Abstract
Free full text

Analysis of Sex Chromosome Evolution in the Clade Palaeognathae from Phased Genome Assembly
Abstract
Birds in the clade Palaeognathae, excluding Tinamiformes, have morphologically conserved karyotypes and less differentiated ZW sex chromosomes compared with those of other birds. In particular, the sex chromosomes of the ostrich and emu have exceptionally large recombining pseudoautosomal regions (PARs), whereas non-PARs are classified into two strata according to the date of their origins: stratum 0 and stratum 1 (S1). However, the construction and analysis of the genome sequences in these regions in the clade Palaeognathae can be challenging because assembling the S1 region is difficult owing to low sequence diversity between gametologs (Z-linked and W-linked sequences). We addressed this issue by applying the Platanus-allee assembler and successfully constructed the haplotype-resolved (phased) assembly for female emu, cassowary, and ostrich using only sequence read data derived from the Illumina platform. Comparative genomic and phylogenetic analyses based on assembled Z-linked and W-linked sequences confirmed that the S1 region of emu and cassowary formed in their common ancestor. Moreover, the interspersed repetitive sequence landscapes in the S1 regions of female emu showed an expansion of younger repetitive elements in the W-linked S1 region, suggesting an interruption in homologous recombination in the S1 region. These results provide novel insights into the trajectory of sex chromosome evolution in the clade Palaeognathae and suggest that the Illumina-based phased assembly method is an effective approach for elucidating the evolutionary process underlying the transition from homomorphic to differentiated sex chromosomes.
Introduction
Similar to mammals, sex in avian species is determined by the chromosome set (Witschi 1959). In contrast to mammals, birds have a ZW-type sex chromosome pattern, in which the homogametic sex (ZZ) is male, and the heterogametic sex (ZW) is female. Aves comprise two large clades, Neognathae and Palaeognathae, which are distinguished by plate shape. This classification is also supported by the results of molecular phylogenetic analyses (Sibley and Ahlquist 1990; van Tuinen et al. 1998). The Z and W chromosomes are homologous between Neognathae and Palaeognathae, and the fundamental sex-determining system based on sex chromosomes is conserved in these two clades, indicating that they share a common ancestor. However, there are marked differences in the sex chromosomes of each clade. According to the latest estimate, Neognathae, which comprises more than 16,000 species, including chicken (Barrowclough et al. 2016), has a W chromosome containing a small heterochromatic region that undergoes extensive degradation, similar to that of the Y chromosome in mammals (Tone et al. 1982; Ellegren 2000). In contrast, palaeognathous birds have morphologically conserved karyotypes and less differentiated ZW sex chromosomes (Ogawa et al. 1998; Shetty et al. 1999; Tsuda et al. 2007) with exceptionally large recombining pseudoautosomal regions (PARs). Moreover, recent sequencing analyses of ostrich and emu genomes have revealed that at least two-thirds of the Z chromosome consists of PARs, with the rest of the sequence represented by two strata: stratum 0 (S0) and stratum 1 (S1) (Vicoso et al. 2013; Zhou et al. 2014). S0 is the more evolutionarily ancient stratum; the putative male sex-determining gene DMRT1 is located in the S0 region, suggesting that this region was formed in the common avian ancestor about 120 Ma (van Tuinen and Hedges 2001; Smith et al. 2009). In contrast, S1 is relatively younger than S0 and consists of sequences approximately 5–10 Mb long that exhibit higher levels of sequence similarity (approximately 90–95% in genome level) between the Z and W chromosomes (Zhou et al. 2014; Xu, Auer, et al. 2019; Xu and Zhou 2020) than those in the S0 region.
In general, sex chromosomes are considered to have originated from regular autosomes (Fridolfsson et al. 1998); therefore, genome analysis of palaeognathous birds, which have relatively fewer differentiated sex chromosomes than those of neognathous birds, could provide insights into the evolutionary trajectory of sex chromosomes. However, the low degree of differentiation between gametologous sex chromosomes makes it difficult to construct continuous sequences in the S1 and S0 regions. In particular, little success has been achieved in constructing Z-linked and W-linked phased full-length sequences, hampering the comparative genomic analysis of sex chromosomes across the clade Palaeognathae.
De novo assembly programs generally attempt to merge haplotype sequences and construct a single contig/scaffold as a mosaic of these sequences. However, if the sequence difference between the haplotypes exceeds a certain level, only the regions that are differentiated above this level will be recognized as separate sequences, resulting in a mixture of merged and separate output regions and making it difficult to obtain a continuous assembly (Li et al. 2010; Kajitani et al. 2014). In the chicken genome, the Z and W chromosomes are well differentiated and could be assembled into separate sequences. Although the S1 and S0 regions are more differentiated than the PAR in the clade Palaeognathae, there is only a small percentage difference in the S1 region and large accumulation of repetitive sequences that are not sufficient to clearly distinguish the Z-linked and W-linked sequences with conventional assembly tools. Therefore, fragmented scaffolds are the typical output for the S0 and S1 regions of female (ZW) individuals. The genome sequence of the ostrich Z chromosome, derived from a female (ZW) individual, has been widely used as a representative genome of palaeognathous birds (Zhang et al. 2015; Yazdi and Ellegren 2018). This sequence was improved using optical mapping data, showing that the entire PAR was covered by only four continuous super-scaffolds, whereas the S1 and S0 regions included several fragmented scaffolds (Zhang et al. 2015; O’Connor et al. 2018).
To elucidate the evolutionary history of Palaeognathae sex chromosomes, many efforts have been made to retrieve the sequences of these recombination-repressed S1 and S0 regions, particularly in the Z-linked and W-linked phased forms (Vicoso et al. 2013; Zhou et al. 2014; Zhang et al. 2015; Yazdi and Ellegren 2018; Xu, Wa Sin, et al. 2019). The most successful attempt to date was achieved by extracting scaffolds from the female genome assembly exhibiting a certain degree of homology with the Z-chromosome sequence constructed from the male genome with approximately half the sequence coverage compared with that of the autosomes. The extracted scaffolds were classified into Z-linked and W-linked groups according to their homology with the Z-chromosome sequence (Xu and Zhou 2020). However, whether the Z-linked and W-linked sequences are assembled separately or merged depends on the behavior of the assembler due to the local homology between gametologous sequences. Therefore, although Illumina sequencers were mainstream, no analyses used full-length phased Z-linked and W-linked genome sequences to clarify the accumulation of mutations in each lineage or gametologous sequences after differentiation.
The female emu genome sequence was constructed in which the S1 region was consecutively assembled into a single scaffold in a phased manner, with the Z and W chromosomes separated (Liu et al. 2021). This assembly appears to be highly promising for facilitating comparative analyses between gametologous pairs as well as between the S1 regions of different species, which have been highly fragmented using other assembly approaches. However, this assembly was constructed from a large amount of data produced by expensive platforms such as PacBio, 10× Genomics-linked reads, Dovetail Chicago data, and Hi-C data; therefore, this method is not easily applied to other species.
In this study, we phased an Illumina-based assembly of a female palaeognathous bird genome using the recently developed genome assembly program Platanus-allee (Kajitani et al. 2019). This allowed us to overcome cost limitations of long-read single-molecule sequencing and recover haplotypes relatively inexpensively. It is difficult to make a simple comparison of the sequencing costs because they can vary annually, but the cost of our method was estimated to be approximately 10–20% of that of PacBio and the other platform based method. Platanus-allee is an assembler that is mainly designed for heterozygous diploid genomes to conduct “phased” assembly of homologous chromosomes. We verified the effectiveness of the assembly protocol by performing phased assembly of the newly sequenced female emu genome as well as reassembly on published Illumina read data for cassowary (Sackton et al. 2019) and ostrich (Zhang et al. 2015). Through various analyses based on newly obtained long-phased gametologous sequences, including comparative genome analysis and precise synteny analysis of the S1 region, we aimed to confirm that the S1 region was formed in ostriches and the common ancestor of the Casuariiformes. We also sought to obtain novel insights exemplified by the difference of interspersed repetitive sequence landscapes between the Z-linked and W-liked S1 regions. Moreover, this study highlights the advantages of phased genome assembly and its potential to reveal the evolutionary process underlying the differentiation of homomorphic sex chromosomes in broad taxa with young chromosomes, such as in fish and amphibians (Ezaz et al. 2006; Lambert et al. 2016; Furman and Evans 2018; Takehana et al. 2020).
Results
De Novo Assembly of the Male Emu Genome
The draft genome of the male emu had a total length of 1.20 Gb that was slightly shorter than the k-mer-based estimation of 1.35 Gb, an N50 scaffold length of 13.3 Mb, and an L50 of 23 scaffolds (table 1). The newly assembled draft genome showed a longer continuous N50 scaffold than that reported previously (droVov1; GCA_003342905.1; 13.3 Mb vs 3.3 Mb), which was assembled using data obtained from the Illumina platform (Sackton et al. 2019). The read data sequenced in this study are shown in supplementary table 1, Supplementary Material online, and the detailed comparison of our results with other assemblies are shown in supplementary results and supplementary table 2, Supplementary Material online.
Table 1.
Assembly Statistics for the Emu, Cassowary, and Ostrich Scaffolds
| Common Name | Sex | Assembler | Output | Total (Gb) | N50 (bp) | #N50 | Gap (%) |
|---|---|---|---|---|---|---|---|
| Emu | Male | Platanus v1.2.4 | Consensus | 1.200 | 13,304,504 | 23 | 1.13 |
| Female | Platanus-allee v2.2.2 | Consensus | 1.297 | 8,506,830 | 44 | 4.56 | |
| All phased | 1.975 | 59,020 | 5,022 | 6.90 | |||
| Primary bubble | 0.682 | 128,006 | 1,046 | 7.49 | |||
| Cassowary | Female | Platanus-allee v2.2.2 | Consensus | 1.218 | 2,886,591 | 151 | 1.10 |
| All phased | 1.333 | 34,317 | 8,138 | 1.96 | |||
| Primary bubble | 0.132 | 15,055 | 2,037 | 6.44 | |||
| Ostrich | Female | Platanus-allee v2.2.2 | Consensus | 1.294 | 3,315,423 | 104 | 7.89 |
| All phased | 2.042 | 22,231 | 40,124 | 10.72 | |||
| Primary bubble | 0.770 | 31,536 | 9,268 | 9.62 |
Calculated for scaffolds of ≥500 bp.
bp.
Z Chromosome Detection from the Male Emu Genome (ZZ) Assembly
As shown in figure 1, 51 previously reported scaffolds from the ostrich genome, with a total length of 88.8 Mb, were used as scaffolds for the Z chromosome (Zhang et al. 2015; O’Connor et al. 2018; Xu, Wa Sin, et al. 2019). We constructed a draft of the entire Z chromosome for the cassowary that consisted of 106 scaffolds with a total length of 85.6 Mb (Sackton et al. 2019). Similarly, we constructed the counterpart region for the emu, which consisted of 13 scaffolds with a total length of 82.4 Mb. Using data from an emu male individual (ZZ) for the assembly, we succeeded in constructing six continuous scaffolds with a total of 55.3 Mb (PAR), one scaffold of 4.9 Mb (S1 region), and eight scaffolds of 22.2 Mb (S0 region). Moreover, one long scaffold, labeled scaffold1724899_cov79, covered part of the PAR (39.1 Mb), the entire S1 region (4.9 Mb), and part of the S0 region (7.7 Mb).
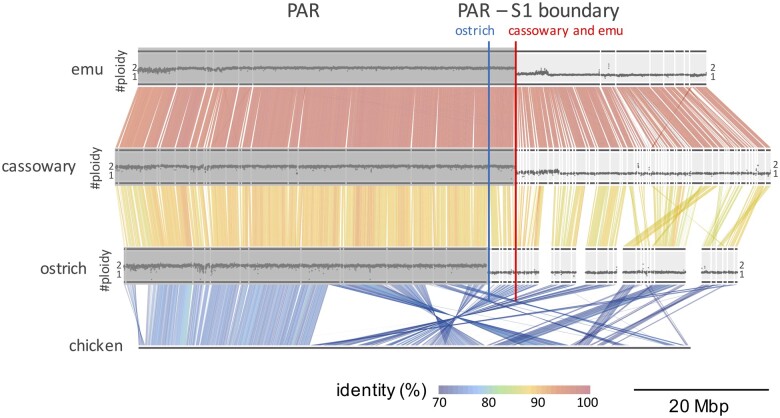
Overview of the Z chromosome with the boundary between a pseudoautosomal region (PAR) and stratum 1 (S1) region. The whole Z chromosomes of emu, cassowary, ostrich, and chicken are illustrated. Gray horizontal lines represent the scaffolds derived from the Z chromosome of each species. The order of the scaffolds in the ostrich is based on previous data (Zhang et al. 2015), and that in the cassowary and emu is based on an alignment with the chicken and ostrich Z chromosomes. The color map represents sequence alignments and their identities between the two species. The histograms shown on the top of each emu, cassowary, and ostrich scaffold represent normalized read coverage based on the sequence depth of the Illumina paired-end (PE) reads. Dark gray boxes on the histograms indicate the pseudoautosomal region (PAR) for each species. The red vertical line indicates the boundary between the PAR and S1 region of the emu and cassowary, and the blue line indicates the boundary between the PAR and S1 region in the ostrich. These graphs were plotted using an in-house python script with Matplotlib.
PAR Estimation from Read Mapping Data
The boundaries between the PARs and S1 regions were detected using the normalized read depth for each female (ZW) emu, cassowary, and ostrich. The normalized read depth along the Z chromosome clearly changed from two to one, suggesting that the upper region corresponded to the PAR (Z-W recombination); in contrast, the downstream region corresponded to the W-degenerated (Z-specific) region (fig. 1). Therefore, we defined the breakpoint of the read depth changes as the boundary between the PAR and S1 region in each species.
Comparisons of the loci of the breakpoints and synteny among the three species showed that the boundary was consistent between the emu and cassowary (Casuariiformes; indicated by the red vertical line in fig. 1), whereas the boundary in the ostrich chromosome (blue vertical line) was in a distinct position. We further confirmed that the ostrich Z chromosome had a relatively large S1 region that expanded approximately 2–3 Mb toward the PAR side compared with those of the emu and cassowary, consistent with previously studies (Zhou et al. 2014; Xu, Wa Sin, et al. 2019; Xu and Zhou 2020).
Phased De Novo Assembly of the Female Genomes
To construct phased S1 sequences, we initially applied Platanus-allee v2.2.2, a de novo haplotype assembler, to one individual genome for each female emu, cassowary, and ostrich (table 1). Read data from the female emu are shown in supplementary table 1, Supplementary Material online, and those of cassowary and ostrich were downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database and are presented in supplementary table 3, Supplementary Material online. In the female emu, the total length of the assembled consensus sequences was 1.30 Gb, which was almost equal to the estimated genome size. The total length of the primary haplotype-phased sequences (referred to as “bubble” scaffolds in Platanus-allee) corresponded to the longer haplotype-phased sequences in a pair, which was 682 Mb with an N50 scaffold length of 128 kb. In contrast, the total length of the cassowary consensus sequences was 1.22 Gb, and the total length of the primary bubble sequences was shorter than that of the emu at 131 Mb, with an N50 scaffold length of 15 kb. The estimated heterozygosity rate in the cassowary genome was lower than that in the emu, suggesting a link between heterozygosity and total size of the phased regions (supplementary table 4, Supplementary Material online). Additionally, this indicated that the libraries used for de novo assembly contributed to the continuity of the scaffolds. Specifically, in the emu, four mate-pair (MP) libraries with an insert size of 3–15 kb were used for de novo assembly, whereas only the 3-kbp MP library was used for the cassowary assembly (supplementary tables 1 and 3, Supplementary Material online). By contrast, the total length of the ostrich consensus sequences was 1.29 Gb and that of the primary bubble sequences was 770 Mb, which were slightly longer than that in the emu and had an N50 scaffold length of 31 kb. Detailed assembly results and comparison of our results with other assemblies are shown in supplementary results and supplementary table 2, Supplementary Material online.
Construction of Phased S1 Regions
Following the haplotype assembly for female emu, cassowary, and ostrich, we detected bubble scaffolds derived from the S1 region and assigned each of the phased scaffolds to the Z or W chromosome (table 2 and supplementary fig. 1, Supplementary Material online). We verified the accuracy of our haplotype-resolved assembly in female individuals using two methods. First, we confirmed the high sequence identity between the male emu (ZZ) S1 region and the female emu Z-linked S1 region according to dot-plot alignments (supplementary fig. 2, Supplementary Material online). Second, we confirmed that both the Z-linked and W-linked S1 regions had high sequence identity with those of the PacBio-based female emu assembly (fig. 2A; ZJU1.0; Liu et al. 2021). The results of dot-plot with another Illumina-based study (ASM1339679v1; Feng et al. 2020; Xu and Zhou 2020) are shown in figure 2B. However, it is clear that this Illumina-based study did not provide sufficient phased assembly, as only a few regions were constructed as separate phased sequences into Z-linked and W-linked scaffolds (fig. 2B, enlarged image), and most regions were constructed as only one sequence. The sum length of the previously reported assembly was only approximately 5.5 Mb, whereas the sum length of our six scaffolds was approximately 9.9 Mb (4.9 Mb consisting of three Z-linked scaffolds and 5.0 Mb consisting of three W-linked scaffolds), which was the same as the length of the PacBio-based assembly results in the S1 region. In addition, the published Illumina-based assembly had approximately 96–98% homology with both of our W-linked and Z-linked sequences compared with PacBio assembly, which had almost 100% homology between the same phased sequences and approximately 94–96% homology between the opposite phased sequences. This result strongly indicated that both our assembly and PacBio-based assembly succeeded in obtaining fully phased S1 sequences, whereas the previous Illumina-based assembly consisted of a mosaic-like mixture of Z-linked and W-linked sequences. Similar results were obtained from analyses of the assembly results from cassowary compared with previous studies (supplementary fig. 3, Supplementary Material online). These results confirmed that our assembly has high accuracy and comprehensiveness (table 2 and fig. 2).
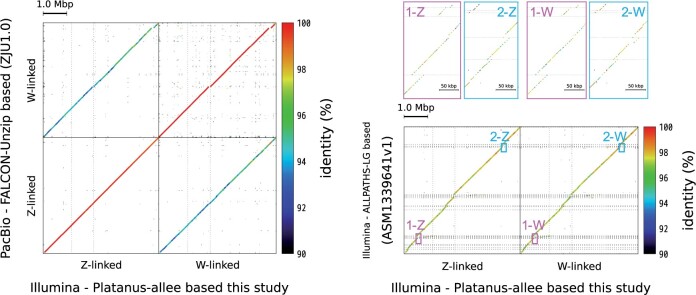
Dot-plot alignment of the female emu S1 region. (A) Dot-plot alignment of the female emu S1 regions between the Platanus-allee assembly constructed in this study and the PacBio-based assembly (ZJU1.0) (Liu et al. 2021). The horizontal axis shows the Z-linked and W-linked S1 region sequences from the female emu that was constructed by Platanus-allee assembler in this study. The vertical axis shows the Z-linked and W-linked S1 region sequences of the female emu constructed from the PacBio-based data (ZJU1.0). (B) Dot-plot alignment of female emu S1 regions between Platanus-allee and Illumina-based (droNov1) assemblies (Sackton et al. 2019). The horizontal axis shows the Z-linked and W-linked S1 region sequences in female emu constructed by the Platanus-allee assembler. The vertical axis shows the S1 region sequences in female emu constructed from Illumina-based data by the ALLPATHS-LG assembler (droNov1). Magnified views of the two regions are shown in the dot-plot alignments framed in pink and light blue, respectively.
Table 2.
Total Size of Constructed S1 Regions and Sequence Identity between Gametologs
| Common Name | No. of Phased Scaffolds | Z-Linked (Mb) | W-Linked (Mb) | Sequence Identity between Z and W Chromosomes (%) |
|---|---|---|---|---|
| Emu | 3 | 4.91 | 5.00 | 95.85 |
| Cassowary | 6 | 4.91 | 4.89 | 96.15 |
| Ostrich | 9 | 7.16 | 6.88 | 89.07 |
Comparisons of Phased S1 Regions
Sequence alignments between the Z-linked S1 and W-linked S1 regions within species (gametologous sequences) are presented in figure 3A. We noted that these results were based on the alignment of corresponding regions to show the sequence homology between Z-linked S1 and W-linked S1 regions, which were not anchored to chromosomes and do not necessarily represent their positions on chromosomes. The detected S1 regions of the emu and cassowary were 4.9–5.1 Mb in length and nearly in the same loci in both species. Conversely, the S1 region constructed from phased scaffolds in the ostrich was approximately 7.2 Mb, with nine scaffolds in total that were approximately 2.2 Mb longer toward the PAR side than that in emu and cassowary (fig. 3B). Moreover, within the same species, average sequence differences between gametologous sequences in the S1 region were markedly higher than the heterozygosity rates (i.e., the average sequence difference rate of the whole genome), which were estimated with GenomeScope (supplementary table 4, Supplementary Material online). The sequence identities between gametologous sequences in the S1 regions of the emu and cassowary were approximately 95.9% and 96.2%, respectively, which were slightly higher than that reported previously (Xu and Zhou 2020). Gametologous sequence divergence, described as the density of heterozygous sites in the S1 region, slightly increased from the S0 region side toward the PAR side (fig. 3C). Whereas the sequence identity between gametologous sequences in the ostrich S1 region was 89.1%. These results suggested that there was a higher level of differentiation in the S1 region in the ostrich than in the emu and cassowary. Interspersed repetitive sequence landscapes in the Z-linked and W-linked S1 regions of emu and cassowary females showed that the expansion of the repetitive elements, especially for LTR families, occurred in the emu W-linked region after Z/W differentiation and emu/cassowary differentiation. The same insertion pattern of young repetitive sequences was also observed in the ostrich, however, no traces of recent repetitive sequence accumulation in the W-linked region of the cassowary were observed. (fig. 3D and supplementary figs. 4 and 5, Supplementary Material online).
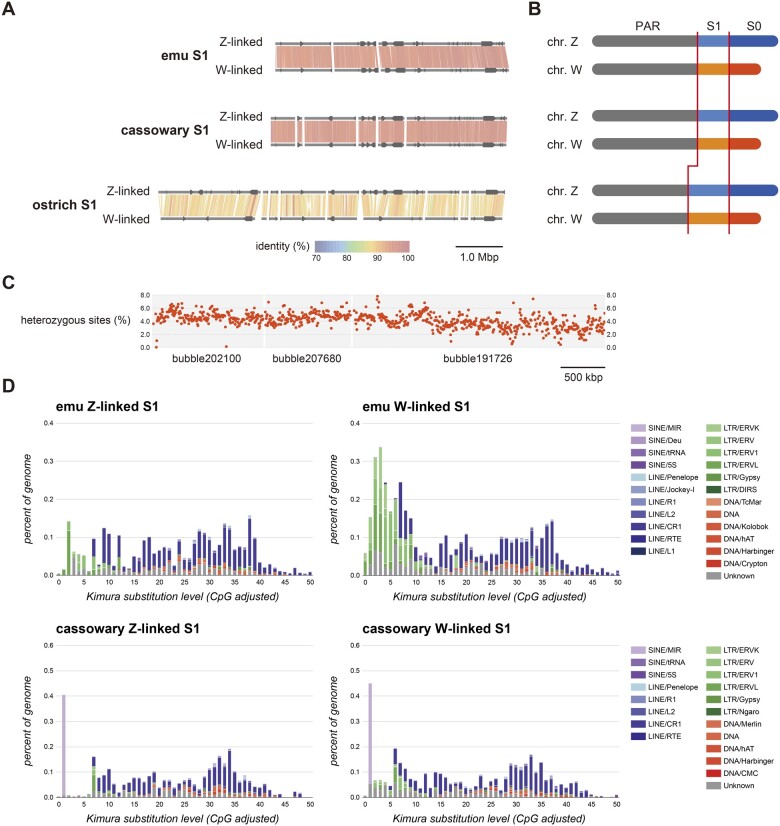
Overview of the phased Z-linked and W-linked S1 regions and their features. (A) Phased S1 scaffolds and gene synteny. Gray horizontal lines represent phase scaffolds derived from the S1 region from each species. The color map indicates sequence identity between Z-linked and W-linked S1 gametologs. The arrows on the scaffolds indicate genes annotated in the S1 regions. (B) Schematic diagram of sex chromosomes from each species. The pseudoautosomal (PAR), Z-linked S1, and W-linked S1 regions are labeled in gray, light blue, and light orange, respectively. The Z-linked S0 and W-linked S0 regions are labeled in blue and orange, respectively. Red vertical lines show the boundaries between the PAR and S1 regions, and between the S1 and S0 regions. (C) Distribution of gametologous heterozygous sites in the S1 region of a female emu individual. (D) Interspersed repetitive sequence landscapes in the Z-linked S1 region (left) and W-linked S1 region (right) of a female emu individual. The horizontal axis represents the Kimura’s substitution rate from the consensus sequences of each repetitive unit. Older copies are located on the right of the graphs, whereas more recent copies are located on the left.
Phylogenetic Analysis of W/Z Gametologs in the S1 and S0 Regions
To reveal the evolutionary trajectory of sex chromosomes in the clade Palaeognathae, phylogenetic trees were estimated for the S0 and S1 regions using annotated genes (fig. 4). The gametologous genes used for the phylogenetic analysis of the S0 and S1 regions are shown in supplementary tables 5 and 6, Supplementary Material online, with all annotated genes for emu S0 and S1 regions shown in supplementary tables 7 and 8, Supplementary Material online.
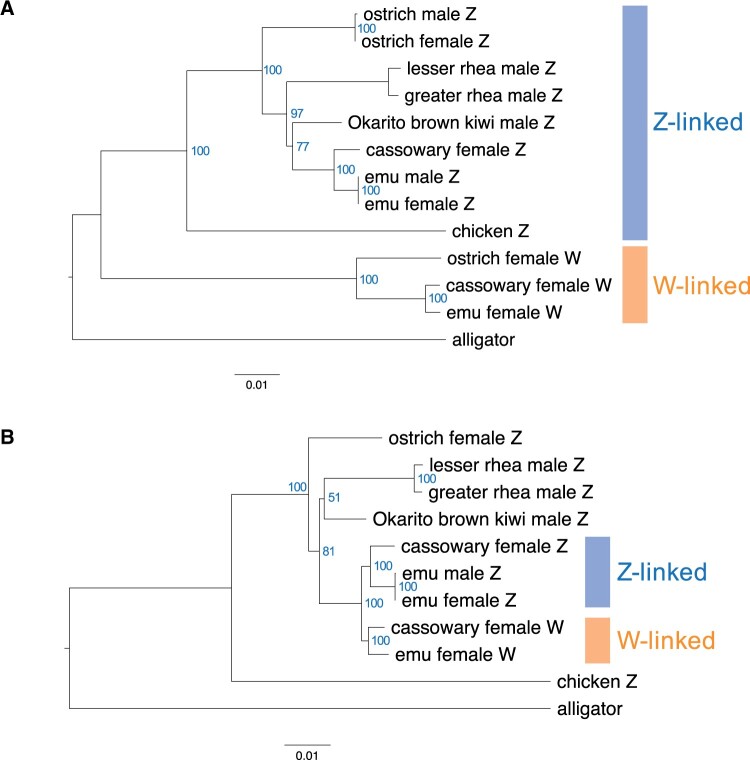
Phylogenetic trees constructed using gametologs in the S0 (A) and S1 (B) regions. We used the chicken (Gallus gallus) as a representative species of Neognathae and the American alligator (Alligator mississippiensis) as an outgroup for the phylogenetic analysis. Bootstrap values of 100 trials are indicated in blue. In Palaeognathae species, we used Z-linked genes derived from male individuals (emu, rheas, and kiwi) and both Z-linked and W-linked genes from female individuals (emu, cassowary, and ostrich).
As reported previously (Zhou et al. 2014; Xu, Wa Sin, et al. 2019; Xu and Zhou 2020), in the S0 phylogenetic tree that corresponds to the most evolutionarily ancient stratum, the Aves, including species in both Neognathae and Palaeognathae, were divided into two clades that corresponds to Z-linked and W-linked sequences. Specifically, the Z-linked clade contained Z-derived operational taxonomic units (OTUs; i.e., concatenated Z-linked or W-linked gene sequences of each species) obtained from both male and female Palaeognathae species and chicken, whereas the W-linked clade contained only female W-derived OTUs. The tree topologies in each clade were in agreement with previously published data that suggested the ostrich was placed in the basal lineage, with the rhea and then the kiwi in the next basal lineages (fig. 4A) (Yonezawa et al. 2017; Xu, Wa Sin, et al. 2019). We found that the S1 phylogenetic tree exhibited a different topology from that of the S0 tree (fig. 4B). First, the Aves were divided from an outgroup into two clades: one clade contained the chicken and was classified as Neognathae, whereas the other contained birds classified as Palaeognathae. Moreover, two clades were formed that comprised the Casuariiformes. The first was a Z-linked clade that contained male emu (ZZ), female emu Z-linked, and female cassowary Z-linked sequences. The other was a W-linked clade that contained female emu W-linked and female cassowary W-linked sequences. This topology result is supported by the findings of the analysis of the genome sequences. The sequence identity between the emu and cassowary Z-linked (or the emu and cassowary W-linked) genome regions was higher than that between the Z-linked and W-linked genome regions within the same species (supplementary figs. 2 and 6B, Supplementary Material online). Moreover, the phylogenetic tree constructed using core genome alignments in the S1 regions (supplementary fig. 6A, Supplementary Material online) clearly showed that female ostrich Z-linked and W-linked regions formed a clade independent of the Casuariiformes.
Discussion
By applying the Platanus-allee assembler to Illumina sequence reads, we successfully assembled the S1 region of the female emu genome in a two phased Z-linked and W-linked manner. In addition to the emu assembly, by published Illumina reads of female cassowary and ostrich were reassembled using the current method. We also successfully constructed almost all S1 regions in a phased form. By comparing the sequences obtained from these three species, we confirmed the previously proposed hypothesis for S1 region formation at a more precise sequence level.
Comparison of both boundaries of the constructed S1 region confirmed that the S0 region likely formed in the common avian ancestor, whereas the S1 regions arose independently after speciation into Struthioniformes (ostrich lineage) and Casuariiformes (Zhou et al. 2014). Furthermore, the loci of the PAR and S1 region boundaries were common between the emu and cassowary but differed in ostrich (Xu et al. 2019; Xu and Zhou 2020) (figs. 1 and 3A). Using both genes and genomes of the S1 region, we observe a greater differentiation of gametologs in ostrich than in the emu and cassowary. Moreover, the phylogenetic topology showed that the emu Z-linked region was closer to the cassowary Z-linked region than the W-linked region (fig. 4B and supplementary fig. 6, Supplementary Material online). These multiple lines of evidence show that the formation of the S1 region occurred independently in the ostrich lineage and common ancestor of the emu and cassowary, which occurred earlier than that in the ostrich (fig. 5).
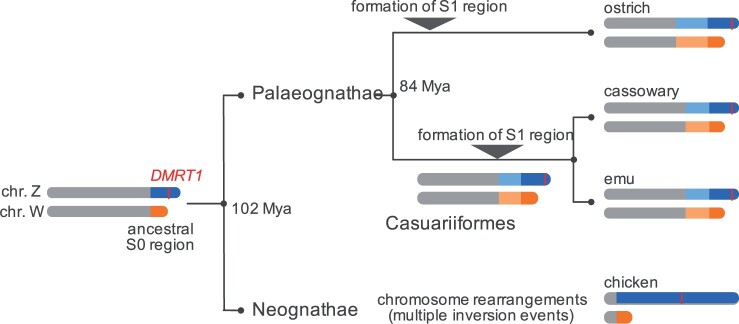
Hypothetical evolutionary trajectory of sex chromosomes in the clade Palaeognathae. Sex chromosome regions are marked as follows: the pseudoautosomal region (PAR), Z-linked S1 region, and W-linked S1 region are labeled in gray, light blue, and light orange, respectively. The Z-linked S0 and W-linked S0 regions are labeled in blue and orange, respectively. The red bar on the Z chromosome represents the locus of DMRT1. Estimated divergence times in the figure are based on previous studies (Handley et al. 2004; Yonezawa et al. 2017).
With our approach, almost the entire length of the S1 region was constructed in a phased manner, facilitating various downstream analyses. For example, more gametologous differentiation in the S1 region was noted from the S0 region side than the PAR side (fig. 3C) (Liu et al. 2021), and interspersed repetitive sequence landscapes in the Z-linked and W-linked S1 regions in female emu suggested a recent expansion of the repetitive elements in the W-linked S1 region (fig. 3D). These phenomena may be caused by the interruption of homologous recombination in the S1 region due to genome inversion, as suggested in a previous study (Liu et al. 2021). The distribution of the repetitive sequences along emu and cassowary genome sequences were examined (supplementary fig. 5, Supplementary Material online); however, this was in conjunction with the age of the repetitive sequences, which showed no significant trend. For a better understanding of the evolutionary processes that occurred in the lineages after speciation, multiple alignments of the four Z-linked and W-linked genome sequences of emu and cassowary should be performed to examine not only insertions of repetitive sequences but also deletions, which would help determine what events occurred in these particular lineages.
In summary, our method provides phased assemblies for the S1 region in the female emu, cassowary, and ostrich genomes using Illumina reads. It is true that phased assemblies have already been obtained for the S1 region of emu in a previous study (Liu et al. 2021); however, the previous assembly was constructed from costly data produced by various platforms, such as PacBio, 10× Genomics-linked reads, Dovetail Chicago data, and Hi-C data, making this method not easily extendable to other species. Similarly, circulating consensus sequencing (CCS) using PacBio sequencers has made it possible to generate highly accurate 10–20 kb high-fidelity (HiFi) reads (Wenger et al. 2019) and combining this technology with Hi-C data has contributed to several published chromosome-scale phased assembly studies (Garg et al. 2021), including the construction of the zig-zag eel Y chromosome (Xue et al. 2021), which is expected to be applied to sex chromosome determination in other species. However, this combined method is less cost-effective than the method described in this study. Indeed, the results presented in this study were constructed only from Illumina reads, which provides us a significant advantage in terms of cost. In addition, various analyses using the almost full-length phased assembly sequences confirmed the evolutionary history of the S1 region formation that was inferred from mapping or partial sequence-based analyses. Notably. we demonstrated that the W-linked and Z-linked emu and cassowary genomes formed a single clade in the phylogenetic analysis, which indicates the usefulness of our method (fig. 4). A recent study performed the same type of phylogenetic analysis using only a single gene (ABHD17B) located in the S1 region, which indicated a single clade formed by the Z-linked and W-linked cassowary genes separated from the Z-linked emu gene, albeit with low bootstrap values and without the W-linked emu gene (Xu and Zhou 2020). This result is not consistent with the hypothesis that the S1 region formed in the common ancestor of emu and cassowary, and this discrepancy may reflect the difficulty previous studies had in extracting sufficient S1 region sequences.
kb high-fidelity (HiFi) reads (Wenger et al. 2019) and combining this technology with Hi-C data has contributed to several published chromosome-scale phased assembly studies (Garg et al. 2021), including the construction of the zig-zag eel Y chromosome (Xue et al. 2021), which is expected to be applied to sex chromosome determination in other species. However, this combined method is less cost-effective than the method described in this study. Indeed, the results presented in this study were constructed only from Illumina reads, which provides us a significant advantage in terms of cost. In addition, various analyses using the almost full-length phased assembly sequences confirmed the evolutionary history of the S1 region formation that was inferred from mapping or partial sequence-based analyses. Notably. we demonstrated that the W-linked and Z-linked emu and cassowary genomes formed a single clade in the phylogenetic analysis, which indicates the usefulness of our method (fig. 4). A recent study performed the same type of phylogenetic analysis using only a single gene (ABHD17B) located in the S1 region, which indicated a single clade formed by the Z-linked and W-linked cassowary genes separated from the Z-linked emu gene, albeit with low bootstrap values and without the W-linked emu gene (Xu and Zhou 2020). This result is not consistent with the hypothesis that the S1 region formed in the common ancestor of emu and cassowary, and this discrepancy may reflect the difficulty previous studies had in extracting sufficient S1 region sequences.
In a previous study (Vicoso et al. 2013), gametologous sequences were extracted from Illumina-based results assembled according to homology with the corresponding S1 region of the male genome. Figure 2B and supplementary figure S3, Supplementary Material online show that only a few regions could be separated into Z-linked and W-linked phased sequences, with most regions constructed as only one sequence that consisted of both Z-linked and W-linked sequences as described previously. The phased regions did not show a high homology with our Z-linked or W-linked sequences, suggesting that these regions consist of a mosaic of both sequences. This may be a cause of the inconsistent topological phylogenetic trees produced in the previous study. By contrast, our female emu assembly had high homology with the previously reported PacBio-based Z-linked and W-linked sequences (fig. 2A). Thus, it is highly likely that the previously reported Illumina-based W-linked and Z-linked sequences were not adequate and that the analyses performed based on these sequences are considered insufficient to accurately describe the sequence homology among these avian species.
De novo assembly programs attempt to merge haplotype sequences and construct a single mosaic sequence. However, if the heterozygosity of a genome exceeds a certain level, only those regions will be provided as separate sequences output, which results in a mixture of merged and separate output regions. The results of the previous study can be attributed to this effect of high heterozygosity when using conventional assemblers. Other problems can be observed in downstream analysis, such as mosaic-like sequences in both alleles that cause false frameshifts. This makes accurate gene prediction difficult, and genes predicted from genome regions constructed separately from both alleles can appear as if they were paralogous genes. The construction of mosaic sequences and incomplete resolution of separate sequences in each haplotype is not a problem unique to Illumina data-based assembly and can occur in long-read data-based assembly. For example, in the long-read data-based emu assembly, the sequences of both alleles were well constructed; however, incomplete separation is likely to occur in cases where gametologous sequences are similar. This is because long reads generally have a high error rate, making it difficult to distinguish between haplotype differences and sequence errors. Therefore, it is expected that using the Platanus-allee assembler for phased assembly of Illumina data is a suitable option, even in species that exhibit high similarity between gametologous sequences. An advantage of Platanus-allee in this regard is that the error rate of Illumina data is low, and Platanus-allee is accurate even when heterozygosity is as low as 1%. The effect of Illumina-based highly accurate Platanus-allee assembly may be reflected by the low number of genes that were annotated as pseudogenes in both the S0 and S1 regions compared with the PacBio-based assembly (Liu et al. 2021) (supplementary tables 7 and 8, Supplementary Material online). The definition of pseudogene is vague and direct comparison is difficult due to the differences in the individuals used; for example, in our assembled genome, the intact annotated genes such as ALDH1A1 and CARNMT1 were identified as pseudogenes in the previous study, even though their expression was confirmed in the same study (Liu et al. 2021).
We believe that adopting a phased assembly approach for other broad taxa with young sex chromosomes, such as those of fish and amphibians. may reveal more hidden events in the history of sex chromosomes. This method could ultimately help elucidate the process of sex chromosome evolution from a homomorphic to differentiated state.
Materials and Methods
Emu Sample Collection and DNA Extraction
Fertilized emu eggs were purchased from Kakegawa Kachouen (Kakegawa Flower and Bird Park, Kakegawa, Shizuoka Prefecture, Japan). Eggs were incubated at 37 °C at 25–30% relative humidity with rotation every hour for 35 days to obtain embryos. Genomic DNA was extracted from the whole brain of each embryo using a Gentra Puregene Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Sexing was performed by polymerase chain reaction (PCR) amplification of the W-specific marker kW1 as described previously (Ishijima et al. 2014).
days to obtain embryos. Genomic DNA was extracted from the whole brain of each embryo using a Gentra Puregene Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Sexing was performed by polymerase chain reaction (PCR) amplification of the W-specific marker kW1 as described previously (Ishijima et al. 2014).
Emu Genome Sequencing
To obtain whole-genome sequencing (WGS) reads from two emu individuals (a male and a female), we constructed paired-end (PE) libraries and four mate-pair (MP) libraries for the Illumina platform (Hayward, CA, USA). PE libraries with an insert size of 600 bp were prepared using a TrueSeq PCR-free DNA Sample Prep kit (Illumina). For MP libraries, we extracted approximately 3, 6, 10, and 15 kb DNA fragments from an agarose gel for each sample and prepared the libraries using a Nextera Mate Pair Library Prep kit (Illumina). We then confirmed the samples by sequencing a small part of the PE libraries with a read length of 250
bp were prepared using a TrueSeq PCR-free DNA Sample Prep kit (Illumina). For MP libraries, we extracted approximately 3, 6, 10, and 15 kb DNA fragments from an agarose gel for each sample and prepared the libraries using a Nextera Mate Pair Library Prep kit (Illumina). We then confirmed the samples by sequencing a small part of the PE libraries with a read length of 250 bp using an Illumina MiSeq platform. We then sequenced all the libraries with a read length of 150
bp using an Illumina MiSeq platform. We then sequenced all the libraries with a read length of 150 bp using an Illumina HiSeq X Ten platform. The sequence data of each sample are presented in supplementary table 1, Supplementary Material online. The basic statistical parameters of the emu genomes (e.g., estimated genome size and heterozygosity) from these sequence data were calculated using the method described in the supplementary methods, and the results are shown in supplementary table 4 and figure S7, Supplementary Material online.
bp using an Illumina HiSeq X Ten platform. The sequence data of each sample are presented in supplementary table 1, Supplementary Material online. The basic statistical parameters of the emu genomes (e.g., estimated genome size and heterozygosity) from these sequence data were calculated using the method described in the supplementary methods, and the results are shown in supplementary table 4 and figure S7, Supplementary Material online.
Preparation of Raw Next-Generation Sequencing Data of Ostrich and Cassowary
Female ostrich and cassowary WGS data were downloaded from the NCBI SRA database. For the ostrich sequence data (NCBI BioProject number PRJNA212875), we used three Illumina PE libraries with insert sizes of 170, 500, and 800 bp and four MP libraries with insert sizes of 2, 5, 10, and 20 kb. For the cassowary sequence data (NCBI BioProject number PRJNA433110), a PE library with an insert size of 220
bp and four MP libraries with insert sizes of 2, 5, 10, and 20 kb. For the cassowary sequence data (NCBI BioProject number PRJNA433110), a PE library with an insert size of 220 bp and an MP library with an insert size of 3 kb were used. The sequence data for each sample are presented in supplementary table 3, Supplementary Material online. The basic genomic features were estimated with the method described in the supplementary methods, and the results are shown in supplementary table 4 and figure 7, Supplementary Material online.
bp and an MP library with an insert size of 3 kb were used. The sequence data for each sample are presented in supplementary table 3, Supplementary Material online. The basic genomic features were estimated with the method described in the supplementary methods, and the results are shown in supplementary table 4 and figure 7, Supplementary Material online.
De Novo Assembly of the Male Emu Genome
The sequence reads of a male emu individual (ZZ) were assembled using Platanus v1.2.4 software (Kajitani et al. 2014). We used an assembler different from that used for the female individuals (ZW), prioritizing long connections because the goal was to not conduct phased assembly. Contig assembly was performed using only the PE library, following which scaffolding and gap closing were performed using all libraries. Additionally, we confirmed that scaffolds ≥500 bp from the contamination were not included by performing alignments with the NCBI nr and nt databases using Diamond v0.9.22 (Buchfink et al. 2015) and Blastn 2.2.27+ (Altschul et al. 1997), respectively. For the subsequent analyses, we used a male emu draft genome with a complete mitochondrial genome. The assembly of the mitochondrial genome is described in the supplementary methods, Supplementary Material online
bp from the contamination were not included by performing alignments with the NCBI nr and nt databases using Diamond v0.9.22 (Buchfink et al. 2015) and Blastn 2.2.27+ (Altschul et al. 1997), respectively. For the subsequent analyses, we used a male emu draft genome with a complete mitochondrial genome. The assembly of the mitochondrial genome is described in the supplementary methods, Supplementary Material online
Z Chromosome Genome Detection
For ostrich, we first downloaded the whole-genome assembly (GCF_000698965.1) and then picked and ordered the scaffolds corresponding to the ostrich pseudo Z chromosome according to previously published reports (Zhang et al. 2015; O’Connor et al. 2018). For emu and cassowary, we initially detected Z chromosome scaffolds based on the similarity with the chicken Z chromosome (Gallus_gallus-5.0, NC_006127.4) independent of the ostrich Z chromosome and then ordered the scaffolds along the ostrich Z chromosome. In emu, the male (ZZ) draft genome was assembled with Platanus v1.2.4 and aligned to the chicken Z chromosome using LASTZ v.1.02.00 (Harris 2007). Then, scaffolds with ≥100 kb and more than 50% of the aligned length were detected as the emu Z chromosome. Finally, scaffolds were ordered and oriented according to the alignment with the ostrich Z chromosome. For the cassowary, we used the draft genome GCA_003342895.1 and subsequent methods described above for the emu genome construction.
PAR Estimation from Read Mapping Data
To estimate the PAR of each species, we calculated read depths normalized to one per haploid set for every 10-kb segment on the Z chromosome using female PE reads. Given that female individuals have Z and W chromosomes, the PAR is expected to be present in two copies (diploid), whereas the W degenerated region is expected to be present in one copy (haploid). The PAR was estimated by detecting changes in the copies along the Z chromosome. PE reads were mapped on each draft genome using BWA aligner v.0.7.12 (Li and Durbin 2009), and the sequence read depth was calculated using SAMtools v.1.8 (Li et al. 2009) based on the reads mapped with ≥98% identity and ≥80% alignment cover rates. Average read depth per haploid set was obtained from the distribution of whole-genome read depth. Lastly, normalized read depth for every 10 kb of scaffold was calculated by dividing the read depth of the 10 kb segment by the average depth per haploid set.
Phased De Novo Assembly of Female Individuals
To construct the phased S1 region, which was expected to have highly divergent W/Z gametologous sequences, we used Platanus-allee v2.2.2 as a de novo haplotype assembler (Kajitani et al. 2019) for female emu, cassowary, and ostrich individuals. The “assemble” command was performed using only the PE library, whereas the “phase” and “consensus” commands were performed using all libraries. For the subsequent comparative analyses, we used “allPhasedScaffold” FASTA files that contained phased (“bubble scaffolds”) and unphased (“nonbubble scaffolds”) sequences. In Platanus-allee, the longer sequence of a pair of phased bubble sequences was defined as the “primary bubble,” whereas the remaining sequence was defined as the “secondary bubble.” A schematic explanation of the Platanus-allee result is shown in supplementary figure 8, Supplementary Material online. Repetitive sequence analysis is described in the supplementary methods, Supplementary Material online.
Construction and Comparison of Phased S1 Regions
We obtained whole genome phased scaffolds that were expected to be haplotype sequences from female WGS data using Platanus-allee. Then, we detected bubble scaffolds derived from the S1 region and assigned each of the phased scaffolds to the original Z or W chromosome. In the pairs of bubbles derived from the S1 region, with each bubble derived from either the Z or W chromosome. We assigned each of the bubble sequences to either the Z or W chromosome according to sequence identity to the male-derived genome or transcriptome such that each of the bubble sequences with high identity to the male-derived sequence was attributed to the Z chromosome. Specifically, for emu and cassowary, we aligned bubble sequences to the male emu draft genome using LASTZ. For the ostrich female individual, we used male-derived transcript contigs for the assignment of the origin.
To obtain transcripts from a male ostrich individual, we assembled RNA-seq reads (SRR1619445) from the adult cerebellum using Trinity v2.2.2 (Haas et al. 2013). Additionally, we aligned the transcript contigs ≥500 bp to bubble sequences using GMAP 2018-07-04 (Wu and Watanabe 2005). Then, average alignment identities for each phased S1 scaffold were calculated using transcript contigs with ≥80% alignment cover rate to both primary and secondary bubble scaffolds (supplementary table 9, Supplementary Material online). One bubble sequence of a pair was aligned with more than 99% sequence identity to male transcript contigs, whereas the other bubble sequence was aligned with 86.5–92.0% identity. According to the respective sequence identities of the nine pairs of phased scaffolds, the bubble with the higher identity to the male transcript contig was defined as Z-linked, whereas the other was defined as W-linked.
bp to bubble sequences using GMAP 2018-07-04 (Wu and Watanabe 2005). Then, average alignment identities for each phased S1 scaffold were calculated using transcript contigs with ≥80% alignment cover rate to both primary and secondary bubble scaffolds (supplementary table 9, Supplementary Material online). One bubble sequence of a pair was aligned with more than 99% sequence identity to male transcript contigs, whereas the other bubble sequence was aligned with 86.5–92.0% identity. According to the respective sequence identities of the nine pairs of phased scaffolds, the bubble with the higher identity to the male transcript contig was defined as Z-linked, whereas the other was defined as W-linked.
We calculated the sequence identity between phased scaffolds in the S1 region to define the sequence identity between gametologous sequences. First, secondary bubble scaffolds were divided into 1-kb segments and the segments were aligned to primary bubble scaffolds using LASTZ (–step = 5, –notransition, –nochain). Next we obtained each of the sequence identities of the alignment following synteny detection within the gametolous sequences and measured distributions of the sequence identity for each species.
Phylogenetic Analysis of W/Z Gametologs in the S1 and S0 Regions
We conducted phylogenetic analysis of the S0 and S1 regions based on W/Z gametologs. We used the following species: emu (male and female), cassowary (female), ostrich (male and female), lesser rhea (male), greater rhea (male), Okarito brown kiwi (male), chicken (female), and American alligator as an outgroup. We used the draft genomes assembled in this study for the male and female emus, female cassowary, and female ostrich. Transcript contigs assembled with RNA-seq reads were used for male ostrich. For other species, the accession IDs of the draft or complete genomes used are listed in supplementary table 10, Supplementary Material online.
For the S0 region, we used eight Z- and W-linked gametologs (Zhou et al. 2014). To predict these genes in Palaeognathae species, the chicken genes were aligned to S0 scaffolds using Exonerate v.2.2.0 (–model protein2genome) (Slater and Birney 2005). The alignments were manually corrected and predicted genes, including the in-frame stop codon, were filtered out from the phylogenetic analysis. Multiple alignments with each of the genes were performed using MAFFT 7.429 (Katoh et al. 2002) converted to amino acid sequences. After removing gapped loci from the alignments, we conducted phylogenetic estimation based on concatenated sequences using the maximum-likelihood method (Kozlov et al. 2019) in RAxML-ng v.0.9.0 (–all, –bs-trees 100, –model HIVB+I+G4+F). The model was selected as the best-fit model under the Akaike’s Information Criterion using ModelTest-NG v.0.1.6 (Darriba et al. 2020). Phylogenetic analysis of the S1 region using the core genome is described in the supplementary methods, Supplementary Material online. Finally, the phylogenetic trees were plotted using FigTree.
Supplementary Material
evab242_Supplementary_Data
Acknowledgments
This work was supported by JSPS KAKENHI (15H04401, 16H06279 [PAGS], 19H03267, and 19H03206). We thank the staff of the Itoh Laboratory at Tokyo Tech for supporting genome sequencing and their valuable participation in scientific discussions.
Data Availability
The raw Illumina sequence reads have been deposited in the NCBI Sequence Read Archive (DRR233936-DRR233947). The assembled genomes are available in DDBJ/ENA/GenBank under accession numbers BLZG01000000 (a male emu), BMAD01000000 (primary bubble-based pseudohaplotype of a female emu), BMAE01000000 (secondary bubble-based pseudohaplotype of a female emu), EAAC01000000 and EAAD01000000 (a female cassowary), and EAAE01000000 and EAAF01000000 (a female ostrich). The scripts we used in this article, including estimate of copy number and a visualization tool (used in figs. 1 and and3),3), are available at GitHub (https://github.com/mokuno3430/emu).
Literature Cited
- Altschul SF, et al.1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [Europe PMC free article] [Abstract] [Google Scholar]
- Barrowclough GF, Cracraft J, Klicka J, Zink RM.. 2016. How many kinds of birds are there and why does it matter? PLoS One. 11(11):e0166307. [Europe PMC free article] [Abstract] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [Abstract] [Google Scholar]
- Darriba D, et al.2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294. [Europe PMC free article] [Abstract] [Google Scholar]
- Ellegren H. 2000. Evolution of the avian sex chromosomes and their role in sex determination. Trends Ecol Evol. 15(5):188–192. [Abstract] [Google Scholar]
- Ezaz T, , StiglecR, , VeyrunesF, , Marshall Graves JA.. 2006. Relationships between vertebrate ZW and XY sex chromosome systems. Curr Biol. 16(17):R736–R743. [Abstract] [Google Scholar]
- Feng S, et al.2020. Dense sampling of bird diversity increases power of comparative genomics. Nature 587(7833):252–257. [Europe PMC free article] [Abstract] [Google Scholar]
- Fridolfsson A-K, et al.1998. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci U S A. 95(14):8147–8152. [Europe PMC free article] [Abstract] [Google Scholar]
- Furman BLS, , Evans BJ.. 2018. Divergent evolutionary trajectories of two young, homomorphic, and closely related sex chromosome systems. Genome Biol Evol. 10(3):742–755. [Europe PMC free article] [Abstract] [Google Scholar]
- Garg S, et al.2021. Chromosome-scale, haplotype-resolved assembly of human genomes. Nat Biotechnol. 39(3):309–312. [Europe PMC free article] [Abstract] [Google Scholar]
- Haas BJ, et al.2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [Europe PMC free article] [Abstract] [Google Scholar]
- Handley LL, Ceplitis H, Ellegren H.. 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167(1):367–376. [Europe PMC free article] [Abstract] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignment of genomic DNA [PhD thesis]. State College (PA): Pennsylvania State University.
- Ishijima J, Uno Y, Nishida C, Matsuda Y.. 2014. Genomic structures of the kW1 loci on the Z and W Chromosomes in ratite birds: structural changes at an early stage of W chromosome differentiation. Cytogenet Genome Res. 142(4):255–267. [Abstract] [Google Scholar]
- Kajitani R, et al.2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [Europe PMC free article] [Abstract] [Google Scholar]
- Kajitani R, et al.2019. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat Commun. 10(1):1–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [Europe PMC free article] [Abstract] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A.. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35(21):4453–4455. [Europe PMC free article] [Abstract] [Google Scholar]
- Lambert MR, , SkellyDK, , Ezaz T.. 2016. Sex-linked markers in the North American green frog (Rana clamitans) developed using DArTseq provide early insight into sex chromosome evolution. BMC Genomics. 17(1):844. [Europe PMC free article] [Abstract] [Google Scholar]
- Li H, et al.2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [Europe PMC free article] [Abstract] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu J, et al.2021. A new emu genome illuminates the evolution of genome configuration and nuclear architecture of avian chromosomes. Genome Res. 31(3):497–511. [Europe PMC free article] [Abstract] [Google Scholar]
- Li R, et al.2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20(2):265–272. [Europe PMC free article] [Abstract] [Google Scholar]
- O’Connor RE, et al.2018. Chromosome-level assembly reveals extensive rearrangement in saker falcon and budgerigar, but not ostrich, genomes. Genome Biol. 19(1):171. [Europe PMC free article] [Abstract] [Google Scholar]
- Ogawa A, Murata K, Mizuno S.. 1998. The location of Z- and W-linked marker genes and sequence on the homomorphic sex chromosomes of the ostrich and the emu. Proc Natl Acad Sci U S A. 95(8):4415–4418. [Europe PMC free article] [Abstract] [Google Scholar]
- Sackton TB, et al.2019. Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364(6435):74–78. [Abstract] [Google Scholar]
- Shetty S, Griffin DK, Graves JAM.. 1999. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 7(4):289–295. [Abstract] [Google Scholar]
- Sibley CG, Ahlquist JE.. 1990. Phylogeny and classification of birds: a study in molecular evolution. New Haven (CT): Yale University Press. [Google Scholar]
- Slater GSC, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6(1):31. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith CA, et al.2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461(7261):267–271. [Abstract] [Google Scholar]
- Takehana Y, et al. . 2020. Genome sequence of the euryhaline Javafish Medaka, Oryzias javanicus: A small aquarium fish model for studies on adaptation to salinity. G3 (Bethesda) 10(3):907–915. [Europe PMC free article] [Abstract] [Google Scholar]
- Tone M, Nakano N, Takao E, Narisawa S, Mizuno S.. 1982. Demonstration of W chromosome-specific repetitive DNA sequences in the domestic fowl, Gallus g. domesticus. Chromosoma 86(4):551–569. [Abstract] [Google Scholar]
- Tsuda Y, Nishida-Umehara C, Ishijima J, Yamada K, Matsuda Y.. 2007. Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma 116(2):159–173. [Abstract] [Google Scholar]
- van Tuinen M, Hedges SB.. 2001. Calibration of avian molecular clocks. Mol Biol Evol. 18(2):206–213. [Abstract] [Google Scholar]
- van Tuinen M, Sibley CG, Hedges SB.. 1998. Phylogeny and biogeography of ratite birds inferred from DNA sequences of the mitochondrial ribosomal genes. Mol Biol Evol. 15(4):370–376. [Abstract] [Google Scholar]
- Vicoso B, Kaiser VB, Bachtrog D.. 2013. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc Natl Acad Sci U S A. 110(16):6453–6458. [Europe PMC free article] [Abstract] [Google Scholar]
- Wenger AM, et al.2019. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 37(10):1155–1162. [Europe PMC free article] [Abstract] [Google Scholar]
- Witschi E. 1959. Age of sex-determining mechanisms in vertebrates: distribution of differentiation patterns indicates the evolutionary path of genes and chromosomes. Science 130(3372):372–375. [Abstract] [Google Scholar]
- Wu TD, Watanabe CK.. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21(9):1859–1875. [Abstract] [Google Scholar]
- Xu L, Auer G, et al.2019. Dynamic evolutionary history and gene content of sex chromosomes across diverse songbirds. Nat Ecol Evol. 3(5):834–844. [Abstract] [Google Scholar]
- Xu L, Wa Sin SY, Grayson P, Edwards SV, Sackton TB.. 2019. Evolutionary dynamics of sex chromosomes of paleognathous birds. Genome Biol Evol. 11(8):2376–2390. [Europe PMC free article] [Abstract] [Google Scholar]
- Xu L, Zhou Q.. 2020. The female-specific W chromosomes of birds have conserved gene contents but are not feminized. Genes 11(10):1126. [Europe PMC free article] [Abstract] [Google Scholar]
- Xue L, et al.2021. Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 22(1):203. [Europe PMC free article] [Abstract] [Google Scholar]
- Yazdi HP, Ellegren H.. 2018. A genetic map of ostrich Z chromosome and the role of inversions in avian sex chromosome evolution. Genome Biol Evol. 10(8):2049–2060. [Europe PMC free article] [Abstract] [Google Scholar]
- Yonezawa T, et al.2017. Phylogenomics and morphology of extinct paleognaths reveal the origin and evolution of the ratites. Curr Biol. 27(1):68–77. [Abstract] [Google Scholar]
- Zhang J, Li C, Zhou Q, Zhang G.. 2015. Improving the ostrich genome assembly using optical mapping data. Gigascience 4(1):4–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou Q, et al.2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346(6215):1246338. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genome Biology and Evolution are provided here courtesy of Oxford University Press
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/115911306
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/gbe/evab242
Article citations
Sex Chromosome Evolution: Hallmarks and Question Marks.
Mol Biol Evol, 41(11):msae218, 01 Nov 2024
Cited by: 0 articles | PMID: 39417444 | PMCID: PMC11542634
Review Free full text in Europe PMC
Sex chromosome cycle as a mechanism of stable sex determination.
J Biochem, 176(2):81-95, 01 Jul 2024
Cited by: 1 article | PMID: 38982631 | PMCID: PMC11289310
Review Free full text in Europe PMC
Avian Introgression Patterns are Consistent With Haldane's Rule.
J Hered, 113(4):363-370, 01 Jul 2022
Cited by: 3 articles | PMID: 35134952 | PMCID: PMC9308041
Review Free full text in Europe PMC
The Length Polymorphism of the 9th Intron in the Avian CHD1 Gene Allows Sex Determination in Some Species of Palaeognathae.
Genes (Basel), 13(3):507, 12 Mar 2022
Cited by: 3 articles | PMID: 35328061 | PMCID: PMC8954394
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject (2)
- (1 citation) BioProject - PRJNA212875
- (1 citation) BioProject - PRJNA433110
GCA - NCBI genaome assembly (2)
- (1 citation) GCA - GCA_003342905.1
- (1 citation) GCA - GCA_003342895.1
Nucleotide Sequences (Showing 6 of 6)
- (1 citation) ENA - BLZG01000000
- (1 citation) ENA - BMAD01000000
- (1 citation) ENA - SRR1619445
- (1 citation) ENA - DRR233936
- (1 citation) ENA - DRR233947
- (1 citation) ENA - BMAE01000000
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Phylogeny and sex chromosome evolution of Palaeognathae.
J Genet Genomics, 49(2):109-119, 13 Jul 2021
Cited by: 10 articles | PMID: 34872841
Evolution of bird sex chromosomes: a cytogenomic approach in Palaeognathae species.
BMC Ecol Evol, 24(1):51, 23 Apr 2024
Cited by: 2 articles | PMID: 38654159 | PMCID: PMC11036779
Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds.
Chromosoma, 116(2):159-173, 12 Jan 2007
Cited by: 53 articles | PMID: 17219176
About PAR: the distinct evolutionary dynamics of the pseudoautosomal region.
Trends Genet, 27(9):358-367, 01 Sep 2011
Cited by: 115 articles | PMID: 21962971
Review
Funding
Funders who supported this work.
JSPS KAKENHI (4)
Grant ID: 19H03206
Grant ID: 15H04401
Grant ID: 19H03267
Grant ID: 16H06279



