Abstract
Free full text

Vesicle-Mediated Transfer of Virulence Genes from Escherichia coli O157:H7 to Other Enteric Bacteria
Abstract
Membrane vesicles are released from the surfaces of many gram-negative bacteria during growth. Vesicles consist of proteins, lipopolysaccharide, phospholipids, RNA, and DNA. Results of the present study demonstrate that membrane vesicles isolated from the food-borne pathogen Escherichia coli O157:H7 facilitate the transfer of genes, which are then expressed by recipient Salmonella enterica serovar Enteritidis or E. coli JM109. Electron micrographs of purified DNA from E. coli O157:H7 vesicles showed large rosette-like structures, linear DNA fragments, and small open-circle plasmids. PCR analysis of vesicle DNA demonstrated the presence of specific genes from host and recombinant plasmids (hly, L7095, mobA, and gfp), chromosomal DNA (uidA and eaeA), and phage DNA (stx1 and stx2). The results of PCR and the Vero cell assay demonstrate that genetic material, including virulence genes, is transferred to recipient bacteria and subsequently expressed. The cytotoxicity of the transformed enteric bacteria was sixfold higher than that of the parent isolate (E. coli JM109). Utilization of the nonhost plasmid (pGFP) permitted the evaluation of transformation efficiency (ca. 103 transformants μg of DNA−1) and demonstrated that vesicles can deliver antibiotic resistance. Transformed E. coli JM109 cells were resistant to ampicillin and fluoresced a brilliant green. The role vesicles play in genetic exchange between different species in the environment or host has yet to be defined.
Many gram-negative bacteria produce membrane vesicles, suggesting that vesicle production is not purposeless; indeed, studies during the last two decades have presented strong evidence supporting the importance of vesicles. Typical vesicles released from the surfaces of gram-negative bacteria are 50 to 250 nm, spherical, and made up of outer membrane and encapsulated periplasmic components (4, 26). Vesicle components include outer membrane proteins, lipopolysaccharide, periplasmic proteins, phospholipids, DNA, and RNA (9, 12, 15, 22, 34, 40). Vesicles from gram-negative bacteria were reported to fuse to both gram-positive and gram-negative bacteria and in some instances to promote lysis of the target cell (28). Moreover, vesicles may function as an alternative secretory pathway (3, 23) and promote adherence of the parent cell to host cells (17, 32). By virtue of their small size, bilayer protecting envelope, and ability to integrate into the membranes of foreign bacteria and to adhere to or be engulfed by eukaryotic cells, a potential role of vesicles in delivery of virulence factors, including enzymes and toxins, is not unlikely (23). In fact, virulence factors associated with the parent strain, including proteases, phospholipases, autolysin, hemolysins, and Shiga toxins, have been isolated from vesicles (3, 22, 26, 28).
Aside from toxic compounds, DNA has also been isolated from vesicles. Vesicles produced by Pseudomonas aeruginosa were reported to contain DNA (22). Vesicles released by Neisseria gonorrhoeae harbor both linear and circular DNA, including 4.2- and 7.1-kb plasmids (12). Chromosomal and bacteriophage-associated virulence genes were detected in Escherichia coli O157:H7 vesicles (26). Moreover, this research demonstrated that DNA was protected from digestion by DNase, suggesting that DNA is packaged within vesicles (26).
Bacterial evolution often proceeds by horizontal gene transfer between different genera and species (1, 7). Antibiotic resistance genes and pathogenicity islands have been acquired by a variety of pathogens, including E. coli, Salmonella enterica serovar Typhimurium, Yersinia pestis, Dichelobacter nodosis, and Helicobacter pylori (19). Virulence factors contributing to the pathogenicity of E. coli O157:H7, including Shiga toxins (45, 46) and intimin (31, 44), are encoded on pathogenicity islands in the O157 chromosome and are thought to have been acquired by horizontal transfer. Results of previous studies suggest that vesicles may be involved in the transfer of genetic material among similar bacterial species (8, 12, 26). The hypothesis has been put forth that vesicles influence antibiotic resistance in other bacteria in two ways: by physical dissemination of preformed antibiotic-inactivating enzymes into the recipient periplasm and by delivery of antibiotic resistance plasmids (3, 12). Competent Haemophilus influenzae produces vesicles which are released into the medium when cells are returned to normal growth conditions or a noncompetent state (8). Specific DNA-binding peptides were reported to be present on the surfaces of H. influenzae vesicles (24, 25) and to be associated with vesicles from N. gonorrhoeae (11).
Previously, it was reported that vesicles released by E. coli O157:H7 into culture medium contain virulence genes and Shiga toxin (26). In the present study, we demonstrate that E. coli O157:H7 vesicles mediate the transfer of virulence genes, which are subsequently expressed by recipient enteric bacteria. Moreover, the origin of the DNA in E. coli O157:H7 vesicles is elucidated. Observations show that in addition to bacteriophage-associated genes, E. coli O157:H7 vesicles contain plasmids and fragments of chromosomal DNA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli O157:H7 (ATCC 43895), E. coli JM109 (Promega, Madison, Wis.), and Salmonella enterica serovar Enteritidis (ATCC 13076) were grown in Luria-Bertani (LB) broth (Difco, Detroit, Mich.) at 37°C with shaking (200 rpm). Transformation of pGFP (Clontech, Palo Alto, Calif.), which encodes green fluorescent protein (GFP), was performed using the calcium chloride method (43). Following transformation, E. coli O157:H7(pGFP) cells were grown on LB agar containing 100 μg of ampicillin ml−1.
Vesicle isolation.
Vesicles were isolated from early-stationary-phase (14-h) cultures of E. coli O157:H7 and E. coli O157:H7(pGFP) according to the method of Kadurugamuwa and Beveridge (22). In brief, tryptic soy broth (150 ml) was inoculated with 106 E. coli cells and incubated at 37°C for 15 h with shaking (150 rpm). Following incubation, cells were pelleted by centrifugation (10,000 × g, 10 min, 4°C), and the supernatant was decanted and passed through a 0.22-μm-pore-size filter (Micron Separations Inc., Westboro, Mass.) to remove remaining cells and cellular debris. Vesicles were collected by centrifugation (150,000 × g, 3 h, 4°C) (Ti 45 rotor; Beckman Instruments, Inc., Fullerton, Calif.), washed, resuspended in 50 mM HEPES (Fisher Scientific, Pittsburgh, Pa.) supplemented with 0.5 mM dithiothreitol (Sigma Chemical Co., St. Louis, Mo.), and stored at −20°C until needed. Vesicle preparations were checked for the presence of E. coli by surface plating of the vesicle suspension on Trypticase soy agar and by electron microscopy. Vesicle preparations were treated with DNase (DNase I from bovine pancreas; Sigma Chemical Co.), as described by Kolling and Matthews (26).
Electron microscopy.
DNA associated with E. coli O157:H7 vesicles was visualized using DNA shadowing as described by Inman and Schnos (21). Briefly, DNA isolated from vesicles was adsorbed onto collodion-coated copper grids (400 mesh) and stained. Rotary shadowing was performed using platinum-palladium. For negative staining, 5 μl of vesicle suspension was layered onto a Formvar-coated copper grid (300 mesh) and wicked off after 10 s of contact. A 5-μl aliquot of 2% uranyl acetate was used to stain vesicles. Samples were visualized using a JEM-1230 electron microscope (JEOL USA, Inc., Peabody, Mass.).
DNA isolation.
Total cellular DNA was isolated from E. coli and Salmonella serovar Enteritidis cells using guanidium thiocyanate according to the method of Pitcher et al. (41). Prior to DNA isolation from vesicle preparations, samples (200 μl) were treated with DNase to hydrolyze surface-associated DNA and free DNA in the suspension. Reactions were stopped by heat treatment at 80°C for 10 min. DNase-treated vesicles were lysed with 0.125% Triton X-100 solution (30 min at 37°C), and DNA was purified using a glass fiber matrix binding and elution technique according to the manufacturer's directions (Amersham Pharmacia Biotech, Piscataway, N.J.). DNA was resuspended in 20 μl of TE (10 mM Tris, 1 mM EDTA; pH 8.0).
PCR.
All PCRs were performed in a total volume of 50 μl containing 4 μg of each primer ml−1, a 0.2 mM concentration of each deoxynucleoside triphosphate (New England Biolabs, Inc., Beverly, Mass.), 10× PCR buffer (Sigma), nucleotide-free water (Promega), and 2.5 U of Taq polymerase (Sigma). DNA primers used for amplification of genes are listed in Table Table1.1. Primers were not designed to have a common melting temperature. The DNA template was either whole transformed E. coli JM109 cells, DNase-treated vesicles (~4 ng of DNA), or purified vesicle DNA (~20 ng). An E. coli O157:H7 whole-cell suspension or cellular DNA was used as a positive control, and E. coli JM109 was used as a negative control. Cell pellets were resuspended in TE to a concentration of 108 cells ml−1, and 4 μl was used for PCR. PCRs were carried out in a GeneAmp PCR system 2400 thermocycler (Perkin-Elmer Corp., Foster City, Calif.). Reactions were started with 1 cycle of 5 min at 94°C, 2 min at 50°C, and 3 min at 72°C and continued with 25 cycles of 45 s at 94°C, 90 s at 50°C, and 120 s at 72°C. The reaction was completed with an extension step of 10 min at 72°C. PCR products were separated on agarose gels by electrophoresis, stained with ethidium bromide, and visualized by UV transillumination.
TABLE 1
DNA primers used for amplification of specific genes
genes
| Gene | Primer name | Primer sequence (5′→3′) | Gene source (size [kb]) | Product size (bp) | Reference or source |
|---|---|---|---|---|---|
| mobA | n-mobA | CGCGCAGGCGGGCTATCAGTC | p4821 (3.3) | 576 | 18 |
| c-mobA | CCAAAGCCACATCATTTGC | ||||
| cdaA | n-cdaA | CTCGATGGTCGTGTGGCCG | pColD157 (6.7) | 632 | 20 |
| c-cdaA | GGAGTATTCATCTGACCCACAC | ||||
| L7095 | n-L7095 | CAGTTCGCTCGTAAAGCAG | pO157 (92) | 668 | 6, 29 |
| c-L7095 | GTATAAATGGCTCTATGTCTAC | ||||
| hlyCAa | n-hlyCA | CTTTTGACGTCATGGGGAAGG | pO157 (92) | 792 | 6, 29 |
| c-hlyCA | CGAATATTGCAACACCACGTTCAG | ||||
| stx1 | stx-1F | ACACTGGATGATCTCAGTGG | Phage 933J | 614 | 16 |
| stx-1R | CTGAATCCCCCTCCATTATG | ||||
| stx2 | stx-2F | CCATGACAACGGACAGCAGT | Temperate phage 933W | 779 | 16 |
| stx-2R | CCTGTCAACTGAGCACTTTG | ||||
| eaeA | n-eae | CCCGAATTCGGCACAAGCATAAGC | Chromosomal DNA | 863 | 44 |
| c-eae | CCCGAATCCGTCTCGCCAGTATTCG | ||||
| uidA | n-uidA | CTCTACACCACGCCGAACAC | Chromosomal DNA | 922 | 26 |
| c-uidA | CCTTCTCTGCCGTTTCCAAAT | ||||
| 933W | W61311F | GTCAGTATGCGATAGCTCTG | DNA endpoints of 933W | 660 | 42 |
| W300R | GCGACATGGGATTCATTCAGC | ||||
| gfp | n-gfp | ATGAGTAAAGGAGAAGAACTTTTC | pGFP | 800 | Clontech |
| c-gfp | CGAAAGGGCCCGTACGGCCG |
Vesicle-mediated transformation.
E. coli JM109 and Salmonella serovar Enteritidis were cultured in LB broth at 37°C with shaking (200 rpm) for 3 to 4 h to an optical density at 600 nm (OD600) of 0.6. Cells were pelleted and resuspended in cold SOC medium (Bio 101, Vista, Calif.) to a concentration of approximately 107 cells ml−1 (for GFP transformation, cells were concentrated to approximately 1011 cells ml−1). The cell suspension (100 μl) was mixed with 800 μl of SOC medium and 100 μl of vesicle suspension (approximately 1.65 ng of DNA ml−1) and DNase (final concentration, 1 ng ml−1). The suspension was incubated statically at 37°C for 1 h and then for an additional 2 h with shaking (150 rpm). Ten milliliters of LB broth was added to the suspension, and incubation in LB broth continued at 37°C for 20 h with shaking (200 rpm). Samples (2 ml) for the Vero cell assay (as described below) were collected after 5 and 20 h of incubation after the addition of LB broth. Samples were centrifuged, and the supernatant was collected and passed through a 0.22-μm-pore-size filter (Micron Separations Inc.) to remove remaining cells. Samples were either used immediately or stored (at −20°C) for later use. Controls included E. coli O157:H7, E. coli O157:H7 vesicles alone (no cells), Salmonella serovar Enteritidis, and E. coli JM109 either alone or with total DNA isolated from E. coli O157:H7. For PCR experiments, cells were harvested, pelleted, washed twice, and resuspended in TE buffer. All experiments were done in duplicate and repeated twice. Bacteriophages, if present, may contaminate vesicle preparations during isolation. To ensure that vesicles were responsible for DNA transformation and not bacteriophages (2, 37), vesicle samples were treated with 50 μg of proteinase K (Sigma) ml−1 for 30 min at 37°C to hydrolyze phage coats and release phage contents. Prior to transformation, proteinase K was removed from treated samples using microconcentrators that exclude molecules with an Mr of <30,000 (Millipore Corp., Bedford, Mass.). DNase was added to treated vesicle preparations to degrade released DNA. To further demonstrate that DNA transfer was vesicle mediated, separate experiments were conducted using vesicles that were hydrolyzed with 50 mg of lysozyme ml−1 at 37°C for 30 min. A control experiment was conducted using 20 ng of purified total E. coli O157:H7 DNA.
DNA packaging.
Vesicles were isolated from E. coli O157:H7(pGFP) to investigate packaging and transfer of nonhost (the term “nonhost” is used to underscore the idea that pGFP is not naturally associated with E. coli O157:H7) DNA. Transformation was completed as described above, and E. coli JM109 cells were plated onto LB agar supplemented with 100 μg of ampicillin ml−1 and incubated at 37°C for 24 h. Ampicillin-resistant colonies, which fluoresce a brilliant green upon exposure to UV light, were considered transformants.
Vero cell assay.
The Vero cell assay was completed as described previously (26, 33). In short, Vero cells suspended in growth medium (Basal Medium Eagle, 15% fetal bovine serum; pH 7.1) (Sigma) were seeded into wells (2 × 104 cells/well) of a 96-well microtiter plate and incubated for 24 h at 37°C. Growth medium was removed by aspiration and replaced with fresh medium, and 100 μl of the filtered supernatants (obtained as described above) was added to the first row of wells. Serial dilutions (1:2) were made, and the plates were incubated for 48 h. After 48 h, 25 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; 5 mg ml−1) was added to each well without removal of supernatants. Controls included E. coli O157:H7 (ATCC 43895), E. coli JM109, and Salmonella serovar Enteritidis (ATCC 13076) supernatants and vesicle preparations (treated like the transformed-cell supernatants), bacterial growth medium (LB) alone, and buffer (phosphate-buffered saline) alone. Results are based on cell survival using the following formula: percent live cells = (ODtreated cells/ODcontrol cells) × 100. The assay was performed in triplicate and repeated twice.
Plaque assay.
The presence of phage in vesicle preparations was examined using a plaque assay. Samples (200 μl) of transformed E. coli JM109 cells, either immediately after vesicle transformation or after overnight growth, were diluted (1:10) in buffer (20 mM Tris, 10 mM NaCl, 10 mM MgSO4; pH 7.4), mixed with 3 ml of molten (45°C) TB top agar (1% trypton, 0.5% NaCl, 10 mM MgSO4, and 0.6% agarose), and poured onto the surfaces of LB plates. Plates were incubated at 37°C for 24 h and observed for plaque formation. In plaque assays, phage 933W is capable of forming plaques on E. coli O157:H7 (ATCC 43895) (also called EDL933).
RESULTS
Detection of genetic material within vesicles.
Electron microscopy was performed on DNA isolated from vesicles to determine its origin, i.e., plasmid, chromosome, or phage. Electron photomicrographs show that various forms of DNA are associated with vesicles (Fig. (Fig.1).1). Large rosette-like structures, linear DNA fragments, and supercoiled plasmids were evident; small open-circle plasmids were also visualized (ca. 2 to 4 kb, based on comparisons to known plasmids). The rosette-like structures were frequently observed in vesicle samples.
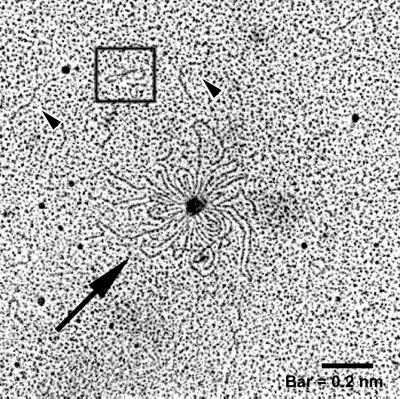
Electron micrograph of DNA isolated from E. coli O157:H7 vesicles. Purified DNase-treated vesicles were lysed using 0.125% Triton X-100, and DNA was isolated and prepared for electron microscopy. Linear DNA (arrowheads), a plasmid (box), and rosette-like structures (arrow) were present.
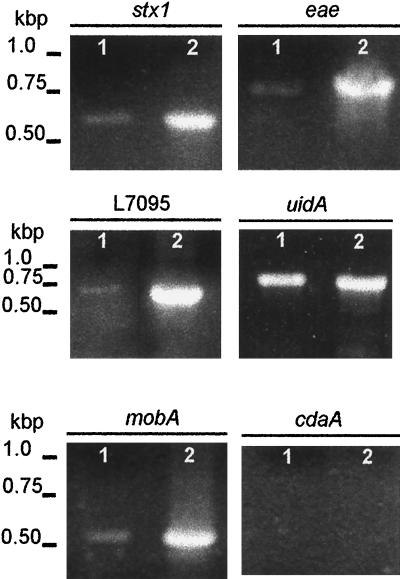
PCR amplification of specific genes provided additional information relating to the origin of vesicle-associated DNA. Ten primer pairs (Table (Table1)1) were used to amplify genes of chromosomal, phage, and plasmid origins. Genes of interest may not be associated with all E. coli O157:H7 isolates; therefore, cellular DNA isolated from strain ATCC 43895 was used as a template in PCRs to determine whether the strain harbors the selected genes (Fig. (Fig.22 and and3).3). The chromosomal genes eaeA (863 bp) and uidA (992 bp), the bacteriophage-related genes stx1 (614 bp) and stx2 (779 bp), and three of the four plasmid-related genes were amplified. Amplification of hlyCA (792 bp), L7095 (668 bp), and mobA (576 bp) suggests that strain ATCC 43895 harbors plasmids pO157 (92 kb) and p4821 (3.3 kb). The primer set n-hlyCA and c-hlyCA was designed to amplify the C- and N-terminus-encoding portions of hlyC and hlyA, respectively, from the pO157 hemolysin operon, hlyCABD. No PCR product was obtained using primers for cdaA, suggesting that the 6.7-kb plasmid pColD157, which is responsible for the colicinogenic phenotype of some E. coli O157 strains, is not carried by strain ATCC 43895 (5, 20). Two primers homologous to phage 933W DNA endpoints that correlate to circular phage DNA were used to amplify a 660-bp fragment from vesicle-associated DNA (42). Vesicles released by E. coli O157:H7 were screened for genes that gave positive results with whole-cell DNA analysis (Fig. (Fig.22 and and3).3). PCR products associated with stx1, eaeA, uidA, mobA, and L7095 were obtained using purified, concentrated (10-fold) vesicle DNA (Fig. (Fig.3).3). Other genes that were screened for were detected using nonconcentrated vesicle suspensions.
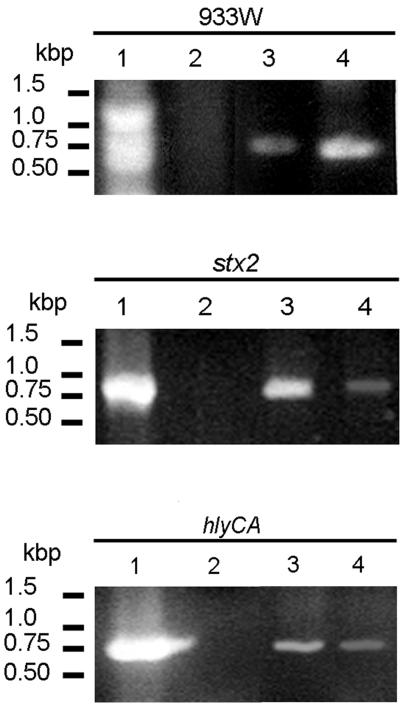
PCR-detected genes transferred by E. coli O157:H7 vesicles to recipient JM109 cells. Lanes 1, E. coli O157:H7 cells; lanes 2, E. coli JM109 cells; lanes 3, intact vesicles; lanes 4, E. coli JM109, vesicle transformed. All genes listed in Table Table11 were screened for, but only positive profiles are shown.
Vesicle-mediated transformation.
Vesicle-mediated transfer of genetic material was determined qualitatively by PCR amplification of the selected E. coli O157:H7 genes using recipient E. coli JM109 cells (Fig. (Fig.3).3). Following transformation and overnight growth in LB broth, cells were concentrated (10,000 × g, 5 min) and washed, and PCR analysis was performed using primers specific for eaeA, stx1, stx2, hlyCA, L7095, mobA, and the endpoints of phage 933W. PCR products corresponding to stx2 and hlyCA were visualized in ethidium bromide-stained agarose gels. The genes listed above were not amplified using nontransformed E. coli JM109. Two fragments of phage 933W were amplified using primers homologous to the phage endpoints, likely indicating the presence of the entire circular phage DNA.
pGFP uptake, transfer, and expression in recipient cells.
E. coli O157:H7 was transformed with pGFP, a derivative of the high-copy-number plasmid pUC18, to determine whether nonhost DNA would be entrapped in vesicles. Vesicles isolated from the transformed strain were analyzed by PCR for the presence of gfp. A single band of the appropriate size (800 bp) was amplified, indicating that plasmid DNA of foreign origin was packaged in the vesicles (Fig. (Fig.4).4). Vesicles (gfp positive) were used to transform E. coli JM109, which was plated onto LB agar alone or supplemented with ampicillin, and the transformation frequency was calculated as the number of transformants in the total cell count. The frequency of transformation of pGFP by vesicles was 3 × 10−10, and transformation efficiency was 103 transformants per μg of DNA based on the assumption that 0.83 ng of DNA is associated with 10 μg of vesicle proteins (26). In fact, the efficiency is higher since pGFP is not the only DNA present in vesicles. No colonies formed in the following controls on LB agar supplemented with ampicillin: GFP-positive vesicles without cells, E. coli JM109 cells without vesicles, and E. coli JM109 cells incubated with 20 ng of purified plasmid ml−1.
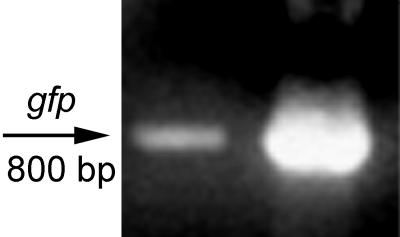
PCR analysis of vesicle DNA for the presence of gfp. Vesicles isolated from E. coli O157:H7 harboring pGFP were treated with DNase and used as a template for PCR analysis with gfp primers. Left lane, vesicles containing gfp; right lane, purified pGFP (control). The molecular size of the expected fragment was 800 bp.
Cytotoxicity of supernatants from transformed enteric bacteria.
The expression of virulence genes by recipient cells was analyzed using the Vero cell assay. Cytotoxic activity was sixfold greater for supernatants from transformed E. coli JM109 than for supernatants from parental E. coli JM109, indicating that Stx is expressed by the recipient cells (Fig. (Fig.5).5). Vesicles were also evaluated in the Vero cell assay to eliminate the possibility that vesicles were solely responsible for cytotoxicity. Cytotoxic activity was greater for supernatants from transformed cells than for supernatants from wild-type cells or vesicles alone. Transformation experiments were also conducted using total DNA from E. coli O157:H7 in place of vesicles; the cytotoxicities of supernatants from these experiments were similar to those for controls (Fig. (Fig.5).5). Treatment of vesicles with proteinase K did not affect the results; however, cytotoxicity decreased when lysozyme-treated vesicles were used, suggesting that vesicles and not phages are responsible for gene transfer (Fig. (Fig.5).5).
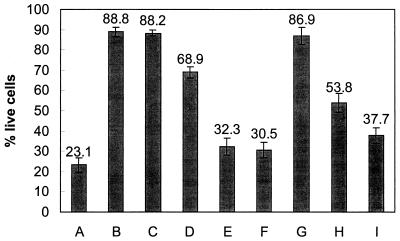
Vero cell cytotoxicity of 20-h supernatants from transformed enteric bacteria. A, E. coli O157:H7 (positive control); B, E. coli JM109 (negative control); C, Salmonella serovar Enteritidis (negative control); D, E. coli O157:H7 vesicles; E, E. coli JM109, vesicle transformed; F, Salmonella serovar Enteritidis, vesicle transformed; G, E. coli JM109 transformed with total DNA purified from E. coli O157:H7; H, E. coli JM109 transformed with lysozyme-treated vesicles; I, E. coli JM109 transformed with proteinase K-treated vesicles. Values were determined as described in Materials and Methods. Error bars indicate the standard errors of the means. The assay was performed in triplicate and repeated twice. Values are from a representative experiment.
Vesicle-mediated transformation of enteric pathogens other than E. coli was determined using Salmonella serovar Enteritidis, a food-borne pathogen. Cell-free supernatants from transformants were used in the Vero cell assay. The cytotoxic activities of the supernatants from transformed Salmonella serovar Enteritidis and E. coli JM109 increased in 20-h supernatants compared to 5-h culture supernatants, suggesting continued expression of Stx by transformants (Table (Table2).2).
TABLE 2
Effect of 5- and 20-h supernatants of transformants in Vero cell cytotoxicity assay
assay
| Incubation time (h)a | Vero cell survival (mean % live cells ± SEM)b with:
| ||
|---|---|---|---|
| Transformed E. coli JM109c | Transformed Salmonella serovar Enteritidisc | Vesiclesd | |
| 5 | 47.3 ± ± 2.8 2.8 | 46.0 ± ± 3.0 3.0 | 64.9 ± ± 3.2 3.2 |
| 20 | 36.4 ± ± 4.1 4.1 | 34.6 ± ± 3.9 3.9 | 68.9 ± ± 2.6 2.6 |
Plaque assay.
E. coli O157:H7 strain ATCC 43895 (also called EDL933) is reported to spontaneously release phage 933W particles (42). The authors of the study indicate that propagation of the phage was unsuccessful in broth culture (as used in the present study); regardless, experiments were conducted to verify that the transfer of genetic material was indeed mediated by vesicles and not by lysogenic bacteriophages. Plunkett et al. (42) reported that phage 933W titers fell more than 20-fold after overnight storage at 4°C despite supplementation with CaCl2, MgCl2, or gelatin. In the present study, vesicle samples were stored for extended periods without supplementation of the above chemicals, and indeed, plaque assay results for putative transformants were negative.
DISCUSSION
In this study, the origin of DNA within membrane vesicles released by the food-borne pathogen E. coli O157:H7 was identified, and vesicle-mediated transformation of enteric pathogens other than E. coli was demonstrated. Vesicles harbored genes of different origins: chromosomal, phage, and plasmid. Electron micrographs of vesicle-associated DNA show the presence of plasmids and linear DNA. Transformation experiments demonstrated that vesicles can export and transfer genetic material to other enteric bacteria. The mechanism(s) by which DNA is entrapped or packaged into vesicles is not yet known. E. coli JM109 transformed with pGFP-positive vesicles also exhibited antibiotic resistance. The β-lactamase gene encoding resistance to ampicillin is carried on pGFP as a selectable marker.
Based on PCR, chromosomally integrated genes are associated with vesicles. The uidA gene is located in the chromosome, and its product, β-d-glucuronidase, is expressed in virtually all E. coli isolates (13, 14). The eaeA gene is located in a 35-kb chromosomal pathogenicity element and encodes intimin, a protein involved in the attachment of E. coli O157:H7 to epithelial cells (10, 30). The temperate bacteriophage 933W contains DNA encoding Stx2, while Stx1 is encoded by 933J, a putative cryptic prophage, and both genes are integrated into the chromosome (35, 36, 42). Analysis of the data set obtained during sequencing of phage 933W indicated that the assembled sequence is probably a circular permutation of prophage DNA (42). Amplification of the putative ends of phage 933W arms (int and attR) indicated that both arms are incorporated into vesicles and likely the circular form of the phage is present in vesicles (Fig. (Fig.22).
E. coli O157:H7, like most gram-negative pathogens, contains plasmids, which generally carry genes encoding virulence determinants. Three known plasmids are associated with E. coli O157:H7; a large plasmid, of approximately 92 kb (pO157), is present in virtually all clinical isolates, and two small plasmids, 6.7 and 3.3 kb in size, are present less frequently (27, 38, 39). Based on the results of PCR analysis, E. coli O157:H7 (ATCC 43895) harbors both pO157 and the 3.3-kb plasmid but not the 6.7-kb plasmid. The complete DNA sequence of pO157 isolated from E. coli O157:H7 ATCC 43895 and RIMD 0509952 has recently been determined (6, 29). In this study, screening was done for the presence of three genes on pO157, hlyC, hlyA, and L7095. A protein encoded by L7095 has a putative cytotoxin active site and sequence homology to the large clostridial toxin family. The genes hlyC and hlyA are part of the hlyCABD operon, which encodes a pore-forming cytolysin and its secretion apparatus (6). All three genes were detected in vesicles (Fig. (Fig.22 and and3).3). The hly genes were amplified using template DNA from samples that were not concentrated, suggesting that pO157 is frequently contained in vesicles.
The 3.3-kb plasmid contains all the information necessary for its replication, stability, and mobilization (including origin of transfer and mobA, which encodes a mobility protein), but it lacks the tra genes, which mediate close physical contact of bacteria and are important for efficient conjugation. The lack of these genes is indicative of a nonconjugative but mobile plasmid (18). Nucleotide sequence analysis showed that the plasmid is extremely similar (>98%) to an antibiotic-resistant plasmid, NTP16, derived from Salmonella serovar Typhimurium, with the exception of antibiotic resistance transposons (18, 29). The presence of mobA (Fig. (Fig.3)3) indicates that the 3.3-kb plasmid is found within vesicles. Vesicles may play a role in the transfer of the nonconjugative 3.3-kb plasmid.
Based on the presence of chromosomal (derived from bacterial genomes and pathogenicity islands) and bacteriophage genes, large and small plasmids, and a foreign recombinant plasmid (pGFP), DNA entrapment likely occurs randomly. Although all genes that were screened for were detected within vesicles, results suggest that phage 933W and plasmid pO157 are more commonly associated with vesicles, since they were detected consistently without using concentrated vesicle DNA. Several ideas may explain these results, since the stability of genes is influenced by various factors. Assuming random DNA entrapment in vesicles, one would expect to find a similar distribution of genes in parental cells and vesicles, i.e., a higher frequency in cases of genes harbored by high-copy-number plasmids (like pGFP) or propagated phage. However, the mechanism by which DNA is entrapped in vesicles is not yet known. DNA-binding proteins were observed in vesicle lysates of H. influenzae (8) and N. gonorrhoeae (11). Vesicles released from N. gonorrhoeae sedimented into two fractions on a sucrose density gradient, termed BI and BII. Distinct profiles of DNA-binding proteins observed within the two fractions suggests that vesicles may play different roles and that specific mechanisms may exist for determining the association between DNA and vesicles (11).
The source of genetic material entrapped in vesicles was determined through PCR analysis. Electron microscopy was done to determine whether entire plasmids or intact phage was present in vesicles. Micrographs revealed that vesicles harbor intact plasmids; however, phage or phage components were not observed in micrographs of negative stained vesicle preparations. Large rosette structures observed in micrographs may be pO157 or the entire DNA of phage 933W. Although many small DNA fragments were evident, linear DNA fragments approximately a nanometer in length were also observed.
The intent of this study was also to investigate whether DNA contained within vesicles can be transferred to recipient bacteria and whether the gene products expressed will ultimately be active. A recent report indicates that vesicles derived from either Shigella flexneri or P. aeruginosa rapidly fused with the outer membrane of Salmonella enterica serovar Typhi, Salmonella serovar Typhimurium, or E. coli DH5α (22). The authors suggested that the integration of vesicles from a donor bacterium into the membrane of a recipient would introduce constituents from the donor directly into the recipient. In the present study, the results of PCR, antibiotic resistance selection, and the Vero cell assay demonstrate that genetic material is exported by E. coli O157:H7 vesicles and transferred to recipient enteric bacteria (E. coli JM109 and Salmonella serovar Enteritidis). Simply combining vesicles with noncompetent cells in vitro resulted in the transfer of genetic material and subsequent expression of active compounds (e.g., Shiga toxin). Although the experiments related to transformation and expression were restricted to replicons that do not require recombination, this does not preclude transformation and subsequent expression of chromosomal genes.
Three main mechanisms of gene transfer have been identified in bacteria: transformation, involving the uptake and incorporation of naked DNA; conjugation, a cell contact-dependent DNA transfer mechanism; and transduction, whereby host DNA is encapsidated into a bacteriophage which acts as the vector for its injection into a recipient cell. Perhaps vesicles constitute an alternative mode of gene transfer among bacteria. Based on the results of the present study, genes encoding virulence factors and antibiotic resistance can be transferred by vesicles. The role vesicles play in genetic exchange between different species and genera in the environment or host has yet to be defined.
ACKNOWLEDGMENTS
We thank Helen Ricalde for her technical assistance. Processing of electron microscopy samples was conducted by Gloria Binkowski.
Funding from NJAES supported this work.
REFERENCES
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.66.10.4414-4420.2000
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc92318?pdf=render
Citations & impact
Impact metrics
Article citations
Horizontal gene transfer and beyond: the delivery of biological matter by bacterial membrane vesicles to host and bacterial cells.
Curr Opin Microbiol, 81:102525, 26 Aug 2024
Cited by: 0 articles | PMID: 39190937
Review
Outer membrane vesicles secreted from Actinobacillus pleuropneumoniae isolate disseminating the floR resistance gene to Enterobacteriaceae.
Front Microbiol, 15:1467847, 05 Sep 2024
Cited by: 0 articles | PMID: 39301187 | PMCID: PMC11410613
Assessing the Public Health Implications of Virulent and Antibiotic-Resistant Bacteria in Côte d'Ivoire's Ready-to-Eat Salads.
Int J Microbiol, 2024:3264533, 06 Aug 2024
Cited by: 0 articles | PMID: 39139471 | PMCID: PMC11321884
Outer membrane vesicles of Acinetobacter baumannii DS002 carry circular DNA similar to bovine meat and milk factors (BMMFs) and SPHINX 2.36 and probably play a role in interdomain lateral gene transfer.
Microbiol Spectr, 12(9):e0081724, 05 Aug 2024
Cited by: 0 articles | PMID: 39101807 | PMCID: PMC11370262
The mouse epididymal amyloid matrix is a mammalian counterpart of a bacterial biofilm.
iScience, 27(6):110152, 31 May 2024
Cited by: 0 articles | PMID: 38974467
Go to all (188) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7.
Appl Environ Microbiol, 65(5):1843-1848, 01 May 1999
Cited by: 193 articles | PMID: 10223967 | PMCID: PMC91264
Semi-automated fluorogenic PCR assays (TaqMan) forrapid detection of Escherichia coli O157:H7 and other shiga toxigenic E. coli.
Mol Cell Probes, 13(4):291-302, 01 Aug 1999
Cited by: 76 articles | PMID: 10441202
Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7.
Infect Immun, 71(9):4908-4916, 01 Sep 2003
Cited by: 67 articles | PMID: 12933832 | PMCID: PMC187321
Molecular Profiling: Catecholamine Modulation of Gene Expression in Escherichia coli O157:H7 and Salmonella enterica Serovar Typhimurium.
Adv Exp Med Biol, 874:167-182, 01 Jan 2016
Cited by: 5 articles | PMID: 26589218
Review