Abstract
Background
Insulin's discovery 100 years ago and its ongoing use since that time to treat diabetes belies the molecular complexity of its structure and that of its receptor. Advances in single-particle cryo-electron microscopy have over the past three years revolutionized our understanding of the atomic detail of insulin-receptor interactions.Scope of review
This review describes the three-dimensional structure of insulin and its receptor and details on how they interact. This review also highlights the current gaps in our structural understanding of the system.Major conclusions
A near-complete picture has been obtained of the hormone receptor interactions, providing new insights into the kinetics of the interactions and necessitating a revision of the extant two-site cross-linking model of hormone receptor engagement. How insulin initially engages the receptor and the receptor's traversed trajectory as it undergoes conformational changes associated with activation remain areas for future investigation.Free full text

Understanding insulin and its receptor from their three-dimensional structures
Abstract
Background
Insulin's discovery 100 years ago and its ongoing use since that time to treat diabetes belies the molecular complexity of its structure and that of its receptor. Advances in single-particle cryo-electron microscopy have over the past three years revolutionized our understanding of the atomic detail of insulin-receptor interactions.
Scope of review
This review describes the three-dimensional structure of insulin and its receptor and details on how they interact. This review also highlights the current gaps in our structural understanding of the system.
Major conclusions
A near-complete picture has been obtained of the hormone receptor interactions, providing new insights into the kinetics of the interactions and necessitating a revision of the extant two-site cross-linking model of hormone receptor engagement. How insulin initially engages the receptor and the receptor's traversed trajectory as it undergoes conformational changes associated with activation remain areas for future investigation.
1. Introduction
The 100-year history of insulin intimately intertwines with the quest to determine insulin's three-dimensional atomic structure [[1], [2], [3], [4], [5], [6]] and, through the last few decades, with the quest to understand the molecular details of insulin's interactions with its cell surface receptor [[7], [8], [9], [10], [11], [12], [13], [14], [15], [16]]. This century of endeavor spans the development of protein- and DNA-sequencing technologies [17,18], protein X-ray crystallography [19], molecular biology and recombinant protein expression [20], and—most recently—high-resolution cryo-electron microscopy (cryoEM) [[21], [22], [23]]. These technologies have enabled three-dimensional atomic-level imaging of insulin engaging its receptor, revealing a complexity of information that would have astonished a century ago.
I begin with an overview of the three-dimensional structure of insulin in its receptor-free form. This structure— now known for more than 50 years—revealed how the hormone is packaged into a hexamer for storage in β-cell granules and how it disassembles into monomers in plasma circulation. The structure led to an understanding, at least in part, of the surfaces of the hormone responsible for its physiological action. I then describe the three-dimensional structure of the extra-cellular (“ectodomain”) region of the insulin receptor in its insulin-free (“apo”) form, determined progressively over the past two decades. The structure reveals how the receptor's tyrosine kinase domains are held apart but, at the same time, the ectodomain is poised to engage an incoming insulin molecule. I then discuss the intricate engagement of insulin with the receptor, the structural detail of which has been revealed only over the past few years. The three-dimensional views of the insulin-receptor complex reveal the multifaceted conformational transitions that the respective molecules undergo as they engage and how these transitions ultimately effect receptor activation.
2. Insulin's three-dimensional structure
The amino acid sequences of human insulin polypeptides were determined by Sanger et al. in the early 1950s [[24], [25], [26], [27], [28]]. The hormone in its mature form comprises two polypeptide chains: a 30-residue B chain and a 21-residue A chain, interlinked by two disulfide bonds (CysB7 to CysA7 and CysB19 to CysA21), with a further disulfide bond located within the A chain (CysA6 to CysA11). The mature form of insulin arises from a “proinsulin” precursor—a single-chain polypeptide wherein the C terminus of the B chain is linked to the N terminus of the A chain by a so-called “C peptide” [29]. Maturation involves the enzymatic removal of the C peptide as well as trimming of the nascent B- and C-peptide C termini by carboxypeptidases [30,31]. The three-dimensional atomic structure of insulin was published in 1969 by Hodgkin et al. [6], insulin being the sixth protein to have its structure determined.1 That structure, as determined by X-ray crystallography at 2.8 Å resolution, revealed that the B chain contains a central α helix (spanning residues B9 to B19) and the A chain contains two α helices (spanning residues A1 to A8 and A12 to A20, respectively) (Figure 1A). Residues B20 to B23 form a type-II β turn and residues B24 to B30 form an extended strand that runs anti-parallel to the B-chain α helix. Within the crystal, the 51-residue molecule is assembled into dimers related by a two-fold (non-crystallographic) axis, and the dimers then turn into trimers related by a three-fold (crystallographic) axis (Figure 1B). Assembly of monomers into dimers is mediated largely by their respective B24-to-B30 strands forming an anti-parallel, two-stranded β sheet (Figure 1C). Two zinc ions are located on the hexamer's three-fold symmetry axis, each ion being coordinated by individual triplets of residue HisB10 (Figure 1B). Insulin's spatial resolution has since improved progressively to 1.00 Å (PDB 1MSO) [32].
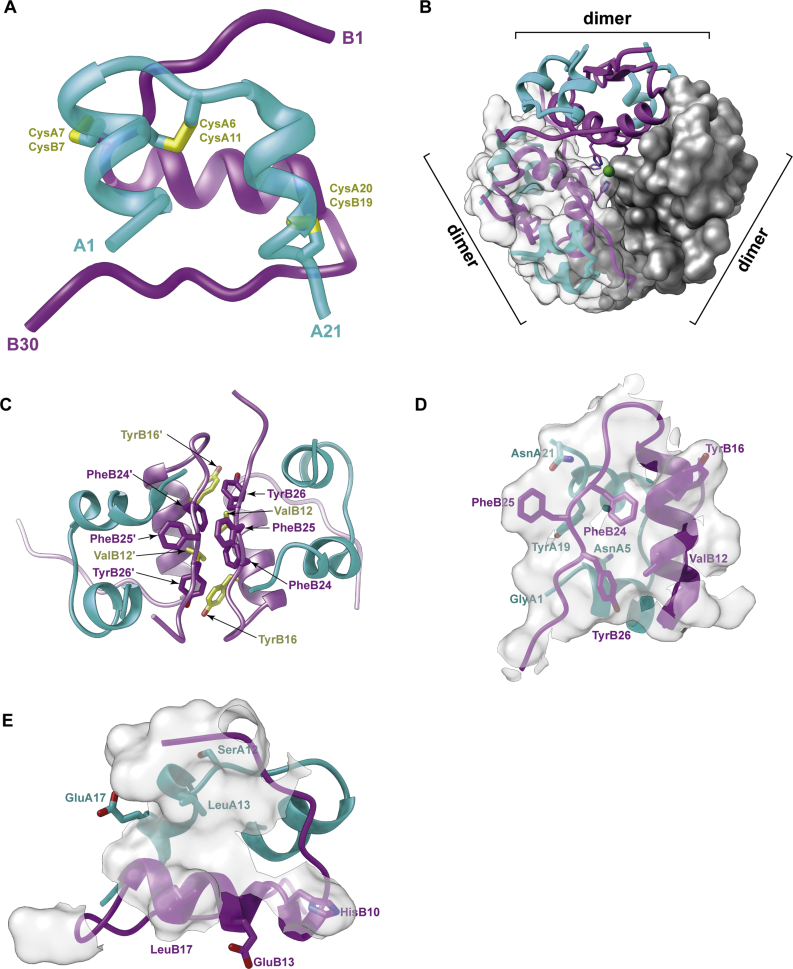
Three-dimensional structure of insulin and its predicted receptor-binding surfaces. (A) Structure of the insulin monomer showing chains A (cyan) and B (magenta) together with two inter-chain disulfide bonds and one intra-chain disulfide bond. (B) Assembly of the insulin dimer and three dimers into the insulin hexamer. The two zinc atoms (green) lie on the hexamer axis. The three dimers are shown in varying representations (ribbon, ribbon plus transparent surface, and opaque surface, respectively). (C) Detail of the insulin dimer interface, with selected residues highlighted. (D) The so-called “classical” set of receptor-binding residues of insulin (stick representation); these overlap insulin's dimer-forming surface (transparent white). (E) Predicted additional set of receptor-binding residues of insulin (stick representation); these overlap insulin's hexamer-forming surface (transparent white). Panels are based on PDB 1MSO [32].
Insulin's three-dimensional structure allowed the immediate interpretation of a wealth of biochemical and biophysical data regarding the hormone [33]. The hexameric assembly within the crystal was observed to correspond to the 36 kDa form adopted by the hormone at neutral pH in the presence of zinc ions, and dimeric subcomponents corresponded to the 12 kDa form observed for the hormone under dilute conditions. Disassembly of the hexamer upon loss of the zinc ions was subsequently rationalized to result from the mutual repulsion of the six centrally located residues GluB13 [34,35].
Further insight arose from analyzing the structural location of insulin residues conserved across species [36]. Conservation across species of particular amino acids within proteins typically occurs at sites required either for structural integrity or function; more subtle structural conservation occurs in protein cores, reflecting conservation of the shape and hydrophobicity. Conservation of surface residues is related more likely to function, although surface conservation may also be related to the need to maintain solubility or multimeric assembly. The striking examples in insulin of residues conserved for structural integrity are the six disulfide-bond-forming cysteines—all known insulins contain these six residues. In insulins, the conserved hydrophobic core includes residues LeuB6, GlyB8, LeuB11, LeuB15, ValB18, GlyB23, IleA2 and LeuA16. Strictly conserved surface residues include ValB12 and PheB24—these residues are involved in hormone dimerization (Figure 1C), suggesting in turn that dimerization is a property conserved across species. The salient exception to surface residue conservation is in hystricomorph insulins, which have substitutions (a) at the zinc-chelating residue HisB10 (asparagine in guinea pigs and glutamine in coypu), (b) at residue GlyB20 (a residue involved in the β turn that enables the B-chain C-terminal segment to fold back anti-parallel to the B-chain helix; glutamine in guinea pigs and arginine in coypu), and (c) at the dimer interface-forming residues PheB25 and TyrB26 (tyrosine and arginine, respectively, in coypu) [33]. Taken together, these substitutions suggested that the hystricomorph insulins likely do not assemble into hexamers and possibly not into dimers. A recent analysis of sequence conservation across the broader insulin family is in Reference [37].
Whereas it ultimately required the determination of the structure of the insulin-complexed insulin receptor to provide a full picture, extensive clues arose beforehand as to how insulin and its receptor might engage (reviewed in [[38], [39], [40]]). In particular, insulin's receptor engagement was deduced to be mediated by what was called the hormone's “classical” receptor-binding region—residues ValB12, TyrB16, GlyB23, PheB24, PheB25, TyrB26, GlyA1, GlnA5, TyrA19 and AsnA21. This region overlaps insulin's dimer-forming surface (Figure 1D), and suggestions of its involvement in receptor binding arose initially from its conservation across species [41] but was subsequently supported by a wealth of data derived from mutant insulins (see, for example, [[42], [43], [44], [45], [46], [47], [48]]). Confirmation also arose from chemical cross-linking studies, wherein it was shown that residue PheB25 (and ValA3) of insulin could be cross-linked to the C-terminal region of the receptor α chain [49,50] and that residues TyrB16 and PheB24 of insulin could be cross-linked to the N-terminal region of the receptor α chain [51,52] (see below for the definition of the receptor α and β chains).
Integral to the emerging picture of how insulin might engage its receptor were detailed kinetic studies of its receptor binding. Insulin binding to its receptor showed curvilinear Scatchard plots initially interpreted to reflect two independent binding sites with different affinities [53]. However, the detection of accelerated dissociation of bound radio-labeled insulin upon added unlabeled insulin showed that the curvilinear plots resulted instead from negative cooperativity between binding sites [54]. Intriguingly, whereas the degree of accelerated dissociation increased with the concentration of added cold insulin, it reduced at very high concentrations of added cold insulin [54]. Germane to this observation was the fact that such reduction in negative cooperativity was not observed for hagfish insulin [55], despite the “classical” receptor-binding surface of hagfish insulin being highly similar to that of human insulin. The conundrum was resolved by proposing that there was a further receptor-binding surface on insulin. This second surface was deduced to include residues LeuB17 and LeuA13, as mutation of these residues in human insulin led to insulins that also fail to show reduced negative cooperativity at high insulin concentrations [56]. Taken together, these observations then led to a model wherein it was proposed that (a) the receptor monomers are arranged in a two-fold symmetric fashion, (b) insulin binding cross-links two distinct sites (sites 1 and 2′) on the receptor, one provided by each receptor monomer, (c) such cross-linking of one site 1-site 2′ pair reduces insulin's affinity for the alternate site 1′-site 2 pair (negative cooperativity), and (d) at high insulin concentrations, the receptor becomes saturated with three bound insulins, one insulin cross-linking the site 1-site 2′ pair and the other two binding individually to site 1′ and site 2, respectively [57]. Within this model, one of these sites was proposed to be engaged by the classical binding surface of insulin as described above and the other by a surface that included the insulin residues LeuB17 and LeuA13. Subsequent studies extended insulin's second receptor-binding surface to include residues HisB10, GluB13, LeuB17, SerA12, LeuA13, and GluA17 [39,58]—this subset of residues overlaps the hormone's hexamer-forming surface (Figure 1E).
It also became apparent that receptor engagement requires the C-terminal segment of the insulin B chain to disengage from the hormone's core: in other words, insulin must undergo conformational change upon receptor binding. The evidence for this was as follows. First, a single-chain insulin constructed by linking the B-chain's C terminus directly to the A-chain's N terminus had no detectable activity despite the resultant polypeptide retaining the canonical dimer mode of assembly of the native hormone (PDB 6INS) [59]. Such linking stabilizes the packing of the B-chain C-terminal segment against the hormone's core and prevents it from disengaging. Second, the clinical ValA3Leu mutant (insulin Wakayama) has less than 1% of native insulin's affinity for the receptor [60] despite having a crystal structure effectively identical to that of the native hormone (PDB 1XW7) [61]. However, the side chain of the mutant residue LeuA3 is largely buried within the hormone's core, the burial being effected by the residues at the B chain's C terminus. The implication is hence that, for the mutation to effect a reduction in receptor affinity, the mutated residue must become exposed to engage the receptor. Third, the clinical PheB24Ser mutant (insulin Los Angeles) retains a native-like solution structure but lacks activity (PDB 1HIQ) [62, 63]. The serine side chain is mostly buried against the mutant hormone's core; again, the implication is that for the substitution to effect a reduction in receptor affinity, the residue needs to be exposed. These conclusions were indirectly supported in turn by data from a solution structure of PheB24Gly insulin, wherein the B20–B30 segment was found to be highly mobile, despite the fact that GlyB24 insulin retains 78% of its native insulin affinity for receptor (PDB 1HIT) [64].
An intriguing observation is that insulin can also adopt within crystals a so-called “R-state” form, wherein its B-chain helix is extended N-terminally to include most of residues B8 to B1, the hexamer under these circumstances coordinating four—rather than two—zinc ions (PDB 1ZNI) [65]. The R-state structure is presumably of lower energy than the form previously described (the so-called “T state”) and was tentatively suggested to reflect a receptor-bound form (see [66] for a review). However, no R-state form of insulin has been found in any of the structures of the insulin-complexed receptor determined to date. The R-state form is hence likely an artifact of crystallization or perhaps an intermediate along the receptor-engagement pathway.
In summary, insulin's three-dimensional structure is that of a compact globular protein but one that has to fulfill many functions—storage in the pancreatic β cells, transport in the plasma, receptor engagement and, ultimately, to be available for proteolytic degradation within the endosome. Most of the hormone's surface is thus likely involved in more than one role and, indeed, complete functionality appears to require the generation of additional surfaces through conformational change that exposes the hormone's core. Understanding this complexity necessitates atomic-level imaging of the receptor in both insulin-free and insulin-bound form.
3. The insulin receptor’s three-dimensional structure
Insulin's role in promoting glucose transport across cell membranes was discovered in 1949 [67], and an insulin-binding fraction of rat liver membrane was first isolated in 1970 [68], establishing that insulin acted via a cell-surface receptor. Competing results were published by two other groups in 1971 [[69], [70], [71], [72]]. Insulin's receptor was first isolated in 1972 from liver and fat cell membranes [73] and subsequently shown to be an (αβ)2 disulfide-linked homodimer [74,75]. The receptor's cDNA was sequenced in 1985, revealing that the α chain lay N-terminal to the β chain and that these chains were separated by a proteolytic cleavage site [76,77]. The multidomain organization of both the α and β chains was also evident: the receptor was seen to comprise (from the N terminus) a leucine-rich-repeat domain (L1), a cysteine-rich region (CR), a second leucine-rich region (L2), and three predicted fibronectin type III domains (denoted FnIII-1, FnIII-2, and FnIII-3, or F1, F2, and F3, respectively), followed by a transmembrane (TM) domain, an intra-cellular juxtamembrane segment (JM), a tyrosine kinase (TK) domain, and a C-terminal tail (C-tail) [76,77,[80], [81], [82], [83]] (Figure 2A). The α/β cleavage site lies within a largely disordered region—called the insert domain (ID)—that in turn lies within domain FnIII-2 (see below). The protein was found to be expressed as two splice mutants differing by the presence (in isoform B) or absence (in isoform A) of the product of exon 11 [84] (Figure 2A). Inter-α-chain disulfides were located at residue Cys524 (within domain FnIII-1) [85] and at one or more sites within the Cys682, Cys683, and Cys685 triplet [86] (Figure 2A). An α-to β-chain disulfide bond was also shown to be formed between residues Cys647 and Cys872 (A isoform numbering) [86]. The receptor sequence was found to have fifteen potential N-linked glycosylation sites within its α chain and four within its β chain [77]. All but one of these sites (Asn78) subsequently proved to be glycosylated [78], though their role in receptor biosynthesis and function remains only partly understood [79]. Insulin binding was shown to result in the auto-phosphorylation of two tyrosine residues in the JM segment, three in the TK domain, and two in the C-tail segment [[87], [88], [89], [90]]. It should be noted that the insulin receptor together with its homologs—the type 1 insulin like growth factor receptor and the insulin receptor related receptor—are unique among the receptor tyrosine kinases (RTKs) in that they are pre-formed disulfide-linked dimers, the remainder of the RTKs being monomers [91]. Discussion of how the mechanism of activation of the insulin receptor sub-family aligns with or differs from that of other RTK sub-families is beyond the scope of this review.
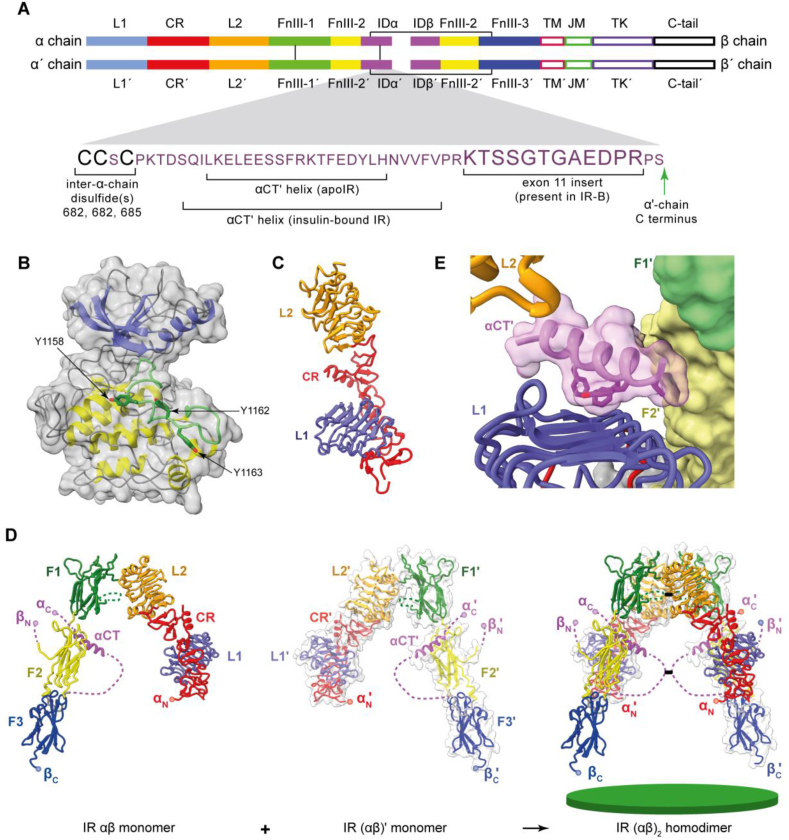
Three-dimensional structure of the human apo insulin-receptor (IR) ectodomain. (A) Domain layout of the disulfide-linked (αβ)2 homodimer; close-up view shows the sequence detail of the α chain's C-terminal region. The domain nomenclature is defined in the main text. Interchain disulfide bonds are shown as green lines. (B) Bi-lobal structure of the IR TK domain (N-terminal lobe in blue, C-terminal lobe in yellow, and the activation loop in green). (C) Structure of the IR L1-CR-L2 module. (D) Structure of the αβ monomer showing its pairing with the (αβ)′ monomer to form the (αβ)2 homodimer. The green disc (bottom right) represents the cell membrane. In the panel, the N and C termini of the α chain and β chain are denoted as αN, αC, βN, and βC, respectively. Dashed connectors indicate residue segments disordered in the crystal structure. (E) Detail of the engagement of the αCT′ helix with the L1-β2 surface. Panels are based on PDB 1IRK [92], PDB 2HR7 [7] and PDB 4ZXB [95].
The three-dimensional structure of the receptor itself was revealed in stages. The first structure was obtained in 1994 and was that of the intra-cellular tyrosine kinase domain in its unphosphorylated form, determined at 2.1 Å resolution (PDB 1IRK) [92]. This structure was also the first to be determined of any tyrosine kinase domain and was shown to be similar to those of protein serine/threonine kinases. In particular, the TK domain displayed the characteristic two-domain structure of the latter kinases, comprising an N-terminal lobe with five anti-parallel β strands and a single α helix, followed a C-terminal lobe with eight α helices and four β strands (Figure 2B). In this structure, the so-called activation loop (residues 1149–1170, which span two short strands of the C-terminal lobe and contain the phosphorylation sites Tyr1158, Tyr1162, and Tyr1163) traverses the cleft between the N- and C-terminal lobes (Figure 2B). Although residue Tyr1162 is poised for phosphorylation within the structure, the ATP-binding site is blocked by this residue, preventing Tyr1162 cis-phosphorylation. Further structural study of the non-phosphorylated TK domain revealed that residue Tyr984 (lying in the immediate upstream region of the juxtamembrane segment and not a phosphorylation site) stabilizes a catalytically non-productive position of the canonical αC helix of the domain's N-terminal lobe. Alanine mutation of Tyr984 results in a marked increase in receptor phosphorylation levels in cells, and phenylalanine mutation of Tyr984 results in a four-fold increase in the enzyme's kcat [93]. How insulin activates the TK domains will be discussed later in this review.
The second structure of an insulin receptor fragment to be obtained was that of the receptor's isolated L1-CR-L2 module determined at 2.32 Å resolution (PDB 2HR7) [7]. In this structure, the receptor domains L1 and L2 have β-barrel fold topologies, with the constituent β strands assembling into three distinct sheets (denoted β1, β2, and β3). Domain L1 is intimately associated with the downstream CR region, which itself has an irregular structure consisting of eight modules, each containing either one or two disulfide bonds (Figure 2C). The connection between the CR region and domain L2 is flexible as evidenced by the two distinct conformations of the respective receptor L1-CR-L2 modules within the crystallographic asymmetric unit. The L1-CR-L2 module crystal structure was followed the same year by that of the intact receptor ectodomain in apo form, crystallization of the latter being facilitated by the attachment of four antibody fragments (“Fabs”) as well as by removal of a highly glycosylated segment near the β chain's N terminus (PDB 2DTG) [8]. Within this structure (determined at 3.8 Å resolution), the L1-CR-L2 module has an extended conformation, followed by domains FnIII-1, FnIII-2, and FnIII-3 in a folded-over linear arrangement. The second αβ monomer (the domains of which are conventionally indicated with a prime symbol ′) packs with its L1′-CR′-L2′ module adjacent to the first monomer's [FnIII-1]-[FnIII-2]-[FnIII-3] module, the entire assembly then exhibiting a Λ-shaped two-fold-symmetric structure (Figure 2D). Subsequent refinement of this structure (PDB 3LOH, PDB 4ZXB) [94,95] revealed that the C-terminal region (“αCT”) of the receptor α chain is assembled as an α helix on the surface of the L1-β2 sheet, inter-digitated with it by a series of hydrophobic residues (Figure 2E). The remainder of the insert domain is largely disordered within the structure.
A salient feature of the apo ectodomain structure is that the membrane entry points (the respective C termini of domains FnIII-3 and FnIII-3′) are separated by ~120 Å. This separation arguably holds the intra-cellular TK domains apart and prevents their transphosphorylation (transphosphorylation being the receptor's mode of activation [96]). Insulin binding was thus proposed to alter the Λ-shaped conformation, effecting the uniting of the FnIII modules and concomitantly transphosphorylation of the intra-cellular TK domains. It could be asked whether the Λ-shaped conformation of the apo ectodomain is a crystallographic artifact caused by the removal of the TM and intra-cellular domains and/or by the attachment of the four Fabs. This is a legitimate question, as many of the observed domain–domain interfaces in the ectodomain structure are sparse, indicative of conformational flexibility. Also, the isolated receptor ectodomain does not display high-affinity hormone binding nor does it display negative cooperativity [97], and hence its three-dimensional structure may not fully reflect that of the holo-receptor. Nevertheless, evidence argues against the ectodomain structure being artifactual. First, low-resolution imaging of apo holo-receptor embedded in lipid nanodiscs reveals a similar Λ-shaped conformation for the ectodomain region [11]. Second, a similar structure has since been obtained for the apo ectodomain of the type 1 insulin-like growth factor receptor (IGF-1R) in complex with a qualitatively distinct combination of antibody fragments (two Fv modules) (PDB 5U8R) [98]. IGF-1R is very closely related in sequence to the insulin receptor [99]. What must be recognised, however, is that the crystal structures are static, they do not reveal the degree of conformational flexibility that occurs in vivo. Conformational flexibility may allow the apo insulin receptor to adopt an active conformation for a percentage of time, which could in turn be responsible for the 5–10% basal signaling level of the receptor system reported in [90,100]. We will return to this issue later.
The ectodomain structure's second salient feature is the interaction of domain L1 with an αCT segment [8,94]. The structure in itself does not—due to the disorder of most of the insert domain residues—resolve whether these elements belong to the same α chain. However, chemical cross-linking data showed that insulin binding cross-links receptor α chains [101], with one receptor monomer contributing domain L1 and the second its αCT′ segment [94]. As both the receptor domain L1 and receptor αCT segment harbor residues critical for insulin binding [102,103], it thus appeared natural that the observed L1 plus αCT tandem element of the apo receptor was the likely insulin-binding site and that the observed L1 and αCT elements arose from different monomers [94]. However, the structure did not reveal how insulin engaged the tandem element nor the location of further insulin-binding sites.
4. Putting it all together: How insulin engages its receptor
Whereas the above structures provided clues on how insulin and its receptor might engage, they ultimately disguised the complexity of the hormone's interaction with the receptor. I begin by describing the discovery of how insulin opens up to engage the L1+αCT′ tandem element, the latter now called the receptor's “primary” binding site. The nature of this engagement was revealed through crystal structures (to 2.9 Å resolution) of insulin in co-complex with a domain-minimized version of the insulin receptor comprising the two-domain L1-CR receptor fragment complemented with an exogenous αCT′ peptide (the so-called insulin “micro-receptor” or “μIR”; Figure 3A) (PDB 3W11, PDB 4OGA, PDB 6VEP) [9,10,16]. Despite such drastic domain minimization, details on the hormone μIR complex are found to be conserved within the more recent insulin-bound ectodomain and insulin-bound holo-receptor structures obtained by cryoEM.
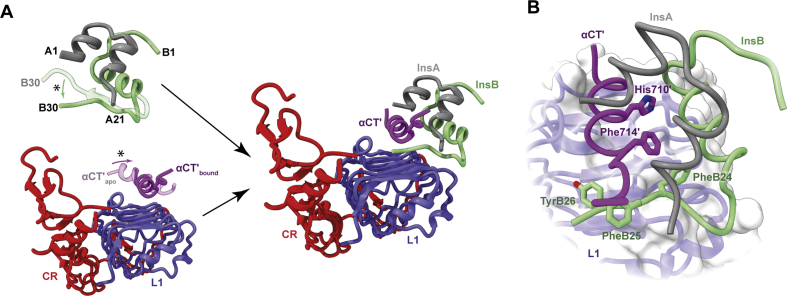
Insulin's engagement with its primary binding site on the receptor. (A) Schematic diagram illustrating both the folding out of the B-chain C-terminal region of insulin from the hormone's core (asterisk, top left), the reconfiguration upon insulin binding of the αCT′ helix on the L1-β2 surface (asterisk, bottom left), and insulin's engagement with the L1+αCT′ tandem element (right). (B) Detail of the packing of αCT′ residues His710′ and Phe714′ into the hormone's core as well as the reconfiguration of the three aromatic residues PheB24, PheB25, and TyrB26 within insulin's B-chain C-terminal region. Panels are based on PDB 1MSO [32], PDB 4ZXB [95] and PDB 6VEP [16].
The insulin-μIR complex shows that, upon engagement, the C-terminal segment (residues B24 to B30) of the insulin B chain folds away from the hormone's core, rotating from being oriented anti-parallel to the B-chain helix to lying approximately perpendicular to it. The observed folding out of insulin's B-chain C-terminal segment thus aligned completely with the earlier predictions described in Section 2 above. The concomitantly exposed surface of the hormone's B-chain helix docks onto the receptor's αCT′ helix, with the two helices aligning parallel (Figure 3A). In this assembly, the insulin B-chain helix engages with the C-terminal ends of the L1-β2 sheet strands, whereas the insulin A chain engages solely the αCT′ helix and not with receptor domain L1 (Figure 3B). Insulin residue PheB24 appears as a hinge point in the folding out of the B-chain C-terminal segment, its side chain remaining docked with the hormone's core by virtue of a compensatory rotameric change in its phenyl ring. The αCT′ helix also undergoes conformational changes upon insulin docking: it rotates about an axis perpendicular to the L1-β2 surface to align approximately perpendicular to the direction of the L1-β2 strands (Figure 3B). The C-terminal part of the αCT′ helix extends to include a further turn, bringing receptor residue Phe714′ into engagement with the hormone's core. In μIR crystal structures, the N-terminal (non-insulin-engaged) part of the αCT′ helix disassembles; however, such disassembly was later seen to be an artifact of the μIR platform [12].
The structural features of the μIR complex explain a wealth of biochemical data regarding both insulin and its receptor. First, the long-predicted folding out of insulin's B-chain C-terminal segment was confirmed. Second, insulin residues engaging the receptor tandem element include GlyB8, SerB9, LeuB11, ValB12, GluB13, LeuB15, TyrB16, ArgB22, GlyB23, PheB24, PheB25, TyrB26, GlyA1, IleA2, ValA3, GluA4, AsnA18, and TyrA19. This set coincides with the hormone's “classical” binding surface, but includes additional residues within the insulin core that are exposed on the folding out of the B chain's C-terminal strand. Third, residues on the surface of the L1-β2 sheet identified prior as important for insulin binding—on the basis of either clinical mutation or alanine scanning mutagenesis (reviewed in [7])—are seen in this structure to engage either insulin (L1-β2 residues Asp12, Asn15, Leu37, Phe39, Lys40, and Arg65), the reconfigured αCT′ segment (L1-β2 residues Leu36, Leu37, Phe88, Phe89, Tyr91, Val94, Phe96, Arg118, and Glu120), or both (L1-β2 residues Arg14, Phe64, and Glu97). Likewise, key residues within the αCT′ segment identified prior as important for insulin binding (Thr704′, Phe705′, Glu706′, Tyr708′, Leu709′, His710′, Asn711′, Val713′, Phe714′, and Val715′) [103] are seen in this structure to engage either insulin, the L1-β2 sheet, or both. αCT′ residues that upon mutation to alanine do not affect insulin binding [103] are likewise seen to have very limited engagement with insulin or the L1-β2 surface (αCT′ residues Asp707′, Val712′, Pro716′, and Arg717′). In interpreting these results, it is important to note that the L1-CR module is incapable of binding insulin in the absence of an αCT′ element [104,105].
A critical deduction from the insulin-bound μIR structure is that when its L1-CR module is overlaid onto the corresponding module of the apo ectodomain structure, extensive steric overlap occurs between the μIR-bound insulin and the adjacent domains FnIII-1′ and FnIII-2′ [9]. Insulin binding thus requires either prior separation of the apo L1+αCT′ element away from the adjacent FnIII′ module, an induced fit mode of the hormone's engagement with these elements, or both.
As intriguing as the insulin-complexed μIR structures are, they did not foreshadow the large-scale domain rearrangements that occur in the ectodomain as a whole upon insulin binding. These rearrangements' complexities were ultimately revealed by cryoEM [[12], [13], [14], [15]], a technology that has recently enabled 3D structures of large macromolecular complexes to be generated with considerably more ease than via the X-ray crystallographic route, its enablers being awarded the Nobel Prize in Chemistry [[21], [22], [23]]. CryoEM structures of the insulin-complexed receptor have been obtained from a repertoire of receptor constructs, including (a) the insulin holo-receptor (the full-length receptor inclusive of its transmembrane and cytoplasmic elements) [14], (b) the isolated receptor ectodomain [12,15], and (c) an ectodomain construct that has a leucine-zipper element attached at the C terminus of each β chain, leucine-zipper attachment arguably providing a mimic of the holo-receptor's membrane anchoring, which ostensibly reduces the conformational mobility of the receptor extracellular domains [13]. These receptor constructs (Figure 4) have different insulin-binding kinetics: the insulin holo-receptor displays sub-nanomolar affinity for insulin and negative cooperativity of insulin binding [54], whereas the isolated ectodomain displays nanomolar insulin affinity for insulin and no negative cooperativity [97], and the leucine-zippered receptor ectodomain sub-nanomolar affinity for insulin, but its cooperativity of insulin binding has not been explicitly investigated [13,106]. It is thus not immediately clear whether differences in the details of the cryoEM structures reflect intrinsic differences in the constructs or differences in cryoEM sample preparation/methodology. The structures will be described here in increasing order of hormone-to-receptor stoichiometry, beginning with the cryoEM structure of a single insulin molecule bound to the leucine-zippered receptor ectodomain (Figure 5A; PDB 6HN4, PDB 6HN5) [13]—all the other structures (bar one of significantly lower resolution; PDB 6CE7 [12]) have a higher stoichiometric ratio of bound insulin. The zippered structure's average resolution is 3.2 Å in its membrane-distal portion and 4.2 Å in its membrane-proximal region, and its salient features are as follows [13]:
- (a)
The single insulin molecule is bound to one of the two L1+αCT′ elements within the construct and in a manner closely similar to that seen in the μIR-based structure described above. In particular, the B-chain C-terminal segment is folded out away from the hormone's core, the insulin B-chain helix docks parallel to that of αCT′, and the αCT′ element is reconfigured on the L1-β2 sheet surface (Figure 5A).
- (b)
Within the construct, the insulin-bound L1-CR-L2 module undergoes marked conformational changes with respect to its location within the apo receptor ectodomain. In particular, domain L2 is folded out of the four-domain [L2+FnIII-1]2 “head” component of the receptor, and the L1-CR module is disengaged from domain FnIII-2′ and then rotated further about the CR-to-L2 junction (Figure 5B)—these changes combine to place the [L1+αCT′]-bound insulin in interaction with the membrane-distal loops of domain FnIII-1′ (Figure 5C). Insulin residues involved in this latter interaction include AsnB3, HisB5, HisB10, and GluB13, these four residues reside in the N-terminal portion of the insulin B chain. Receptor residues involved in the interaction include Asp 496′, Phe497′, Arg498′, and Arg539′ and possibly further residues with the loop formed by FnIII-1′ domain residues 540′-545′ (the latter loop is disordered within the structure).
- (c)
The insulin-bound αCT′ helix extends N-terminally with respect to its apo form—the αCT′ helix now spans receptor residues 688 to 714, i.e., to within four residues of the cysteine triplet (Cys682, Cys683, and Cys685) that forms inter-α-chain disulfide bond(s) (Figure 5D).
- (d)
These conformational changes also cause extensive interactions between (i) receptor domain L2 and the extended αCT′ helix, (ii) receptor domain FnIII-1′ and the extended αCT′ helix, and (iii) receptor domains L1 and L2 (Figure 5E). These intra-receptor interactions are absent in the apo ectodomain structure. Residues involved include αCT′ residues Lys687′, Asp696′, Glu697′, Ser700′, Phe701′, Lys703′, Glu706′, and Asp707′, domain L2 residues Ser323, Val324, Thr325, Gln328, Arg345, Gly346, Leu350, and Glu353, domain FnIII-2′ residues Asp496′, Arg498′, Asp499′, Asp533′, and Thr571′, and domain L1 residues Leu87, Phe89, Asn90, Arg114, and Asp142 (residues selected are based on an assessment of PDB 6HN5 [13] as well on those reported in the further insulin-complexed receptor structures discussed below). Of interest in this set is residue Ser323, which upon clinical mutation to leucine causes severe insulin resistance [107,108], and Asp707, which upon clinical mutation to alanine results in receptors being unable to bind insulin [109]. Uchikawa et al. [14] showed that disruption of the salt bridge between domain L2 residue Arg345 and αCT′ residue Glu697′ diminishes receptor activation.
- (e)
The receptor fibronectin-domain modules hinge inwards towards the receptor ectodomain's pseudo-symmetry axis, with a contact forming between domains FnIII-3 and FnIII-3′ (Figure 5F).
- (f)
The insulin-free L1′+αCT element remains in an apo-like association with domain FnIII-2 (Figure 5F), although the αCT helix appears poorly defined on the L1′ domain surface [13].
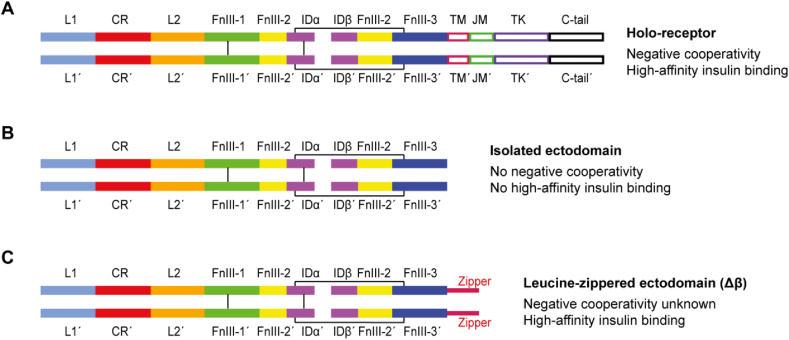
Insulin receptor (IR) constructs used in structures determined by cryoEM. (A) Holo-receptor IR construct employed by Uchikawa et al. [14]. (B) IR-A ectodomain constructs employed by Scapin et al. [12] and Gutmann et al. [15]. (C) Leucine-zippered ectodomain construct employed by Weis et al. [13] including the so-called Δβ modification that removes the highly glycosylated segment near the β chain's N terminus [8].
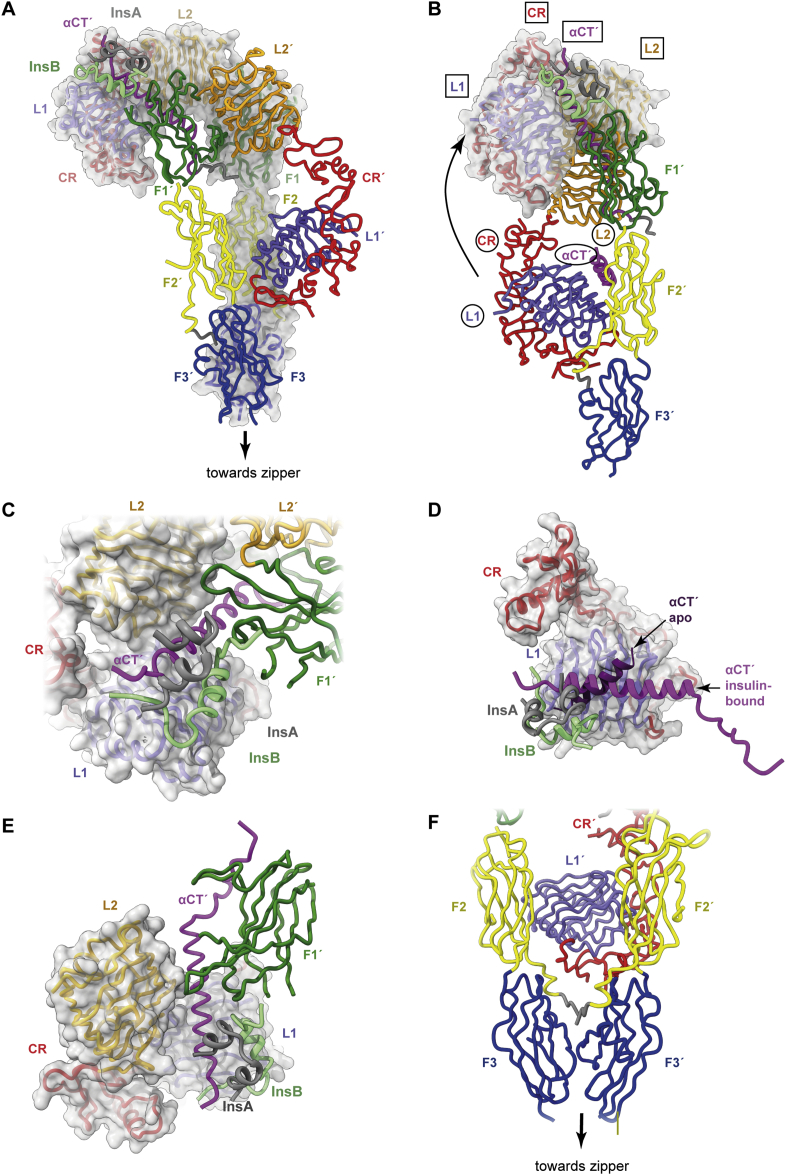
Three-dimensional structure of the single-insulin-bound insulin receptor ectodomain. (A) Overall domain configuration of the single-insulin-bound insulin receptor ectodomain determined in the context of the leucine-zippered construct. (B) Schematic demonstrating how the L1, CR, and L2 domains and the αCT′ segment relocate to the receptor “head” upon insulin binding. Domain names are circled for the insulin-free elements and boxed for the insulin-bound elements. Only a single receptor “leg” is shown (that is, domains L1, CR, and L2 from one monomer and domains FnIII-1, FnIII-2, FnIII-3, and αCT′ from the alternate monomer). (C) Detail of (A) showing engagement of insulin with domain FnIII-1′. (D) Detail of (A) showing the reconfiguration of αCT′ upon the L1 surface upon insulin binding. (E) Detail of (A) showing engagement of domains L2 and FnIII-1′ with the extended αCT′ helix and engagement of domain L1 with domain L2 upon insulin binding. (F) Detail of (A) showing how the domains FnIII-2, FnIII-2, FnIII-3, and FnIII-3′ fold inwards with respect to the apo receptor ectodomain structure, forming an interaction between domains FnIII-3 and FnIII-3′ while retaining an apo-like association of domain L1′ with domain FnIII-2. Panels are based on PDB 6HN5 [13], PDB 6HN4 [13] and PDB 4ZXB [95].
Relevant to this structure's interpretation is the way in which the cryoEM sample was prepared—namely, by elution from an insulin-affinity column upon application of a stoichiometric excess of insulin. Excess insulin was then removed by size-exclusion chromatography and the complex then further combined with the Fv module of monoclonal antibody 83-7 and repurified by size-exclusion chromatography, with the final sample concentration being 0.09 mg mL−1 [110, 111].2 Critically, the retention of only a single insulin molecule under these conditions suggests that the two possible binding sites for insulin (formed by L1+αCT′ plus FnIII-1′ in one instance and by L1′+αCT plus FnIII-1 in the other) are non-equivalent, arguably reflecting the known negative cooperativity of insulin binding to holo-receptor [54].
The interaction of the FnIII-3 domains in this structure is also informative. The juxtaposition of these domains is not a hard constraint of zipper attachment, as there is an approximately 10-residue segment within each receptor β chain between the C-terminal exit of domain FnIII-3 and the start of the zipper sequence [13,106]. The juxtaposition of the receptor legs is thus likely a consequence of insulin binding, compatible with the paradigm of insulin binding enabling juxtaposition of the tyrosine kinase domains and their transphosphorylation. The interaction between domains FnIII-3 and FnIII-3′ is not extensive, suggesting that their interaction does not play a major role in stabilizing the signaling-active conformation of the receptor ectodomain [13]. This point will be returned to later.
A key issue raised by this structure is that it does not display the predicted engagement of insulin's hexamerization surface with the receptor (Figure 1E), the latter surface surmised to be involved in the formation of the site 1-site 2′ cross-link proposed necessary for high-affinity insulin binding. Indeed, the only additional interaction of insulin with receptor observed in this structure is that of insulin with domain FnIII-1′ (Figure 5C, see (b) above); this interaction is sparse and does not involve the hexamerization surface [13], implying that there was likely yet a further binding surface on the receptor to be discovered.
The second cryoEM structure to be discussed is that published by Scapin et al. [12], shortly before the appearance of the leucine-zippered receptor complex in the literature. The sample comprised the isolated receptor ectodomain combined with insulin at a 10:1 insulin-to-receptor ratio and prepared at a concentration of 0.3 mg mL−1 The structure displays two insulins bound in an apparently symmetric fashion at a resolution of 4.3 Å (PDB 6CE9, PDB 6CEB). The insulins are bound to the respective receptor L1+αCT′ and L1′+αCT elements in a fashion similar to that described above, with the L1+αCT′ and L1′+αCT modules detaching from their respective adjacent domains FnIII-2′ and FnIII-2 and both undergoing similar conformational changes to bring their respective insulins into contact with the membrane-distal loops of the respective domains FnIII-1′ and FnIII-1 (Figure 6). Both domains L2 and L2′ fold out of the receptor head and the αCT′ and αCT helices both undergo N-terminal extension, these changes again being individually similar to those in the single-insulin-bound leucine-zippered receptor construct described above. Domains FnIII-2, FnIII-3, FnIII-2′, and FnIII-3′ are, however, unresolved, suggesting their conformational flexibility (domain FnIII-2′ could, however, be partially resolved when two-fold symmetry was relaxed [12]). Scapin et al. also presented a lower-resolution structure (7 Å resolution) of the single-insulin-bound receptor ectodomain [12], with domains FnIII-2, FnIII-3, and FnIII-3′ likewise unresolved. The overall conformation of this lower-resolution structure appears compatible with that described for the single-insulin bound leucine-zippered ectodomain, but its (insulin-free) L1′-CR′ module is detached from domain FnIII-2.
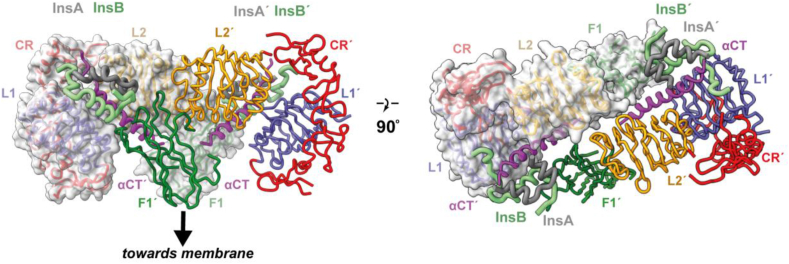
CryoEM structure of the two-insulin-bound receptor “head.” The left- and right-hand panels show orthogonal views of the isolated insulin receptor ectodomain prepared in complex with two insulins. Within this structure, domains FnIII-2, FnIII-2′, FnIII-3, and FnIII-3′ are disordered. The mode of interaction of the first insulin with the receptor domains L1, αCT′, and FnIII-1′ and of the second insulin with receptor domains L1′, αCT, and FnIII-1 are both effectively identical to that of the single insulin bound to the leucine-zippered construct shown in Figure 5. Panel is based on PDB 6CE9 [12].
The final cryoEM structures to be described are two that each display four bound insulins. The first of these two was obtained using an insulin receptor ectodomain construct prepared at 40 μM concentration and at a 28:1 insulin-to-receptor ratio and was obtained at an average resolution of 4.3 Å (PDB 6SOF) [15]3. The second was obtained using detergent-solubilized insulin holo-receptor prepared at 7 mg mL−1 at a 4:1 insulin-to-receptor ratio and yielded an average resolution of 3.1 Å for the head part of the insulin-bound receptor and 3.2 Å for the structure as a whole (PDB 6PXW, PDB 6PXV) [14]. The TM- and cytoplasmic domains were absent from the latter model. Whereas these two structures differ in detail, the overall conformational changes caused by insulin binding are closely similar across the two structures. Single insulin molecules are bound to the respective primary binding sites formed by the L1+αCT′ and L1′+αCT elements. The L1-CR-L2 modules undergo similar conformational changes to those seen in the structure of Scapin et al. [12], bringing their two bound insulins into contact with the membrane-distal loops of domains FnIII-1′ and FnIII-1, respectively, again in a fashion similar to that described above.
The striking difference in these two structures with respect to the earlier structures of Scapin et al. [12] and Weis et al. [13] is the presence of the two additional insulins, attached to the outward-facing β-sheet surfaces of domains FnIII-1 and FnIII-1′, respectively (Figure 7A). The additional insulins bind the receptor via residues GlnB4, GluB13, LeuB17, ValB18, GluB21, LeuA13, TyrA14, and GluA17 (Figure 7B). This residue set aligns closely with the hexamer-forming set of insulin residues (HisB10, GluB13, LeuB17, SerA12, LeuA13, and GluA17; Figure 1E) predicted to engage receptor site 2′ [58,112]. The two additional insulins display the receptor-free conformation of insulin, that is, their B-chain C-terminal segment remains engaged with the hormone's core. Residues of domain FnIII-1 involved in the interaction include Tyr477, Arg479, Lys484, Leu486, Arg488, Pro537, Leu552, and Arg554 (similar to the interaction of domain FnIII-1′ with its insulin). Mutation of residues Lys484 and Leu552 individually to alanine were previously shown to reduce receptor IC50 for insulin two- and five-fold, respectively [112], and in Uchikawa et al.’s study [14], the authors showed that the individual mutation of Tyr477Ala, Arg479Glu, Lys484Glu, Arg488Glu, Pro536Ala, Pro537Ala, Leu552Ala, and Arg554Glu each led to decreased insulin-dependent activation, with the Lys484 and Leu552 mutations having the greatest effect. However, these [FnIII-1]- and [FnIII-1′]-bound insulins do not form inter-monomer cross-links to sites 1 or 1′ (apart from relatively minor contacts with domains L1 and with part of the insert domain [15]). Their binding sites presumably have lower affinity for insulin than the primary [L1+αCT′]- and [L1′+αCT]-binding sites as they appear to have been populated only under insulin-saturating conditions.
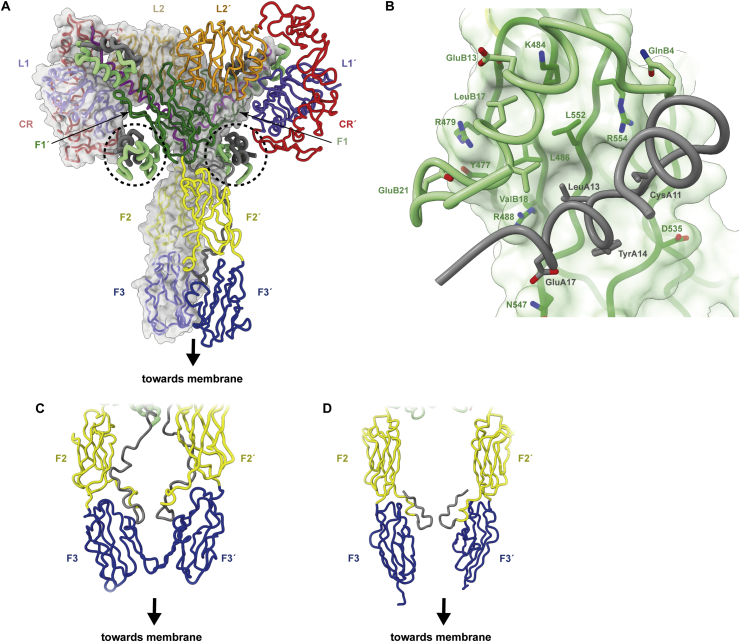
Three-dimensional structure of the four-insulin-bound insulin receptor ectodomain. (A) Overall pseudo-symmetric structure of the four-insulin-bound insulin receptor ectodomain structure determined by Gutmann et al. [15]. The binding sites of the additional bound insulins are enclosed in dashed circles. (B) Detail of insulin's binding to the surface of domain FnIII-1′ in the structure, as seen in the structure determined by Uchikawa et al. [14]. (C) Interaction between domains FnIII-3 and FnIII-3′ in the structure determined by Gutmann et al. [15]. (D) Interaction between insert domain segments IDα and IDα′ in the structure determined by Uchikawa et al. [14]. Panels are based on PDB 6SOF [15] and PDB 6PXV [14].
Intriguingly, in the four-insulin-bound ectodomain-only structure [15], domains FnIII-3 and FnIII-3′ contact each other, whereas in the four-insulin-bound holo-receptor structure, a contact forms instead between α- and α′-chain segments immediately downstream of the respective α-to-β and α′-to-β′ disulfide bonds, with none occurring between domains FnIII-3 and FnIII-3′ (Figure 7C). Both of these inter-monomer contacts differ from that in the single-insulin-bound leucine-zippered ectodomain structure described above. The lack of a common interaction across these structures' membrane-proximal elements suggests that the insulin-bound form of the receptor ectodomain is not stabilized by an interaction between its membrane-proximal elements; these interactions are likely secondary to the domain rearrangements that occur within the receptor's “head” region.
There are further differences between the two four-insulin-bound structures. The structure presented by Uchikawa et al. [14] had an “almost perfect” two-fold (C2) symmetry and was hence refined with the symmetry enforced. Focused refinement of the head region alone allowed further improvement to the 3.1 Å resolution. However, relaxation of C2 symmetry allowed the detection of a subset of particles that possibly had only three bound insulins, one each to [L1+αCT′] and [L1′+αCT] and one to one of the domain FnIII-1 sites. By contrast, the structure presented by Gutmann et al. [15] displayed only pseudo-two-fold symmetry. Notably, the insulin bound to site 2′ (that is, to the site on domain FnIII-1′) was seen to interact with the segment Gln672-Ser673 (within the insert domain of the alternate monomer to domain FnIII-1′) and with the segment Glu676′-Cys682′ (within the insert domain of the same monomer as that contributing domain FnIII-1′), whereas the site 2-bound insulin displayed no interaction with either monomer's insert domain. These insulins also differed in the extent of their interaction with the respective domains L1 and L1′. The reasons for these asymmetries were unclear. However, it should be noted that the interactions of the site 2,2′-bound insulins with the insert domains and with domains L1 and L1′ were sparse and may lack physiological relevance in a holo-receptor context. Gutmann et al. also detected subsets of particles that displayed marked asymmetry of the head, apparently arising from two- or three-insulin-bound particles. Molecular dynamics analysis of their four-insulin-bound particles also indicated that one of the two site 1-bound insulins was more mobile than the other. Whereas it is possible that these asymmetries reflected binding subtleties, it is also possible that they reflected asymmetries induced by asymmetric association of the particles with the air–water interface.
The role of the additional insulin-binding sites is not well understood. It is possible that they are involved in the initial interaction of insulin with its receptor (individual mutation of insulin residues LeuA13 and GluA17 to alanine reduces the receptor on-rate by nearly 20-fold [113]). It is also possible that insulin binding to these site(s) indirectly destabilizes the interaction between the apo L1+αCT′ tandem element and the adjacent domain FnIII-2′—this destabilization may allow domain L1 to disassociate from domain FnIII-2′, resulting in the L1+αCT′ element then being accessible to an incoming insulin molecule. One such mechanism for destabilization may be interactions between one of these insulins and domain L2 upon binding the apo-receptor: superimposition of the insulin-bound domain FnIII-1′ onto its counterpart in the apo-receptor results in steric overlap with an N-terminal loop segment of domain L2 [14]. Destabilization of domain L2 will likely destabilize the interaction of the [L1-CR]+αCT′ module with domain FnIII-2′, making the module accessible to insulin. Alternatively, it is possible that the insulin molecule bound to domain FnIII-1′ itself becomes “captured” by the adjacent insulin-free L1+αCT′ element, binding the latter by an induced fit mechanism and in the process freeing domain L1 from adjacent domain FnIII-2′. The existence and location of these additional insulin-binding sites may thus provide a hint as to how insulin gains access to the L1+αCT′ element. A further point is that if the receptor sites bound by the additional insulins are indeed sites 2 and 2′ within the kinetic model, then the assignment of the interaction of the [L1+αCT′]-bound insulin with domain FnIII-1′ as that of site 2′ (as in [12]) is incorrect.
As noted, the cryoEM structures of the insulin-bound insulin receptor described above display different hormone-to-receptor stoichiometry—one, two, and four insulin(s) per (αβ)2 receptor. It is instructive to consider a recent study by Gutmann et al. [11] wherein the authors reported a low-resolution negative-stain electron microscopy study of the insulin-complexed holo-receptor embedded in a lipidic nanodisc, with insulin added at discrete concentrations (0 nM, 0.8 nM, 12 nM, and 1000 nm, respectively). Under these experimental conditions, three-dimensional reconstruction was precluded, but image analysis yielded a set of two-dimensional class averages that broadly concurred with the structures described above (Figure 8). In particular, a Λ-shaped conformation of the apo ectodomain was observed. Addition of insulin then resulted in progressive appearance of T-shaped structures wherein the legs of the Λ-shaped structure transitioned to a single stalk, although the low resolution precluded detection of individual insulin molecules. Furthermore, it appeared that a single insulin molecule was sufficient to effect the transition to the T-shaped structure, compatible with data indicating that the receptor could be activated by a single insulin molecule [114,115]. The paucity of class averages that reflected the shape of the structure reported by Weis et al. (Figure 5; [13]) suggested that the association of the insulin-free [L1-CR]′ module with the adjacent domain FnIII-2 within that structure may have been weak, leading to their separation under negative-stain experimental conditions. This weak attachment may also explain the low-resolution structure reported by Scapin et al. of the single insulin-bound receptor ectodomain [12]. The T-shaped structures did not appear to display precise two-fold symmetry, suggesting that the insulin-complexed receptor “head” region may be somewhat mobile with respect to the “stalk” or that it may be complexed asymmetrically with insulin or by a varying number of insulins.
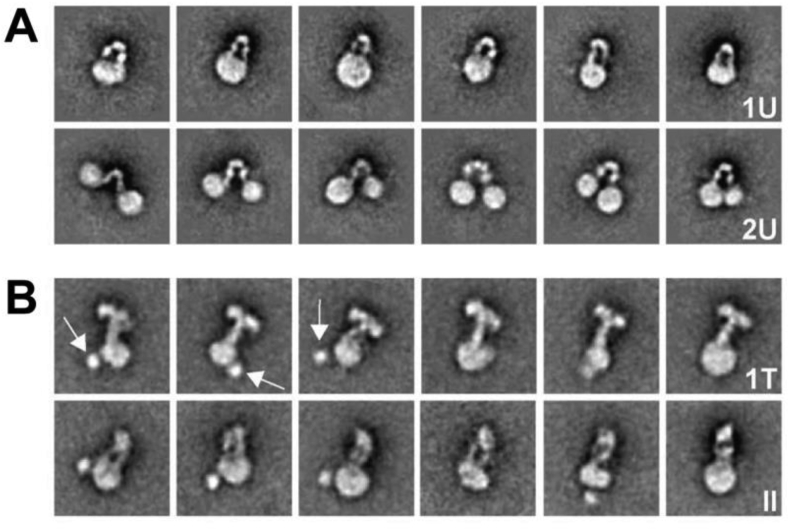
Two-dimensional class averages obtained from negative-stain images of insulin-bound nanodisc-embedded holo insulin receptor. (A) Apo holo-receptor showing the characteristic Λ-shaped conformation assembled either into one or two nanodiscs (1U and 2U, respectively). (B) Insulin-complex holo-receptor assembled into nanodiscs after complexation with insulin. All display incorporation into a single nanodisc, suggesting that insulin binding permits or directs the combination of the ectodomain's membrane proximal elements. Arrows are the blobs putatively corresponding to the tyrosine kinase domain(s). The class averages reflect either a T-shaped conformation (1T) or a narrower conformation with two legs visible (II), the latter may in some instances be a side view of the 1T conformation. Based on Figure 3 in Gutmann et al. (2018) J Cell Biol 217, 1643-9 (https://doi.org/10.1083/jcb.201711047) and used under a Creative Commons License.
5. An alternative model of the kinetics of insulin's engagement with its receptor
Taken together, the cryoEM studies of the insulin-bound receptor ectodomain and insulin-bound holo-receptor provide a set of structures that can be interpreted in a mostly unified fashion. However, it is not possible to map these structures directly onto states within the kinetic models of insulin binding proposed by De Meyts [57] and Schäffer [56]. In particular,
- (a)
no insulin molecule is seen to form an inter-monomer crosslink wherein the insulin interacts with one receptor monomer via its classical binding surface and with the other receptor monomer via its hexamer-forming surface.
- (b)
Instead, the classical binding surface of insulin is seen itself to cross-link monomers (that is, a single insulin cross-links domain L1 of one monomer to the αCT′ segment of the other), even in the isolated ectodomain context. This cross-link cannot in itself reflect high-affinity (sub-nM) binding as its affinity is nM [104,105].
- (c)
An additional receptor-binding surface of insulin is detected spanning residues AsnB3, HisB5, HisB10, and GluB13 and interacting with membrane-distal loops of domain FnIII-1′. These receptor-binding residues lie largely outside of both the dimerization and hexamerization surface of insulin. However, their interaction with the receptor is not extensive and it is reasonable to group them together with those that bind the L1+αCT′ element. As this insulin interaction is present in both holo-receptor and ectodomain-only structures, it also does not in itself appear responsible for high-affinity binding. High-affinity binding must therefore result from the additional enthalpic contributions that arise from the extensive reorganization of the receptor domains.
- (d)
However, whereas the receptor sites bound by the two additional insulins do not correspond to sites within the cross-linking model, they do correspond to sites that become populated at high concentrations and hence might be responsible for the reduced negative cooperativity at high insulin concentrations.
Some clues emerge to the source of negative cooperativity (that is, the non-equivalence of the [L1+αCT′] and [L1′+αCT] insulin-binding sites). Weis et al. [13] showed that negative cooperativity may result from the coupling of the αCT′ and αCT elements via the upstream disulfide bond(s) at residues Cys682, Cys683, and/or Cys685, proposing that the αCT′ and αCT elements cannot both simultaneously adopt configurations compatible with high-affinity binding to the respective [L1+αCT′] tandem elements.
A revised model is thus proposed herein for insulin's receptor engagement. In the apo state, the receptor ectodomain adopts the Λ shape seen in both its crystal structure [8,94,95] and in negative-stain imaging of nanodisc-embedded apo holo-receptor [11]. In this state, the primary insulin-binding sites are partly occluded as a consequence of domain L1 engaging domain FnIII-2′ and domain L1′ engaging domain FnIII-2.4 Receptor activation is then initiated due to insulin engaging via its hexamer-forming surface, the outward-facing β-sheet surface of domain FnIII-1′, this interaction being of low affinity. Such insulin binding then releases domain L1 from domain FnIII-2′, enabling the L1+αCT′ element to then bind an insulin molecule with higher (nM) affinity. The disengagement of the L1 domain from domain FnIII-1′ is likely facilitated by an interaction with domain L2 of the insulin bound to domain FnIII-1′ [14]. Whether the insulin molecule that binds L1+αCT′ is the same as that bound to FnIII-1′ is an open question—if it is, then the binding of that insulin to the L1+αCT′ element via its initial engagement of domain FnIII-1′ is an induced fit process.5 The [L1-CR]+αCT′ module then undergoes conformational changes to transport its bound insulin to the “head” of the receptor whereupon the insulin docks onto the membrane-distal loops of domain FnIII-1′. Significant rearrangement of receptor domains occurs in this process, which in turn enables the bringing together of the receptor ectodomain's membrane-proximal regions. These conformational changes also alter the affinity of insulin for the alternate [L1-CR]′+αCT module, either increasing it through destabilizing its attachment to domain FnIII-2 or decreasing it through the destabilization of the αCT element on the surface of the L1′-β2 sheet. Whether a second insulin molecule is able to bind to this site will be a function of the local insulin concentration, but were it free to bind insulin, its affinity for insulin would be greater than that of the surrounding receptors, which have only (low-affinity) sites 2 and 2′ free to bind insulin.
These processes do not preclude domain L1 detaching transiently from domain FnIII-2′ to allow insulin access to site 1. As noted above, such receptor domain mobility may underlie the receptor's basal activity (i.e., in the absence of insulin), perhaps in a fashion similar to that proposed by Kiselyov et al. (i.e., “harmonic oscillation”) [117]. In this regard, it is worth noting that attempts at cryoEM reconstruction of the apo insulin receptor ectodomain discern considerable heterogeneity of conformation, precluding high-resolution reconstruction [12,15].
Several alternative schemes have been proposed. Scapin et al. [12] proposed that the αCT segment is disengaged from the L1 domain prior to insulin binding and that the L1-CR-L2 modules to not engage the FnIII modules prior to insulin binding, i.e., the apo receptor does not have the Λ-shaped conformation found in its crystal structure but instead has a T-shaped conformation prior to insulin binding. However, this seems very unlikely given that the Λ-shaped conformation is confirmed by images of the apo holo-receptor in nanodiscs [11]. The Λ-shaped conformation is also consistent with that adopted by the homologous ectodomain of IGF-1R in the absence of ligand and wherein the αCT segments are also seen docked on the respective L1 domains [98]. Uchikawa et al. proposed that one insulin binds first to site 1 [14], although they did not specify how this occurs. They posited that a second insulin binds to site 2, enabling insulin's engagement with site 1′ and a transition to the T-shaped structure. In this model, the receptor is only partially active when bound with a single insulin, but fully active when bound by three (or four) insulins. Whether insulin receptors with multiple bound insulins occur under physiological conditions remains an open question.
6. The final step: receptor activation
The structures discussed above align with the paradigm of insulin activating its receptor by intra-receptor transphosphorylation: in the Λ-shaped apo receptor, the TK domains are kept apart, whereas in the insulin-bound receptor, these domains are brought together. Two additional structural studies are germane to these events. The first is that co-obtained in the cryoEM study of detergent-solubilized insulin-bound holo-receptor [14]. In that study, a three-dimensional reconstruction was also obtained at 6.7 Å that showed a tentative interaction between the receptor's two TM domains (Figure 9A). Such an interaction may assist in stabilizing the insulin-bound holo-receptor, in turn aiding transphosphorylation of the intra-cellular TK domains [14]. It should be noted, however, that the low-resolution map precluded any accurate modeling of the TM domains themselves, although a solution structure of the latter has been determined in isolation (and shown to be helical [118]; Figure 9B). The second is the crystal structure of the receptor's tyrosine kinase domains extended N terminally to include residues of the juxtamembrane region and in co-complex with AMPPCP and Mg2+, with the juxtamembrane residue Tyr972 and TK activation-loop residues Tyr1158, Tyr1162, and Tyr1163 being phosphorylated prior to crystallization [119]. Within this structure, the C-terminal segment of the juxtamembrane region associates in trans across the TK domains, stabilizing the N-terminal lobe's regulatory αC helix; the activation loops have a folded-out active conformation (Figure 9C). Such trans interaction likely stabilizes the activated conformation of the TK domains and promotes its kinase activity with respect to downstream substrates.
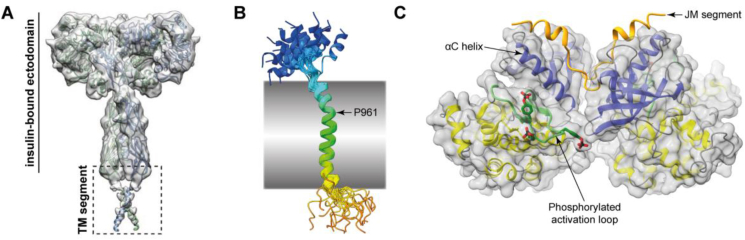
Transmembrane and cytoplasmic domains of the activated insulin receptor. (A) Putative association of the TM domains of the insulin-activated holo-receptor within a detergent micelle as detected by single-particle cryoEM [14]. The panel was extracted from Figure 1, Supplementary Figure 4 from Uchikawa et al. (2019) eLife 8, e48630 (https://doi.org/10.7554/eLife.48630) and used under a Creative Commons License. (B) Solution structure of the insulin receptor TM domain (backbone trace only, 20 structures superimposed [118]). The traces are rainbow-colored representations from blue (Met939) to red (Glu988). The kink at residues Gly960/Pro961 [118] is arrowed; the shaded box represents the membrane's approximate span. (C) Trans-association of upstream juxtamembrane segment with the respective TK domains (residues Tyr972, Tyr1158, Tyr1162, and Tyr1163 are phosphorylated) showing the interaction of the JM segment with the αC helix of the opposing TK domain [119]. Blue ribbons: N-terminal lobes of the respective TK domains; yellow ribbons: C-terminal lobes of the respective TK domains; green ribbons: phosphorylated activation loops; orange ribbons: respective upstream JM segments; gray surfaces: respective molecular surfaces of the TK domains. The labeled segments are from the same TK domain.
7. Conclusion
The first diffraction patterns from insulin were obtained in 1935 by Dorothy Hodgkin (née Crowfoot) [3]; 34 years then elapsed before insulin's atomic structure emerged [6] and a further 44 years then elapsed before atomic structures of the insulin-bound receptor began to emerge [9]. These structures illuminate decades of corresponding biochemical investigation but at the same time display a molecular complexity that remains understood only in part. The difficulty is that current structural biology techniques provide only static images and, consequently, little is understood of the dynamics of insulin insulin-receptor engagement. Two pathways forward emerge. The first is to increase the number of static images, with the aim of generating a set of structures that populate the conformational trajectory traversed by insulin as it activates the receptor. CryoEM is ideally suited to this task given its ability to discern subpopulations (three-dimensional “classes”) of particles within a single sample. Better sample preparation techniques will aid this process as well as the judicious use of mutant insulins or receptor constructs that could lead to the trapping of stable intermediates. The second is via in silico simulation. Molecular dynamics simulation has been used to improve or validate the atomistic detail of insulin-complexed insulin receptor models derived from cryoEM as well as to understand the degree of mobility of domains within these complexes [15]. It has also been used to investigate the engagement of novel insulins with the receptor sites described above [120]. Computational techniques to model the trajectories of the large-scale domain motion that occurs within the receptor as whole upon ligand binding may also become within reach [[121], [122], [123]]. The next step forward is likely to occur within years, not decades.
Acknowledgments
I thank my many colleagues and collaborators who have contributed to our structural studies of the insulin receptor system over the past two decades. I pay particular tribute to the late Drs Colin Ward and Neil McKern, whose prior insulin receptor studies formed the basis for my laboratory's current research. My research was made possible at WEHI through the Victorian State Government Operational Infrastructure Support and the Australian NHMRC Independent Research Institutes Infrastructure Support Scheme, with part of my salary being derived from sub-awards of NIH R01 grants DK127761, DK124401, and DK040949.
Footnotes
1The insulin crystallized was of porcine origin; porcine insulin differs slightly from human insulin, having an alanine in place of the human threonine at position B30.
2The attachment of the Fv 83-7 module [16,111] was originally intended to aid crystallographic study of the complex; in the cryoEM experiment; here, it potentially aided particle classification and three-dimensional refinement. The Fv module is attached to the receptor's CR domain, distal to the insulin-binding site.
3Published in advance of peer-review as https://www.biorxiv.org/content/10.1101/679233v1.
4These inter-monomer interactions (between domain L1 and domain FnIII-2′ and between domain L1′ and domain FnIII-2) have been shown in IGF-1R to be essential for the auto-inhibition of the receptor: an IGF-1R construct devoid of domains L1 and L1′ is constitutively active [116].
5Indirect evidence of an induced fit mechanism arises from the soaking insulin-like growth factor I (IGF-I) into crystals of the apo ectodomain of IGF-1R. In that study, IGF-I was seen to engage the occluded L1+αCT′ element of IGF-1R despite the constraints of lattice embedding [98].
Conflict of interest
MCL's laboratory has a funded agreement with Eli Lilly and Company (USA) as well as past collaborations with Sanofi-Aventis (Germany). MCL is a co-inventor of an insulin-related patent licensed to Monolog LLC.
References
Articles from Molecular Metabolism are provided here courtesy of Elsevier
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/105834424
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.molmet.2021.101255
Article citations
Insulin oxidation and oxidative modifications alter glucose uptake, cell metabolism, and inflammatory secretion profiles.
Redox Biol, 77:103372, 05 Oct 2024
Cited by: 0 articles | PMID: 39378614 | PMCID: PMC11492613
Glucose-sensitive insulin with attenuation of hypoglycaemia.
Nature, 634(8035):944-951, 16 Oct 2024
Cited by: 0 articles | PMID: 39415004 | PMCID: PMC11499270
Spontaneous Dimerization and Distinct Packing Modes of Transmembrane Domains in Receptor Tyrosine Kinases.
Biochemistry, 63(20):2692-2703, 25 Sep 2024
Cited by: 0 articles | PMID: 39322977 | PMCID: PMC11483822
Advancements in strategies for overcoming the blood-brain barrier to deliver brain-targeted drugs.
Front Aging Neurosci, 16:1353003, 26 Aug 2024
Cited by: 0 articles | PMID: 39253614 | PMCID: PMC11381257
Review Free full text in Europe PMC
The Pathophysiological Mechanism and Clinical Treatment of Polycystic Ovary Syndrome: A Molecular and Cellular Review of the Literature.
Int J Mol Sci, 25(16):9037, 20 Aug 2024
Cited by: 0 articles | PMID: 39201722 | PMCID: PMC11354688
Review Free full text in Europe PMC
Go to all (46) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Protein structures in PDBe (Showing 21 of 21)
-
(4 citations)
PDBe - 4ZXBView structure
-
(3 citations)
PDBe - 6HN5View structure
-
(3 citations)
PDBe - 1MSOView structure
-
(2 citations)
PDBe - 6HN4View structure
-
(2 citations)
PDBe - 6PXVView structure
-
(2 citations)
PDBe - 1IRKView structure
-
(2 citations)
PDBe - 2HR7View structure
-
(2 citations)
PDBe - 6SOFView structure
-
(2 citations)
PDBe - 6VEPView structure
-
(2 citations)
PDBe - 6CE9View structure
-
(1 citation)
PDBe - 6PXWView structure
-
(1 citation)
PDBe - 6INSView structure
-
(1 citation)
PDBe - 5U8RView structure
-
(1 citation)
PDBe - 6CE7View structure
-
(1 citation)
PDBe - 1HIQView structure
-
(1 citation)
PDBe - 4OGAView structure
-
(1 citation)
PDBe - 1XW7View structure
-
(1 citation)
PDBe - 1ZNIView structure
-
(1 citation)
PDBe - 6CEBView structure
-
(1 citation)
PDBe - 1HITView structure
-
(1 citation)
PDBe - 3W11View structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556(7699):122-125, 28 Feb 2018
Cited by: 107 articles | PMID: 29512653 | PMCID: PMC5886813
The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9(1):4420, 24 Oct 2018
Cited by: 65 articles | PMID: 30356040 | PMCID: PMC6200814
Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain.
J Cell Biol, 219(1):e201907210, 01 Jan 2020
Cited by: 53 articles | PMID: 31727777 | PMCID: PMC7039211
The Activation Mechanism of the Insulin Receptor: A Structural Perspective.
Annu Rev Biochem, 92:247-272, 31 Mar 2023
Cited by: 18 articles | PMID: 37001136 | PMCID: PMC10398885
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIDDK NIH HHS (3)
Grant ID: R01 DK040949
Grant ID: R01 DK124401
Grant ID: R01 DK127761
National Health and Medical Research Council
National Institutes of Health (3)
Grant ID: DK127761
Grant ID: DK040949
Grant ID: DK124401




