Abstract
Free full text

Sex Chromosome Evolution: Hallmarks and Question Marks
Abstract
Sex chromosomes are widespread in species with separate sexes. They have evolved many times independently and display a truly remarkable diversity. New sequencing technologies and methodological developments have allowed the field of molecular evolution to explore this diversity in a large number of model and nonmodel organisms, broadening our vision on the mechanisms involved in their evolution. Diverse studies have allowed us to better capture the common evolutionary routes that shape sex chromosomes; however, we still mostly fail to explain why sex chromosomes are so diverse. We review over half a century of theoretical and empirical work on sex chromosome evolution and highlight pending questions on their origins, turnovers, rearrangements, degeneration, dosage compensation, gene content, and rates of evolution. We also report recent theoretical progress on our understanding of the ultimate reasons for sex chromosomes’ existence.
Introduction
In many species with separate sexes, whether an organism becomes male or female is controlled by sex chromosomes. These peculiar chromosomes stand out in many ways, first due to their role in sex determination and also because they often undergo extensive changes following their evolution from a normal autosomal pair. Sex chromosomes determine the sex of individuals in various ways (Fig. 1). Most often, sex is expressed at the diploid phase with males and females carrying different combinations of sex chromosomes. In some cases, sex is determined by the presence/absence of a dominant gene, such as the masculinizing SRY gene carried by the Y chromosome of placental mammals (Koopman et al. 1991). Sex can also be determined in a dosage-dependent manner, as, for example, in Drosophila, where individuals with a single X chromosome are males and individuals with two or more Xs are females (Bridges 1925). Different types of sex chromosomes exist; females can be the heterogametic sex (ZW) and males the homogametic sex (ZZ) as in birds (Bachtrog et al. 2014). However, in some clades, the W chromosome is absent, leading to a Z0 system, with ZZ males and Z females that are otherwise diploid for autosomes, as in Psychidae moths (Hejníčková et al. 2019). Most therian mammals and many other species harbor a male heterogametic system with XY males and XX females. In the grape phylloxera (insects related to aphids), an X0 sex-determining system is observed with XX females and X males that are otherwise diploid for autosomes (Li et al. 2023). In some species, multiple X and Y chromosomes are found and form chains during meiosis, as seen, for example, in the platypus, where males have five Xs and five Ys (Veyrunes et al. 2008). Finally, in species with substantial development of the haploid phase, sex can be determined at that phase instead of the diploid phase: females carry a sex chromosome called U and males carry a sex chromosome called V (Bachtrog et al. 2014). On top of this variety in sex chromosome types (XY, X0, ZW, Z0, UV, and chains, Fig. 1), sex chromosomes are also very diverse in size. Some sex chromosome pairs can be distinguished cytologically and are called heteromorphic. Y and W chromosomes are often smaller than their counterparts, but it is not always the case. For instance, the plant Silene latifolia giant Y chromosome is larger than the X and any autosome in that species (Matsunaga et al. 1994). In many cases, however, the sex chromosome pair is homomorphic and distinguishing the X and Y (or Z and W or U and V) often requires sequencing. In some clades, chromosomal rearrangements have occurred between sex chromosomes and autosomes, leading to the evolution of neo-sex chromosomes. For example, in the plant Rumex hastatulus, an autosome has fused to the X, leading to XY1Y2 males and XX females, where Y1 is homologous to the ancestral X and Y2 is derived from the autosome that fused with the X (Sacchi et al. 2024). Sex chromosomes have evolved many times independently and are therefore not all homologous, sometimes even in closely related species (Bachtrog et al. 2014).
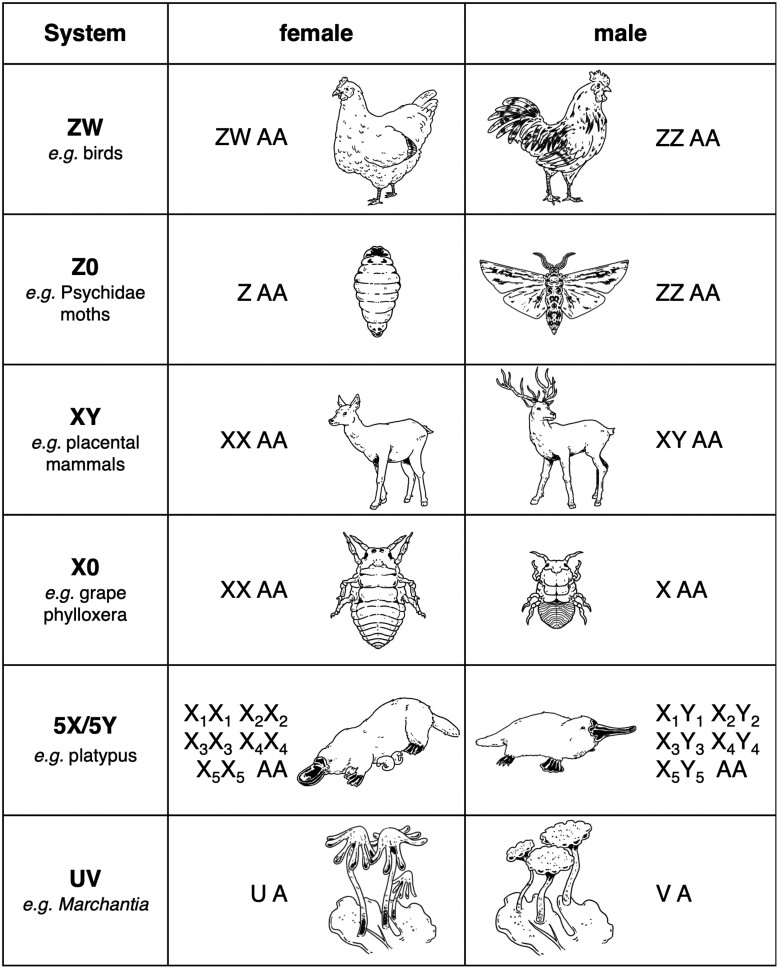
Sex chromosome types. A stands for an autosome (AA when diploid). Please refer to the introduction for more details and references. Custom illustrations by Alice Mazel.
Sex chromosomes, in spite of their diversity, share some common features and evolutionary processes (Fig. 2a). The sex-specific chromosome (the Y, W, U, or V) most often has a nonrecombining region (hereafter abbreviated NRR). This absence of recombination triggers divergence at the sequence level between the X and the Y (or Z-W or U-V), meaning that sex-linked homologous X/Y (or Z/W or U/V) gene pairs (gametologs) accumulate substitutions over time. The size of the NRR varies substantially across species, ranging from small regions containing only a few genes to large regions encompassing almost the entire sex chromosome, with all possible intermediate situations (Bachtrog et al. 2014). The recombining region(s) of sex chromosomes are called pseudoautosomal region(s) (hereafter PAR). The absence of recombination of sex-specific chromosomes causes them to degenerate. In particular, the NRR loses genes and accumulates deleterious mutations and repeats (such as transposable elements , hereafter TEs). These nonrecombining regions can therefore become extremely gene poor, repeat rich, and heterochromatinized.
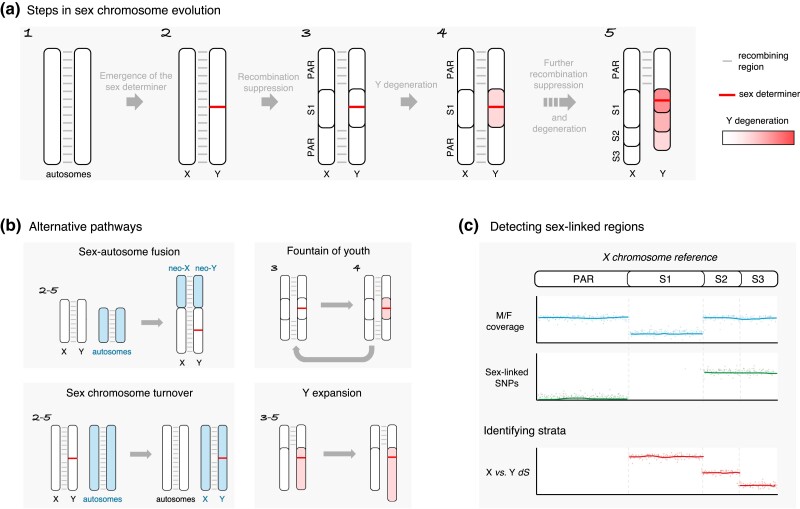
Overview of the steps in sex chromosome evolution and how to detect them using sequencing data. Sex chromosomes depicted here are XY chromosomes, all information shown is also valid for ZW chromosomes. a) The classical pathway of sex chromosome evolution, starting when a pair of autosomes acquires a sex-determining gene/locus. Recombination suppression around the sex-determining locus forms a first evolutionary stratum S1, and PARs. Because of a lack of recombination, stratum S1 starts to degenerate on the Y chromosome. Following multiple successive events of recombination suppression, old sex chromosomes typically show multiple strata (here S1–S3), with different degrees of X-Y degeneration shown in shades of red. b) Some alternative pathways of sex chromosome evolution. Numbers on the top-left of each diagram refer to the steps of (a) (1–5) at which each alternative scenario can branch off from the classical model. Sex-autosome fusion can involve the fusion of either X or Y or both sex chromosomes to autosomes, forming neo-sex chromosomes. In the fountain of youth model, nonrecombining, poorly differentiated X-Y chromosomes undergo a rare recombination event, reducing X-Y differentiation and Y degeneration. A sex chromosome turnover is the change in the genomic location (and sometimes identity) of the sex-determining gene. Y expansion usually results from a strong accumulation of repeats on the Y. c) Main methods to detect sex-linked regions and identify strata using sequencing data mapped to the X chromosome. Analyzing male-to-female depth of coverage ratio (M/F coverage) allows detecting old and highly divergent strata. Contrasting genotypes from multiple males and females allows identifying sex-linked SNPs to detect younger strata with less divergence. Measuring synonymous divergence (dS) between X and Y linked genes is the most commonly used method to identify and delimitate strata of different ages.
Here, we review the evolutionary mechanisms that give rise to sex chromosomes and summarize open questions in the field. The fantastic diversity observed in sex chromosomes, sometimes even among closely related species, is a direct demonstration of how dynamic sex chromosome evolution can be (Furman and Evans 2018; Furman et al. 2020).
Origins of Sex Chromosomes
Sex chromosomes derive from a simple pair of autosomes following the emergence of one or multiple sex-determining genes (Fig. 2a). A classical theory for the origin of sex chromosomes posits that their emergence coincides with the evolutionary transition from hermaphroditism to separate sexes (Westergaard 1958; Charlesworth and Charlesworth 1978). In this model, sex chromosomes carry two closely linked sex-determining genes: one responsible for female sterility and the other for male sterility. Crossover events in-between these two genes lead to sterile individuals, which may select for initial recombination suppression between young proto-sex chromosomes. The “two-gene model” therefore accounts for both the emergence of sex chromosomes and initial recombination suppression. This model has been validated in a number of species, including the plant S. latifolia, where deletion mutants revealed the existence of at least two Y regions involved in sex determination (Moraga et al. 2023). One of the two S. latifolia sex-determining genes was recently proposed to be CLAVATA3, a Y-encoded regulator of stem cell maintenance (Kazama et al. 2022).
This two-gene model is not universal, as sex chromosomes can originate in species with preexisting separate sexes or environmental sex determination (rather than hermaphroditism). This is the case in the majority of animals. In Daphnia magna, a small freshwater crustacean, such a transition has occurred recently. Sex determination is mostly controlled by environmental cues in this species, but some populations have evolved ZW sex chromosomes, most likely driven by the spread of a single feminizing mutation (Reisser et al. 2017). Another alternative route to the evolution of sex chromosomes could come from cases where the W or Y chromosome evolves from B chromosomes, as has been hypothesized in various insects, including Drosophila (Carvalho et al. 2009) and butterflies (Fraïsse et al. 2017; Lewis et al. 2021); though it was also proposed that the butterfly ZW pair originated from an autosomal pair and that the W evolves too fast to allow a proper identification of its origin (Berner et al. 2023). Even in plants, the two-gene model is likely not universal, because some sex chromosomes seem to carry only one sex-determining gene (Renner 2016). This raises the question of the other mechanisms involved in the initial arrest of recombination between sex chromosomes.
Regardless of their origin, recombination suppression is a hallmark of sex chromosome evolution. Initial recombination suppression between proto-sex chromosomes seems to often start in a small region, as suggested by studies of evolutionary young sex chromosomes, such as those found in some houseflies in which X-Y divergence is limited to only a few genes (Son and Meisel 2021). However, in various cases, recombination suppression has spread across a much larger region, sometimes encompassing the near entirety of the sex chromosome pair (Sacchi et al. 2024). The analysis of X-Y gene divergence reveals whether recombination suppression is recent (low X-Y divergence) or old (high X-Y divergence). Interestingly, divergence is not always homogeneous across the whole NRR. In S. latifolia, for instance, some sex-linked genes have higher X-Y divergence than others (Atanassov et al. 2001), and in birds, various genomic regions of the Z have different Z-W divergence levels (Gu et al. 2023). This phenomenon reflects that recombination suppression along sex chromosomes has happened independently at different time points in different regions, forming strata. Strata are genomic regions of homogeneous X-Y (or Z-W or U-V) divergence, usually measured with the synonymous divergence rate dS (Fig. 2c). Comparative genomics of related species allows to identify strata that are specific to some lineages. For example, in paleognaths (ostrich and emu), the W only has a small NRR, while other birds have multiple additional strata (Yazdi and Ellegren 2014, 2018).
The ultimate (why?) and proximate (how?) mechanisms of recombination suppression have been studied extensively. We have dedicated a separate section below to thoroughly discuss the ultimate mechanisms of recombination suppression. Here we rapidly discuss the proximate mechanisms. Inversions and other types of chromosomal rearrangements are thought to be major contributors to recombination suppression on sex chromosomes. In humans, the two most recent strata seem to have arisen following Y inversions (Lemaitre et al. 2009), and inversions are also likely to have contributed to bird sex chromosome evolution (Yazdi and Ellegren 2018). However, in many other organisms, no inversions are found to coincide with strata boundaries, as observed for one stratum of the black-spotted stickleback (Sardell et al. 2021). Therefore, other mechanisms must exist that suppress X-Y recombination. Given recombination rates are not homogenous in genomes, and sometimes even vary between sexes, recombination suppression may occur simply because the sex-determining gene(s) emerge in a region with preexisting low recombination levels, such as a pericentromeric region (Charlesworth 2023). In the plant genus Rumex, the comparative study of species with and without sex chromosomes suggested that sex chromosomes had ancestrally low rates of recombination, even before becoming sex chromosomes (Rifkin et al. 2021). Large differences in recombination landscapes between males and females (heterochiasmy) can also trigger recombination suppression between sex chromosomes. For instance, the emergence of a male sex determiner in a species with lower rates of recombination in males can lead to immediate recombination suppression (Charlesworth 2023). In R. hastatulus as well as in anuran amphibians, recombination in males is restricted to chromosomal tips, explaining Y chromosome recombination suppression in the absence of chromosome inversions (Dufresnes et al. 2021; Rifkin et al. 2022). This phenomenon likely applies to a variety of groups with sex chromosomes since heterochiasmy occurs in various clades (Sardell and Kirkpatrick 2020). An extreme case of this phenomenon, achiasmy, is found in a few organisms where recombination is altogether absent in one sex, as in Drosophila (with male-specific achiasmy) and lepidopterans (with female-specific achiasmy). In those taxa, newly evolved Ys (in Drosophila) or Ws (in lepidopterans) instantly become nonrecombining on their entire length. Alternatively, it has also been proposed that recombination suppression could be a gradual process involving a progressive expansion of the nonrecombining region outwards from the sex-determining region (Natri et al. 2013), meaning that the PAR boundary is progressively moved further toward the telomere end. This could be caused by a gradual reduction of crossover frequencies, which could theoretically be due to genetic modifiers of recombination rates (Brooks 1988) or the accumulation of TEs close to the NRR (Kent et al. 2017). Note that a recombination arrest caused by an ancestral low recombination rate, or gradual recombination suppression cannot alone account for the formation of evolutionary strata. Indeed, stratum formation requires an abrupt event of recombination suppression over a genomic region. Although, as time goes by, different strata that originated at slightly different time points can appear as a single stratum, because the synonymous X-Y divergence (dS) reaches saturation due to accumulated substitutions.
Diversity of Sex Chromosomes
The rate at which new sex chromosomes evolve is remarkably variable among taxa. In certain lineages, the same sex chromosomes have persisted for tens to hundreds of millions of years, while others experience high rates of sex chromosome turnover, i.e. changes in the identity and/or location of the sex-determing gene or region (Fig. 2b). Old and conserved sex-determining systems with sex chromosomes that have evolved over 100 Ma have been described in groups as diverse as therian mammals (Veyrunes et al. 2008), birds (Nam and Ellegren 2008), some lizards (e.g. Anguinomorpha; Rovatsos et al. 2019), and stick insects (Stuart et al. 2023). It has even recently been proposed that the X chromosome of many insects is shared from a common ancestor and has persisted in some lineages for more than 450 My (Toups and Vicoso 2023). At the other end of the spectrum are taxa with highly dynamic sex determination. The recent advances in sequencing technologies have made it possible to characterize sex chromosomes in an increasing number of nonmodel species, revealing an astonishing diversity in sex-determining mechanisms even across closely related species in certain lineages. For instance, at least 13 turnover events are necessary to account for the diversity of sex chromosome systems found among 28 species of ranid frogs (Jeffries et al. 2018). African cichlids also show an extraordinary rate of turnover, even exhibiting divergence across populations within single species (Böhne et al. 2019; Behrens et al. 2024). In plants, high turnover rates have been shown to occur in Salicaceae and Silene for instance (Martin et al. 2019; Yang et al. 2021).
From a proximate point of view, sex chromosome turnovers can be due to the emergence and spread of a new sex-determining gene replacing the old one (Fan et al. 2021), the translocation of the existing sex determiner to a new location in the genome (Yang et al. 2021), or even through hybridization, as in the nine-spined stickleback, in which the Y chromosome was acquired through an introgression event from a sister species (Dixon et al. 2019). Multiple evolutionary mechanisms have been proposed as potential drivers, with support from theoretical models. In most models, the new sex determiner will spread to fixation if it provides a fitness advantage to its bearers. A first model shows that transitions can be driven by the accumulation of deleterious mutations on the ancestral Y or W chromosomes, as the spread of a new sex chromosome can result in the elimination of the old degenerated one (mutation load model, Blaser et al. 2013). In a second model, an emergent sex-determining gene can benefit from an indirect fitness advantage if it is tightly linked to a gene experiencing sexually antagonistic (hereafter SA) selection. For instance, if a masculinizing allele arises in linkage with a male-beneficial—female-detrimental allele, it will have a selective advantage, as the SA allele will be more likely to be inherited by sons than daughters (van Doorn and Kirkpatrick 2007). In a third type of models, sex chromosome turnovers are caused by genetic conflicts over sex ratio and the transmission of sex chromosomes. For instance, selfish genetic elements that skew the transmission of sex chromosomes (i.e. through meiotic drive) can draw the sex ratio away from its optimal value. In response, selection for a balanced sex ratio will favor mutants that allow the production of more of the rarer sex, including mutant sex-determining genes (Kozielska et al. 2010). Finally, a last type of model shows that transitions in sex determination systems can occur under the sole action of genetic drift (Bull and Charnov 1977), and that with certain types of transitions, the fixation of a new sex determiner occurs at a much higher rate than regular autosomal neutral mutations (Veller et al. 2017; Saunders et al. 2018). What evolutionary forces are actually responsible for sex chromosome transitions is very hard to establish empirically. The driving forces are only detectable during the transition phase and sex chromosome turnovers are assumed to occur rapidly, so any clues are rapidly erased (Saunders 2019).
Why certain lineages never experience turnovers is also puzzling. Early observations that sex chromosomes in lineages with old and conserved sex determination tend to be highly differentiated and heteromorphic, while sex chromosomes in lineages with diverse sex-determining systems tend to be poorly differentiated and homomorphic led to the idea that sex chromosomes act as evolutionary traps, precluding changes in the sex-determining mechanism once enough divergence has accumulated (Pokorná and Kratochvíl 2009). Changing the inheritance of highly differentiated and specialized sex-linked regions should be strongly counter-selected, because their specialized features would be deleterious in an autosomal context. This model remains a subject of debate, as transitions have been described even in groups with very old sex chromosomes, such as some Drosophila, therian mammals and crocodile lizards (Vicoso and Bachtrog 2015; Saunders and Veyrunes 2021; Pinto et al. 2024). In contrast, some lineages have retained sex chromosomes over long evolutionary periods despite a lack of substantial differentiation (e.g. the homomorphic ZW chromosomes of some Paleognathous birds; Yazdi and Ellegren 2014; Okuno et al. 2021), which further indicates that the degree of differentiation does not necessarily correlate with sex chromosome lifespan.
Even in the absence of turnovers, sex chromosomes remain dynamic entities throughout their evolutionary life. The process of sex chromosome evolution is far from linear, and sex chromosomes can assume radically different evolutionary trajectories. This is true for young and poorly differentiated sex chromosomes for which interspecies or interpopulation divergence has repeatedly been observed (e.g. Furman and Evans 2018; Hill et al. 2018). This is also true for older sex chromosomes, which continue evolving even in the most differentiated systems, such as the mammalian XY system. In placental mammals, the small Y chromosome gene repertoire and expression are largely shared across species due to shared ancestry. Nevertheless, all lineages show specificities and no two Y chromosomes are identical across species (Cortez et al. 2014; Martínez-Pacheco et al. 2020). At a smaller taxonomic scale, the comparison of Y chromosomes in three mice species of the genus Mus also revealed significant variation, including in copy-number variation of multicopy Y genes (Morgan and Pardo-Manuel de Villena 2017), demonstrating that old sex chromosomes are far from being static entities.
Another source of diversity comes from fusions between sex chromosomes and autosomes, generating so-called neo-sex chromosomes (Fig. 2b). Such fusions are not frequent, but have been described in many lineages. Neo-sex chromosomes, resulting from recent fusion with otherwise old sex chromosomes, have been recognized as prime models to study the very first steps of sex chromosome evolution, and are studied in a great range of species (e.g. Li et al. 2021; Sardell et al. 2021; Sigeman et al. 2021; Sacchi et al. 2024). Just as SA selection was proposed to explain sex chromosome turnovers, it has also been suggested as a driver of sex-autosome fusions (Charlesworth and Charlesworth 1980), yet this remains to be confirmed.
Diversity can also arise through chromosomal rearrangements inside the sex chromosome pair. Indeed, in some complex sex-determining systems, inversions on the X chromosome can cause the evolution of an X* that does not recombine with the normal X over a portion of its length. The X* can influence sex determination, with, for example, X*Y females found in the African pygmy mice Mus minutoides (Veyrunes et al. 2010) or entirely female progeny for females carrying the X* in Sciaridae (Baird et al. 2023). In both cases, the molecular mechanisms through which the X* impact sex determination remain unknown, but interestingly, the Sciaridae X* evolutionary trajectory is similar to a W chromosome, with an NRR that degenerates due to its lack of recombination.
Evolution of Y Degeneration
Population genetics theory has long predicted that the absence of recombination would cause the degeneration of the sex-specific NRRs, due to processes such as Muller's ratchet, genetic hitchhiking, and background selection (reviewed in Bachtrog 2006). These effects result in increased fixation of deleterious mutations, decreased rates of adaptation, and a lower effective population size of NRRs as demonstrated by their observed lower genetic diversity compared with their recombining homologs (Hellborg and Ellegren 2004; Morgan and Pardo-Manuel de Villena 2017). The predicted degeneration has been verified on various characteristics of sex chromosomes: (i) Y allele expression is usually lower than on the X (Muyle et al. 2012; Beaudry et al. 2017); (ii) Y genes are more often lost than X genes (Wilson et al. 2013; Blavet et al. 2015; Charlesworth et al. 2021); (iii) repeats accumulate on the Y chromosome (Schield et al. 2022; Wang et al. 2024); (iv) Y proteins degenerate due to relaxed selection, as shown by their increased dN/dS ratio (Filatov 2005), and (v) the proportion of optimal codons is reduced on the Y compared with the X (Qiu et al. 2011; Carpentier et al. 2022). These hallmarks of NRR degeneration can be observed even in young sex chromosomes of a few million years of age or less (Martin et al. 2019; Wang et al. 2024). In the most extreme cases, it has even been proposed that degeneration could ultimately end with the complete loss of the Y in some species, as observed in the Ryukyu spiny rat (Li et al. 2024). In this species, males and females are both X0. Some ancestrally Y-linked genes were translocated to the X or autosomes, and others were lost, including the male-determiner SRY. Comparison of male and female genomes in the Ryukyu spiny rat led to the identification of a male-specific duplication of a Sox9 enhancer located on an autosome, which could act as the new sex-determining gene (Terao et al. 2022).
Though the sex-limited Y or W sex chromosome is often smaller than its counterpart due to degeneration, the accumulation of TEs can lead to the evolution of a larger Y/W (Fig. 2, Matsunaga et al. 1994). Y/W enlargement can have another origin, as, for example, in cichlid fish of the tribe Oreochromini, in which fusion of the Y with a highly repetitive B chromosome led to a giant Y chromosome three times the size of any autosome (Conte et al. 2021). Some TEs accumulate specifically on the Y or the X, revealing that they are only active in the male or female germline (Puterova et al. 2017). The epigenetic silencing of accumulated TEs on NRRs of sex chromosomes can have dramatic consequences for their evolution (reviewed in Muyle et al. 2021). TE silencing on the Y even has consequences on the rest of the genome in Drosophila melanogaster, causing the Y to sequester silencing epigenetic marks and depleting them from the rest of the male genome (Brown et al. 2020). This imbalance of silencing epigenetic marks between males and females may contribute to sex-biased gene expression in that species.
The extent and patterns of degeneration of sex-limited chromosomes vary substantially across lineages, and even sometimes among closely related species (Sardell et al. 2021). Multiple factors seem to influence these differences. First and foremost, the extent of the Y/W degeneration should reflect the age of the nonrecombining region(s), because old sex chromosomes have had more time to accumulate deleterious mutations and degenerate (Fig. 2). Second, sex chromosomes with larger NRRs and/or carrying more genes should be more degenerated because selective interference is stronger when an NRR contains more genes (Bachtrog 2008). In other words, advantageous mutations are less likely to fix in larger NRRs because they carry a higher number of linked deleterious mutations than small NRRs. In the Silene genus, sex chromosomes have evolved independently at least twice (Bernasconi et al. 2009). In the Silene Otites group, sex chromosomes are about 2.3 My old (Balounova et al. 2019) and NRRs in the Silene Otites group are smaller and less degenerated than NRRs in the Silene Melandrium group (Bergero et al. 2015; Martin et al. 2019), where sex chromosomes evolved around 11 Ma (Krasovec et al. 2018). The age of the sex chromosomes could be one of the factors explaining the difference in NRR degeneration level between these two Silene groups of species. However, other factors may contribute, such as the size of the NRR.
Other factors are likely to influence the speed of NRR degeneration, such as variation in TE activity, mutation rate, and effective population size. The intensity of Muller's ratchet is expected to increase with reduced effective population size, because deleterious mutations accumulate at a higher rate with increased genetic drift, as shown by Bachtrog (2008) using simulations of Y chromosomes. On the other hand, the rate of degeneration caused by genetic hitchhiking increases with increasing population size, since selection for advantageous Y mutations is stronger in larger populations (Bachtrog 2008). Indeed, selective sweeps have drastically reduced Y genetic diversity in mice (Morgan and Pardo-Manuel de Villena 2017). The multifaceted effects of effective population size on the speed of sex chromosome degeneration make it difficult to predict whether species with varying population sizes will differ in terms of the intensity of degeneration of their NRRs. More empirical and theoretical work is required to address this question.
Comparing young sex chromosomes in species that inherited them from a common ancestor allows us to control for the age of the NRR and gene content and test whether other factors such as population size actually influence the speed of degeneration. This was recently done in three closely related stickleback species that share the same ancestral XY chromosomes but display various degrees of X-Y differentiation and Y degeneration (Sardell et al. 2021). The ancestral stratum common to all three species shows substantial interspecific differences, suggesting that the speed of Y degeneration is variable, though the factors responsible could not be identified.
Though degeneration appears to be an inevitable fate of recombination suppression, some lineages with ancient and ancestral separation of sexes carry nonrecombining sex chromosomes displaying extremely low levels of degeneration. Two alternative scenarios can explain this observation. Frequent sex chromosome turnover events (Fig. 2b; see section “Diversity of Sex Chromosomes” for details) will act against degeneration as suggested in the Salicaceae family (Yang et al. 2021). Alternatively, occasional recombination between the X and the Y (or the Z and W or the U and the V) can prevent degeneration in the absence of turnover (Fig. 2b). In some systems, X-Y (or Z-W) recombination events occur in sex-reversed individuals (XY females or ZW males), a model called “the fountain of youth” (Perrin 2009). For instance, the frog Rana temporaria has XY chromosomes with a very low level of differentiation despite recombination suppression across most of the Y chromosome. It has been shown that the occasional emergence of fertile XY females allows “rejuvenation” of the Y via recombination with the X in those individuals (Rodrigues et al. 2018). These occasional X-Y recombination events in sex-reversed XY females may contribute to the low levels of Y degeneration observed in many frogs.
Rare Z-W recombination events have also been suggested to occur on the sex chromosomes of the butterfly Melanargia ines. In this species, the neo-W chromosome was expected to show uniform Z-W divergence due to the absence of crossover in females. However, Decroly et al. (2024) observed three plateaux of Z-W divergence, highlighting at least two ancient events of Z-W recombination.
Occasional recombination between sex chromosomes can also take the form of gene conversion events, a process where two alleles undergo recombination without crossover and one allele is copy-pasted onto the other. For instance, in humans, occasional events of X-Y gene conversion have been identified in the youngest stratum (Trombetta et al. 2014). These examples suggest that rare recombination events (including gene conversion events) may be a common feature of young sex chromosomes (or strata) and likely contribute to slow down Y/W degeneration in some cases.
Evolution of Dosage Compensation
Following the degeneration of the sex-specific nonrecombining region, the heterogametic sex should experience decreased expression levels of its sex-linked genes compared with the homogametic sex, if it was not for the evolution of a phenomenon called dosage compensation (hereafter DC). DC patterns are very diverse and have been classified into categories depending on two expression ratios (Gu and Walters 2017). The first expression ratio is XY male/XX female (or ZW female/ZZ male). If this ratio is equal to 1, males and females have equal expression levels and dosage balance is achieved. Conversely, if this ratio is <1, Y/W degeneration is not compensated for and the heterogametic sex has lower expression than the homogametic sex. The second expression ratio is XY male/AA (or ZW female/AA), where AA stands for autosomal expression levels. If this second expression ratio is equal to 1, ancestral expression levels have been recovered for sex-linked genes in the heterogametic sex. This type of DC is called DC sensu stricto. If Y genes have been lost, to obtain DC sensu stricto, X expression has to be upregulated to the extent it has doubled relative to its ancestral state. If the XY male/AA expression ratio is <1, Y degeneration causes males to have lower expression levels than ancestrally, before the evolution of the Y chromosome.
DC patterns are categorized into four broad types that reflect levels of dosage balance and DC sensu stricto (Gu and Walters 2017). Type I DC corresponds to perfect DC, meaning cases where both dosage balance and DC sensu stricto are achieved (Gu and Walters 2017), as in the guppy fish Poecilia picta and Poecilia parae, in which X gene expression is upregulated in males (Metzger et al. 2021). Similar patterns of equal expression of Z-linked and autosomal genes in males and females are observed in the crustacean Artemia franciscana (Huylmans et al. 2019). Mosquito XY sex chromosomes also show patterns consistent with complete DC (Jiang et al. 2015).
Type II DC groups patterns similar to placental mammal DC, where dosage balance is achieved but not DC sensu stricto (Gu and Walters 2017). Indeed, in humans and other placental mammals, the X/AA expression ratio in males is close to 0.5 in both transcriptomic and proteomic data (Chen and Zhang 2015, 2016). These results suggest that ancestral expression levels have not been recovered in males. However, it is still debated whether DC sensu stricto is achieved in placental mammals thanks to a posttranscriptional upregulation of gene expression (Wang et al. 2020; Cecalev et al. 2024). Dosage balance is achieved in placental mammals thanks to the inactivation of one X chromosome in females (Barr and Bertram 1949). Interestingly, X chromosome inactivation does not affect all X genes equally. In particular, about 25% of X genes escape X inactivation in humans and were found to be under strong selective constraint, suggesting that escaping X chromosome inactivation is selected for in those genes (Park et al. 2010; Slavney et al. 2016). Most of these X inactivation escapees are found in recent strata, where Y degeneration is less advanced and therefore DC is not as necessary as in older strata (Carrel and Willard 2005).
Type III DC stands for absence of DC altogether, i.e. the absence of dosage balance and absence of DC sensu stricto (Gu and Walters 2017), as observed, for example, in the Komodo dragon (Rovatsos et al. 2019).
Finally, type IV DC corresponds to DC sensu stricto but absence of dosage balance (Gu and Walters 2017). This rather rare pattern of X upregulation in both males and females was observed in S. latifolia (Muyle et al. 2018), in the flour beetle Tribolium castaneum (Prince et al. 2010), in stratum 2 of the three-spined sticklebacks (Schultheiß et al. 2015) and in schistosome parasites, which have increased Z expression in both sexes (Picard et al. 2018; Chen et al. 2020). However, follow-up studies led to contradictory results both in S. latifolia (Krasovec et al. 2019) and beetles (Mahajan and Bachtrog 2015; Whittle et al. 2020; Bracewell et al. 2023). Type IV DC could correspond to an intermediary stage of DC evolution where expression levels are corrected in XY males (or ZW females) but not yet in XX females (or ZZ males).
Even though some species lack global mechanisms of DC, few species seem to have a complete absence of DC for all genes, and many species show mixed DC patterns, with some genes that are dosage compensated (type I or II) and others not at all (type III). These mixed patterns, also called “gene-by-gene DC” were observed, for example, in gila monster (Webster et al. 2024), in the lizard Anolis carolinensis (Rupp et al. 2017), in various plant species (Crowson et al. 2017; Martin et al. 2019; reviewed in Muyle et al. 2022), in chicken (Zimmer et al. 2016), and in butterflies and moths (Walters and Hardcastle 2011; Harrison et al. 2012; Walters et al. 2015; Gu et al. 2017; Catalán et al. 2018). Some butterfly species and specific tissues in certain species indeed seemed to have incomplete DC, but the removal from analyses of genes with differential expression between sexes (hereafter sex-biased genes) led to reevaluate DC as being complete for multiple species (Huylmans et al. 2017; Höök et al. 2019). Indeed, the presence of genes with sex-biased expression on sex chromosomes can bias the analysis of global DC patterns. For instance, an excess of female-biased genes on the X will cause the global XY male/XX female expression ratio to be <1, suggesting a lack of dosage balance. Disentangling the effect of sex-biased gene expression from Y/W gene silencing is challenging. Indeed, a loss of Y expression looks like female-biased expression in the absence of DC. However, at the gene level, loss of Y expression in the absence of DC leads to XY male/XX female gene expression ratios of 0.5 at the lowest. Female-biased gene expression at the gene level can lead to XY male/XX female gene expression ratios much <0.5. We therefore recommend removing all sex-biased genes with a >2-fold expression ratio for DC analyses, to avoid biasing global sex chromosome patterns of expression ratios. Sex-biased genes likely play roles in sex-specific functions and are therefore not expected to require equal dosage between males and females.
Dosage-sensitive genes, unlike sex-biased genes, are genes whose function is impaired by dosage imbalance, as, for example, genes involved in protein complexes, where it is crucial to have stoichiometric quantities of each partner. Genes forming protein complexes were shown to be fully dosage compensated in human sex chromosomes (Pessia et al. 2012, 2014). A better characterization of dosage-sensitive genes may help us understand why in some species only a few genes are dosage compensated, while in others, complete DC is observed.
DC patterns not only vary among gene types but also among tissues and developmental stages, as observed in Drosophila (Nozawa et al. 2014; Huylmans and Parsch 2015). DC patterns can even vary between stages of the life cycle, as in Schistosoma mansoni (Vicoso and Bachtrog 2011; Picard et al. 2019). The causes behind this variation of DC patterns among tissues, developmental stages, and life forms are still poorly understood. However, different abundances of sex-biased genes in different tissues could bias patterns (Huylmans and Parsch 2015).
Most of the above-mentioned studies were performed on transcriptomic data. However, RNA sequencing (RNA-seq) data can only provide an incomplete view on DC, since regulatory mechanisms can also act at the translation stage to balance protein quantity between males and females. In birds, for example, most genes are not dosage compensated when looking at RNA-seq data (Uebbing et al. 2013). However, when measuring protein quantity in embryos, some genes showed complete DC at the protein level (Uebbing et al. 2015).
The past years of research have unfolded the diversity of DC patterns in species with sex chromosomes; however, the exact molecular actors of DC regulation are only known in a few species: the MSL complex in Drosophila (Lucchesi and Kuroda 2015), the long noncoding RNA XIST in placental mammals (Marahrens et al. 1997), the convergently evolved long noncoding RNA RSX in marsupials (Grant et al. 2012), the sex-specific factor SOA in Anopheles mosquitoes (Kalita et al. 2023), and the long noncoding RNA MAYEX in the green anole lizard (Tenorio et al. 2024). These DC regulators modify epigenetic marks such as DNA methylation and histone modifications on sex chromosomes in a sex-specific manner, allowing to balance expression between sexes and/or sex chromosomes and autosomes.
In the absence of identified molecular actors for DC regulation in most species, it remains unclear whether DC patterns are due to bona fide DC or simple buffering mechanisms. Buffering occurs when any gene present in a hemizygous state (only one gene copy in an otherwise diploid individual) is more highly expressed than the expected 50% of gene expression in the diploid state. Buffering was observed in Drosophila (Malone et al. 2012) and yeast (Hose et al. 2015), when one of the two copies of an autosomal gene was artificially deleted. Buffering differs from DC bona fide because it can affect any gene (on autosomes and sex chromosomes), while DC is a phenomenon limited to sex chromosomes with degenerated Y/W expression. Because buffering automatically upregulates gene expression in the hemizygous state, it might contribute to DC patterns on sex chromosomes, without the need for a specific mechanism to evolve (such as the MSL complex in Drosophila). However, buffering is unlikely to explain full compensation patterns because, at least in Drosophila autosomal genes, buffering was shown to be heterogeneous across genes and sexes, with some genes being fully buffered when hemizygous, while other genes had a complete absence of buffering when hemizygous (Malone et al. 2012). In addition, buffering is expected to affect expression levels only in the heterogametic sex (the sex with Y/W degeneration), meaning that buffering cannot explain DC patterns where the homogametic sex expression has changed. For instance, X chromosome imprinting in S. latifolia, with a higher maternal X expression in both males and females does not fit well with buffering (Muyle et al. 2018, 2022), nor does lower X expression of some sex-linked genes in females of Rumex rothschildianus (Crowson et al. 2017).
We still have a lot to learn about how DC evolves. The effects of effective population size, size and age of the NRR should be further explored empirically and theoretically. In particular, it is still unclear why some sex chromosomes have incomplete DC even after hundreds of millions of years of evolution, while others have evolved complete DC. It also remains to be discovered how DC can transition from being local to global. DC evolution on the Drosophila miranda neo-X gives a partial answer to this question. Indeed, some D. miranda neo-X genes had their expression doubled in males, while other neo-X genes still maintain ancestral expression (Nozawa et al. 2018). The status of each X gene depends on whether the Y copy is fully degenerated or not. This gradual spread of DC on the D. miranda neo-X was facilitated by the acquisition of cis-regulators through TE transpositions on the neo-X (Ellison and Bachtrog 2013). However, the mechanism regulating DC was already present in the ancestor of D. miranda, before the evolution of the neo-X, which likely facilitated the gradual spread of DC onto the neo-X. Different mechanisms might be at play on young sex chromosomes evolving global DC for the first time.
Evolution of Sex Chromosome Gene Content and Evolutionary Rates
On top of changes in expression levels linked to Y degeneration and DC evolution, sex chromosomes can also evolve different proportions of sex-biased genes compared with the ancestral autosomal pair from which they evolved. For instance, the X chromosome spends two-thirds of its time in females, which can drive its enrichment in female-biased genes, the so-called X feminization effect (Rice 1984; Bachtrog 2006). In ZW sex chromosomes, a similar process can lead to Z masculinization, as observed in Lepidoptera (Huylmans et al. 2017). An opposite phenomenon is called X masculinization and consists in enrichment of the X in male-biased genes when the X is mostly hemizygous in males, as is the case in humans (Lercher et al. 2003). Indeed, when Y genes are lost, recessive male-beneficial alleles are directly visible to selection in X-hemizygous males and can become fixed. If these male-beneficial X alleles are deleterious for females, male-biased expression can evolve to resolve sexual antagonism (Rice 1984; Bachtrog 2006).
The Y chromosome, on the other hand, is expected to become enriched in genes related to male functions, since it is male specific. For example, in D. miranda, the neo-Y male-biased genes are retained longer than female-biased genes and poorly expressed genes, which are more likely to be lost due to degeneration (Kaiser et al. 2011). Although most Y genes are involved in spermatogenesis in placental mammals, slightly different gene sets were retained on the Y of mice and humans (Subrini and Turner 2021).
The gene content of sex chromosomes can also evolve through gene translocations, as described in S. latifolia for the SlCyt gene whose movement to the sex chromosomes is supposed to have been selected for (Kaiser et al. 2009). A similar phenomenon occurred in the seed beetle Callosobruchus maculatus, where males carry an additional TOR copy on the Y chromosome. TOR is a regulator of body size and its presence on the Y likely contributes to sexual dimorphism. Duplication and translocation of the autosomal TOR gene onto the Y chromosome may have helped resolve sexual conflict over body size in that species (Kaufmann et al. 2023). On the other hand, some Y genes escape from the Y by translocating to autosomes (Bracewell and Bachtrog 2020). These translocation events out of the Y are likely favored to avoid gene degeneration.
In animals, the male germline induces a higher mutation rate than the female germline due to the higher number of cell divisions that occur in the testes compared with the ovaries. Because the Y chromosome is only transmitted through the male germline, it has a higher mutation rate than the autosomes. The X chromosome spends two-thirds of its time in females and is not subject to this increased mutation rate. Autosomes spend equal amounts of time in males and females, causing the X chromosome to have a lower mutation rate than autosomes (Malcom et al. 2003). This has important implications for the rate of degeneration of Y vs. W chromosomes, because Ws are female specific and, therefore, expected to degenerate more slowly than Y chromosomes.
The gene content of the Y chromosome also evolves through duplication of genes into multiple copies that are called amplicons, often forming palindromes that can recombine through gene conversion (Geraldes et al. 2010; Oetjens et al. 2016; Vegesna et al. 2020; Tomaszkiewicz et al. 2023). It has been proposed that palindromic Y gene conversion maintains the sequence integrity of Y genes and prevents Y degeneration in the absence of X-Y recombination (Marais et al. 2010). Another hypothesis for the origin of Y ampliconic gene clusters is that they arise because of transmission distorters (genes that favor their own transmission during meiosis), as observed in mice (Moretti et al. 2020). Palindromes are also observed on the mouse X and similarly undergo gene conversion, which may contribute to their accelerated evolutionary rates (Swanepoel et al. 2020).
Following the degeneration of NRRs, sex-linked genes can become hemizygous (without a Y/W homolog). Hemizygosity reveals recessive alleles to selection, which potentially triggers higher levels of positive selection. Therefore, hemizygosity may lead to accelerated levels of X/Z protein evolution. On the other hand, the X/Z chromosomes have three quarters the effective population size of autosomes, which may lead to relaxed selection and higher rates of X/Z protein evolution. Therefore, the so-called faster-X (or faster-Z) evolution may be due to two phenomena: (i) relaxed selection causing faster-X divergence or (ii) increased adaptation rates resulting in faster-X adaptation (Meisel and Connallon 2013). In Dr. miranda, X-hemizygous genes evolve faster than X genes that retained a functional Y homolog, suggesting that higher adaptation in X-hemizygous genes is responsible for the faster-X effect (Zhou and Bachtrog 2012). However, in many species, the role of adaptation in faster-X evolution is less clear than in Drosophila. The analysis of two Stegodyphus spider species with contrasting life-history traits (inbreeding social vs. subsocial outcrossing) showed a prominent role for drift in faster-X evolution, although some patterns were consistent with positive selection acting on the X (Bechsgaard et al. 2019). In birds, faster-Z evolution at the sequence level was attributed to the reduced effective population size of the Z compared with autosomes (Mank et al. 2010). Also in birds, Z-gene expression levels evolve faster than autosomal gene expression levels (Dean et al. 2015). This faster-Z effect at the gene expression level was linked to selection acting on recessive beneficial alleles of Z-hemizygous genes in females. A similar faster-X effect was observed in Drosophila at the gene expression level and was linked to a combination of neutral processes and also natural selection fixing advantageous cis-regulatory changes in male-biased genes on the X (Coolon et al. 2015).
While faster-X effect is expected in degenerated sex chromosomes that are mostly hemizygous due to degeneration and gene loss, another effect called the slower-X effect has been described on recently evolved sex chromosomes (Mrnjavac et al. 2023). Because of the smaller effective population size of the X and its biased transmission through females, it is predicted that selection is less efficient on diploid X loci than on autosomal and X-hemizygous loci, especially for genes related to male functions. This slow-X effect may cause higher rates of nonsynonymous substitutions and demasculinization of the X in young sex chromosomes.
Therefore, sex chromosomes do not necessarily maintain their ancestral gene content over evolutionary time, can differentially accumulate female- or male-biased genes, and experience changes in their evolutionary rates.
The Ultimate Causes of Sex Chromosome Recombination Suppression
Over the past years, the field of sex chromosome evolution witnessed a burst of theoretical developments related to the question of why sex chromosomes stop recombining (reviewed in Käfer 2022; Charlesworth 2023; Jay et al. 2024; Veltsos et al. 2024). Please refer to the above section on sex chromosome origins for details on the proximate mechanisms explaining how recombination becomes suppressed. It is important to understand why recombination suppression occurs, given that it triggers all other evolutionary processes involved in sex chromosome evolution. At any given time, selection could favor mechanisms reestablishing recombination between sex chromosomes to reduce the deleterious mutation load of the NRR. There must, therefore, be one or multiple balancing evolutionary forces that explain why, in many cases, recombination suppression between sex chromosomes is maintained over evolutionary time.
Until recently, the literature had been dominated by a single theory proposing that recombination suppression of sex chromosomes is driven by SA selection (Rice 1984, 1987). This model posits that SA mutations (advantageous in one sex and deleterious in the other) are selected if they are tightly linked to the sex-specific region, providing advantage to one sex without damaging the other. SA selection may therefore favor recombination suppression between the sex-specific NRR and a closely linked SA loci. So far, no direct evidence of this theory has been observed. It is particularly difficult to prove the SA theory, because SA genes are also predicted to accumulate on sex chromosomes after recombination suppression, and identifying genes under SA selection in the recombining part of sex chromosomes is challenging (Dagilis et al. 2022). Nevertheless, SA selection is still a good candidate to explain why recombination suppression is maintained in the long run, and likely contributes to prevent reestablishment of recombination on sex chromosomes.
Various patterns indirectly consistent with the expectations of the SA theory have been described in different species. For instance, PAR genes show increased diversity and increased male–female single nucleotide polymorphism (SNP) differentiation in S. latifolia (Qiu et al. 2016). This pattern is consistent with SA alleles being maintained polymorphic at partially sex-linked loci. But other scenarios could explain the pattern, such as demographic changes. In another study, it was shown that the NRR of monkeys and apes is larger than that of lemurs and lorises (Shearn et al. 2020). It was proposed that SA selection may have driven further recombination suppression in the former, given they display stronger sexual dimorphism.
A recent test of the SA theory in guppies was a source of debate. These fish are particularly interesting when it comes to SA selection, because they are known to harbor SA color patterns (Endler 1980). Bright coloration alleles are advantageous in males due to sexual selection, but disadvantageous in females due to predation (Houde and Endler 1990). In theory, if encoded on the sex chromosomes, these SA color patterns could impact recombination suppression (Charlesworth 2018). Wright et al. (2017) studied sex chromosomes in various populations of the guppy Poecilia reticulata experiencing different predation rates, and presumably different intensity of SA selection. The authors used male over female SNP density to annotate NRRs and concluded that recombination suppression affected 40% of the sex chromosome pair in populations with low predation, as opposed to only 12% in populations with high predation. They concluded that SA selection triggered further recombination suppression in populations with low predation where males are more colorful (Wright et al. 2017). Further studies by the same research team using other data and closely related guppy species claimed similar results, i.e. that recombination suppression affects a large part of the Y chromosome and that the NRR has a common ancestry among multiple guppy species (Darolti et al. 2019; Wright et al. 2019; Almeida et al. 2021; Fong et al. 2023). In the meantime, an independent research team concluded that recombination is focused at the tips of all chromosomes in guppy males, but rare recombination events occur on almost the entire length of the sex chromosome pair, with only about 4% of the Y chromosome that is completely nonrecombining (Bergero et al. 2019; Bergero and Charlesworth 2019; Charlesworth et al. 2020a, 2020b; Qiu et al. 2022; Charlesworth et al. 2024). A third research team assembled a male guppy genome and corroborated that the NRR is small in P. reticulata (Fraser et al. 2020). Finally, a fourth research team reanalyzed data from the aforementioned teams and found no Y-specific SNPs in “stratum II” identified by Wright et al. (2017), suggesting again that this region recombines from time to time (Kirkpatrick et al. 2022). Genetic mapping in different P. reticulata populations recently confirmed that recombination patterns are similar among populations, regardless of whether populations undergo more or less intense SA selection (Charlesworth et al. 2024). Therefore, as of today, there is no consensual evidence that SA selection is involved in recombination suppression in guppies.
A few years ago, a second theory explaining why recombination becomes suppressed between sex chromosomes proposed that gradual neutral sequence divergence between the X and the Y (or Z-W or U-V) may arise in the PAR near the boundary with the NRR due to partial sex linkage, until divergence reaches levels that entirely prevent recombination (Jeffries et al. 2021). This model predicts that the NRR might expand gradually, which should be reflected by a pattern of gradual increase in X-Y sequence divergence when moving away from the PAR-NRR boundary. This model relies on the idea that X-Y divergence in the partially sex-linked region of PAR close to the boundary should reduce the rate of recombination, which remains to be demonstrated. As raised by Lenormand and Roze (2024), current data on meiotic recombination rates in species without sex chromosomes suggest that higher heterozygosity can actually promote meiotic crossovers rather than repress them. This model does not propose a mechanism to explain the long-term maintenance of recombination suppression once deleterious mutations start accumulating in the NRR, but it makes clear predictions which should allow to test whether it is responsible for recombination suppression in some sex chromosome systems (gradual increase in X-Y differentiation away from the PAR-NRR boundary and elevated genetic diversity in the PAR in the region adjacent to the boundary; Jeffries et al. 2021).
A third recent theory relies on regulatory evolution to explain recombination suppression on sex chromosomes, and accounts for both the establishment and the long-term maintenance of recombination suppression (Lenormand and Roze 2022). This theory starts with a “lucky inversion” on the Y that carries fewer deleterious mutations than noninverted haplotypes and can be fixed by chance in a population thanks to this initial fitness advantage. Once recombination is suppressed, cis-regulators diverge between the X and the Y (Lenormand et al. 2020). A stochastic decrease in Y allele expression fires a feedback loop where Y alleles accumulate mutations in cis-regulators that lower their expression level, which in turn lowers the efficacy of selection on Y proteins, causing the accumulation of deleterious mutations, that consequently select for adaptive silencing of Y alleles, eventually leading to complete Y gene loss. This phenomenon cannot occur on the X because deleterious recessive mutations are not sheltered in a heterozygous state on the X, unlike on the Y. Importantly, the evolution of Y silencing through X/Y cis-regulator divergence is independent from Y degeneration due to selective interference. If X/Y genes are dosage sensitive, the silencing of Y alleles will quickly be compensated by the evolution of DC, either through cis or trans effects. Once DC has evolved, X-Y recombination reestablishment is counter selected, because it would lead to unfavorable combinations of upregulating or silencing cis and trans effects that would cause expression issues for dosage-sensitive genes. In other words, the evolution of DC results in SA effects, because the presence of two X alleles is only advantageous in a female background, while the combination of X and Y alleles is only advantageous in a male background.
The regulatory evolution theory for recombination suppression predicts that Y degeneration and DC will both evolve early during sex chromosome evolution. This differs from the classical theory where Y degeneration increases progressively due to selective interference. In particular, the classical theory fails to explain why small nonrecombining regions should degenerate, because selective interference is limited when only a few genes are trapped in an NRR. Interestingly, DC has evolved for a few genes in Silene otites ZW sex chromosomes which are extremely young (~0.6 My old), even though the NRR is small (Martin et al. 2019), consistent with predictions made by the regulatory evolution theory. In another study, putative enhancers of Y expression exhibited elevated substitution rates and decreased polymorphism in stickleback and were associated with lower Y expression compared with the X (Shaw et al. 2024). These results suggest that cis-regulatory regions are under positive selection to become silenced on the Y, consistent with expectations of the model by Lenormand and Roze (2022).
A fourth theory was published in parallel and coined the “sheltering of deleterious mutations” model. This theory focuses on what corresponds to the first step of the regulatory evolution theory (the fixation of a lucky inversion), but no regulatory evolution was included in models. The sheltering theory proposes that Y inversions that prevent recombination between the X and the Y have a higher probability to become fixed in populations compared to autosomal inversions (Jay et al. 2022). The first step of the theory is the occurrence of a Y inversion that initially carries fewer deleterious mutations than other noninverted haplotypes in the population. This initial lower load advantage can lead to an increase in frequency of the inversion (at this stage, the sheltering is not happening yet). The next step is a race against time for the inversion to become fixed before it accumulates too many nonrecessive deleterious mutations due to Y degeneration in the absence of recombination. The Y inversion's recessive deleterious mutations are sheltered from selection due to the heterozygous state of the Y chromosome (i.e. mutations arising on the Y inverted haplotype are never in a homozygous state). This sheltering of recessive deleterious mutations does not occur for autosomal inversions, explaining why Y inversions have a higher probability to fix than autosomal inversions (Jay et al. 2022).
The validity and relevance of the sheltering theory is under ongoing debate (Olito and Charlesworth 2023; Lenormand and Roze 2024; Jay et al 2024; Charlesworth and Olito 2024). Olito and Charlesworth (2023) initially criticized Jay et al. (2022) for (i) excluding inversions that were lost in the first 20 generations of their simulations, which they claim biased the results towards higher fixation probabilities for Y inversions, and (ii) not including simulations with neutral inversions (i.e. inversions carrying no deleterious mutations), which are necessary to establish whether an inversion carrying deleterious mutations has a net selective advantage. Olito and Charlesworth (2023) reran Jay et al. (2022)’s simulations in the light of this missing data. As expected, including the first 20 generations resulted in a decrease in fixation probabilities across all simulations and parameter values. Nevertheless, the qualitative results of Jay et al. (2022) remained largely unchanged: under some conditions, inversions carrying deleterious mutations are more likely to become fixed on Y chromosomes than autosomes (as shown by Fig. 2 in Olito and Charlesworth 2023). However, Charlesworth and Olito (2024) argue that Y inversion fixation probability should be compared to neutral inversions, to test for any selective advantage of Y inversions that carry mutations. When comparing Y inversions to neutral inversions, Charlesworth and Olito (2024) showed that Y inversions are selectively favored only under restricted conditions that the authors judge unrealistic (when mutations are fully recessive, or when selection is very weak). These conclusions were corroborated by Olito et al. (2024), after correcting an error in their retracted paper (Olito el al. 2022). Jay et al. (2024) argue that comparisons with a neutral model is not as informative as comparisons to autosomes, given the sheltering effect is not a standard directional selective force but rather a protection against a detrimental effect (the expression of recessive mutations). Olito et al. (2024) highlighted other constraints faced by lucky Y inversions which they claim make their probability of fixation negligible: (i) recessive or partially recessive deleterious mutations captured by Y inversions can also be segregating on X chromosomes, so they will be expressed as homozygous in XY individuals at the frequency with which they occur on the X (ii) the advantage of lucky inversions will gradually erode as new mutations accumulate over time. Both phenomena can occur in simulations carried by Jay et al. (2022). To summarize the current state of the controversy, both groups seem to agree that the sheltering of deleterious mutations can cause Y inversions to fix at a higher rate than autosomal ones, under some conditions. A debate remains on how general or specific the sheltering theory is, and what are the parameter ranges where it may apply.
Unlike the regulatory evolution theory, the sheltering theory does not propose an explanation for the long term maintenance of recombination suppression (Lenormand and Roze 2024). Indeed, Y inversions accumulate deleterious load due to degeneration and any reversion to a recombining state should be selected for because it increases the fitness of the heterogametic sex. Experimental data are currently lacking to know how plausible it is for recombination suppression to be reverted. Jay et al. (2022) argue that it is unlikely to revert a Y inversion to a recombining state because a second inversion would have to occur at the exact same breakpoints as the initial inversion. However, in a new simulation study, Lenormand and Roze (2024) showed that very low rates of recombination reestablishment are sufficient to prevent the sheltering theory from acting. The sheltering theory may still explain initial recombination suppression. Reversion to a recombining state may be prevented by the rapid evolution of a complex combination of various chromosomal rearrangements on the Y (Jay et al. 2022, 2024). However, a sex chromosome turnover leading to the loss of the Y chromosome is another plausible solution to restore recombination, regardless of how rearranged the Y chromosome is (Lenormand and Roze 2024). Jay et al. (2024) still argue that the sheltering theory may allow Y inversions to fix in the short term and be observed in nature, regardless of their long-term fate, similar to other evolutionary phenomena such as self-fertilization, which have short-term advantages but are deleterious in the long-run.
Finally, preexisting low recombination rates have sometimes been listed as a possible cause for recombination suppression in newly evolved sex chromosomes (Rifkin et al. 2021; Charlesworth 2023). However, we would like to highlight that this factor is not directly comparable with the models mentioned above. Heterochiasmy and achiasmy can indeed trigger recombination suppression on sex chromosomes (as explained above in the “Origins of Sex Chromosomes” section), but we believe those factors explain how recombination becomes suppressed rather than addressing the evolutionary mechanisms causing recombination suppression. In this case, recombination suppression on sex chromosome is a by-product of a preexisting genomic feature that evolved independently. Why heterochiasmy and achiasmy have evolved in some lineages and not others is a topic that is still open to debate (Lenormand et al. 2016; Sardell and Kirkpatrick 2020). In addition, heterochiasmy and achiasmy cannot explain the evolution of strata, but only initial recombination suppression of sex chromosomes.
Identification of Sex Chromosomes and Sex-Linked Sequences
To study their evolution and diversity, sex chromosomes and sex-linked sequences first need to be properly identified. Historically, sex chromosomes used to be detected cytologically by analyzing karyotypes and identifying chromosome pairs displaying morphological differences in one sex. Though efficient for species with heteromorphic chromosomes, this approach does not allow to characterize sex chromosomes in species with homomorphic sex chromosomes, common in fish and reptiles for instance. This deficit was recently addressed with the development of multiple bioinformatic approaches based on sequencing data. The high TE content and heterozygosity level of sex chromosomes makes assembling their sequences a challenge and specific methods had to be developed. These new methods have played a determinant role in shaping our current view of sex chromosome diversity. Palmer et al. (2019) and Muyle et al. (2017) provided comprehensive reviews of these methods; here, we will focus on the most widely utilized approaches.
The “depth of coverage” approach is suited to detect sex-linked regions with a substantial level of divergence. The principle behind this method involves conducting genomic sequencing on one or more individuals of each sex from the same species, aligning sequenced reads to a reference genome, and searching for regions with unbalanced male:female coverage ratio along assembled sequences (Fig. 2c). If enough divergence has accumulated between a pair of X and Y (or Z and W), X sequences will exhibit approximately one half the sequencing depth in males compared with females (and vice versa for Z sequences), while Y or W sequences will exhibit, respectively, male- or female-limited coverage. This approach has been successfully applied across a massive range of taxa, including insects, trematodes, plants, reptiles and many more (e.g. Fraïsse et al. 2017; Müller et al. 2020; Elkrewi et al. 2021; Kostmann et al. 2021). Its main limitation is that it does not work well on sex chromosome pairs with poor differentiation.
A second type of approach, coined the “subtraction approach” aims specifically at identifying sex-specific sequences in species with known sex-determining systems. In its most classic form, if a reference genome is available for the homogametic sex, RNA-seq is performed for the heterogametic sex, raw reads are mapped to the reference, and unmapped reads are assembled to obtain putative Y- or W-limited transcripts (Cortez et al. 2014). In the absence of a reference genome, similar results can be achieved by sequencing RNA and assembling transcripts for the heterogametic sex. Mapping genomic reads from both sexes to those transcripts allows identifying putative Y or W transcripts, by filtering transcripts with no coverage from the homogametic sex (Cornejo-Páramo et al. 2020). Because they require that sex chromosomes are sufficiently diverged for reads from sex chromosomes to not match to each other, subtraction approaches are also most suited for species with high sex chromosome divergence.
So-called k-mer-based approaches are quite similar in essence and yield similar results to subtraction approaches. One approach consists in mapping k-mers of whole-genome sequencing reads from an individual of the homogametic sex to an assembled genome of the heterogametic sex. Assembled sequences with no hits are likely to be part of the Y/W chromosome (Carvalho and Clark 2013). An extension of this approach, which does not require a reference genome, uses DNA/RNA data from both sexes. Following the identification of k-mers specific to the heterogametic sex, reads carrying these k-mers are extracted and assembled into W- or Y-specific genetic sequences or transcripts (Tomaszkiewicz et al. 2016; Elkrewi et al. 2021).
The third type of approach relies on SNPs and is suited to detect sex-linked regions with a low level of divergence. It is based on population genetics and the comparison of male and female genotypic polymorphisms, using either wild-caught or bred specimens. The core principle is to sequence short genomic or transcriptomic reads for multiple males and females, and scan the data to identify genomic signatures of recombination suppression in one sex, usually following mapping to a reference genome or transcriptome and performing variant calling. An elevated SNP density, an enrichment in heterozygous markers or sex-limited variants are all expected in the heterogametic sex for regions with low sex chromosomes divergence (i.e. assuming that X and Y or Z and W sequenced reads are close enough to map on the same reference, Fig. 2c). Male vs. female Fst (an indicator of genetic differentiation) can also be used to detect sex-linked regions. Different implementations of this approach have been used to identify sex chromosomes and sex-linked sequences in plants, fish, and reptiles among others (Pucholt et al. 2017; Hill et al. 2018; El Taher et al. 2021; Käfer et al. 2022). Variations of this approach allow characterizing the sex-determining system (XY or ZW) and identifying sex-linked sequences or genes without using a reference genome, following the assembly of raw reads (RAD-seq reads assembled into RADtags, e.g. Brelsford et al. 2017, or RNA-seq reads assembled into transcripts, e.g. Saunders et al. 2024). Instead of sampling males and females in natural populations, it is also possible to analyze segregation patterns of polymorphic markers in a cross, which involves sequencing parents and a few offspring of each sex (Michalovova et al. 2015; Muyle et al. 2016). This provides a much higher sensitivity and specificity to analyses, but is only applicable to species that can be bred in controlled conditions.
A multitude of programs or R packages has been developed to facilitate and streamline analyses for all types of approaches (Muyle et al. 2016; Rangavittal et al. 2018; Feron et al. 2021; Käfer et al. 2021; Cabrera et al. 2022; Grayson et al. 2022; Nursyifa et al. 2022; Sigeman et al. 2022; Devloo-Delva et al. 2024). The best program or approach to use depends on multiple factors. Key elements to consider are the degree of sex chromosome differentiation, the existence of a reference genome, the type of data, and the number of samples available (Muyle et al. 2017). Because the degree of sex chromosome differentiation is difficult to ascertain prior to analysis, a good strategy is to combine different approaches to maximize the chances to detect sex-linked markers (Elkrewi et al. 2021; Käfer et al. 2021). This also provides the opportunity to detect regions with various degrees of differentiation in the same genome (i.e. young and old strata). Most approaches can work with different data types. Whole-genome sequencing data provides the best coverage of the genome and allows detecting small/poorly differentiated sex-linked regions. Reduced-representation genome sequencing (e.g. RAD-seq) is a cheap way to sequence many individuals, but the patchiness of sequenced regions might overlook small sex-linked regions, and the absence of a reference genome significantly limits mapping capacities for enrichment or comparative analyses (Hill et al. 2018). This issue can be mitigated with RNA-seq, via the assembly of transcripts, which size and degree of genetic conservation allows mapping to genomes of distantly related species (Saunders et al. 2024). Limited sample sizes make some approaches prone to errors, especially if recombination is reduced but not halted (e.g. contrasting male vs. female Fst), leading to erroneous inference of recombination suppression. This was partially responsible for the debates regarding guppy sex chromosome evolution, detailed in the “The Ultimate Causes of Sex Chromosome Recombination Suppression” section. The program SDpop (Käfer et al. 2021) includes a probabilistic framework to conduct statistical analyses of sex linkage to minimize these types of errors.
Conclusions and Perspectives
Sex chromosomes have fascinated researchers for over a century because of their involvement in fundamental aspects of life, namely sex determination and sexual reproduction, and because they are among the most dynamic and fast-evolving regions of the genome. The classical model of sex chromosome evolution, gradually elaborated since the start of the 20th century mostly based on the study of sex chromosomes in model species (human, Drosophila, chicken, Caenorhabditis elegans) is widely regarded as a fundamental paradigm and one of the great achievements of evolutionary biology. In the past decade, the field of sex chromosome evolution has extended to a plethora of new nonmodel species, thanks to the development and increased accessibility of DNA sequencing. Genomic analyses in these species uncovered a truly unexpected diversity of sex chromosome systems, revealing that many species do not follow classical models and highlighted that important aspects of the biology of sex chromosome evolution remain unclear. In particular, in the past few years, the importance of some central aspects of the classical model was questioned, and novel theoretical models were developed to account for new empirical observations, notably to explore the mechanisms responsible for recombination suppression. Despite continual refinement of theoretical models, and inclusion of novel species in empirical studies bringing novel insights in our understanding of sex chromosome evolution, many questions remain unanswered. What is the role of sexually antagonistic selection in their evolution? What are the proximate and ultimate causes of recombination suppression? What is the pace of genetic divergence and degeneration of sex chromosomes? Why do rates of sex chromosome turnover vary so drastically across taxa? To summarize, understanding why, how, and when sex chromosomes evolve is still a major goal in evolutionary biology.
Acknowledgments
The authors thank the Molecular Biology and Evolution Editors-in-Chief for inviting them to write this Perspective. The authors also acknowledge Thomas Lenormand, Tatiana Giraud and referees for their feedback on the manuscript. The authors are grateful to Alice Mazel for the drawings of Fig. 1.
Contributor Information
Paul A Saunders, CEFE, University of Montpellier, CNRS, EPHE, IRD, Montpellier, France.
Aline Muyle, CEFE, University of Montpellier, CNRS, EPHE, IRD, Montpellier, France.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
References
- Almeida P, Sandkam BA, Morris J, Darolti I, Breden F, Mank JE. Divergence and remarkable diversity of the Y chromosome in guppies. Mol Biol Evol. 2021:38(2):619–633. 10.1093/molbev/msaa257. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Atanassov I, Delichère C, Filatov DA, Charlesworth D, Negrutiu I, Monéger F. 2001. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol Biol Evol. 18(12):2162–2168. 10.1093/oxfordjournals.molbev.a003762. [Abstract] [CrossRef] [Google Scholar]
- Bachtrog D. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 2006:16(6):578–585. 10.1016/j.gde.2006.10.007. [Abstract] [CrossRef] [Google Scholar]
- Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008:179(3):1513–1525. 10.1534/genetics.107.084012. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, Hahn MW, Kitano J, Mayrose I, Ming R, et al. . Sex determination: why so many ways of doing it? PLoS Biol. 2014:12(7):e1001899. 10.1371/journal.pbio.1001899. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Baird RB, Urban JM, Mongue AJ, Jaron KS, Hodson CN, Grewoldt M, Martin SH, Ross L. Recent evolution of a maternally acting sex-determining supergene in a fly with single-sex broods. Mol Biol Evol. 2023:40(7):msad148. 10.1093/molbev/msad148. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Balounova V, Gogela R, Cegan R, Cangren P, Zluvova J, Safar J, Kovacova V, Bergero R, Hobza R, Vyskot B, et al. . Evolution of sex determination and heterogamety changes in section Otites of the genus Silene. Sci Rep. 2019:9(1):1045. 10.1038/s41598-018-37412-x. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949:163(4148):676–677. 10.1038/163676a0. [Abstract] [CrossRef] [Google Scholar]
- Beaudry FEG, Barrett SCH, Wright SI. Genomic loss and silencing on the Y chromosomes of Rumex. Genome Biol Evol. 2017:9(12):3345–3355. 10.1093/gbe/evx254. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bechsgaard J, Schou MF, Vanthournout B, Hendrickx F, Knudsen B, Settepani V, Schierup MH, Bilde T. Evidence for faster X chromosome evolution in spiders. Mol Biol Evol. 2019:36(6):1281–1293. 10.1093/molbev/msz074. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Behrens KA, Koblmueller S, Kocher TD. Diversity of sex chromosomes in vertebrates: six novel sex chromosomes in basal haplochromines (Teleostei: Cichlidae). Genome Biol Evol. 2024:16(7):evae152. 10.1093/gbe/evae152. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bergero R, Charlesworth D. 2019. Reply to Wright et al.: how to explain the absence of extensive Y-specific regions in the guppy sex chromosomes. Proc Natl Acad Sci U S A. 116(26):12609–12610. 10.1073/pnas.1906633116. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bergero R, Gardner J, Bader B, Yong L, Charlesworth D. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc Natl Acad Sci U S A. 2019:116(14):6924–6931. 10.1073/pnas.1818486116. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bergero R, Qiu S, Charlesworth D. Gene loss from a plant sex chromosome system. Curr Biol. 2015:25(9):1234–1240. 10.1016/j.cub.2015.03.015. [Abstract] [CrossRef] [Google Scholar]
- Bernasconi G, Antonovics J, Biere A, Charlesworth D, Delph LF, Filatov D, Giraud T, Hood ME, Marais GB, McCauley D, et al. . Silene as a model system in ecology and evolution. Heredity (Edinb). 2009:103(1):5–14. 10.1038/hdy.2009.34. [Abstract] [CrossRef] [Google Scholar]
- Berner D, Ruffener S, Blattner LA. Chromosome-level assemblies of the Pieris mannii butterfly genome suggest Z-origin and rapid evolution of the W chromosome. Genome Biol Evol. 2023:15(6):evad111. 10.1093/gbe/evad111. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution. 2013:67(3):635–645. 10.1111/j.1558-5646.2012.01810.x. [Abstract] [CrossRef] [Google Scholar]
- Blavet N, Blavet H, Muyle A, Kafer J, Cegan R, Deschamps C, Zemp N, Mousset S, Aubourg S, Bergero R, et al. . Identifying new sex-linked genes through BAC sequencing in the dioecious plant Silene latifolia. BMC Genomics. 2015:16(1):1–8. 10.1186/s12864-015-1698-7. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Böhne A, Weber AA-T, Rajkov J, Rechsteiner M, Riss A, Egger B, Salzburger W. Repeated evolution versus common ancestry: sex chromosome evolution in the haplochromine cichlid Pseudocrenilabrus philander. Genome Biol Evol. 2019:11(2):439–458. 10.1093/gbe/evz003. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bracewell R, Bachtrog D. Complex evolutionary history of the Y chromosome in flies of the Drosophila obscura species group. Genome Biol Evol. 2020:12(5):494–505. 10.1093/gbe/evaa051. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bracewell R, Tran A, Chatla K, Bachtrog D. Sex chromosome evolution in beetles. bioRxiv 524646. 10.1101/2023.01.18.524646, 20 January 2023, preprint: not peer reviewed. [CrossRef]
- Brelsford A, Lavanchy G, Sermier R, Rausch A, Perrin N. Identifying homomorphic sex chromosomes from wild-caught adults with limited genomic resources. Mol Ecol Resour. 2017:17(4):752–759. 10.1111/1755-0998.12624. [Abstract] [CrossRef] [Google Scholar]
- Bridges CB. Sex in relation to chromosomes and genes. Am Nat. 1925:59(661):127–137. 10.1086/280023. [CrossRef] [Google Scholar]
- Brooks L. The evolution of recombination rates. In: Michod RE, Levin BR, editors. The evolution of sex. Sunderland (MA): Sinauer; 1988. p. 87–105. [Google Scholar]
- Brown EJ, Nguyen AH, Bachtrog D. The Drosophila Y chromosome affects heterochromatin integrity genome-wide. Mol Biol Evol. 2020:37(10):2808–2824. 10.1093/molbev/msaa082. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bull JJ, Charnov EL. Changes in the heterogametic mechanism of sex determination. Heredity (Edinb). 1977:39(1):1–14. 10.1038/hdy.1977.38. [Abstract] [CrossRef] [Google Scholar]
- Cabrera AA, Rey-Iglesia A, Louis M, Skovrind M, Westbury MV, Lorenzen ED. How low can you go? Introducing SeXY: sex identification from low-quantity sequencing data despite lacking assembled sex chromosomes. Ecol Evol. 2022:12(8):e9185. 10.1002/ece3.9185. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Carpentier F, Rodríguez de la Vega RC, Jay P, Duhamel M, Shykoff JA, Perlin MH, Wallen RM, Hood ME, Giraud T. Tempo of degeneration across independently evolved nonrecombining regions. Mol Biol Evol. 2022:39(4):msac060. 10.1093/molbev/msac060. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005:434(7031):400–404. 10.1038/nature03479. [Abstract] [CrossRef] [Google Scholar]
- Carvalho AB, Clark AG. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013:23(11):1894–1907. 10.1101/gr.156034.113. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Carvalho AB, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009:25(6):270–277. 10.1016/j.tig.2009.04.002. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Catalán A, Macias-Muñoz A, Briscoe AD. Evolution of sex-biased gene expression and dosage compensation in the eye and brain of Heliconius butterflies. Mol Biol Evol. 2018:35(9):2120–2134. 10.1093/molbev/msy111. [Abstract] [CrossRef] [Google Scholar]
- Cecalev D, Viçoso B, Galupa R. Compensation of gene dosage on the mammalian X. Development. 2024:151(15):dev202891. 10.1242/dev.202891. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. Am Nat. 1978:112(988):975–997. 10.1086/283342. [CrossRef] [Google Scholar]
- Charlesworth B, Olito C. Making sense of recent models of the “sheltering” hypothesis for recombination arrest between sex chromosomes. Evolution. 2024. 10.1093/evolut/qpae119. [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D. The guppy sex chromosome system and the sexually antagonistic polymorphism hypothesis for Y chromosome recombination suppression. Genes (Basel). 2018:9(5):264. 10.3390/genes9050264. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D. Why and how do Y chromosome stop recombining? J Evol Biol. 2023:36(3):632–636. 10.1111/jeb.14137. [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D, Bergero R, Graham C, Gardner J, Yong L. Locating the sex determining region of linkage group 12 of guppy (Poecilia reticulata). G3 (Bethesda). 2020a:10(10):3639–3649. 10.1534/g3.120.401573. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D, Charlesworth B. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res. 1980:35(2):205–214. 10.1017/S0016672300014051. [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D, Graham C, Trivedi U, Gardner J, Bergero R. PromethION sequencing and assembly of the genome of Micropoecilia picta, a fish with a highly degenerated Y chromosome. Genome Biol Evol. 2021:13(9):evab171. 10.1093/gbe/evab171. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D, Qiu S, Bergero R, Gardner J, Keegan K, Yong L, Hastings A, Konczal M. Has recombination changed during the recent evolution of the guppy Y chromosome? Genetics. 2024:226(1):iyad198. 10.1093/genetics/iyad198. [Abstract] [CrossRef] [Google Scholar]
- Charlesworth D, Zhang Y, Bergero R, Graham C, Gardner J, Yong L. Using GC content to compare recombination patterns on the sex chromosomes and autosomes of the guppy, Poecilia reticulata, and its close outgroup species. Mol Biol Evol. 2020b:37(12):3550–3562. 10.1093/molbev/msaa187. [Abstract] [CrossRef] [Google Scholar]
- Chen J, Wang M, He X, Yang J-R, Chen X. The evolution of sex chromosome dosage compensation in animals. J Genet Genomics. 2020:47(11):681–693. 10.1016/j.jgg.2020.10.005. [Abstract] [CrossRef] [Google Scholar]
- Chen X, Zhang J. No X-chromosome dosage compensation in human proteomes. Mol Biol Evol. 2015:32(6):1456–1460. 10.1093/molbev/msv036. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chen X, Zhang J. The X to autosome expression ratio in haploid and diploid human embryonic stem cells. Mol Biol Evol. 2016:33(12):3104–3107. 10.1093/molbev/msw187. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Conte MA, Clark FE, Roberts RB, Xu L, Tao W, Zhou Q, Wang D, Kocher TD. Origin of a giant sex chromosome. Mol Biol Evol. 2021:38(4):1554–1569. 10.1093/molbev/msaa319. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Coolon JD, Stevenson KR, McManus CJ, Yang B, Graveley BR, Wittkopp PJ. Molecular mechanisms and evolutionary processes contributing to accelerated divergence of gene expression on the Drosophila X chromosome. Mol Biol Evol. 2015:32(10):2605–2615. 10.1093/molbev/msv135. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cornejo-Páramo P, Dissanayake DSB, Lira-Noriega A, Martínez-Pacheco ML, Acosta A, Ramírez-Suástegui C, Méndez-de-la-Cruz FR, Székely T, Urrutia AO, Georges A, et al. . Viviparous reptile regarded to have temperature-dependent sex determination has old XY chromosomes. Genome Biol Evol. 2020:12(6):924–930. 10.1093/gbe/evaa104. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014:508(7497):488–493. 10.1038/nature13151. [Abstract] [CrossRef] [Google Scholar]
- Crowson D, Barrett SCH, Wright SI. Purifying and positive selection influence patterns of gene loss and gene expression in the evolution of a plant sex chromosome system. Mol Biol Evol. 2017:34(5):1140–1154. 10.1093/molbev/msx064. [Abstract] [CrossRef] [Google Scholar]
- Dagilis AJ, Sardell JM, Josephson MP, Su Y, Kirkpatrick M, Peichel CL. Searching for signatures of sexually antagonistic selection on stickleback sex chromosomes. Philos Trans R Soc B Biol Sci. 2022:377(1856):20210205. 10.1098/rstb.2021.0205. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Darolti I, Wright AE, Sandkam BA, Morris J, Bloch NI, Farré M, Fuller RC, Bourne GR, Larkin DM, Breden F, et al. . Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc Natl Acad Sci U S A. 2019:116(38):19031–19036. 10.1073/pnas.1905298116. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Dean R, Harrison PW, Wright AE, Zimmer F, Mank JE. Positive selection underlies faster-Z evolution of gene expression in birds. Mol Biol Evol. 2015:32(10):2646–2656. 10.1093/molbev/msv138. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Decroly T, Vila R, Lohse K, Mackintosh A. Rewinding the ratchet: rare recombination locally rescues neo-W degeneration and generates plateaus of sex-chromosome divergence. Mol Biol Evol. 2024:41(7):msae124. 10.1093/molbev/msae124. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Devloo-Delva F, Gosselin T, Butcher PA, Grewe PM, Huveneers C, Thomson RB, Werry JM, Feutry P. An R-based tool for identifying sex-linked markers from restriction site-associated DNA sequencing with applications to elasmobranch conservation. Conservation Genet Resour. 2024:16:11–16. 10.1007/s12686-023-01331-5. [CrossRef] [Google Scholar]
- Dixon G, Kitano J, Kirkpatrick M. The origin of a new sex chromosome by introgression between two stickleback fishes. Mol Biol Evol. 2019:36(1):28–38. 10.1093/molbev/msy181. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Dufresnes C, Brelsford A, Baier F, Perrin N. When sex chromosomes recombine only in the heterogametic sex: heterochiasmy and heterogamety in hyla tree frogs. Mol Biol Evol. 2021:38(1):192–200. 10.1093/molbev/msaa201. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Elkrewi M, Moldovan MA, Picard MAL, Vicoso B. Schistosome W-linked genes inform temporal dynamics of sex chromosome evolution and suggest candidate for sex determination. Mol Biol Evol. 2021:38(12):5345–5358. 10.1093/molbev/msab178. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ellison CE, Bachtrog D. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science. 2013:342(6160):846–850. 10.1126/science.1239552. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- El Taher A, Ronco F, Matschiner M, Salzburger W, Böhne A. Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci Adv. 2021:7(36):eabe8215. 10.1126/sciadv.abe8215. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Endler JA. Natural selection on color patterns in Poecilia reticulata. Evolution. 1980:34(1):76–91. 10.2307/2408316. [Abstract] [CrossRef] [Google Scholar]
- Fan B, Xie D, Li Y, Wang X, Qi X, Li S, Meng Z, Chen X, Peng J, Yang Y, et al. . A single intronic single nucleotide polymorphism in splicing site of steroidogenic enzyme hsd17b1 is associated with phenotypic sex in oyster pompano, Trachinotus anak. Proc R Soc Lond B Biol Sci. 2021:288(1963):20212245. 10.1098/rspb.2021.2245. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Feron R, Pan Q, Wen M, Imarazene B, Jouanno E, Anderson J, Herpin A, Journot L, Parrinello H, Klopp C, et al. . RADSex: a computational workflow to study sex determination using restriction site-associated DNA sequencing data. Mol Ecol Resour. 2021:21(5):1715–1731. 10.1111/1755-0998.13360. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Filatov DA. Substitution rates in a new Silene latifolia sex-linked gene, SlssX/Y. Mol Biol Evol. 2005:22(3):402–408. 10.1093/molbev/msi003. [Abstract] [CrossRef] [Google Scholar]
- Fong LJM, Darolti I, Metzger DCH, Morris J, Lin Y, Sandkam BA, Mank JE. Evolutionary history of the Poecilia picta sex chromosomes. Genome Biol Evol. 2023:15:evad030. 10.1093/gbe/evad030. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fraïsse C, Picard MAL, Vicoso B. The deep conservation of the Lepidoptera Z chromosome suggests a non-canonical origin of the W. Nat Commun. 2017:8(1):1486. 10.1038/s41467-017-01663-5. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fraser BA, Whiting JR, Paris JR, Weadick CJ, Parsons PJ, Charlesworth D, Bergero R, Bemm F, Hoffmann M, Kottler VA, et al. . Improved reference genome uncovers novel sex-linked regions in the guppy (Poecilia reticulata). Genome Biol Evol. 2020:12(10):1789–1805. 10.1093/gbe/evaa187. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Furman BLS, Evans BJ. Divergent evolutionary trajectories of two young, homomorphic, and closely related sex chromosome systems. Genome Biol Evol. 2018:10(3):742–755. 10.1093/gbe/evy045. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, Shu JJ, Mank JE. Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol. 2020:12(6):750–763. 10.1093/gbe/evaa081. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Geraldes A, Rambo T, Wing RA, Ferrand N, Nachman MW. Extensive gene conversion drives the concerted evolution of paralogous copies of the SRY gene in European rabbits. Mol Biol Evol. 2010:27(11):2437–2440. 10.1093/molbev/msq139. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, et al. . Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012:487(7406):254–258. 10.1038/nature11171. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Grayson P, Wright A, Garroway CJ, Docker MF.. SexFindR: a computational workflow to identify young and old sex chromosomes. bioRxiv 481346. 10.1101/2022.02.21.481346, 22 February 2022, preprint: not peer reviewed. [CrossRef]
- Gu H, Wen J, Zhao X, Zhang X, Ren X, Cheng H, Qu L. Evolution, inheritance, and strata formation of the W chromosome in duck (Anas platyrhynchos). Genome Biol Evol. 2023:15(11):evad183. 10.1093/gbe/evad183. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gu L, Walters JR. Evolution of sex chromosome dosage compensation in animals: a beautiful theory, undermined by facts and bedeviled by details. Genome Biol Evol. 2017:9(9):2461–2476. 10.1093/gbe/evx154. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gu L, Walters JR, Knipple DC. Conserved patterns of sex chromosome dosage compensation in the Lepidoptera (WZ/ZZ): insights from a moth neo-Z chromosome. Genome Biol Evol. 2017:9(3):802–816. 10.1093/gbe/evx039. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012:4(11):1118–1126. 10.1093/gbe/evs086. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hejníčková M, Koutecký P, Potocký P, Provazníková I, Voleníková A, Dalíková M, Visser S, Marec F, Zrzavá M. Absence of W chromosome in Psychidae moths and implications for the theory of sex chromosome evolution in Lepidoptera. Genes (Basel). 2019:10(12):1016. 10.3390/genes10121016. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hellborg L, Ellegren H. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol Biol Evol. 2004:21(1):158–163. 10.1093/molbev/msh008. [Abstract] [CrossRef] [Google Scholar]
- Hill PL, Burridge CP, Ezaz T, Wapstra E. Conservation of sex-linked markers among conspecific populations of a viviparous skink, Niveoscincus ocellatus, exhibiting genetic and temperature-dependent sex determination. Genome Biol Evol. 2018:10(4):1079–1087. 10.1093/gbe/evy042. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Höök L, Leal L, Talla V, Backström N. Multilayered tuning of dosage compensation and Z-chromosome masculinization in the wood white (Leptidea sinapis) butterfly. Genome Biol Evol. 2019:11(9):2633–2652. 10.1093/gbe/evz176. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hose J, Yong CM, Sardi M, Wang Z, Newton MA, Gasch AP. Dosage compensation can buffer copy-number variation in wild yeast. Elife. 2015:4:e05462. 10.7554/eLife.05462. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Houde AE, Endler JA. Correlated evolution of female mating preferences and male color patterns in the guppy Poecilia reticulata. Science. 1990:248(4961):1405–1408. 10.1126/science.248.4961.1405. [Abstract] [CrossRef] [Google Scholar]
- Huylmans AK, Macon A, Vicoso B. Global dosage compensation is ubiquitous in Lepidoptera, but counteracted by the masculinization of the Z chromosome. Mol Biol Evol. 2017:34(10):2637–2649. 10.1093/molbev/msx190. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Huylmans AK, Parsch J. Variation in the X: autosome distribution of male-biased genes among Drosophila melanogaster tissues and its relationship with dosage compensation. Genome Biol Evol. 2015:7(7):1960–1971. 10.1093/gbe/evv117. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Huylmans AK, Toups MA, Macon A, Gammerdinger WJ, Vicoso B. Sex-biased gene expression and dosage compensation on the Artemia franciscana Z-chromosome. Genome Biol Evol. 2019:11(4):1033–1044. 10.1093/gbe/evz053. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jay P, Jeffries D, Hartmann FE, Véber A, Giraud T. Why do sex chromosomes progressively lose recombination? Trends Genet. 2024:40(7):564–579. 10.1016/j.tig.2024.03.005. [Abstract] [CrossRef] [Google Scholar]
- Jay P, Tezenas E, Véber A, Giraud T. Sheltering of deleterious mutations explains the stepwise extension of recombination suppression on sex chromosomes and other supergenes. PLoS Biol. 2022:20(7):e3001698. 10.1371/journal.pbio.3001698. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jeffries DL, Gerchen JF, Scharmann M, Pannell JR. A neutral model for the loss of recombination on sex chromosomes. Philos Trans R Soc B Biol Sci. 2021:376(1832):20200096. 10.1098/rstb.2020.0096. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN, Canestrelli D, Crochet P-A, Dufresnes C, et al. . A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat Commun. 2018:9(1):4088. 10.1038/s41467-018-06517-2. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jiang X, Biedler JK, Qi Y, Hall AB, Tu Z. Complete dosage compensation in Anopheles stephensi and the evolution of sex-biased genes in mosquitoes. Genome Biol Evol. 2015:7(7):1914–1924. 10.1093/gbe/evv115. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Käfer J. How sex chromosomes get trapped into nonrecombination. PLoS Biol. 2022:20(7):e3001718. 10.1371/journal.pbio.3001718. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Käfer J, Bewick A, Andres-Robin A, Lapetoule G, Harkess A, Caïus J, Fogliani B, Gâteblé G, Ralph P, dePamphilis CW, et al. . A derived ZW chromosome system in Amborella trichopoda, representing the sister lineage to all other extant flowering plants. New Phytol. 2022:233(4):1636–1642. 10.1111/nph.17662. [Abstract] [CrossRef] [Google Scholar]
- Käfer J, Lartillot N, Marais GAB, Picard F. Detecting sex-linked genes using genotyped individuals sampled in natural populations. Genetics. 2021:218(2):iyab053. 10.1093/genetics/iyab053. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kaiser VB, Bergero R, Charlesworth D. Slcyt, a newly identified sex-linked gene, has recently moved onto the X chromosome in Silene latifolia (Caryophyllaceae). Mol Biol Evol. 2009:26(10):2343–2351. 10.1093/molbev/msp141. [Abstract] [CrossRef] [Google Scholar]
- Kaiser VB, Zhou Q, Bachtrog D. Nonrandom gene loss from the Drosophila miranda neo-Y chromosome. Genome Biol Evol. 2011:3:1329–1337. 10.1093/gbe/evr103. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kalita AI, Marois E, Kozielska M, Weissing FJ, Jaouen E, Möckel MM, Rühle F, Butter F, Basilicata MF, Keller Valsecchi CI. The sex-specific factor SOA controls dosage compensation in Anopheles mosquitoes. Nature. 2023:623(7985):175–182. 10.1038/s41586-023-06641-0. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kaufmann P, Wiberg RAW, Papachristos K, Scofield DG, Tellgren-Roth C, Immonen E. Y-linked copy number polymorphism of target of rapamycin is associated with sexual size dimorphism in seed beetles. Mol Biol Evol. 2023:40(8):msad167. 10.1093/molbev/msad167. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kazama Y, Kitoh M, Kobayashi T, Ishii K, Krasovec M, Yasui Y, Abe T, Kawano S, Filatov DA. A CLAVATA3-like gene acts as a gynoecium suppression function in white campion. Mol Biol Evol. 2022:39(10):msac195. 10.1093/molbev/msac195. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kent TV, Uzunović J, Wright SI. Coevolution between transposable elements and recombination. Philos Trans R Soc B Biol Sci. 2017:372(1736):20160458. 10.1098/rstb.2016.0458. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kirkpatrick M, Sardell JM, Pinto BJ, Dixon G, Peichel CL, Schartl M. Evolution of the canonical sex chromosomes of the guppy and its relatives. G3 (Bethesda). 2022:12(2):jkab435. 10.1093/g3journal/jkab435. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991:351(6322):117–121. 10.1038/351117a0. [Abstract] [CrossRef] [Google Scholar]
- Kostmann A, Augstenová B, Frynta D, Kratochvíl L, Rovatsos M. Cytogenetically elusive sex chromosomes in scincoidean lizards. Int J Mol Sci. 2021:22(16):8670. 10.3390/ijms22168670. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kozielska M, Weissing FJ, Beukeboom LW, Pen I. Segregation distortion and the evolution of sex-determining mechanisms. Heredity (Edinb). 2010:104(1):100–112. 10.1038/hdy.2009.104. [Abstract] [CrossRef] [Google Scholar]
- Krasovec M, Chester M, Ridout K, Filatov DA. The mutation rate and the age of the sex chromosomes in Silene latifolia. Curr Biol. 2018:28(11):1832–1838.e4. 10.1016/j.cub.2018.04.069. [Abstract] [CrossRef] [Google Scholar]
- Krasovec M, Kazama Y, Ishii K, Abe T, Filatov DA. Immediate dosage compensation is triggered by the deletion of Y-linked genes in Silene latifolia. Curr Biol. 2019:29(13):2214–2221.e4. 10.1016/j.cub.2019.05.060. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lemaitre C, Braga MDV, Gautier C, Sagot M-F, Tannier E, Marais GAB. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol Evol. 2009:1:56–66. 10.1093/gbe/evp006. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lenormand T, Engelstädter J, Johnston SE, Wijnker E, Haag CR. Evolutionary mysteries in meiosis. Philos Trans R Soc Lond B Biol Sci. 2016:371(1706):20160001. 10.1098/rstb.2016.0001. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lenormand T, Fyon F, Sun E, Roze D. Sex chromosome degeneration by regulatory evolution. Curr Biol. 2020:30(15):3001–3006.e5. 10.1016/j.cub.2020.05.052. [Abstract] [CrossRef] [Google Scholar]
- Lenormand T, Roze D. Y recombination arrest and degeneration in the absence of sexual dimorphism. Science. 2022:375(6581):663–666. 10.1126/science.abj1813. [Abstract] [CrossRef] [Google Scholar]
- Lenormand T, Roze D. Can mechanistic constraints on recombination reestablishment explain the long-term maintenance of degenerate sex chromosomes? Peer Community J. 2024:4. 10.24072/pcjournal.373. [CrossRef] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol Biol Evol. 2003:20(7):1113–1116. 10.1093/molbev/msg131. [Abstract] [CrossRef] [Google Scholar]
- Lewis JJ, Cicconardi F, Martin SH, Reed RD, Danko CG, Montgomery SH. The Dryas iulia genome supports multiple gains of a W chromosome from a B chromosome in butterflies. Genome Biol Evol. 2021:13(7):evab128. 10.1093/gbe/evab128. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li J, Song S, Zhang J. Where are the formerly Y-linked genes in the Ryukyu spiny rat that has lost its Y chromosome? Genome Biol Evol. 2024:16(3):evae046. 10.1093/gbe/evae046. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li M, Zhang R, Fan G, Xu W, Zhou Q, Wang L, Li W, Pang Z, Yu M, Liu Q, et al. . Reconstruction of the origin of a neo-Y sex chromosome and its evolution in the spotted Knifejaw, Oplegnathus punctatus. Mol Biol Evol. 2021:38(6):2615–2626. 10.1093/molbev/msab056. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li Z, Xue AZ, Maeda GP, Li Y, Nabity PD, Moran NA. Phylloxera and aphids show distinct features of genome evolution despite similar reproductive modes. Mol Biol Evol. 2023:40(12):msad271. 10.1093/molbev/msad271. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lucchesi JC, Kuroda MI. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 2015:7(5). 10.1101/cshperspect.a019398. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mahajan S, Bachtrog D. Partial dosage compensation in Strepsiptera, a sister group of beetles. Genome Biol Evol. 2015:7(2):591–600. 10.1093/gbe/evv008. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Malcom CM, Wyckoff GJ, Lahn BT. Genic mutation rates in mammals: local similarity, chromosomal heterogeneity, and X-versus-autosome disparity. Mol Biol Evol. 2003:20(10):1633–1641. 10.1093/molbev/msg178. [Abstract] [CrossRef] [Google Scholar]
- Malone JH, Cho D-Y, Mattiuzzo NR, Artieri CG, Jiang L, Dale RK, Smith HE, McDaniel J, Munro S, Salit M, et al. . Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012:13(4):r28. 10.1186/gb-2012-13-4-r28. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mank JE, Nam K, Ellegren H. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 2010:27(3):661–670. 10.1093/molbev/msp282. [Abstract] [CrossRef] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997:11(2):156–166. 10.1101/gad.11.2.156. [Abstract] [CrossRef] [Google Scholar]
- Marais GAB, Campos PRA, Gordo I. Can intra-Y gene conversion oppose the degeneration of the human Y chromosome? A simulation study. Genome Biol Evol. 2010:2(0):347–357. 10.1093/gbe/evq026. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Martin H, Carpentier F, Gallina S, Godé C, Schmitt E, Muyle A, Marais GA, Touzet P. Evolution of young sex chromosomes in two dioecious sister plant species with distinct sex determination systems. Genome Biol Evol. 2019:11(2):350–361. 10.1093/gbe/evz001. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Martínez-Pacheco M, Tenorio M, Almonte L, Fajardo V, Godínez A, Fernández D, Cornejo-Páramo P, Díaz-Barba K, Halbert J, Liechti A, et al. . Expression evolution of ancestral XY gametologs across all major groups of placental mammals. Genome Biol Evol. 2020:12(11):2015–2028. 10.1093/gbe/evaa173. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Matsunaga S, Hizume M, Kawano S, Kuroiwa T. Cytological analyses in Melandrium album: genome size, chromosome size and fluorescence in situ hybridization. Cytologia (Tokyo). 1994:59(1):135–141. 10.1508/cytologia.59.135. [CrossRef] [Google Scholar]
- Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends Genet. 2013:29(9):537–544. 10.1016/j.tig.2013.05.009. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Metzger DCH, Sandkam BA, Darolti I, Mank JE. Rapid evolution of complete dosage compensation in Poecilia. Genome Biol Evol. 2021:13(7):evab155. 10.1093/gbe/evab155. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Michalovova M, Kubat Z, Hobza R, Vyskot B, Kejnovsky E. Fully automated pipeline for detection of sex linked genes using RNA-seq data. BMC Bioinformatics. 2015:16(1):78. 10.1186/s12859-015-0509-0. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Moraga C, Branco C, Rougemont Q, Veltsos P, Jedlička P, Muyle A, Hanique M, Tannier E, Liu X, Mendoza-Galindo E, et al. The Silene latifolia genome and its giant Y chromosome. bioRxiv 558754. 10.1101/2023.09.21.558754, 22 September 2023, preprint: not peer reviewed. [CrossRef]
- Moretti C, Blanco M, Ialy-Radio C, Serrentino M-E, Gobé C, Friedman R, Battail C, Leduc M, Ward MA, Vaiman D, et al. . Battle of the sex chromosomes: competition between X and Y chromosome-encoded proteins for partner interaction and chromatin occupancy drives multicopy gene expression and evolution in muroid rodents. Mol Biol Evol. 2020:37(12):3453–3468. 10.1093/molbev/msaa175. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Morgan AP, Pardo-Manuel de Villena F. Sequence and structural diversity of mouse Y chromosomes. Mol Biol Evol. 2017:34(12):3186–3204. 10.1093/molbev/msx250. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mrnjavac A, Khudiakova KA, Barton NH, Vicoso B. Slower-X: reduced efficiency of selection in the early stages of X chromosome evolution. Evol Lett. 2023:7(1):4–12. 10.1093/evlett/qrac004. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Müller NA, Kersten B, Leite Montalvão AP, Mähler N, Bernhardsson C, Bräutigam K, Carracedo Lorenzo Z, Hoenicka H, Kumar V, Mader M, et al. . A single gene underlies the dynamic evolution of poplar sex determination. Nat Plants. 2020:6(6):630–637. 10.1038/s41477-020-0672-9. [Abstract] [CrossRef] [Google Scholar]
- Muyle A, Bachtrog D, Marais GAB, Turner JMA. Epigenetics drive the evolution of sex chromosomes in animals and plants. Philos Trans R Soc Lond B Biol Sci. 2021:376(1826):20200124. 10.1098/rstb.2020.0124. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Muyle A, Käfer J, Zemp N, Mousset S, Picard F, Marais GA. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biol Evol. 2016:8(8):2530–2543. 10.1093/gbe/evw172. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Muyle A, Marais GAB, Bačovský V, Hobza R, Lenormand T. Dosage compensation evolution in plants: theories, controversies and mechanisms. Philos Trans R Soc Lond B Biol Sci. 2022:377(1850):20210222. 10.1098/rstb.2021.0222. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Muyle A, Shearn R, Marais GA. The evolution of sex chromosomes and dosage compensation in plants. Genome Biol Evol. 2017:9(3):627–645. 10.1093/gbe/evw282. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Muyle A, Zemp N, Deschamps C, Mousset S, Widmer A, Marais GAB. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 2012:10(4):e1001308. 10.1371/journal.pbio.1001308. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Muyle A, Zemp N, Fruchard C, Cegan R, Vrana J, Deschamps C, Tavares R, Hobza R, Picard F, Widmer A, et al. . Genomic imprinting mediates dosage compensation in a young plant XY system. Nat Plants. 2018:4(9):677–680. 10.1038/s41477-018-0221-y. [Abstract] [CrossRef] [Google Scholar]
- Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008:180(2):1131–1136. 10.1534/genetics.108.090324. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Natri HM, Shikano T, Merilä J. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 2013:30(5):1131–1144. 10.1093/molbev/mst035. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nozawa M, Fukuda N, Ikeo K, Gojobori T. Tissue- and stage-dependent dosage compensation on the neo-X chromosome in Drosophila pseudoobscura. Mol Biol Evol. 2014:31(3):614–624. 10.1093/molbev/mst239. [Abstract] [CrossRef] [Google Scholar]
- Nozawa M, Ikeo K, Gojobori T. Gene-by-gene or localized dosage compensation on the neo-X chromosome in Drosophila miranda. Genome Biol Evol. 2018:10:1875–1881. 10.1093/gbe/evy148. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nursyifa C, Brüniche-Olsen A, Garcia-Erill G, Heller R, Albrechtsen A. Joint identification of sex and sex-linked scaffolds in non-model organisms using low depth sequencing data. Mol Ecol Resour. 2022:22(2):458–467. 10.1111/1755-0998.13491. [Abstract] [CrossRef] [Google Scholar]
- Oetjens MT, Shen F, Emery SB, Zou Z, Kidd JM. Y-Chromosome structural diversity in the bonobo and chimpanzee lineages. Genome Biol Evol. 2016:8(7):2231–2240. 10.1093/gbe/evw150. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Okuno M, Mizushima S, Kuroiwa A, Itoh T. Analysis of sex chromosome evolution in the clade palaeognathae from phased genome assembly. Genome Biol Evol. 2021:13(11):evab242. 10.1093/gbe/evab242. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Olito C, Charlesworth B. Do deleterious mutations promote the evolution of recombination suppression between X and Y chromosomes? bioRxiv 568803. 10.1101/2023.11.27.568803, 27 November 2023, preprint: not peer reviewed. [CrossRef]
- Olito C, Ponnikas S, Hansson B, Abbott JK. Consequences of partially recessive deleterious genetic variation for the evolution of inversions suppressing recombination between sex chromosomes. Evolution. 2022:76(6):1320–1330. 10.1111/evo.14496. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Olito C, Ponnikas S, Hansson B, Abbott JK. Consequences of partially recessive deleterious genetic variation for the evolution of inversions suppressing recombination between sex chromosomes. Evolution. 2024:78(8):1499–1510. 10.1093/evolut/qpae060. [Abstract] [CrossRef] [Google Scholar]
- Palmer DH, Rogers TF, Dean R, Wright AE. How to identify sex chromosomes and their turnover. Mol Ecol. 2019:28(21):4709–4724. 10.1111/mec.15245. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Park C, Carrel L, Makova KD. Strong purifying selection at genes escaping X chromosome inactivation. Mol Biol Evol. 2010:27(11):2446–2450. 10.1093/molbev/msq143. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Perrin N. Sex reversal: a fountain of youth for sex chromosomes? Evolution. 2009:63(12):3043–3049. 10.1111/j.1558-5646.2009.00837.x. [Abstract] [CrossRef] [Google Scholar]
- Pessia E, Engelstädter J, Marais GAB. The evolution of X chromosome inactivation in mammals: the demise of Ohno's hypothesis? Cell Mol Life Sci. 2014:71(8):1383–1394. 10.1007/s00018-013-1499-6. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012:109(14):5346–5351. 10.1073/pnas.1116763109. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Picard MAL, Cosseau C, Ferré S, Quack T, Grevelding CG, Couté Y, Vicoso B. Evolution of gene dosage on the Z-chromosome of schistosome parasites. Elife. 2018:7:e35684. 10.7554/eLife.35684. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Picard MAL, Vicoso B, Roquis D, Bulla I, Augusto RC, Arancibia N, Grunau C, Boissier J, Cosseau C. Dosage compensation throughout the Schistosoma mansoni lifecycle: specific chromatin landscape of the Z chromosome. Genome Biol Evol. 2019:11(7):1909–1922. 10.1093/gbe/evz133. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pinto BJ, Nielsen SV, Sullivan KA, Behere A, Keating SE, van Schingen-Khan M, Nguyen TQ, Ziegler T, Pramuk J, Wilson MA, et al. . It's a trap?! Escape from an ancient, ancestral sex chromosome system and implication of Foxl2 as the putative primary sex-determining gene in a lizard (Anguimorpha; Shinisauridae). Evolution. 2024:78:355–363. 10.1093/evolut/qpad205. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linn Soc. 2009:156(1):168–183. 10.1111/j.1096-3642.2008.00481.x. [CrossRef] [Google Scholar]
- Prince EG, Kirkland D, Demuth JP. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linked genes in the flour beetle. Genome Biol Evol. 2010:2:336–346. 10.1093/gbe/evq024. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pucholt P, Wright AE, Conze LL, Mank JE, Berlin S. Recent sex chromosome divergence despite ancient dioecy in the willow Salix viminalis. Mol Biol Evol. 2017:34(8):1991–2001. 10.1093/molbev/msx144. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Puterova J, Razumova O, Martinek T, Alexandrov O, Divashuk M, Kubat Z, Hobza R, Karlov G, Kejnovsky E. Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biol Evol. 2017:9:197–212. 10.1093/gbe/evw303. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Qiu S, Bergero R, Guirao-Rico S, Campos JL, Cezard T, Gharbi K, Charlesworth D. RAD mapping reveals an evolving, polymorphic and fuzzy boundary of a plant pseudoautosomal region. Mol Ecol. 2016:25(1):414–430. 10.1111/mec.13297. [Abstract] [CrossRef] [Google Scholar]
- Qiu S, Bergero R, Zeng K, Charlesworth D. Patterns of codon usage bias in Silene latifolia. Mol Biol Evol. 2011:28(1):771–780. 10.1093/molbev/msq251. [Abstract] [CrossRef] [Google Scholar]
- Qiu S, Yong L, Wilson A, Croft DP, Graham C, Charlesworth D. Partial sex linkage and linkage disequilibrium on the guppy sex chromosome. Mol Ecol. 2022:31(21):5524–5537. 10.1111/mec.16674. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rangavittal S, Harris RS, Cechova M, Tomaszkiewicz M, Chikhi R, Makova KD, Medvedev P. Recovery: k-mer-based read classification for Y-chromosome-specific sequencing and assembly. Bioinformatics. 2018:34(7):1125–1131. 10.1093/bioinformatics/btx771. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Reisser CMO, Fasel D, Hürlimann E, Dukic M, Haag-Liautard C, Thuillier V, Galimov Y, Haag CR. Transition from environmental to partial genetic sex determination in daphnia through the evolution of a female-determining incipient W chromosome. Mol Biol Evol. 2017:34:575–588. 10.1093/molbev/msw251. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Renner SS. Pathways for making unisexual flowers and unisexual plants: moving beyond the “two mutations linked on one chromosome” model. Am J Bot. 2016:103(4):587–589. 10.3732/ajb.1600029. [Abstract] [CrossRef] [Google Scholar]
- Rice W. Sex-chromosomes and the evolution of sexual dimorphism. Evolution. 1984:38(4):735–742. 10.2307/2408385. [Abstract] [CrossRef] [Google Scholar]
- Rice W. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution. 1987:41(4):911–914. 10.2307/2408899. [Abstract] [CrossRef] [Google Scholar]
- Rifkin JL, Beaudry FEG, Humphries Z, Choudhury BI, Barrett SCH, Wright SI. Widespread recombination suppression facilitates plant sex chromosome evolution. Mol Biol Evol. 2021:38(3):1018–1030. 10.1093/molbev/msaa271. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rifkin JL, Hnatovska S, Yuan M, Sacchi BM, Choudhury BI, Gong Y, Rastas P, Barrett SCH, Wright SI. Recombination landscape dimorphism and sex chromosome evolution in the dioecious plant Rumex hastatulus. Philos Trans R Soc Lond B Biol Sci. 2022:377(1850):20210226. 10.1098/rstb.2021.0226. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rodrigues N, Studer T, Dufresnes C, Perrin N. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Mol Biol Evol. 2018:35(4):942–948. 10.1093/molbev/msy008. [Abstract] [CrossRef] [Google Scholar]
- Rovatsos M, Rehák I, Velenský P, Kratochvíl L. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol Biol Evol. 2019:36(6):1113–1120. 10.1093/molbev/msz024. [Abstract] [CrossRef] [Google Scholar]
- Rupp SM, Webster TH, Olney KC, Hutchins ED, Kusumi K, Wilson Sayres MA. Evolution of dosage compensation in Anolis carolinensis, a reptile with XX/XY chromosomal sex determination. Genome Biol Evol. 2017:9:231–240. 10.1093/gbe/evw263. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sacchi B, Humphries Z, Kružlicová J, Bodláková M, Pyne C, Choudhury BI, Gong Y, Bačovský V, Hobza R, Barrett SCH, et al. . Phased assembly of neo-sex chromosomes reveals extensive Y degeneration and rapid genome evolution in Rumex hastatulus. Mol Biol Evol. 2024:41(4):msae074. 10.1093/molbev/msae074. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sardell JM, Josephson MP, Dalziel AC, Peichel CL, Kirkpatrick M. Heterogeneous histories of recombination suppression on stickleback sex chromosomes. Mol Biol Evol. 2021:38(10):4403–4418. 10.1093/molbev/msab179. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sardell JM, Kirkpatrick M. Sex differences in the recombination landscape. Am Nat. 2020:195(2):361–379. 10.1086/704943. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Saunders PA. Sex chromosome turnovers in evolution. eLS. John Wiley & Sons, Ltd; 2019. 10.1002/9780470015902.a0028747. [CrossRef] [Google Scholar]
- Saunders PA, Ferre-Ortega C, Hill PL, Simakov O, Ezaz T, Burridge CP, Wapstra E. Using a handful of transcriptomes to detect sex-linked markers and develop molecular sexing assays in a Species with homomorphic sex chromosomes. Genome Biol Evol. 2024:16(4):evae060. 10.1093/gbe/evae060. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Saunders PA, Neuenschwander S, Perrin N. Sex chromosome turnovers and genetic drift: a simulation study. J Evol Biol. 2018:31(9):1413–1419. 10.1111/jeb.13336. [Abstract] [CrossRef] [Google Scholar]
- Saunders PA, Veyrunes F. Unusual mammalian sex determination systems: a cabinet of curiosities. Genes (Basel). 2021:12(11):1770. 10.3390/genes12111770. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Schield DR, Perry BW, Card DC, Pasquesi GIM, Westfall AK, Mackessy SP, Castoe TA. The rattlesnake W chromosome: a GC-rich retroelement refugium with retained gene function across ancient evolutionary strata. Genome Biol Evol. 2022:14(9):evac116. 10.1093/gbe/evac116. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Schultheiß R, Viitaniemi HM, Leder EH. Spatial dynamics of evolving dosage compensation in a young sex chromosome system. Genome Biol Evol. 2015:7(2):581–590. 10.1093/gbe/evv013. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Shaw DE, Naftaly AS, White MA. Positive selection drives cis-regulatory evolution across the threespine stickleback Y chromosome. Mol Biol Evol. 2024:41(2):msae020. 10.1093/molbev/msae020. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Shearn R, Wright AE, Mousset S, Régis C, Penel S, Lemaitre J-F, Douay G, Crouau-Roy B, Lecompte E, Marais GA. Evolutionary stasis of the pseudoautosomal boundary in strepsirrhine primates. Elife. 2020:9:e63650. 10.7554/eLife.63650. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sigeman H, Sinclair B, Hansson B. Findzx: an automated pipeline for detecting and visualising sex chromosomes using whole-genome sequencing data. BMC Genomics. 2022:23(1):328. 10.1186/s12864-022-08432-9. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sigeman H, Strandh M, Proux-Wéra E, Kutschera VE, Ponnikas S, Zhang H, Lundberg M, Soler L, Bunikis I, Tarka M, et al. . Avian neo-sex chromosomes reveal dynamics of recombination suppression and W degeneration. Mol Biol Evol. 2021:38(12):5275–5291. 10.1093/molbev/msab277. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Slavney A, Arbiza L, Clark AG, Keinan A. Strong constraint on human genes escaping X-inactivation is modulated by their expression level and breadth in both sexes. Mol Biol Evol. 2016:33(2):384–393. 10.1093/molbev/msv225. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Son JH, Meisel RP. Gene-level, but not chromosome-wide, divergence between a very young house fly proto-Y chromosome and its homologous proto-X chromosome. Mol Biol Evol. 2021:38(2):606–618. 10.1093/molbev/msaa250. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Stuart OP, Cleave R, Magrath MJL, Mikheyev AS. Genome of the Lord Howe Island stick insect reveals a highly conserved phasmid X chromosome. Genome Biol Evol. 2023:15(6):evad104. 10.1093/gbe/evad104. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Subrini J, Turner J. Y chromosome functions in mammalian spermatogenesis. eLife. 2021:10. 10.7554/eLife.67345. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Swanepoel CM, Gerlinger ER, Mueller JL. Large X-linked palindromes undergo arm-to-arm gene conversion across Mus lineages. Mol Biol Evol. 2020:37(7):1979–1985. 10.1093/molbev/msaa059. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tenorio M, Cruz-Ruiz S, Encarnación-Guevara S, Hernández M, Corona-Gomez JA, Sheccid-Santiago F, Serwatowska J, López-Perdomo S, Flores-Aguirre CD, Arenas-Moreno DM, et al. . MAYEX is an old long noncoding RNA recruited for X chromosome dosage compensation in a reptile. Science. 2024:385(6715):1347–1354. 10.1126/science.adp1932. [Abstract] [CrossRef] [Google Scholar]
- Terao M, Ogawa Y, Takada S, Kajitani R, Okuno M, Mochimaru Y, Matsuoka K, Itoh T, Toyoda A, Kono T, et al. . Turnover of mammal sex chromosomes in the Sry-deficient Amami spiny rat is due to male-specific upregulation of Sox9. PNAS [Internet]. 2022:119(49):e2211574119. 10.1073/pnas.2211574119. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tomaszkiewicz M, Rangavittal S, Cechova M, Campos Sanchez R, Fescemyer HW, Harris R, Ye D, O’Brien PCM, Chikhi R, Ryder OA, et al. . A time- and cost-effective strategy to sequence mammalian Y chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 2016:26(4):530–540. 10.1101/gr.199448.115. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tomaszkiewicz M, Sahlin K, Medvedev P, Makova KD. Transcript isoform diversity of ampliconic genes on the Y chromosome of great apes. Genome Biol Evol. 2023:15(11):evad205. 10.1093/gbe/evad205. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Toups MA, Vicoso B. The X chromosome of insects likely predates the origin of class Insecta. Evolution. 2023:77(11):2504–2511. 10.1093/evolut/qpad169. [Abstract] [CrossRef] [Google Scholar]
- Trombetta B, Sellitto D, Scozzari R, Cruciani F. Inter- and intraspecies phylogenetic analyses reveal extensive X-Y gene conversion in the evolution of gametologous sequences of human sex chromosomes. Mol Biol Evol. 2014:31(8):2108–2123. 10.1093/molbev/msu155. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Uebbing S, Konzer A, Xu L, Backström N, Brunström B, Bergquist J, Ellegren H. Quantitative mass spectrometry reveals partial translational regulation for dosage compensation in chicken. Mol Biol Evol. 2015:32(10):2716–2725. 10.1093/molbev/msv147. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Uebbing S, Künstner A, Mäkinen H, Ellegren H. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol. 2013:5(8):1555–1566. 10.1093/gbe/evt114. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007:449(7164):909–912. 10.1038/nature06178. [Abstract] [CrossRef] [Google Scholar]
- Vegesna R, Tomaszkiewicz M, Ryder OA, Campos-Sánchez R, Medvedev P, DeGiorgio M, Makova KD. Ampliconic genes on the great ape Y chromosomes: rapid evolution of copy number but conservation of expression levels. Genome Biol Evol. 2020:12(6):842–859. 10.1093/gbe/evaa088. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Veller C, Muralidhar P, Constable GWA, Nowak MA. Drift-induced selection between male and female heterogamety. Genetics. 2017:207(2):711–727. 10.1534/genetics.117.300151. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Veltsos P, Shinde S, Ma W-J. 2024. Sex chromosome evolution: the classical paradigm and so much beyond. Encyclopedia of Evolutionary Biology. In press.
- Veyrunes F, Chevret P, Catalan J, Castiglia R, Watson J, Dobigny G, Robinson TJ, Britton-Davidian J. A novel sex determination system in a close relative of the house mouse. Proc Biol Sci. 2010:277:1049–1056. 10.1098/rspb.2009.1925. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grützner F, Deakin JE, Whittington CM, Schatzkamer K, et al. . Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008:18(6):965–973. 10.1101/gr.7101908. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vicoso B, Bachtrog D. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 2011:3:230–235. 10.1093/gbe/evr010. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vicoso B, Bachtrog D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015:13(4):e1002078. 10.1371/journal.pbio.1002078. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Walters JR, Hardcastle TJ. Getting a full dose? Reconsidering sex chromosome dosage compensation in the silkworm, Bombyx mori. Genome Biol Evol. 2011:3:491–504. 10.1093/gbe/evr036. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Walters JR, Hardcastle TJ, Jiggins CD. Sex chromosome dosage compensation in heliconius butterflies: global yet still incomplete? Genome Biol Evol. 2015:7(9):2545–2559. 10.1093/gbe/evv156. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang T, Gong G, Li Z, Niu J-S, Du W-X, Wang Z-W, Wang Y, Zhou L, Zhang X-J, Lian Z-Q, et al. . Genomic anatomy of homozygous XX females and YY males reveals early evolutionary trajectory of sex-determining gene and sex chromosomes in silurus fishes. Mol Biol Evol. 2024:41(8):msae169. 10.1093/molbev/msae169. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang Z-Y, Leushkin E, Liechti A, Ovchinnikova S, Mößinger K, Brüning T, Rummel C, Grützner F, Cardoso-Moreira M, Janich P, et al. . Transcriptome and translatome co-evolution in mammals. Nature. 2020:588(7839):642–647. 10.1038/s41586-020-2899-z. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Webster TH, Vannan A, Pinto BJ, Denbrock G, Morales M, Dolby GA, Fiddes IT, DeNardo DF, Wilson MA. Lack of dosage balance and incomplete dosage compensation in the ZZ/ZW gila monster (Heloderma suspectum) revealed by de novo genome assembly. Genome Biol Evol. 2024:16(3):evae018. 10.1093/gbe/evae018. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Westergaard M. The mechanism of sex determination in dioecious flowering plants. Adv Genet. 1958:9:217–281. 10.1016/S0065-2660(08)60163-7. [Abstract] [CrossRef] [Google Scholar]
- Whittle CA, Kulkarni A, Extavour CG. Absence of a faster-X effect in beetles (Tribolium, Coleoptera). G3. 2020:10:1125–1136. 10.1534/g3.120.401074. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wilson S, Melissa A, Makova KD. Gene survival and death on the human Y chromosome. Mol Biol Evol. 2013:30:781–787. 10.1093/molbev/mss267. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wright AE, Darolti I, Bloch NI, Oostra V, Sandkam B, Buechel SD, Kolm N, Breden F, Vicoso B, Mank JE. Convergent recombination suppression suggests role of sexual selection in guppy sex chromosome formation. Nat Commun. 2017:8:14251. 10.1038/ncomms14251. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wright AE, Darolti I, Bloch NI, Oostra V, Sandkam BA, Buechel SD, Kolm N, Breden F, Vicoso B, Mank JE. On the power to detect rare recombination events. Proc Natl Acad Sci U S A. 2019:116:12607–12608. 10.1073/pnas.1905555116. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yang W, Wang D, Li Y, Zhang Z, Tong S, Li M, Zhang X, Zhang L, Ren L, Ma X, et al. . A general model to explain repeated turnovers of sex determination in the Salicaceae. Mol Biol Evol. 2021:38:968–980. 10.1093/molbev/msaa261. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yazdi HP, Ellegren H. Old but not (so) degenerated—slow evolution of largely homomorphic sex chromosomes in ratites. Mol Biol Evol. 2014:31:1444–1453. 10.1093/molbev/msu101. [Abstract] [CrossRef] [Google Scholar]
- Yazdi HP, Ellegren H. A genetic map of ostrich Z chromosome and the role of inversions in avian sex chromosome evolution. Genome Biol Evol. 2018:10:2049–2060. 10.1093/gbe/evy163. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012:337:341–345. 10.1126/science.1225385. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zimmer F, Harrison PW, Dessimoz C, Mank JE. Compensation of dosage-sensitive genes on the chicken Z chromosome. Genome Biol Evol. 2016:8:1233–1242. 10.1093/gbe/evw075. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from Molecular Biology and Evolution are provided here courtesy of Oxford University Press
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/169432000
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sex Chromosome Evolution: So Many Exceptions to the Rules.
Genome Biol Evol, 12(6):750-763, 01 Jun 2020
Cited by: 96 articles | PMID: 32315410 | PMCID: PMC7268786
Review Free full text in Europe PMC
Progress and prospects toward our understanding of the evolution of dosage compensation.
Chromosome Res, 17(5):585-602, 01 Jan 2009
Cited by: 73 articles | PMID: 19626444 | PMCID: PMC2758192
Review Free full text in Europe PMC
Sex chromosome evolution: historical insights and future perspectives.
Proc Biol Sci, 284(1854):20162806, 01 May 2017
Cited by: 60 articles | PMID: 28469017 | PMCID: PMC5443938
Sex chromosome dosage compensation: definitely not for everyone.
Trends Genet, 29(12):677-683, 14 Aug 2013
Cited by: 115 articles | PMID: 23953923
Review

 and
and 

