Abstract
Free full text

SIRT7: a sentinel of genome stability
Associated Data
Abstract
Abstract
SIRT7 is a class III histone deacetylase that belongs to the sirtuin family. The past two decades have seen numerous breakthroughs in terms of understanding SIRT7 biological function. We now know that this enzyme is involved in diverse cellular processes, ranging from gene regulation to genome stability, ageing and tumorigenesis. Genomic instability is one hallmark of cancer and ageing; it occurs as a result of excessive DNA damage. To counteract such instability, cells have evolved a sophisticated regulated DNA damage response mechanism that restores normal gene function. SIRT7 seems to have a critical role in this response, and it is recruited to sites of DNA damage where it recruits downstream repair factors and directs chromatin regulation. In this review, we provide an overview of the role of SIRT7 in DNA repair and maintaining genome stability. We pay particular attention to the implications of SIRT7 function in cancer and ageing.
1. Introduction
The integrity and stability of the genome are constantly challenged by both intrinsic or extrinsic insults such as replication stress, oxidative damage, ultraviolet light, ionizing radiation and various genotoxic reagents, which can ultimately lead to DNA damage [1]. If DNA damage is not properly repaired, it can result in diseases such as cancer, or pathologies associated with ageing [2,3]. To counteract DNA damage, cells have evolved an elaborate mechanism—a tightly regulated DNA damage response (DDR) that detects, signals and repairs DNA lesions. Both normal and malignant cells depend on various DDR pathways to protect their genomes [4]. Depending on the cell cycle stage, genetic background and types of DNA damage, there are five major repair pathways, including non-homologous end joining (NHEJ), homologous recombination (HR), mismatch repair (MMR), base excision repair (BER) and nucleotide excision repair (NER) [5].
Post-translational modifications have a crucial role in mediating the cellular response to DNA damage, providing a means of changing protein activity without the necessity of de novo protein synthesis [6]. The most common post-translational modifications include phosphorylation, ubiquitination, acetylation, methylation and sumoylation [6]. These modifications are reversible due to their regulation by two opposing enzymes. For example, lysine residues are acetylated due to the activity of acetyltransferases that attach acetyl groups and deacetylated due to the activity of histone deacetylases (HDACs) [7].
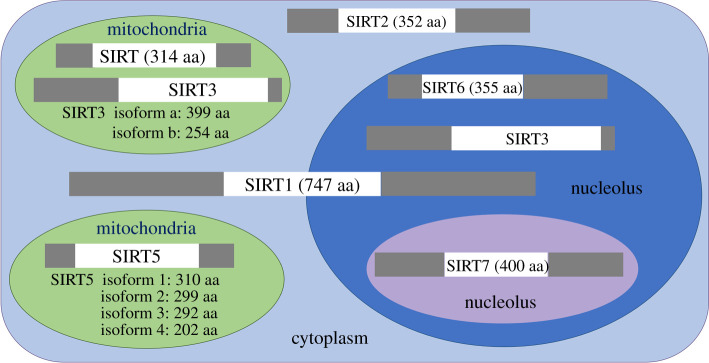
In higher eukaryotes, HDACs can be divided into four classes. Class I Rpd3-like enzymes are comprised of HDAC1, 2, 3 and 8. Class II Hda1-like enzymes are composed of HDAC4, 5, 6, 7, 9 and 10. Class III Sir2-like enzymes consist of seven sirtuins, SIRT1-7, which depend on NAD+ as a coenzyme. Class IV contains only HDAC11 [7–9]. Sirtuins are a class of deacetylases that are homologous to Sir2 (silent information regulator 2), seven members of this family, SIRT1-7, all have a conserved catalytic domain (figure 1). In addition to homology, sirtuins have different types of enzyme catalytic activities, such as ADP ribosyl transferase, desuccinylase and demalonylase, the diverse enzyme activities endow sirtuins with diverse biological functions [10–12].
Among the sirtuins, SIRT7 is the least studied protein, but recent breakthroughs have shown that it is also involved in multiple cellular processes and its biological function is gradually becoming clear. In this review, we outline the current studies regarding the role of SIRT7 in DDR and its potential therapeutic role in disease.
2. SIRT7 structure and function
SIRT7 encodes a 400 amino acid protein and in humans' functions as an NAD+-dependent class III histone deacetylase [13]. Compared with other nuclear-localized sirtuins (SIRT1 and SIRT6), SIRT7 exhibits deacetylase, desuccinylase and deglutarylase activities [14–16]. Over the past two decades, several SIRT7 substrates have been identified (table 1). The wide variety of SIRT7 substrates suggests that SIRT7 participates in diverse biological processes.
Table 1.
SIRT7 targets.
| substrate | activity | functions |
|---|---|---|
| p53 | deacetylation | apoptosis, heart hypertrophy and inflammatory cardiomyopathy [17] |
| H3K18 | deacetylation | oncogenic transformation [14] |
| DAF-16 | deacetylation | stress response [18] |
| PAF53 | deacetylation | pre-rRNA processing [19] |
| NPM1 | deacetylation | ageing [20] |
| PGK1 | deacetylation | glycolysis [21] |
| GABPβ1 | deacetylation | mitochondrial homeostasis [17,22,23] |
| U3-55k | deacetylation | pre-rRNA processing [24] |
| H3K122 | desuccinylation | chromatin compaction [15] |
| FOXO3 | deacetylation | monocyte apoptosis [25] |
| FKBP51 | deacetylation | Akt activity [26] |
| CDK9 | deacetylation | RNA polymerase II transcription [27] |
| DDB1 | deacetylation | activity of the CUL4B/DDB1/DCAF1 E3 ubiquitin ligase complex [28] |
| DDX21 | deacetylation | transcription elongation and genome stability [29] |
| SMAD4 | deacetylation | breast cancer metastasis [30] |
| OSX | deacetylation | bone formation [31] |
| WDR77 | deacetylation | transmethylase activity of the WDR77/PRMT5 complex [32] |
| Fibrillarin | deacetylation | rRNA synthesis [33] |
| H3K36/K37 | deacetylation | heterochromatin silencing [34] |
| ATM | deacetylation | DNA repair [35] |
| Ran | deacetylation | nuclear export of NF-κB p65 [36] |
| H4K91 | deglutarylation | chromatin structure [16] |
| GATA4 | deacetylation | stress-induced cardiac hypertrophy |
| STRAP | deacetylation | p53 activity and stability [37] |
| CRY1 | deacetylation | circadian phase coherence and glucose homeostasis [38] |
| Nfatc1 | deacetylation | hair growth [39] |
| USP39 | deacetylation | hepatocellular carcinoma development [40] |
In chromatin, SIRT7 selectively deacetylates histone H3 lysine 18 (H3K18Ac), which serves to maintain the cellular transformation ability of human cancer cells and tumour formation in vivo [14]. SIRT7 also functions as a desuccinylase of histone H3 lysine 122 and a deglutarylase of histone H4 lysine 91 to promote chromatin compaction [15,16]. Despite its prominent roles regulating chromatin, SIRT7 also deacetylates several non-histone proteins, including U3-specific protein U3-55 k and nucleolar organizer polymerase-associated factor 53 (PAF53) that is involved in the precursor ribose RNA (pre-rRNA) processing [19,24]. SIRT7 also deacetylates GA-binding protein β1 (GABPβ1) to regulate mitochondrion function and phosphoglycerate kinase 1 (PGK1) in regulating glycolysis [21,22]. In addition, SIRT7 participates in ageing processes and breast cancer lung metastasis by deacetylating nucleophosmin (NPM1) and SMAD4 [20,30]. SIRT7 also serves as a key activator of the telogen-to-anagen transition in cycling hair follicles; here, it acts as the deacetylase of NFATc1, which helps activate dynamic hair follicle stem cells [39]. To further widen the range of SIRT7 deacetylation targets, our laboratory conducted stable isotope labelling in SIRT7 knockout cell line coupled with quantitative mass spectrometry. We found a comprehensive list of candidates involved in a variety of functions, ranging from gene regulation to chromatin architecture homeostasis and metabolism [41].
Moreover, multiple studies reveal that SIRT7 regulates proteostasis/endoplasmatic reticulum (ER) stress, mitochondrial protein folding stress and mitochondrial metabolism [17,22,23,42]. SIRT7 is recruited to the promoters of ribosomal protein genes via transcription factor Myc to repress gene expression and to alleviate ER stress [42]. In addition, SIRT7 inactivation caused reduced quiescence, increased mitochondrial protein folding stress, and expression of SIRT7 is reduced in aged haematopoietic stem cells (HSCs) [23]. The same phenomenon was observed in human haematopoietic cells [43]; conversely, SIRT7 upregulation significantly improved the regenerative capacity of aged HSCs [23]. This is the first report linking stem cell ageing and SIRT7, giving the hope for targeting the dysregulated cellular programme to reverse HSC ageing. SIRT7 deacetylates GABPβ1, an important role of regulator of nuclear-encoded mitochondrial genes, which impacts mitochondrial function [22]. SIRT7 arginine methylation, which inhibits its H3K18 deacetylase activity, mediated glucose sensing and signalling with mitochondria biogenesis to maintain energy balance [17].
Most notably, SIRT7 is a crucial player in the DDR: it has histone deacetylase activity at DNA damage sites and exhibits other catalytic activities towards proteins involved in DNA damage and repair [15,35,44]. We discuss these processes in more detail below.
3. SIRT7 in maintaining genome stability
3.1. SIRT7: guardians of genome integrity and stability
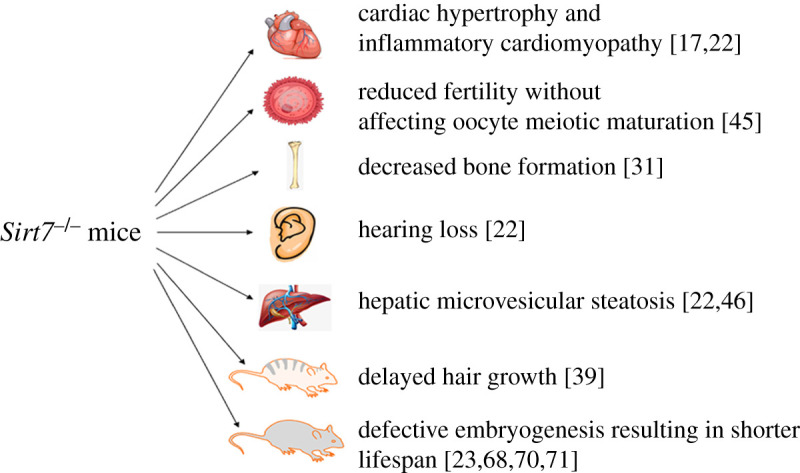
Numerous studies support a role for SIRT7 in genome stability and organismal viability. Much support has come from the use of Sirt7 knockout mice (figure 2). Vakhrusheva et al. [17] found that Sirt7-deficient mice suffer from degenerative heart hypertrophy, accompanied by inflammatory cardiomyopathy and decreased resistance to cytotoxic and oxidative stress. In female Sirt7 knockout mice, Vazquez et al. [45] found that Sirt7−/− females exhibit reduced fertility without an effect on oocyte meiotic maturation. Multisystemic mitochondrial dysfunction is also observed in Sirt7-deficient mice.
SIRT7 functions at chromatin to suppress ER stress and prevents fatty liver disease, and SIRT7-deficient mice develop chronic hepatosteatosis resembling human fatty liver disease, and liver-specific reconstitution of SIRT7-deficient mice reversed the fatty liver phenotype. Strikingly, SIRT7 overexpression in the livers of high-fat, diet-fed mice suppressed ER stress and rescued the fatty liver phenotype [42]. Sirt7−/− pups are born at sub-Mendelian ratios, indicating a defect in embryogenesis. Mutant mice that survive to adulthood exhibit a shortened lifespan with signs of accelerated ageing such as premature (6 months), kyphosis and decreased gonadal fat pad content [44]. Sirt7−/− mice exhibit elevated blood lactate levels, exercise intolerance, cardiac dysfunction, microvesicular steatosis and age-related hearing loss. In addition, in the liver-specific Sirt7 KO (Sirt7hep−/−) mice display the same hepatic mitochondrial dysfunction and represents SIRT7 activity in a cell-autonomous effect on mitochondria function [22]. SIRT7 expression is reduced in aged HSCs, which are characterized by increased apoptosis, loss of quiescence and decreased reconstitution capacity, features resembling those observed in Sirt7−/− mice, and in mice reconstituted with Sirt7−/− HSCs improved their regenerative capacity [23].
Using hair follicle stem cell-specific Sirt7 knockout mice, Li et al. [39] found that loss of Sirt7 impedes the follicle life cycle transition from telogen to anagen phase and delays hair growth. In addition, in response to pressure overload, the cardiomyocyte-specific Sirt7 knockout mice show severe cardiomyocyte hypertrophy [46]. Osteopenia-specific Sirt7 knockout mice showed decreased bone formation that occurred via acylation of SP7/Osterix (OSX)—a transcription factor that activates genes involved in osteoblast differentiation [31]. Finally, Fang et al. [47] reported that Sirt7-deficient mice show increased Sirt1 activity, resulting in inhibited PPARγ expression and thus restrained adipocyte differentiation and diminished white fat accumulation. The phenotypic consequences of SIRT7 deficiency could be explained by the functional link of SIRT7 with the maintenance of genome stability.
3.2. SIRT7 regulates DNA double-strand break repair
DNA double-strand breaks (DSBs) constitute the most toxic type of DNA lesion. As such, they must be efficiently repaired to maintain genome stability. DSBs are mainly repaired by NHEJ, which is predominant in non-cycling cells exposed to genotoxic stress, and HR, which functions in proliferating cells as it requires the pairing of sister chromatids [48]. Regarding HR, data from a previous report suggested that SIRT7 might regulate HR-mediated repair [49]. However, the detailed mechanism remains largely unknown and requires further investigation. For this reason, we explain how SIRT7, which is efficiently recruited to DSBs, is involved in mediating NHEJ. Whether SIRT7 is involved in other forms of DNA repair is largely unknown.
Upon DSBs, driven by a signalling cascade, which is initiated by ataxia-telangiectasia mutated (ATM)-mediated phosphorylation of histone 2A variant H2AX to generate γ-H2AX, this process is followed by the recruitment of the mediator of DNA damage checkpoint protein 1 (MDC1) and activation of RNF8–RNF168-dependent ubiquitination. Following the ubiquitination of H2A at lysine 13 and lysine 15 (H2AK13ub and H2AK15ub), and histone H4 lysine 20 dimethylation (H4K20me2) and histone H4 lysine 16 monomethylation (H4K16me1), 53BP1 is rapidly recruited onto chromatin surrounding the DSBs where it serves as an effector of the NHEJ pathway [50–54].
Interestingly, Vazquez et al. [44] found that 53BP1 foci are remarkably reduced in SIRT7−/− cells, and that DNA damage, mutations and replication stress accumulate. The resulting genome instability leads to compromised NHEJ (figure 3a).
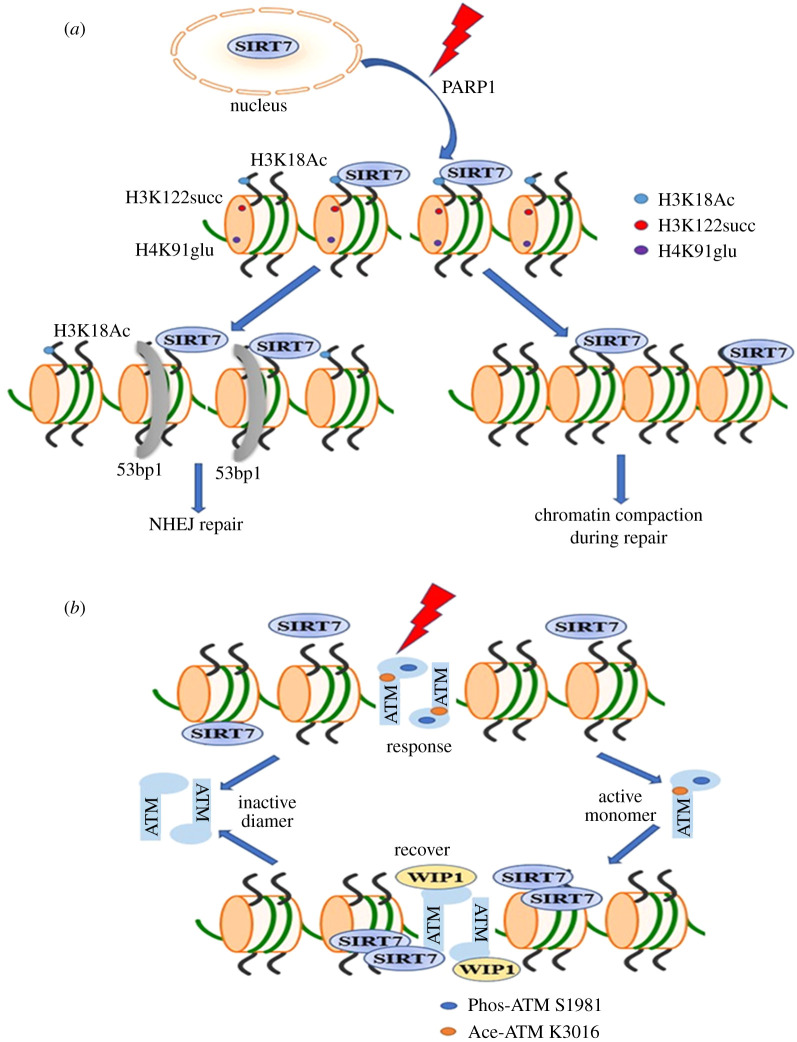
The role of SIRT7 in DNA repair. (a) In response to DNA damage, SIRT7 is recruited to DSBs that depend on PARP1, where it deacetylates H3K18ac and allows 53bp1 recruitment for the repair via the NHEJ pathway. After its recruitment to chromatin, SIRT7 mediates H3K122 desuccinylation and H4K91 deglutarylation for chromatin compaction necessary for DNA repair. (b) In response to DNA damage, ATM is sequentially modified by acetylation and phosphorylation before ATM dissociates into an active monomer. At the late stage of DNA damage and repair, SIRT7 is gradually recruited to DNA damage sites and deacetylates ATM; this process facilitates ATM dephosphorylation by WIP1. After complete repair, ATM dimerizes into an inactive form.
SIRT7 is, in fact, recruited to DSBs, but at a relatively slower rate compared with SIRT1 and SIRT6 [44]; its recruitment depends on poly (ADP-ribose) polymerase (PARP) activity, which ensures the recruitment of several DNA damage repair proteins to damaged sites [44,55,56]. A direct interaction between SIRT7 and PARP1 has been reported [15], but the detailed mechanism and function of SIRT7–PARP1 interplay is unknown.
Consistent with the previous report regarding the effects of SIRT7 on NHEJ, Chen and coworkers identified the Dicer protein in the regulation of SIRT7 localization upon DNA damage. They find that DNA damage agents can induce Dicer expression and results in increased trapping of SIRT7 in the cytoplasm and increases H3K18 acetylation at sites of damaged DNA and facilitates NHEJ repair pathway [57,58]. Growing evidence supports the importance of chromatin modification at or around DNA-damaged sites in DDR [59,60]. Again, data provided by Vazquez et al. [44] showed that SIRT7-mediated H3K18 deacetylation affects 53BP1 recruitment to DNA damage sites. H3K18Ac is directly involved in DNA repair, and H3K18Ac levels are fine-tuned by SIRT7 in response to DNA damage. Meanwhile, Li et al. [15] showed that SIRT7 is recruited to DSBs and catalyses the desuccinylation of histone H3 lysine 122, thereby promoting chromatin condensation and efficient DSB repair. Bao et al. [16] demonstrated that endogenous Sirt7 functions as a histone deglutarylase to regulate histone H4 lysine 91 glutarylation dynamics. In response to DNA damage, Sirt7 depletion hindered the removal of H4K91glu. Similar to SIRT7-mediated H3K122 desuccinylation, the removal of H4K91 glutarylation also aims at promoting chromatin condensation for the DNA repair process [16] (figure 3a). It is of great interest that all three sites—H3K18Ac, H3K122succ and H4K91glu—are mediated by SIRT7 during DNA damage repair. Whether these sites function independently or in a synergistic manner is largely unknown. Further studies are warranted to shed light on how SIRT7-mediated epigenetic regulation collaborates with the functions of other repair proteins recruited to DSBs and the underlying regulatory network. However, based on the above findings, it is clear that SIRT7 is required during the early phase of DNA repair and that a signalling mechanism is deployed that links histone modification to DSB repair.
These findings establish the role of SIRT7 in the early phase of DNA repair and elucidate novel signalling that links histone modification and DSB-related repair. During the process of DNA damage and repair, the proteins recruited to DNA damage sites are gradually displaced and inactivated, which make the cells return to the normal state and ensure faithful DNA repair. Among the numerous key DNA damage response factors, ATM has been reported to be an apical kinase in response to DSBs. Through exposure to DNA damage, ATM is activated through a series of highly organized machineries [61–65]. Acetylation and phosphorylation are two key post-translational modifications involved in activation of ATM in response to DSBs, both are dynamically regulated. Our research fills the gap of the dynamic regulation of ATM acetylation, and we find that SIRT7 is gradually recruited to chromatin in the late phase of repair and deacetylate ATM, which is required for the dephosphorylation of ATM by the phosphatase WIP1, and thus ensure the faithful DNA repair [35] (figure 3b). How SIRT7 regulates the downstream of ATM signalling needs more exploration.
Interestingly, re-localization of SIRT7 from the nucleolus to DNA damage sites affects ribosomal transcriptional repression [44,66,67]. This finding suggests that SIRT7-mediated DNA repair might have consequences on genome-wide transcriptional regulation under conditions of chronic DNA damage, plausibly the restoration of transcriptional profiles.
On the other hand, R-loop is a three-stranded nucleic acid structure; its aberrant formation and persistence cause DNA damage. Song et al. [29] showed that SIRT7-mediated deacetylation of DDX21 deacetylation cooperates which helps to prevent R-loop accumulation and DSBs, thus safeguarding genome integrity.
3.3. Role of SIRT7 in cancer and ageing
Increased genome instability is a common hallmark of both ageing and cancer. Consequently, any defect in DNA repair can contribute to genomic instability and subsequently lead to accelerated ageing or tumorigenesis [68,69]. DNA damage accumulates with age, and defects in DNA repair can cause phenotypes of premature ageing. Below, we describe the emerging data that suggest defects in SIRT7-mediated genome stability can affect ageing.
A longevity function has been proved for mammalian sirtuins. Indeed, Sirt7-deficient mice exhibit a reduction in mean and maximum lifespans, which indicates the role of SIRT7 in the ageing process [17]. By performing a comparative interactomics study associated with DNA repair, chromatin assembly and ageing, Lee et al. [20] found that SIRT6 and SIRT7 regulate NPM1 during the ageing process.
As mentioned earlier, researchers offered insights into the role of SIRT7 in the ageing process, showing that SIRT7 protects adult hair follicle stem cells from ageing by ensuring their progression through the hair growth cycle [39,70]. Bi et al. [71] also delineated the mechanisms of human stem cell ageing, showing that SIRT7 can form a complex with the nuclear lamina and heterochromatin proteins to maintain a repressive heterochromatin state and regulate the innate immune response during stem cell ageing. Moreover, Liu et al. [72] showed that SIRT7 deficiency leads to lowered histone acetyltransferase 1 (HAT1) activity and decreased H4K5 and H4K12 acetylation, which affects chromatin assembly. They also obtained evidence that SIRT7 ablation results in aneuploidy and ageing phenotypes, including senescence and nucleolar expansion [72].
Genomic instability in rDNA repeat sequences is an underlying cause of cell ageing [68]. Paredes et al. [73] uncovered an important role for SIRT7 in guarding against rDNA instability and protecting against senescence through association with SNF2H. Taken together, it seems that SIRT7 serves as an important regulator of mammalian longevity and might act as a molecular bridge between ageing and genome stability, paving the road for use. These preliminary findings offer support to investigate the value of targeting SIRT7 in the treatment of age-related diseases.
Based on the studies of SIRT7 in cancer, Kiran et al. [74] demonstrate that SIRT7 plays an important role in cell survival of osteosarcoma (U2OS) under DNA damage-induced stress. Specifically, the researchers showed that SIRT7 attenuated the effects of genomic stress, as SIRT7 knockdown cells showed increased susceptibility to the DNA damaging agent doxorubicin. Mechanistically, the cell cycle of SIRT7-overexpressing cells is temporarily halted at the G1/S phase when DNA damage is detected, probably to ensure DNA repair. SIRT7 resulted in reduced accumulation of γ-H2AX, p53 and the attenuation of stress-activated protein kinases (p38 and JNK) to maintain the genome integrity [74]. Beside the role of SIRT7-mediated H3K18 deacetylation in maintaining a malignant phenotype, Pandey & Kumar [75] provided evidence that HBx-dependent accumulation of SIRT7 favours H3K18 deacetylation and downregulation of RPS7, which is involved in the DDR and cancer cell transformation. Finally, data from our laboratory support that SIRT7 has degraded in response to 5-fluorouracil treatment and renders colorectal cancer cells sensitive to radiation [76]. The identification of SIRT7 inhibitors could thus be of great importance with respect to cancer treatment.
4. Conclusion
SIRT7 is involved in diverse cellular processes, including energy homeostasis, chromatin regulation, gene regulation and ribosome biogenesis. Here, we have highlighted the roles of SIRT7 in maintaining genome stability through its involvement in the DDR and the repair of DSBs. While it is clear that SIRT7 serves to promote DNA repair and ensure genome stability, how SIRT7 might interact with HR, MMR, NER and BER are still unclear.
While we know that SIRT7 regulates chromatin condensation in response to DNA damage via the desuccinylation of H3K122 and deglutarylation of H4K91 [15,16]. As a master epigenetic regulator, there are no doubt more epigenetic marks regulated by SIRT7 need to be studied for a comprehensive understanding of epigenetic regulation.
The importance of SIRT7 in DNA damage repair suggests that this enzyme might function as a tumour suppressor. However, SIRT7 is overexpressed in various cancers. Thus, SIRT7 might have opposing effects on cancer initiation and progression [14,32,76–78]. More systematic research is necessary to delineate how SIRT7 function might change across cancer evolution and development. A deeper understanding of SIRT7 function in genome stability at the molecular and physiologic levels may enable us to develop novel cancer- or ageing-related therapeutic targets. Such targets will be essential for conceptualizing the translation of SIRT7 biology into clinical applications.
Data accessibility
This article does not contain any additional data.
Authors' contributions
M.T. and H.T. wrote the primary manuscript and revised the manuscript. B.T. and W.-G.Z. conceived and designed the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by grants from the National Key R&D Program of China (2017YFA0503900), National Natural Science Foundation of China (81802811, 81720108027 and 81530074), Science and Technology Program of Guangdong Province in China (2017B030301016), Shenzhen Municipal Commission of Science and Technology Innovation (JCYJ20170818092450901), Discipline Construction Funding of Shenzhen [(2016)1452] and Shenzhen Bay Laboratory (SZBL2019062801011).
References
Articles from Open Biology are provided here courtesy of The Royal Society
Full text links
Read article at publisher's site: https://doi.org/10.1098/rsob.210047
Read article for free, from open access legal sources, via Unpaywall:
https://royalsocietypublishing.org/doi/pdf/10.1098/rsob.210047
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/107767803
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1098/rsob.210047
Article citations
The role of SIRT1 in autophagy and drug resistance: unveiling new targets and potential biomarkers in cancer therapy.
Front Pharmacol, 15:1469830, 30 Sep 2024
Cited by: 0 articles | PMID: 39403142 | PMCID: PMC11471651
Review Free full text in Europe PMC
Sirt1: An Increasingly Interesting Molecule with a Potential Role in Bone Metabolism and Osteoporosis.
Biomolecules, 14(8):970, 08 Aug 2024
Cited by: 0 articles | PMID: 39199358 | PMCID: PMC11352324
Review Free full text in Europe PMC
SIRT7 Inhibits Melanin Synthesis of PIG1 and PIG3V by Suppressing the Succinylation of EZR.
Clin Cosmet Investig Dermatol, 17:1495-1504, 22 Jun 2024
Cited by: 0 articles | PMID: 38933605 | PMCID: PMC11204816
Endothelium-specific SIRT7 targeting ameliorates pulmonary hypertension through Krüpple-like factor 4 deacetylation.
Cardiovasc Res, 120(4):403-416, 01 Mar 2024
Cited by: 1 article | PMID: 38198357 | PMCID: PMC10981524
Sirtuins in intervertebral disc degeneration: current understanding.
Mol Med, 30(1):44, 29 Mar 2024
Cited by: 3 articles | PMID: 38553713 | PMCID: PMC10981339
Review Free full text in Europe PMC
Go to all (21) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability.
Nat Commun, 7:12235, 20 Jul 2016
Cited by: 199 articles | PMID: 27436229 | PMCID: PMC4961794
Sirtuins and DNA damage repair: SIRT7 comes to play.
Nucleus, 8(2):107-115, 17 Feb 2017
Cited by: 39 articles | PMID: 28406750 | PMCID: PMC5403131
Review Free full text in Europe PMC
SIRT7 Is Activated by DNA and Deacetylates Histone H3 in the Chromatin Context.
ACS Chem Biol, 11(3):742-747, 03 Mar 2016
Cited by: 33 articles | PMID: 26907567 | PMCID: PMC4850736
SIRT6, a protein with many faces.
Biogerontology, 14(6):629-639, 10 Nov 2013
Cited by: 53 articles | PMID: 24213807
Review
Funding
Funders who supported this work.
Discipline Construction Funding of Shenzhen (1)
Grant ID: (2016)1452
National Key R&D Program of China (1)
Grant ID: 2017YFA0503900
National Natural Science Foundation of China (1)
Grant ID: 81802811, 81720108027, 81530074
Science and Technology Program of Guangdong Province in China (1)
Grant ID: 2017B030301016
Shenzhen Bay Laboratory (1)
Grant ID: SZBL2019062801011
Shenzhen Municipal Commission of Science and Technology Innovation (1)
Grant ID: JCYJ20170818092450901

 2
2