellular vesicles accumulating at the secretory pores of the cells (Fig. (Fig.11A,11A, B). Tblastn analysis using human MIF as query against the MMETSP database (E-value e-20 cutoff) yielded 92 hits, 28 from dinoflagellates including P. shikokuense, Alexandrium spp., Symbiodiniaceae species, Karenia brevis, Karlodinium veneficum, and Heterocapsa triquetra. Phylogenetic analysis shows it is closely related to a prasinophyte homolog within a clade containing cryptophytes (Fig. (Fig.11C).11C). Most dinoflagellate hits align with human MIF (hsMIF) from position 2 to 115, with some being shorter due to incompleteness (Fig. (Fig.1111D).

Macrophage migration inhibitory factors (MIF) detected in dinoflagellates. A Schematic of MIF structure. B Cell surface localization of MIF in L. polyedra (green fluorescence pointed by arrows). From Jaouannet et al. 2020 [205]. C Phylogenetic tree showing clustering of dinoflagellate MIF with homologs from prasinophytes and other lineages. D Alignment of dinoflagellate MIFs with humans MIF showing high similarity. Homsa, Homo sapiens; Alemo, Alexandrium monilatum; Fugka, Fugacium kawagutii; Linpo, Lingulodinium polyedra; SymbB, Breviolum sp.; Symmi, Symbiodinium microadriaticum
Pfam search confirms these dinoflagellate MIF sequences contain the MIF functional domain. KEGG analysis indicates they are phenylpyruvate tautomerase (EC 5.3.2.1), the second member of the MIF superfamily, involved in Tyrosine and Phenylalanine metabolism. Under biotic stress, L. polyedra shows decreased MIF gene expression and reduced protein levels on the cell surface, suggesting this MIF may be involved in intercellular communication [205] or sensing of environmental stimuli.
RACK1 and Hemerythrin-like protein
Islas-Flores and colleagues constructed yeast-two hybrid system cDNA libraries for Symbiodinium microadriaticum based on DinoSL and the SMART technology [206]. Significantly more cDNAs were retrieved from the DinoSL-based libraries than the SMART-based libraries, demonstrating the advantage of DinoSL for constructing dinoflagellate full-length cDNA libraries. More importantly, using the yeast strain harboring Symbiodinium RACK1 (SmicRACK1) as bait and the DinoSL-based library prepared with the other yeast strain as the prey, the yeast-two hybrid system showed the interaction between SymicRACK1 and hemerythrin-like protein, suggesting the latter being the ligand of the former. RACK1 is a member of the WD-repeat protein family, sharing homology with G-protein β subunit (Gβ) and featuring a seven-bladed β-propeller structure for protein binding [207]. WD-repeat proteins, including RACK1, typically serve as scaffolds for protein complexes. They often interact with multiple partners, coordinating signaling events across various pathways and contributing to the spatiotemporal organization of cellular processes. As such, RACK1 functions in shuttling, anchoring, and stabilizing proteins within the cell, interacting with ribosomal machinery, cell surface receptors, and nuclear proteins, influencing various cellular pathways and functions [207]. Hemerythrin is an oxygen-carrier non-heme protein responsible for oxygen transport in several marine invertebrates and worms [208, 209] but also functions in innate immunity and anterior tissue regeneration in certain worms [210]. The hemerythrin-encoding gene has also been found in numerous bacterial, archaeal, and eukaryotic genomes [209]. Besides O2 transport, hemerythrin proteins can serve in signal transduction, phosphorelay regulation, abiotic resistance, and protein binding [208]. The specific roles of SmicRACK1 and hemerythrin-like protein in S. microadriaticum remain to be determined.
Abscisic acid 8’-hydroxylase
Abscisic acid 8’-hydroxylase (EC 1.14.13.93) belongs to the family of oxidoreductase, and it catalyzes the oxidative degradation of abscisic acid (ABA). As a member of the cytochrome P450 monooxygenase family [211], this enzyme is involved in the regulation of germination and dormancy of plant seeds [212] and participates in carotenoid biosynthesis. In dinoflagellates, the encoding gene of this enzyme was shown to be upregulated in cysts, suggesting its role in encystment and maintenance of dormancy [213]. Besides, upregulated expression of this gene was found during the initial response to heat stress in a Cladocopium species [214].
Small heat shock proteins
sHSPs are ancient, universal molecular chaperones involved in cellular stress response [215, 216]. They have a molecular weight of 12–43 kDa and a tripartite domain architecture, consisting of a conserved alpha-crystallin domain (ACD) and variable C and N terminal extensions [217].
Recent research has revealed new information on sHSP gene sequences and stress-dependent expression profiles in dinoflagellates [218]. Using DinoSL as a selective tool, Deng et al. (2021) [218] constructed dinoflagellate-specific full-length cDNA libraries from marine sediment-derived RNA samples. They found sHSPs with A-X-X-X-D/N/S/H/R-G-V-L in the typical conserved A-X-X-X-N-G-V-L motif. Active expression of these sHSPs in sediment suggests their potential role in maintaining dinoflagellate cyst dormancy.
Meiosis genes
Sexual reproduction is widespread among eukaryotes and serves as a mechanism for chromosome recombination, thereby generating genetic diversity [219–221]. Dinoflagellates typically have a haplontic life cycle, consisting of haploid asexual and diploid sexual stages. Meiosis is one of the hallmarks of sexuality, and its regulating genes only began to be identified in dinoflagellates. From genomes and transcriptomes, a range of genes (31–51) with homology to meiosis genes in model organisms have been identified [18, 222, 223]. These include most of the 12 core meiosis genes previously developed for eukaryotes [224] and homologs of yeast meiosis regulating genes such as MEI2 and several meiotically upregulated genes (Table S8 in Lin et al. 2015 [18]). However, securing, which normally serve in preventing the transition from metaphase to anaphase by inhibiting the protease separin, and some components of the cohesion complex with the function to hold homologous chromosomes together are absent [222]. The absence of cohesion complex elements is consistent with more recent findings suggesting that dinoflagellates may have lost the canonical synaptonemal complex [225].
Syngamy is another important hallmark of sexuality. In a transcriptomic study of P. cordatum, HAP2 was identified, along with MEI2, MutS2, and several other meiosis-associated genes [226]. HAP2 (hapless 2) participates in the fusion of cell membranes [227].
In dinoflagellates, sexual reproduction is known to occur under adverse environmental conditions and lead to encystment for survival (‘sex for encystment’) (review in [228]). However, increasing laboratory observations indicate that sexual reproduction can also produce vegetative cells, bypassing encystment [229]. Recent natural bloom transcriptomic studies suggest that sexual reproduction in dinoflagellates may aid in bloom proliferation or extension (‘sex for proliferation’). Specifically, genes specific to or associated with meiosis were significantly upregulated during blooms, unexpected for the ‘sex for encystment’ scenario [100, 166]. Integrating transcriptomic data from natural blooms and laboratory cultures of P. shikokuense, Scrippsiella acuminata, and Karenia mikimotoi, 28 meiosis genes were identified and found to be highly expressed during blooms or cyst germination. Key genes include MEI2, initially identified in yeast for initiating meiosis. RAD21, SMC3, and MEI2 are linked to bloom-promoting sex, HOP2 and MSH4 to cyst germination, while SPO11, MND1, and DMC1 are common to both cyst-forming and non-encysting sex [100]. Verification of these findings could provide markers for distinguishing different sexual reproduction ecotypes in dinoflagellates.
Response to temperature variation
Temperature responses have not received as much research attention as nutrient responses for molecular understanding. Recently, Zhang et al. (2022) [230] investigated proteomic and metabolomic changes in the HAB species P. shikokuense across four temperatures: optimal (25 °C), supra-optimal (28 °C), and sub-optimal (19 °C and 22 °C). They observed decreases in particulate organic carbon and nitrogen (POC, PON) as temperature increased, with an increased POC/PON ratio at higher temperatures, except at 28 °C. These results suggest a higher cell division rate and photosynthetic carbon fixation at higher temperatures. However, at the supra-optimal temperature, cells increased synthesis of light harvesting, photoreaction, and protein homeostasis related proteins. Sub-optimal temperatures upregulated glutathione (an antioxidant), transcription, and lipid biosynthesis, compensating for decreased translation efficiency and cell membrane fluidity. At 19 °C, there was an increase in nitrate reduction and nitrogen flux towards asparagine, aspartic acid, and glutamine, with the accumulation of glutathione, glutarate semialdehyde, and 5-KETE.
Interestingly, the metabolomic data of the study showed a significant increase in a sulfinpyrazone-like metabolite at 19 °C compared to 25 °C (13-fold) and 28 °C (26-fold). Sulfinpyrazone (C23H20N2O3S) is a medicine to treat gout patients as it inhibits reabsorption (transport) of uric acid and enhances its urine excretion [231]. Whether this compound blocks intracellular cross-membrane trafficking of organic nitrogen molecules or is an accumulated intermediate at lower temperatures remains to be investigated.
Another study on P. cordatum revealed differential gene cluster responses between mRNA editing and alternative splicing (exon usage) to temperature changes between 20 °C and 26 °C [22]. Further verification could indicate a novel mechanism of gene regulation. Additionally, P. glacialis is an ideal model for understanding cold and darkness adaptation, having acquired genes from bacteria for ice-binding proteins. This species shows enhanced transcriptional responses through unidirectional, tandem duplication of single-exon genes involved in cold survival and low-light adaptation [21].
Future prospect
Dinomics has lagged behind the omics research of other major algal groups, but a decade of dedicated effort has produced remarkable insights. Many questions in dinoflagellate biology remain to be explored through broader and deeper omics research complemented by other cutting-edge technologies. Some of the key research gaps are discussed below.
A finished dinoflagellate genome is urgently needed to fully understand genome architecture and coding landscapes. The genomes sequenced so far do not exhibit the previously suspected high complexity, but this may change with the sequencing of larger genomes. Furthermore, accurate gene prediction for dinoflagellates likely requires substantial improvement of algorithms. Additionally, no clear evidence of genome duplication has been found, potentially due to the bias towards smaller genomes. The dual-peak k-mer profile in P. glacialis and chromosomal pairing in P. lunula suggest that genome duplication might be prevalent in large dinoflagellate genomes. Sequencing and achieving high-quality assemblies for ecologically important species like Alexandrium catenella (Group I) would be fruitful.
Tomographical Imaging of Chromosomal Architecture harnessed by combining cutting-edge imaging technology with omics, such as Cryo-EM analyses [158, 173], can address many architectural questions of dinoflagellate chromosomes. It remains unclear how the extreme ‘chimeric’ repertoire of nuclear proteins shapes the constantly condensed chromosomes, regulates DNA duplication, and coordinates gene expression in response to environmental changes. Recent transcriptomic and genome methylation data suggest that dinoflagellates may use nucleosomal arrays to regulate gene transcription [89, 90]. In situ nuclear imaging of samples from different physiological states could shed light on these mysteries.
Functional Genetic Research to accelerate functional characterization of the > 50% unannotated dinoflagellate genes is crucial to advance dinoflagellate biology. Additionally, a substantial fraction of “annotated” genes have only weak matches to known genes, and their exact functions need to be experimentally verified. Accessible functional genetic tools need to be established. Gene transformation began in the 1990s and continued through the 2000s, but reproducible success has been limited [232–234]. Gene knockdown has been explored for a few genes, e.g., Rubisco, rhodopsin [101], cellulose synthase CesA1 [235], and eukaryotic translation initiation factor eIF4e [236]. Gene knockout studies using CRISPR/Cas technology have yet to emerge.
50% unannotated dinoflagellate genes is crucial to advance dinoflagellate biology. Additionally, a substantial fraction of “annotated” genes have only weak matches to known genes, and their exact functions need to be experimentally verified. Accessible functional genetic tools need to be established. Gene transformation began in the 1990s and continued through the 2000s, but reproducible success has been limited [232–234]. Gene knockdown has been explored for a few genes, e.g., Rubisco, rhodopsin [101], cellulose synthase CesA1 [235], and eukaryotic translation initiation factor eIF4e [236]. Gene knockout studies using CRISPR/Cas technology have yet to emerge.
Multi-Omic Approach is necessary to Addressing Gene Regulation
Only by systematic investigation can transcriptional, translational, and post-translational gene regulation be unequivocally determined. The long-held notion of limited transcriptional regulation in dinoflagellates may change, as increasing environment-responsive genes have been reported. Translational and posttranslational regulation, including epigenetic mechanisms, need to be investigated simultaneously. Transcriptomics, proteomics, metabolomics, and epigenomics should be pursued together. Collaboration among experts in these subdisciplines is crucial to tackle the same species in a coordinated manner.
Single-Cell Transcriptomics is widely used for marine prokaryotes, but needs to be explored more extensively for eukaryotes [237]. Applying this technique to sequence the transcriptome of cells from the field has recently led to the discovery of new dinoflagellate species [238]. Single-cell sequencing of sorted cells from different spatial positions in a natural bloom can potentially pinpoint the major drivers of the bloom and the range of physiologies occurring within the population.
Integrative Molecular Ecological Framework should be developed
Translating genomic understanding of dinoflagellates into insights about their ecological behaviors in the natural environment requires an integrative framework. Many fundamental ecological questions cannot be answered with omics data or ecological data alone. For example, understanding how dinoflagellates proliferate to form blooms or what triggers coral bleaching requires a quantitative model relating gene expression to physiological rates or biotic interactions. A standardized approach will enable cross-system comparisons. Data and insights from laboratory cultures are foundational to understanding the dynamics and dominance of dinoflagellate species in natural plankton communities. However, in situ studies should be conducted, and such work should be set on an integrative framework from multiple perspectives such as energy (E) and nutrient (N) acquisition, defense (D) against biotic and abiotic stresses, and sexual and asexual reproduction (S) [101, 178]. This ENDS framework has initially facilitated comparisons of how different phytoplankton lineages perform with various nutritional strategies and understand the factors regulating harmful algal bloom outbreaks [82, 102, 178, 239].
Acknowledgements
I wish to thank my students for assistance in various ways, particularly Xiaoyu Wang, Yifan Gu Jiamin He, Jingtian Wang, and Richard Antosca for gathering information, preparing tables and figures, and organizing the references.
Authors’ contributions
S.L. wrote the manuscript text, prepared the figures, and reviewed the manuscript.
Funding
This work was supported by the Gordon and Betty Moore Foundation through grant GBMF# #4980.01.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Not applicable.
Not applicable.
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
 ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 2013;7(1):13–27.
[Europe PMC free article] [Abstract] [Google Scholar]
ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 2013;7(1):13–27.
[Europe PMC free article] [Abstract] [Google Scholar] +
+ and Mg2
and Mg2 +
+ cations. Eur J Cell Biol. 1983;30(1):33–41.
[Abstract] [Google Scholar]
cations. Eur J Cell Biol. 1983;30(1):33–41.
[Abstract] [Google Scholar] –
– chlorophyll a
chlorophyll a –
– proteins at low temperatures. Biochemistry. 2006;45(47):14052–63.
[Abstract] [Google Scholar]
proteins at low temperatures. Biochemistry. 2006;45(47):14052–63.
[Abstract] [Google Scholar]Articles from BMC Genomics are provided here courtesy of BMC
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/169035316
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Protein Families
- (1 citation) InterPro - IPR052069
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions.
BMC Biol, 18(1):56, 24 May 2020
Cited by: 38 articles | PMID: 32448240 | PMCID: PMC7245778
The enigmatic nucleus of the marine dinoflagellate Prorocentrum cordatum.
mSphere, 8(4):e0003823, 26 Jun 2023
Cited by: 2 articles | PMID: 37358287 | PMCID: PMC10449503
Genomic understanding of dinoflagellates.
Res Microbiol, 162(6):551-569, 07 Apr 2011
Cited by: 134 articles | PMID: 21514379
Genome Evolution of Coral Reef Symbionts as Intracellular Residents.
Trends Ecol Evol, 34(9):799-806, 10 May 2019
Cited by: 16 articles | PMID: 31084944
Review
Funding
Funders who supported this work.
Ann and Gordon Getty Foundation (1)
Grant ID: GBMF# #4980.01.





