Abstract
Free full text

Ubiquitination of stalled ribosomes enables mRNA decay via HBS-1 and NONU-1 in vivo
Abstract
As ribosomes translate the genetic code, they can encounter a variety of obstacles that hinder their progress. If ribosomes stall for prolonged times, cells suffer due to the loss of translating ribosomes and the accumulation of aberrant protein products. Thus to protect cells, stalled ribosomes experience a series of reactions to relieve the stall and degrade the offending mRNA, a process known as No-Go mRNA Decay (NGD). While much of the machinery for NGD is known, the precise ordering of events and factors along this pathway has not been tested. Here, we deploy C. elegans to unravel the coordinated events comprising NGD. Utilizing a novel reporter and forward and reverse genetics, we identify the machinery required for NGD. Our subsequent molecular analyses define a functional requirement for ubiquitination on at least two ribosomal proteins (eS10 and uS10), and we show that ribosomes lacking ubiquitination sites on eS10 and uS10 fail to perform NGD in vivo. We show that the nuclease NONU-1 acts after the ubiquitin ligase ZNF-598, and discover a novel requirement for the ribosome rescue factors HBS-1/PELO-1 in mRNA decay via NONU-1. Taken together, our work demonstrates mechanisms by which ribosomes signal to effectors of mRNA repression, and we delineate links between repressive factors working toward a well-defined NGD pathway.
Author summary
Ribosomes are large molecular machines entrusted with the essential task of reading mRNAs and building proteins. It is critical that ribosomes proceed with their work undeterred, so as to avoid traffic jams between ribosomes on mRNAs; however, they sometimes experience challenges which make traffic jams unavoidable. In these cases, cells have evolved machinery to clean up stalled ribosomes and remove offending mRNAs. Much of this machinery is now identified, but the relationships between factors has yet to be tested. Here, in C. elegans we identify the machinery that responds to stalls, and we order factors relative to one another. We find that ribosomes must be tagged by ubiquitin to signal mRNA decay, as ribosomes lacking ubiquitin sites fail to elicit mRNA decay. Furthermore, we find that these ubiquitin signals recruit an enzyme which cuts mRNA, and we present evidence that ribosome rescue factors are required for the mRNA decay reaction as well. Overall, our work provides new connections between factors orchestrating the elimination of problematic mRNAs that stall ribosomes.
Introduction
An organism’s growth, function, and response to environmental changes rely on ribosomes accurately producing proteins. Subtle errors in mRNAs are often elusive, requiring active translation to detect and trigger downstream repression. If left unchecked, defective mRNAs have the potential to produce toxic proteins resulting in disease phenotypes, such as neurodegeneration [1,2].
Ribosomes that translate problematic mRNAs can elicit various repressive mechanisms. Two such mechanisms are Nonstop mRNA Decay (NSD) and No-Go mRNA Decay (NGD). NSD is triggered by mRNAs lacking stop codons, which can arise from polyadenylation or from mRNA cleavage [3]. NGD occurs on mRNAs containing a variety of elongation-inhibiting features, such as stable secondary structures, stretches of rare codons, polybasic amino acid-encoding sequences, or damaged nucleotides [4]. While triggered by different mRNA species, both of these pathways result in mRNA decay, ribosome rescue, and nascent peptide degradation through the coordinated recruitment of several effectors.
One key event in NGD is the generation of a stalled, collided ribosome species at a problematic site on the mRNA. This species is thought to be unique to NGD and could conceivably recruit downstream effectors [5,6,7]. The interface between collided ribosomes is the site of ubiquitination events on multiple ribosomal proteins near the mRNA’s path [6,7]. The conserved E3 ubiquitin ligase ZNF-598 is thought to deposit ubiquitin marks on at least two ribosomal proteins, including RPS-10 (eS10) and RPS-20 (uS10) [8,9]. Prior work overexpressing ubiquitination-deficient point mutations of eS10 suggests that ribosomal protein ubiquitination is important for NGD [8], though whether additional, ubiquitination-independent mechanisms contribute remains unclear. Our limited understanding of ribosomal ubiquitination is in part due to the difficulty in recovering viable mutants of the target ribosomal proteins, which has thus far complicated a straightforward analysis of individual ubiquitination sites and their relationship to repression via ZNF-598. Harnessing a system to study the functional contributions of ribosomal ubiquitination would clarify its importance.
Early NGD work pointed towards Saccharomyces cerevisiae Dom34 and Hbs1 (Pelota and HBS1 in higher eukaryotes) as being important for mRNA cleavage [4]. Despite an early study claiming nuclease activity of Dom34, it is now thought that this requirement is indirect [10] with subsequent work on Dom34/Hbs1 showing a biochemical role in ribosome rescue [11,12,13,14]. Later work in both Caenorhabditis elegans and S. cerevisiae identified NONU-1/Cue2/YPL199C as endonucleases responsible for cleaving targets of NSD and NGD in the vicinity of stalled ribosomes [15,16]. While NONU-1 is required for efficient NSD and NGD, it remains unclear how and when the factor is targeted to stall-inducing mRNAs. However, clues to its recruitment mechanism exist: NONU-1 and its homologs contain at least two conserved ubiquitin-binding CUE domains [15].
Much of what is known about NGD comes from studying the effect of individual factors and events, and it is unclear how these steps relate to one another to bring about target mRNA repression. Here, we deployed C. elegans to unravel the series of events during NGD. We show that mutation of ribosomal ubiquitination sites on RPS-10 (eS10) and RPS-20 (uS10) phenocopies knock out of ZNF-598. We present data in support of a model in which ZNF-598 first ubiquitinates ribosomes at stall sites, followed by mRNA degradation via NONU-1. Interestingly, we also recovered a role for HBS-1 and PELO-1 in mRNA decay via NONU-1 cleavage, consistent with early northern data in S. cerevisiae, suggesting that ribosome rescue may be an important step that precedes mRNA cleavage.
Results
A novel ribosome stalling screen identifies core NGD machinery
To establish C. elegans as a system to study NGD, we built a ribosome stalling reporter using CRISPR/Cas9 at the unc-54 locus. UNC-54 encodes a major muscle myosin required for animal movement, but dispensable for life [17,18,19]. UNC-54 has also been the starting point for a number of genetic screens, and its expression is relatively well-characterized. To build a NGD reporter, we added a T2A ‘stop-and-go’ peptide, a FLAG tag, twelve rare arginine codons, and GFP to the C-terminus of UNC-54 via CRISPR/Cas9, and we call the resultant allele unc-54(rareArg) (Fig 1A) (reporter sequence available in S1 Table). We selected rare arginine codons for two reasons: (1) arginine is a positively charged amino acid and prior work [20,21] suggests this may induce ribosome stalling via interactions with the peptide exit tunnel, and (2) the Arg-tRNAs decoding two of the rarest codons (CGG, AGG) in C. elegans are very lowly expressed under normal growth conditions (Aidan Manning, Todd Lowe, personal communication, June 2021), providing a second avenue by which stalling may occur.
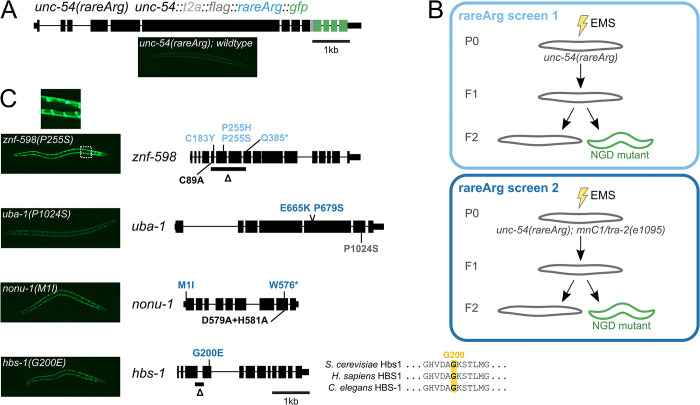
(A) Gene diagram showing annotated exons (black rectangles) of unc-54(rareArg). Colored rectangles represent CRISPR/Cas9 insertions at the endogenous unc-54 locus: T2A sequence (gray), FLAG (dark gray), 12 rare arginine codons (blue), and GFP (green). (B) Schematics of rareArg genetic screens. (C) znf-598, uba-1, nonu-1, and hbs-1 alleles with representative image of one allele per gene on the left. Black rectangles represent exons, thicker rectangles are CDS, and thin lines are introns. Mutations made via EMS in the rareArg screen 1 (light blue) or rareArg screen 2 (dark blue), and via CRISPR/Cas9 (black) or CGC (gray) are shown. For HBS-1, multiple sequence alignment shows conserved glycine (G200) in GTPase domain.
Strains with the unc-54(rareArg) construct showed an uncoordinated (Unc) phenotype consistent with reduction of UNC-54 protein. As other C-terminal tags of UNC-54 are functional [22], the loss of UNC-54 protein suggests that mRNAs produced from the unc-54(rareArg) locus are repressed. unc-54(rareArg) animals also exhibited very low levels of GFP, suggesting that few ribosomes make it to the 3’end of unc-54(rareArg) due to reduced mRNA levels and/or high amounts of stalling during translation elongation.
To initially validate the unc-54(rareArg) reporter as a target of NGD, we crossed it with alleles of two factors known to be required for NGD in C. elegans (nonu-1(AxA)) and other systems (znf-598(Δ)) [8,9,15,23]. In each case we observed de-repression of the reporter, manifest as increased fluorescence and an improvement in animal movement and egg laying (UNC-54 is required in the vulva muscles for egg laying). The nonu-1 result is consistent with our prior work showing that this stretch of twelve rare arginine codons confers nonu-1-dependent mRNA decay [15]. Notably, the phenotypic effects seen upon nonu-1 knockout differ from that seen in S. cerevisiae: knockout of the homologous CUE2 in S. cerevisiae only confers effects upon simultaneous knockout of additional factors [16]. Given that most of our mechanistic understanding of NGD comes from work in S. cerevisiae [4,5,16], and that genetic screens in human K562 cells failed to identify the NONU-1 homolog [24], we reasoned that a genetic screen in C. elegans would prove insightful and augment information gained from other systems.
Using unc-54(rareArg), we performed two genetic screens (Methods) (Fig 1B). In the first of these screens, we EMS-treated unc-54(rareArg) animals, harvested eggs, and screened ~90,000 F1 genomes. Among these, we found six mutants across six plates with increased movement (indicative of de-repression of unc-54(rareArg)) and increased GFP (indicative of translation to the end of the unc-54(rareArg) transcript). Evidence of stall readthrough was visible by the nuclear localization of GFP upon its de-repression (Fig 1C). This observation revealed our serendipitous construction of an N-terminal motif (a run of arginines) matching the sequence requirements for a nuclear localization signal [25]. We genetically mapped and identified variants, and found that four alleles mapped to znf-598 (Fig 1C).
Reasoning that there was more to NGD than znf-598, we repeated the screen using a chromosomal balancer covering the znf-598 locus to preclude recovery of recessive znf-598 mutations. We screened an additional ~105,000 F1 genomes, among which we found an additional 14 mutants across 14 plates. The increase of movement and GFP in these animals were less pronounced than the znf-598 mutants from the first screen. Nevertheless, the phenotypes were robust enough to allow us to genetically map and identify causative loci. Here we report a total of nine alleles in four genes from both screens (Fig 1C). Five alleles in one additional gene were identified (catp-6, S1 Table); mapping and characterization of the remaining six mutants is ongoing.
Seven alleles were split amongst three readily-identifiable NGD components: nonu-1, znf-598, and hbs-1. We found two mutations in nonu-1, as would be expected based on the unc-54(rareArg) reporter validation. We isolated four mutations in znf-598 (all from the first genetic screen), three of which were missense mutations. One mutation (C183Y) was within a region that matches the consensus for a C2H2 zinc finger in C. elegans and Homo sapiens, but is not conserved in S. cerevisiae. Two alleles at P255 (P255S, P255H) mutated a highly conserved proline at the start of a C2H2 zinc finger. One allele was found in the C. elegans’ homolog of HBS1 (k07a12.4, hereafter hbs-1). This allele was in a conserved GTP-binding and active site residue (G200E) (Fig 1C), consistent with a functional requirement for GTP binding and hydrolysis by HBS-1 in NGD.
Two additional mutations mapped in uba-1, the sole E1 ubiquitin-activating enzyme in C. elegans. Both uba-1 mutations (E665K, P679S) exhibited poor viability, as would be expected as complete loss of uba-1 is thought to be lethal [26]. We confirmed the uba-1 result by crossing in a known temperature-sensitive loss-of-function allele of this gene (it129; P1024S), and observed temperature-dependent de-repression of GFP. As ZNF-598 is known to function in other systems as a ubiquitin ligase [8,9], we expect that uba-1 loss-of-function compromised the ubiquitin-conjugation cascade, giving rise to unc-54(rareArg) de-repression.
Cell-specific NGD rescue via overexpression of factors
We also developed a system to analyze the effects of overexpression of particular NGD pathway components using extrachromosomal arrays. Overexpression is a useful means to generate hyperactive alleles of factors to test their relative order in a pathway. Injection of foreign DNA into the germline of C. elegans results in fusion of the DNA sequences into an extrachromosomal array [27,28]. The resultant array can be both meiotically and mitotically inherited, albeit with reduced efficiencies compared to endogenous chromosomes. We created a plasmid vector to overexpress a gene of interest on a transgenic array with the following components: (1) a myo-3 promoter, to drive expression in the body wall muscle where unc-54 is also expressed, (2) a fluorescent mCherry protein, to monitor inheritance and expression of the array, (3) a ‘stop-and-go’ T2A sequence, to separate mCherry from the gene-of-interest, and (4) a site to insert a gene-of-interest. We constructed plasmids encoding fluorescent proteins linked to C. elegans’ znf-598 and nonu-1.
To check the functionality of factors expressed from an array, we made overexpression arrays in each of the cognate mutant backgrounds. Overexpression of the wild-type ZNF-598 and NONU-1 protein restored repression of unc-54(rareArg) and rescued the loss-of-function phenotype of znf-598 (Fig 2A) and nonu-1 (Fig 2B), respectively. Thus the overexpressed factors were functional. Arrays can be stochastically lost or silenced in cell lineages of the animal, allowing us to examine rescue on a cell-by-cell level. For each of znf-598 and nonu-1, we observed an inverse relationship between factor expression (monitored via mCherry) and unc-54(rareArg) (monitored via GFP). Thus the rescue was cell-autonomous, as would be expected under current models of ZNF-598 and NONU-1 acting directly on ribosomes and mRNAs.
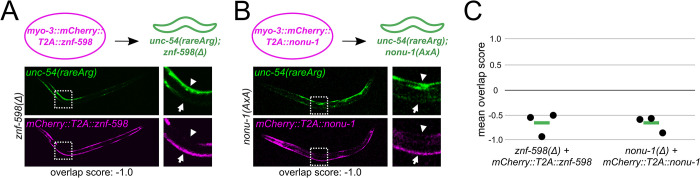
(A) Schematic of znf-598 construct plasmid and znf-598 strain subject to germline microinjection. Below are GFP and mCherry images of a representative animal expressing the above construct, with a zoom in of an area demonstrating the effect of mCherry-marked factor expression on NGD (GFP). (B) As in (A) for nonu-1 construct in nonu-1 strain. (C) Mean overlap score of strains in (A, B). Each black dot represents the mean of one independent isolate (n≥4 animals/isolate), with the mean of all isolates shown as a green bar.
To quantify the inverse relationship of factor overexpression (mCherry) to NGD (GFP), we calculated an overlap score, based on the brightest red and green pixels across multiple, independent animals (Methods) (S1 Fig). If mCherry and GFP are non-overlapping (as would be expected from rescue), the overlap score would be negative. If mCherry and GFP overlap (as would be expected without rescue), the overlap score would be positive. For znf-598 and nonu-1, we observed values close to -1 (Fig 2C), demonstrating the generality of rescue across animals.
With a functional readout of NGD (unc-54(rareArg)), mutants in factors (znf-598, nonu-1, hbs-1, and pelo-1), and the overexpression/rescue system, we set out to characterize the molecular mechanisms by which factors relate and repress gene expression in response to ribosomal stalling.
ZNF-598 is required for ribosomal ubiquitination in C. elegans
In the NGD screen, we recovered several alleles of znf-598. ZNF-598 homologs are thought to recognize ribosomal collisions and ubiquitinate sites on the small subunit, including RPS-10 (eS10) and RPS-20 (uS10) [8,9]. By multiple sequence alignment, we identified a highly conserved cysteine (C89) (Fig 3A) [9] known to be required for ubiquitin conjugation by Hel2/ZNF598, and we generated a point mutation (C89A) via CRISPR/Cas9 at the endogenous locus. The znf-598(C89A) mutant displayed unc-54(rareArg) de-repression indistinguishable from the phenotype of znf-598(Δ) (Fig 3B), supporting a role for ubiquitination in repression of stall-inducing mRNAs in C. elegans, as in other systems.
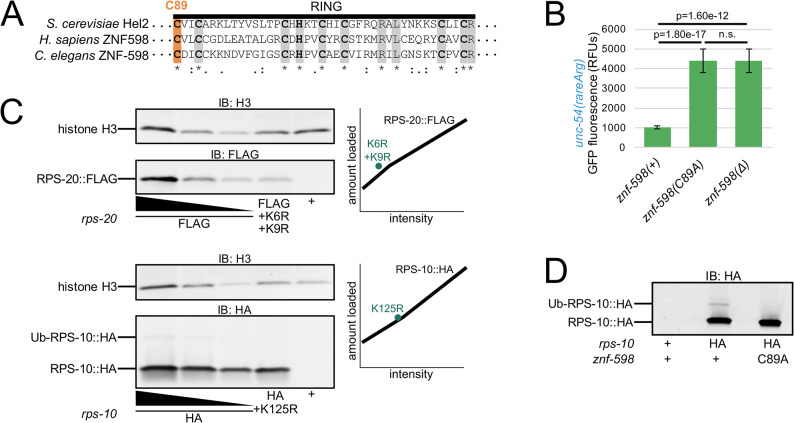
(A) Multiple sequence alignment of S. cerevisiae Hel2, H. sapiens ZNF598, and C. elegans ZNF-598 RING finger domains. Conserved residues (gray) and C89 (orange) are highlighted. Conservation below alignment is as follows: asterisks indicate identity, colons indicate amino acids with strongly similar properties, periods indicate amino acids with weakly similar properties. (B) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test. (C) Western blot of indicated strains to monitor RPS-20 and RPS-10 expression. Dilutions of wild type tagged proteins were loaded as indicated, with two-fold more and two-fold less than other two samples, to generate a standard curve shown as a black line in plots on the right. Lysine mutants of tagged proteins were quantified and plotted as teal points in the plots on the right. (D) Western blot of indicated strains to monitor ubiquitination of HA-tagged RPS-10.
To investigate the ribosomal protein targets of ZNF-598, we tagged both RPS-10 (eS10) and RPS-20 (uS10) at the endogenous locus via CRISPR/Cas9. By immunoblot we observed robust expression of the cognate ribosomal protein in each strain (Fig 3C). We also mutated conserved lysines in RPS-10 and RPS-20 known to be sites of ubiquitination in other systems (S2A and S2B Fig) [8,9]. The K>R substitutions did not adversely impact RPS-10 and RPS-20 expression (Fig 3C) nor animal viability. In the case of RPS-10, we also observed a band corresponding to the expected size of Ub-RPS-10. This band was absent in rps-10(K125R) (Fig 3C) and znf-598(C89A) mutants (Fig 3D). These results show that znf-598 is required for ubiquitination of RPS-10 on K125.
NGD-deficient ribosomes made via ablation of ubiquitination sites
Prior work underscored the importance of ribosomal ubiquitination events in NGD [8,9]. However, the precise functional contributions of individual ubiquitination sites has remained unclear, in part due to the difficulty in obtaining viable mutants in the relevant ribosomal proteins. In prior work, overexpression of RPS-10 with K>R substitutions at ubiquitination sites conferred de-repression of a stalling reporter in between that observed in wild-type and ZNF598 knockout human cells [8]. Interpretation of this experiment is complicated by a number of factors, including residual expression of wild-type RPS-10. Would complete removal of RPS-10 ubiquitination sites mimic loss of ZNF598? Does ZNF598 contribute to functional repression outside of its role in ribosomal ubiquitination? Our work thus far supported a NGD mechanism similar to that of human cells, and therefore our system seemed a useful model to explore these questions. In particular, the viability of ribosomal point substitutions at endogenous loci as well as the ease of making double mutants and overexpression constructs provided us with the means to test models of the functional importance of ribosomal ubiquitination and its relationship to ZNF-598.
To initially test for a functional role of ribosomal ubiquitination in NGD, we crossed rps-10(K125R) and rps-20(K6R+K9R) into unc-54(rareArg). In each case we observed a defect in NGD, manifest as an increase in GFP produced by unc-54(rareArg) (Fig 4A). The double mutant (rps-10(K125R); rps-20(K6R+K9R)) exhibited an even stronger defect in NGD, comparable to that observed in znf-598 mutants. We interpret these data to indicate that ribosomal ubiquitination is required for NGD, and that ablation of these two sites alone is sufficient to prevent functional NGD. Interestingly, these experiments demonstrate that it is possible to make a version of the ribosome deficient in NGD via mutation of a few lysines, highlighting the central role of the ribosome and ubiquitination in NGD.
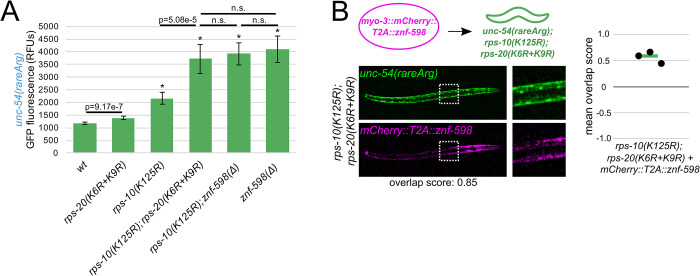
(A) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type. (B) As in Fig 2, with znf-598 construct in rps-10; rps-20 strain.
We also performed additional experiments to clarify the function of ribosomal ubiquitination in relation to znf-598. First, we performed a double mutant analysis. In a model where ribosomal ubiquitination is functionally independent of znf-598, we would expect that a rps-10(K125R); znf-598(Δ) mutant would exhibit a stronger NGD phenotype (more GFP) than either single mutant alone. However, if ZNF-598 and Ub-RPS-10 lie on the same pathway, we would expect the rps-10(K125R); znf-598(Δ) phenotype to resemble one of the single mutants. The rps-10(K125R); znf-598(Δ) phenotype was indistinguishable from the znf-598(Δ) single mutant (Fig 4A), consistent with the latter model. Second, we used our overexpression system. In a model where rps-10(K125R) and rps-20(K6R+K9R) are on a partially redundant pathway to znf-598 (i.e., that znf-598 contains additional repressive functions outside of ubiquitination at these two sites), we would expect that overexpression of ZNF-598 would provide restoration of NGD in the rps-10(K125R); rps-20(K6R+K9R) mutant. However, if ZNF-598 acts through RPS-10 and RPS-20, overexpression of ZNF-598 in a rps-10(K125R); rps-20(K6R+K9R) mutant would yield the same phenotype as rps-10(K125R); rps-20(K6R+K9R). We observed the latter (Fig 4B), again suggesting that ZNF-598 works through ubiquitination of RPS-10 and RPS-20.
Taken together, our experiments suggest that ZNF-598 is a ubiquitin ligase required for ubiquitination of at least RPS-10 and RPS-20, and that these ubiquitination events are essential to NGD. The fact that loss of znf-598 can be mimicked by mutation of ribosomal proteins provides compelling evidence that the primary function of ZNF-598 during NGD is to deposit ubiquitins on ribosomes.
NONU-1 function during NGD requires CUE domains and follows ZNF-598
Having established a requirement for ubiquitination by ZNF-598 in this system, we decided to investigate the relationship of ubiquitination to mRNA decay. A key effector of NGD is NONU-1, which was identified in our screen (Fig 1C) and also in our prior NSD screen [15]. In our prior work, we noted that NONU-1 and its homologs contain CUE domains (Fig 5A) [15,29], which are known to bind ubiquitin, suggesting a mechanism of recruitment for NONU-1 to sites of ribosome stalling. To test a requirement for the CUE domains in nonu-1 function, we deleted the CUE domains and observed a phenotype indistinguishable from other nonu-1 mutants (Fig 5B). This result is consistent with CUE domains being essential to NONU-1 function. We also attempted to examine expression of NONU-1 by tagging the N-terminus (S1 Table), but tagged alleles were non-functional. We did not tag the C-terminus of NONU-1 as it is conserved. We look forward to future work where we can determine whether the requirement of CUE domains in NONU-1 is merely for NONU-1 expression or for its biochemical functions.
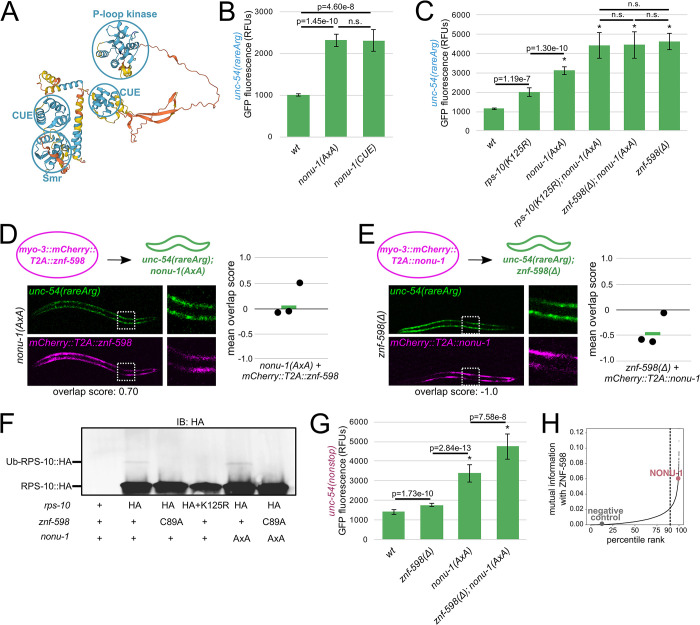
(A) C. elegans NONU-1 protein structure as predicted by AlphaFold [29]. Prediction confidence is as follows: blue regions are confident, yellow regions are low confidence, orange regions are very low confidence. Confident domains are circled in blue and labeled. (B, C) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type. (D) As in Fig 2, with znf-598 construct in nonu-1 strain. (E) As in Fig 2, with nonu-1 construct in znf-598 strain. (F) Western blot of indicated strains to monitor ubiquitination of HA-tagged RPS-10. (G) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(nonstop) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type. (H) ZNF-598 mutual information plot. 90% percentile cutoff is shown as a dashed line, NONU-1 is highlighted in pink, and a negative control protein is highlighted in gray.
To determine whether NONU-1 acts in the same pathway as ZNF-598, we performed a double mutant analysis. If the two factors function in different pathways, we would expect additive phenotypes on the unc-54(rareArg) reporter in a double mutant. However, if NONU-1 and ZNF-598 work in the same pathway of repression, we would expect the double mutant to resemble one of the single mutants. We combined mutations in znf-598 and ribosomal ubiquitination sites with nonu-1(AxA), a catalytic mutation of nonu-1 that is indistinguishable from a deletion of nonu-1 (S3 Fig) [15]. The nonu-1; znf-598 double mutant mimicked the znf-598 single mutant (Fig 5C), consistent with the two factors functioning in the same pathway in NGD.
We next investigated the ordering of ZNF-598 and NONU-1 relative to one another in NGD. In a first pair of experiments, we overexpressed ZNF-598 in a nonu-1 mutant, and also overexpressed NONU-1 in a znf-598 mutant. According to classical logic when ordering genes in functional pathways, we expect that overexpression of an upstream factor will not compensate for loss of a downstream factor, but that overexpression of a downstream factor will compensate for loss of an upstream factor [30,31,32,33]. We observed little effect of the nonu-1 phenotype by ZNF-598 overexpression (Fig 5D), and we saw rescue of the znf-598 phenotype by NONU-1 overexpression (Fig 5E). These results support a model where NONU-1 acts downstream of ZNF-598. In a second set of experiments, we examined RPS-10 ubiquitination by immunoblot in nonu-1(AxA) and observed it to be unchanged (Fig 5F). We interpret this result to indicate that the defect in a nonu-1 mutant is after ribosomal ubiquitination, i.e., in the commitment of ubiquitinated ribosomes to mRNA decay.
We were curious to determine whether nonu-1 and znf-598 function together in NSD as well. Thus, we quantified mutants’ effects on the unc-54(nonstop) reporter, an allele of unc-54 tagged with a C-terminal GFP and lacking all stop codons [15,34]. In contrast to its strong effect on the unc-54(rareArg) reporter, the znf-598 mutant showed mild unc-54(nonstop) de-repression (Fig 5G). This result is consistent with a more modest role for znf-598 in NSD than that observed at unc-54(rareArg), and explains why prior NSD screens failed to identify znf-598 [15,34]. A nonu-1 mutant exhibited a greater de-repression of unc-54(nonstop) than znf-598, and the double mutant expressed higher levels of GFP than either single mutant. The fact that nonu-1 and znf-598 together exhibit additive effects suggests that NONU-1 is recruited via an E3 ubiquitin ligase other than ZNF-598 during NSD. For more on this, see Discussion.
Taken together, our results with the unc-54(rareArg) reporter support a model in which ZNF-598 and NONU-1 function together in NGD. Based on our genetic and molecular analyses, we favor a model in which ribosomal ubiquitination recruits NONU-1 to mRNAs for cleavage.
A conserved role for ZNF-598 and NONU-1 throughout eukaryotes
To determine if the relationship between ZNF-598 and NONU-1 is a broadly conserved phenomenon throughout eukaryotes, we performed phylogenetic profiling (reviewed in [35]). Briefly, phylogenetic profiling scores pairs of proteins using mutual information, which can be interpreted as the tendency of the protein pair to function together, either redundantly or sequentially. High mutual information can be achieved when two proteins are inherited together or lost together, or when two proteins are genetically redundant and at least one protein is maintained.
We carried out phylogenetic profiling by searching 111,921 profile HMMs [36] on genomic and transcriptomic sequences from 473 protists as these represent diverse eukaryotic lifestyles (Methods). To validate this approach, we calculated mutual information between pairs of factors known to function together in a complex, such as PELO-1/HBS-1 (S4A Fig) and LTN-1/RQC-2 (S4B Fig) [11,12,14,37]. PELO-1/HBS-1 and LTN-1/RQC-2 exhibited high mutual information, scoring in each others’ top 99.88% (PELO-1 ranked 19th of 15,909 interactions with HBS-1) and 98.86% (RQC-2 ranked 181th of 15,909 interactions with LTN-1), respectively. Similarly, we observed that ZNF-598 and NONU-1 ranked in each others’ top 99.38% (NONU-1 ranked 99th of 15,909 interactions with ZNF-598) (Fig 5H). Thus ZNF-598 and NONU-1 are a broadly conserved functional pair across diverse eukaryotes, with our work in C. elegans suggesting that this pair functions in NGD to degrade mRNAs that stall ribosomes.
HBS-1 N-terminus resembles a ubiquitin-binding domain and is dispensable for NGD
Several of our NGD suppressor mutants (znf-598, nonu-1, uba-1) encoded factors involved in ubiquitin-dependent processes, so we were initially surprised that this screen also identified the ribosome rescue factor hbs-1. We therefore examined HBS-1 to determine whether it could conceivably function in a ubiquitin-dependent manner.
The HBS-1 protein consists of an N-terminal domain, a disordered linker region, a GTPase domain, and two beta-barrel domains [12,14,29] (Fig 6A). In two published structures, we noticed that the N-terminal domain of HBS-1 is found near known ubiquitination sites on the ribosome (uS10 and uS3) and adopts a distinct triple helix bundle [12,14]. The fold is classified by Pfam as a member of the ubiquitin-binding Uba clan (Fig 6B) [12,14,38]. Notable members of the Uba clan include the Uba and CUE domains, found in S. cerevisiae Rad23 and C. elegans NONU-1, respectively. We also noticed that plant HBS-1 N-term homologs contain a conserved C4-type zinc finger (ZnF) homologous to a known ubiquitin-binding C4 ZnF domain present in the rat Npl4 (Fig 6C) [39]. These observations show that HBS-1 N-termini from diverse organisms lack sequence or structural homology with each other, and yet many N-termini instead share homology with domains that bind ubiquitin. Given this homology to ubiquitin-binding domains, we hypothesized that the N-terminal domain of HBS-1 may bind ubiquitin during NGD.
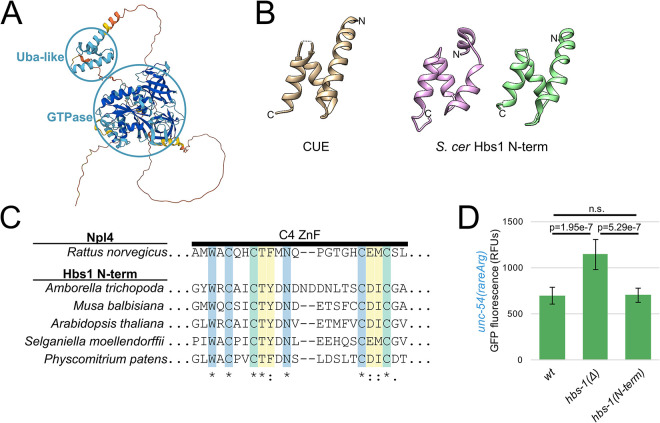
(A) H. sapiens Hbs1 protein structure as predicted by AlphaFold [29]. Prediction confidence is as in Fig 5A, with dark blue showing regions of high confidence. Confident domains are circled in blue and labeled. (B) Structural homology between Hbs1 N-terminal domain and ubiquitin-binding domains. At left is a structure representative of the UBA clan: CUE from [38] (tan, S. cerevisiae Vps9p, 1P3Q). At right are two structures of the Hbs1 N-terminus from [12] (pink, S. cerevisiae, 3IZQ) and [14] (green, S. cerevisiae, 5M1J). Amino and carboxy termini indicated with N and C, respectively. Note overall similarity in topology and fold across structures. (C) The N-terminal zinc finger (ZnF) of plant Hbs1 is homologous to the Ub-binding ZnF of rat Npl4. Multiple sequence alignment of the ZnF of Hbs1 from phylogenetically diverse plants. Residues that are highly conserved among Npl4 homologs are in blue, residues that contact Ub are in yellow, and residues that are both conserved and contact Ub are in green. Coloring and annotation of Npl4 residues from [39]. (D) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test.
To determine whether the N-terminus of HBS-1 is required for its functions in NGD, we generated an N-terminal deletion mutant of hbs-1 by deleting the region spanning the triple helix bundle and much of the linker region, yielding an HBS-1 consisting of only the first few amino acids, a short linker, and the GTPase domain. Surprisingly, when measuring unc-54(rareArg) GFP levels, deleting the N-terminus of HBS-1 had no discernible phenotype, indicating that the N-terminal domain of HBS-1 is dispensable for NGD in C. elegans (Fig 6D).
While our genetic analyses supported a role for HBS-1 in NGD independent of the N-terminal domain, it did not rule out a ubiquitin-binding role for the domain outside of NGD. To test this possibility, we performed an in vitro ubiquitin-binding assay [40]. However, we failed to observe ubiquitin-binding above background (S5 Fig). We speculate that the success of in vitro binding assays may be hindered by a requirement for a ribosome surface to promote HBS-1 binding, as the N-terminus binds to ribosomes in structural studies [12,14]. We look forward to future work to explore the function of the HBS-1 N-terminus on the ribosome.
HBS-1 and PELO-1 are essential for mRNA degradation
HBS-1 functions with PELO-1 in other contexts, and so we decided to test a functional role of pelo-1 in NGD. We crossed a pelo-1 mutant (cc2849, bearing a large deletion) [34] into the unc-54(rareArg) reporter. The pelo-1 mutant exhibited a de-repression of unc-54(rareArg) comparable to that observed in the hbs-1 mutant (S6 Fig). Furthermore, the hbs-1; pelo-1 double mutant exhibited a phenotype similar to the single mutants (S6 Fig), suggesting that the two factors function together in NGD as is known in other systems.
HBS-1 and PELO-1 are predominantly known for their role in ribosome rescue, and we were initially surprised by their requirement for repression of the unc-54(rareArg) reporter. Despite a well-characterized biochemical role in ribosome rescue [11,12,13,14,41], there is also a known but poorly understood requirement for HBS-1/PELO-1 in mRNA degradation in NGD in other systems [4] as well as in NSD in C. elegans [34]. We hypothesized that HBS-1/PELO-1 may facilitate mRNA decay during NGD in C. elegans. To test this hypothesis, we performed RNA-seq on znf-598, nonu-1, and hbs-1 mutants bearing the unc-54(rareArg) reporter (Fig 7A). We observed an increase in unc-54(rareArg) mRNA levels comparable to the level of GFP de-repression observed in each strain, with znf-598 conferring the highest levels and nonu-1 and hbs-1 exhibiting a lesser and similar increase in mRNA levels. Therefore, we conclude that HBS-1 is required for mRNA degradation during NGD in C. elegans.
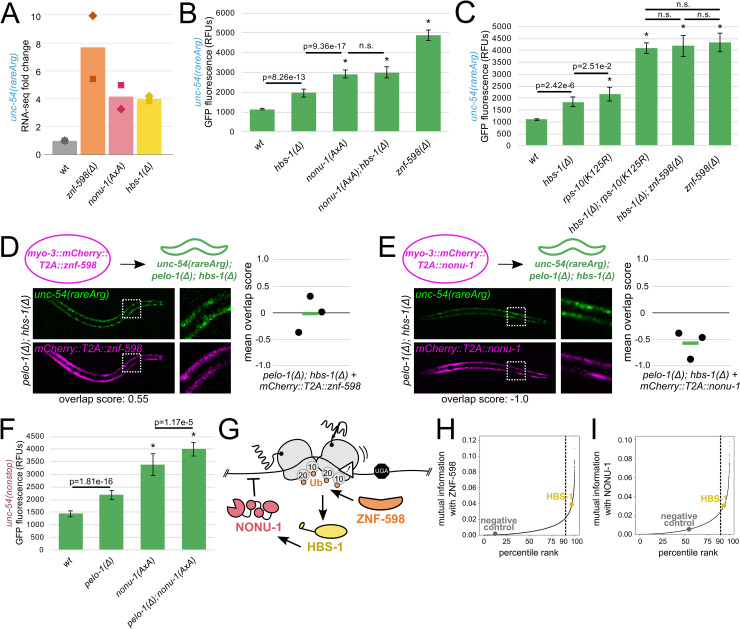
(A) RNA-seq mean fold change of unc-54(rareArg) in the indicated strains from two biological replicates (shown as diamonds and squares). (B, C) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type. (D) As in Fig 2, with znf-598 construct in pelo-1; hbs-1 strain. (E) As in Fig 2, with nonu-1 construct in pelo-1; hbs-1 strain. (F) Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(nonstop) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type. (G) Model for NGD via ZNF-598, HBS-1, and NONU-1. (H) ZNF-598 mutual information plot. 90% percentile cutoff is shown as a dashed line, HBS-1 is highlighted in yellow, and a negative control protein is highlighted in gray. (I) As in (H), showing NONU-1 mutual information with HBS-1.
Studies in S. cerevisiae identified homologs of hbs-1 and pelo-1 (HBS1 and DOM34) as being required for endonucleolytic cleavages during NGD [4,10,42], though at the time of that work, the identity of the nuclease was unknown. Based on this literature and our RNA-seq analysis, we hypothesized that HBS-1/PELO-1 may act in the same pathway as NONU-1. To test this hypothesis, we performed double mutant analysis with hbs-1 and nonu-1. The nonu-1; hbs-1 double mutant was indistinguishable from a nonu-1 single mutant, suggesting that HBS-1 acts in the same pathway as NONU-1 (Fig 7B). We also found HBS-1 to act in the same pathway as ZNF-598 (Fig 7C).
To place HBS-1 relative to ZNF-598 and NONU-1 in NGD, we tested whether excess ZNF-598 or NONU-1 could compensate for the loss of pelo-1 and hbs-1. Overexpression of ZNF-598 resulted in little change to the NGD phenotype (Fig 7D), consistent with a model where HBS-1/PELO-1 act downstream of ZNF-598. In contrast, overexpression of NONU-1 rescued NGD in pelo-1; hbs-1 animals (Fig 7E). This result suggests that NONU-1 acts downstream of HBS-1/PELO-1. Interestingly, analyses using the NSD reporter indicate separate capabilities for NONU-1 and HBS-1/PELO-1 in NSD: a double mutant analysis of pelo-1 and nonu-1 in NSD displayed an additive effect (Fig 7F).
Taken together, our analyses place HBS-1 and PELO-1 on a pathway with ZNF-598 and NONU-1 during NGD, and suggests a model where HBS-1 acts downstream of ZNF-598 to promote mRNA decay by NONU-1 (Fig 7G).
HBS-1 is broadly conserved with NONU-1 and ZNF-598
In light of our genetic analyses, we were curious to measure the conservation of the relationships between HBS-1 and both NONU-1 and ZNF-598. To this end, we carried out phylogenetic profiling of HBS-1 with ZNF-598 homologs, and observed a relatively high level of mutual information falling in each others’ 96.9%/98.3% (Fig 7H), suggesting that HBS-1 and ZNF-598 function together throughout eukarya. NONU-1 and HBS-1 fell in each others’ 93.7% and 96.7%, still exhibiting some conservation, but less so than either factor with ZNF-598 (Fig 7I). Taken together, our conservation analyses support a conserved relationship between ZNF-598 and HBS-1, with a less conserved relationship between HBS-1 and NONU-1.
Discussion
Here, we investigated the functions of and interplay between several factors required for NGD. Overall, our work supports a model where ubiquitination of RPS-10 and RPS-20 by ZNF-598 recruits NONU-1 via ubiquitin-binding to elicit mRNA decay, and HBS-1 acts downstream of ZNF-598 to enable mRNA cleavage by NONU-1.
Our data are consistent with a model in which ZNF-598 directly ubiquitinates RPS-10 and RPS-20. We note that our data could also be explained by a model in which ZNF-598 indirectly affects RPS-10 and RPS-20 ubiquitination, e.g., through an intermediary E3 ligase. While we did not identify other E3 ligases in our screen, it is possible that loss of such E3 ligases would be inviable, precluding their recovery. Additional work will be required to distinguish between these models.
Regardless of whether ZNF-598 acts directly or indirectly on RPS-10 and RPS-20, our data suggest these proteins act in a single pathway to bring about mRNA repression. There may also be additional ubiquitination sites functioning on the ribosome during NGD, as has been suggested in other systems [43,44]. It is notable that we can efficiently block mRNA degradation via ablation of RPS-10 and RPS-20 sites alone. Our analysis of NONU-1’s relationship to ribosomal ubiquitination sites suggests a model that is more complicated than a simple one-ubiquitination-site-one-effector model. This may be expected given NONU-1’s multiple ubiquitin-binding domains. Further genetic work will be required to elucidate the network of ubiquitination sites functioning in NGD, and given the combinatorial possibilities of sites on collided ribosomes, we expect structural studies to be insightful.
While our unc-54(rareArg) reporter indicated that NONU-1 and ZNF-598 largely function in the same pathway during NGD, our NSD data are consistent with flexibility in NONU-1’s recruitment. Given the minor effect of ZNF-598 in NSD, and the absence of znf-598 alleles in NSD screens [15,34], we hypothesize that a factor other than ZNF-598 is used to recognize ribosomes stalled at the 3’end of an mRNA. Other E3 ligases have been implicated in surveillance, and future work will hopefully clarify the recruitment mechanisms of each [7,44,45]. These data could also be explained by differences in the stalling mechanism and/or reporter readout on unc-54(nonstop) vs. unc-54(rareArg). Regardless, the mild NSD de-repression seen in znf-598 indicates that ZNF-598 does act on some ribosomes during NSD. We hypothesize that collisions targeted by ZNF-598 are worsened in a nonu-1 mutant, explaining the greater effect of znf-598 on NSD in a nonu-1 mutant (Fig 5G). These ideas are consistent with other models suggesting multiple types of collisions, with the relative amount of each depending on the genetic background [7].
Here, we discovered that the N-terminal domain of HBS-1 is dispensable for NGD, despite sharing structural homology to ubiquitin-binding domains. Our sequence analyses revealed that HBS-1 N-termini encode domains homologous to ubiquitin-binding domains despite little primary sequence conservation. While this suggests that HBS-1 binds ubiquitin, we failed to observe ubiquitin binding above background, suggesting that additional factors may be required for an interaction. It is also possible that the HBS-1 N-terminus interferes in the ubiquitination reaction. Given that current structures of ribosome-bound HBS-1 lack density for ubiquitin, we expect structural studies with ubiquitinated ribosomes will prove insightful.
Given prior work in S. cerevisiae revealing a role for Hbs1 in NGD cleavage [4,10,42], and in light of our data here, we favor a model where HBS-1 activity greatly enhances mRNA cleavage by NONU-1. We note that we recovered an HBS-1 GTPase mutant in our screen, suggesting that GTP hydrolysis is required for HBS-1 function in mRNA decay. We therefore hypothesize that, in the process of rescuing ribosomes, HBS-1/PELO-1 generate a substrate for NONU-1 or otherwise enable NONU-1 action. In this model it is unclear which ribosomes are rescued, but prior work supports HBS-1/PELO-1 function on internally-stalled ribosomes [41], consistent with a rescue-first-cleavage-second NGD model. These mechanistic ties between HBS-1 and NONU-1 will enable discovery of the molecular mechanisms underlying the known but poorly understood link between ribosome rescue and mRNA decay.
Overall, our work augments information gained from other systems by demonstrating the conservation of NGD factors in a metazoan, and ordering the sequence of events carried out by ZNF-598, HBS-1/PELO-1, and NONU-1 to bring about mRNA repression in response to ribosomal stalls.
Methods
C. elegans strain construction and propagation
C. elegans strains were derived from theVC2010 strain (N2) [46], and the CB4856 (Hawaiian) strain was used for suppressor mutant mapping. Animals were grown at 16C or 20C on NGM plates seeded with OP50 as a food source [47]. Some strains were obtained from the Caenorhabditis Genetic Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). CRISPR/Cas9 was performed to introduce genomic edits as previously described [48]. Multiple independent isolates of each mutation were recovered and observed to have identical phenotypes. Mutant combinations were generated by crossing. A full list of strains, sequences of mutant alleles, PCR primers, and sources is available in S1 Table.
EMS mutagenesis and suppressor mutant screens
EMS mutagenesis was performed essentially as described [34]. Briefly, a large population of each strain was washed with M9 to a final volume of 4mL. EMS was added to a final concentration of ~1mM and animals were incubated at room temperature for 4 hr with nutation. Animals were washed and left overnight at room temperature on NGM plates seeded with OP50. The next day, animals were washed and eggs were collected by hypochlorite treatment. 100–200 eggs were placed on single small NGM+OP50 plates and allowed to propagate. Plates were screened for F2/F3 animals with increased GFP fluorescence and increased movement. Only a single isolate was kept per NGM plate to ensure independence of mutations.
The first rareArg screen was saturated with hits at znf-598, occluding our ability to recover additional alleles at other loci. To address this, we constructed a double-balanced strain covering the region of chromosome II harboring znf-598, similarly to an approach used in an NMD suppressor screen [49]. This strain (WJA 1040) was homozygous on chromosome I for unc-54(srf1004) and heterozygous on chromosome II for mnC1[dpy-10(e128) unc-52(e444) nls190 let-?] (AG226) and tra-2(e1095) (CB2754). Animals homozygous for mnC1 or homozygous for tra-2(e1095) were inviable or Tra/sterile, respectively, allowing us to eliminate recovery of recessive suppressor mutants within the balanced region. This strain was subjected to EMS mutagenesis as described above. Only isolates maintaining the mnC1/tra-2 heterozygous balancer were kept, as seen by myo-2::GFP (marking mnC1), a nonTra phenotype, and subsequent viability.
Suppressor mutant mapping and identification
Following recovery of mutants from EMS screens, we employed a Hawaiian SNP mapping approach as described [50]. We crossed each isolated suppressor mutant to Hawaiian (CB4856) unc-54(cc4112) males (expressing an UNC-54::mCherry fusion engineered by CRISPR/Cas9). Cross progeny were isolated and allowed to self-fertilize. The F2 GFP+ progeny were then backcrossed to the unc-54(cc4112) males at least 2 times.
After rounds of backcrossing, several phenotypically suppressed animals (~20–30) for a given mutant were pooled together onto a single plate and propagated until starvation. Upon starvation, animals were washed off the plate with 1mL EN50, and further washed with EN50 to remove residual E. coli. Genomic DNA was extracted after proteinase K treatment and resuspended in 50uL TE pH7.4. 50ng genomic DNA was used as an input for Nextera (Tn5) sequencing library preparation. Libraries were sequenced at Novogene Corporation Inc. UC Davis Sequencing Center on a NovaSeq 6000.
Reads were mapped to the C. elegans genome using bowtie2 (version 2.3.4.1). Reads were assigned to haplotypes using GATK [51] and a previously published dataset of Hawaiian SNPs [46]. The fraction of reads that were assignable to Hawaiian or N2 animals was calculated across the genome, and linkage was identified by portions of the genome with 0% Hawaiian. Regions of linkage were then manually inspected to identify candidate lesions and loci.
Fluorescence microscopy and image analysis
All animals were maintained at 20C. L4 animals were selected and anesthetized in 3uL EN50 with 1mM levamisole in a microscope well slide with a 0.15mm coverslip.
A Zeiss AxioZoom microscope was used with a 1.0x objective to acquire images for GFP quantification experiments. The following parameters were used for all images: exposure time of 250ms., shift of 50%, and zoom of 80%. 15–25 representative animals were imaged for each strain. All comparisons shown are between images obtained during the same imaging session. We used FIJI to define the area of the animal, subtract the background, and determine mean pixel intensity for the area of each animal.
A Leica SP5 confocal microscope was used with a 10.0x objective to simultaneously acquire images of mCherry and GFP in overexpression experiments. The following parameters were used for all images: pinhole size set to 106.2 μm; bidirectional scan at 400 Hz with a line average of 2; 488 laser to capture GFP and 594 laser to capture mCherry, both at 50% power with a gain of 700 and offset of -0.2%. 4–10 animals were analyzed from a total of 3 independent isolates for each overexpression experiment. A custom python script was used to define the area of the animal, then parse out locations and intensities of green and red pixels. We considered the brightest 1,000 green and brightest 1,000 red pixels and calculated the Jaccard index. The theoretical maximum of the Jaccard index is 1, indicating no overlap of green and red pixels. The observed minimum of the Jaccard index among our strains was ~0.75, higher than the theoretical lower bound of zero, which is the result of expression of mCherry and GFP being driven in the same tissue. To display the image quantification more intuitively, we transformed the Jaccard index into an overlap score. This was done by performing a simple linear transformation of 0.75 (more overlap) to 1.0 (less overlap) in Jaccard space into an overlap score of -1.0 (less overlap) to 1.0 (more overlap). See S1 Fig for images of animals representing various overlap scores.
Immunoblotting
Animals were propagated on NGM plates with OP50 until freshly starved, then washed twice with 1ml M9 and flash frozen in liquid nitrogen. Animal pellets were boiled for 10 min at 99C in 1x SDS loading buffer (0.1M Tris-HCl, 20% glycerol, 4% SDS, 0.1M DTT, 0.05% bromophenol blue). Samples were vortexed and spun to pellet animals and supernatants were collected. Protein was quantified by Qubit and 15ug protein was run on a 4–20% Mini-PROTEAN(R) TGX Stain-Free Protein gel (Bio-Rad). Protein was transferred to a low background fluorescence PVDF membrane (Millipore Sigma) and the membrane was blocked in 5% nonfat milk in 1x TBST. Anti-HA antibody (Enzo Life Sciences 16B12) was used at a 1:2000 dilution to detect HA-tagged RPS-10 protein. Anti-FLAG antibody (Millipore Sigma F1804) was used at a 1:1000 dilution to detect FLAG-tagged RPS-20 protein. Anti-H3 antibody (Millipore Sigma SAB4500352) was used at a 1:2000 dilution to detect the histone H3 protein. Secondary antibody staining was performed with 1:15000 LI-COR goat anti-mouse or LI-COR goat anti-rabbit (LI-COR). Imaging was done using a LI-COR Odyssey Imaging System (LI-COR), with quantification done in ImageStudio.
Mutual information calculation
Phylogenetic profiling was performed using the PANTHER HMM library Version 16.0 on Ensembl Protists Genome (release 51) combined with MMETSP [52]. To identify homologs of proteins from the PANTHER Subfamily HMMs, we used HMMER’s hmmsearch function using the following parameters—noali—notextw—cpu 2. We then parsed each output file from HMMER and determined the presence (1) or absence (0) of a homolog in each organism for a sub-family HMM.
Briefly, if multiple subfamily HMMs matched a given protein in a species, the protein was assigned to the subfamily with the highest bit score, and lesser-scoring subfamilies were ignored. Subfamilies with homologs in <5% or >95% of species were discarded; such subfamilies exhibit too little variation for accurate mutual information calculations. Similarly, species with hits for <10% of subfamilies were discarded; such species may represent poor transcriptome coverage and/or assemblies. We also computed the pairwise hamming distance between all species and discarded species until the pairwise hamming distance between all species was at least 0.1, so as to ensure biodiversity. Mutual information for discrete variables was then calculated as: sum(i = 0,1); sum(j = 0,1) [-log_2(p_ij/(p_i*p_j))].
Negative controls for mutual information calculations were generated as follows. For a given pair of factors A and B, we randomized the binary vector of presence (1) and absence (0) of B in organisms to create a new negative control factor. Mutual information for discrete variables was then calculated between factor A and the newly created negative control.
RNA-seq and analysis
Animals were synchronized by hypochlorite treatment, propagated on NGM plates with OP50 at 16C, and harvested at the L4/young adult stage. Animals were washed off with N50, passed through a 5% sucrose cushion in N50 to remove E. coli, and snap frozen in liquid nitrogen. Animals were lysed by grinding in a mortar and pestle cooled in liquid nitrogen in the presence of frozen PLB (20mM Tris pH8.0, 140mM KCl, 1.5mM MgCl2, 1% Triton) and 100ug/mL cycloheximide. Ground animals were stored as frozen powder at -70C. Total RNA was extracted with trizol, resuspended in TE pH7.4, and quantified by Qubit. Ribosomal RNA was depleted using custom C. elegans-specific rRNA hybridization oligos, similar to a planarian protocol as described [53]. Oligos for this protocol are included in S1 Table. Libraries were prepared using an NEBNext Ultra II Directional RNA Library Prep Kit for Illumina sequencing. Libraries were sequenced at Novogene Corporation Inc. UC Davis Sequencing Center on a NovaSeq 6000 and at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley on a HiSeq 4000.
We generated a custom C. elegans genome (Ensembl, WBCel235) containing unc-54(rareArg) as a separate chromosome and a masked endogenous unc-54 locus. Reads were mapped to this genome including annotated splice junctions using STAR v2.5.4b [54] allowing for six mismatches. All downstream analyses were restricted to uniquely mapping reads.
In vitro ubiquitin-binding
Ubiquitin-binding assay was performed essentially as described in [40]. Briefly, recombinant proteins were induced in E. coli with 1 mM IPTG at 37C for 4 hours. Cells were harvested and suspended in 1x PBS (137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, and 1.8mM KH2PO4), followed by lysis in xTractor buffer (Takara) per the manufacturer’s protocol. Lysates were cleared by centrifugation and immobilized on TALON metal affinity resin (Takara) per the manufacturer’s protocol. Immobilized proteins were washed with 1x PBS and incubated overnight at 4C with clarified lysates containing various GST-fusion constructs. Bound proteins were washed four times with 1x PBS with 5mM imidazole and eluted by heating to 99C for 5 min in 1x SDS loading buffer. Eluted protein was run on a 4–20% Mini-PROTEAN(R) TGX Stain-Free Protein gel (Bio-Rad) and stained with Coomassie brilliant blue.
Supporting information
S1 Fig
Representative animals demonstrating various overlap scores.GFP and mCherry images of representative animals expressing unc-54(rareArg) and an mCherry-tagged array. Above is the calculated overlap score.
(TIF)
S2 Fig
RPS-10 and RPS-20 ubiquitination sites are conserved in C. elegans.(A) Pairwise sequence alignment of H. sapiens eS10 and C. elegans RPS-10. K125 is highlighted in green. Conservation is as shown in Fig 3A. (B) As in (A), showing H. sapiens uS10 and C. elegans RPS-20 with K6 and K9 highlighted in green.
(TIF)
S3 Fig
nonu-1 null mutant displays similar NGD suppression as catalytic mutant.Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type.
(TIF)
S4 Fig
Known surveillance factor partners exhibit high mutual information.(A) HBS-1 mutual information plot. 90% percentile cutoff is shown as a dashed line and PELO-1 is highlighted in purple. (B) As in (A), showing LTN-1 mutual information with RQC-2 in red.
(TIF)
S5 Fig
Ubiquitin-binding assay shows high level of background binding.His6-tagged constructs of S. cerevisiae Cue2 CUE domains, H. sapiens HBS1 N-term domain, H. sapiens HBS1 N-term globular (triple helix) domain, H. sapiens HBS1 N-term globular (triple helix) domain with G93D mutation (Gly in predicted binding site found from [55]), and H. sapiens SMG5 PIN domain. His6 constructs were immobilized on cobalt metal affinity resin and incubated with E. coli lysates expressing GST-Ub or GST. Proteins boiled from resin are shown on Coomassie stained gel, with proteins indicated. Asterisk indicates nonspecific peptide present on resin.
(TIF)
S6 Fig
HBS-1 and PELO-1 exhibit similar phenotypes during NGD in C. elegans.Mean RFUs (relative fluorescence units) of indicated strains (n≥15 animals/strain) in the unc-54(rareArg) background. One standard deviation shown as error bars. p values from Welch’s t-test, with asterisks indicating p<0.01 for all comparisons with wild type.
(TIF)
Acknowledgments
We thank members of the Arribere lab for thoughtful discussions. We thank Benjamin Abrams at the UCSC Life Sciences Microscopy Center for technical support (RRID: SCR_021135). We thank Wormbase.
Funding Statement
This work was supported in part by a T32 Training Grant Fellowship from the National Institute of General Medical Sciences (NIGMS) (5T32GM133391) to P.C.M., an R01 grant from the NIGMS (1R01GM131012) to J.A.A., a Searle Scholars award to J.A.A., and start-up funds from UCSC to J.A.A. The funders did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The data discussed in this publication are accessible through GEO Series accession number GSE218514 and S2 Table.
References
Author response to previous submission
29 Nov 2022
Attachment
Submitted filename: 221129_responseToReviewers.pdf
Decision Letter 0
13 Dec 2022
Dear Dr Arribere,
Thank you very much for submitting your Research Article entitled 'Ubiquitination of stalled ribosomes enables mRNA decay via HBS-1 and NONU-1 in vivo' to PLOS Genetics.
The manuscript was fully evaluated at the editorial level and by independent two peer reviewers. The reviewers are essentially ready to recommend acceptance, but Reviewer #2 has a handful of minor, easily addressable, but nonetheless important issues. After you attend to these I will be able to proceed with an editorial decision without further external evaluation.
We therefore ask you to modify the manuscript according to the review recommendations. Your revisions should address the specific points made by each reviewer.
In addition we ask that you:
1) Provide a detailed list of your responses to the review comments and a description of the changes you have made in the manuscript.
2) Upload a Striking Image with a corresponding caption to accompany your manuscript if one is available (either a new image or an existing one from within your manuscript). If this image is judged to be suitable, it may be featured on our website. Images should ideally be high resolution, eye-catching, single panel square images. For examples, please browse our archive. If your image is from someone other than yourself, please ensure that the artist has read and agreed to the terms and conditions of the Creative Commons Attribution License. Note: we cannot publish copyrighted images.
We hope to receive your revised manuscript within the next 30 days. If you anticipate any delay in its return, we would ask you to let us know the expected resubmission date by email to gro.solp@scitenegsolp.
If present, accompanying reviewer attachments should be included with this email; please notify the journal office if any appear to be missing. They will also be available for download from the link below. You can use this link to log into the system when you are ready to submit a revised version, having first consulted our Submission Checklist.
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email us at gro.solp@serugif.
Please be aware that our data availability policy requires that all numerical data underlying graphs or summary statistics are included with the submission, and you will need to provide this upon resubmission if not already present. In addition, we do not permit the inclusion of phrases such as "data not shown" or "unpublished results" in manuscripts. All points should be backed up by data provided with the submission.
To enhance the reproducibility of your results, we recommend that you deposit your laboratory protocols in protocols.io, where a protocol can be assigned its own identifier (DOI) such that it can be cited independently in the future. Additionally, PLOS ONE offers an option to publish peer-reviewed clinical study protocols. Read more information on sharing protocols at https://plos.org/protocols?utm_medium=editorial-email&utm_source=authorletters&utm_campaign=protocols
Please review your reference list to ensure that it is complete and correct. If you have cited papers that have been retracted, please include the rationale for doing so in the manuscript text, or remove these references and replace them with relevant current references. Any changes to the reference list should be mentioned in the rebuttal letter that accompanies your revised manuscript. If you need to cite a retracted article, indicate the article’s retracted status in the References list and also include a citation and full reference for the retraction notice.
PLOS has incorporated Similarity Check, powered by iThenticate, into its journal-wide submission system in order to screen submitted content for originality before publication. Each PLOS journal undertakes screening on a proportion of submitted articles. You will be contacted if needed following the screening process.
To resubmit, you will need to go to the link below and 'Revise Submission' in the 'Submissions Needing Revision' folder.
Please let us know if you have any questions while making these revisions.
Yours sincerely,
Gregory P. Copenhaver
Editor-in-Chief
PLOS Genetics
Reviewer's Responses to Questions
Comments to the Authors:
Please note here if the review is uploaded as an attachment.
Reviewer #1: The authors successfully addressed my remaining concerns and the manuscript has been greatly improved.
Reviewer #2: This is a very thoroughly revised manuscript that is greatly improved by the addition of new data, removal of data irrelevant to the main point, and by thoroughly revising the text. In this manuscript the authors study no-go mRNA decay in C. elegans. They very effectively use the powerful genetics of this system to identify factors required for no-go decay and then follow up with experiments that provide strong evidence that supports the main conclusion that ribosomal ubiquitination enables mRNA degradation via HBS-1 and NONU-1 in an intact animal. The revised manuscript is a valuable extension of what is known and should be published in PLOS Genetics. I have some very minor comments for the authors that may help them further improve the manuscript detailed below
Minor comments:
1. The authors isolated 20 mutants but only describe 9 alleles. If he authors identified mutations in the other 11 mutants, it would be helpful to the field to add those to table S1, for example if another researcher identifies mutations in the same genes or their orthologs. If the other 11 alleles were not mapped and sequenced, perhaps the authors can clarify that.
2. In line 298 the authors mention creating tagged alleles of NONU-1 that turned out to be nonfunctional. It might be useful to the field to include the details in the supplement. This would prevent future researchers from wasting time trying the same tagging strategy.
3. The use of “phenocopy” in lines 223, 285, 307, 308, and 427 is at best unnecessary jargon but according to the definition of phenocopy in my dictionary or in Wikipedia is actually incorrect. Phenocopy should mean that some specific treatment (e.g. a drug) has the same effect as a mutation and not that two mutations have the same effect. The authors correctly describe mutations with similar phenotypes in lines 296 and 403-406. Even worse is in line 389 where a mutant is described as phenocopying wild type. Much clearer would be to say that deleting the N-terminus of HBS-1 had no discernible phenotype.
4. Line 189 “containing fluorescent protein fusions” is incorrect. The T2A stop and go sequence means that mCherry and ZNF-598 (or NONU-1) are produced as separate proteins and not fusion proteins. In addition, plasmids may encode proteins, but do not “contain” them.
5. Line 429 “acts” should be “act”
**********
Have all data underlying the figures and results presented in the manuscript been provided?
Large-scale datasets should be made available via a public repository as described in the PLOS Genetics data availability policy, and numerical data that underlies graphs or summary statistics should be provided in spreadsheet form as supporting information.
Reviewer #1: Yes
Reviewer #2: Yes
**********
PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: Yes: Seung-Jae V. Lee
Reviewer #2: No
Author response to Decision Letter 0
15 Dec 2022
Attachment
Submitted filename: 221214_responseToReviewers.docx
Decision Letter 1
18 Dec 2022
Dear Dr Arribere,
We are pleased to inform you that your manuscript entitled "Ubiquitination of stalled ribosomes enables mRNA decay via HBS-1 and NONU-1 in vivo" has been editorially accepted for publication in PLOS Genetics. Congratulations!
Before your submission can be formally accepted and sent to production you will need to complete our formatting changes, which you will receive in a follow up email. Please be aware that it may take several days for you to receive this email; during this time no action is required by you. Please note: the accept date on your published article will reflect the date of this provisional acceptance, but your manuscript will not be scheduled for publication until the required changes have been made.
Once your paper is formally accepted, an uncorrected proof of your manuscript will be published online ahead of the final version, unless you’ve already opted out via the online submission form. If, for any reason, you do not want an earlier version of your manuscript published online or are unsure if you have already indicated as such, please let the journal staff know immediately at gro.solp@scitenegsolp.
In the meantime, please log into Editorial Manager at https://www.editorialmanager.com/pgenetics/, click the "Update My Information" link at the top of the page, and update your user information to ensure an efficient production and billing process. Note that PLOS requires an ORCID iD for all corresponding authors. Therefore, please ensure that you have an ORCID iD and that it is validated in Editorial Manager. To do this, go to ‘Update my Information’ (in the upper left-hand corner of the main menu), and click on the Fetch/Validate link next to the ORCID field. This will take you to the ORCID site and allow you to create a new iD or authenticate a pre-existing iD in Editorial Manager.
If you have a press-related query, or would like to know about making your underlying data available (as you will be aware, this is required for publication), please see the end of this email. If your institution or institutions have a press office, please notify them about your upcoming article at this point, to enable them to help maximise its impact. Inform journal staff as soon as possible if you are preparing a press release for your article and need a publication date.
Thank you again for supporting open-access publishing; we are looking forward to publishing your work in PLOS Genetics!
Yours sincerely,
Gregory P. Copenhaver
Editor-in-Chief
PLOS Genetics
Twitter: @PLOSGenetics
----------------------------------------------------
Comments from the reviewers (if applicable):
----------------------------------------------------
Data Deposition
If you have submitted a Research Article or Front Matter that has associated data that are not suitable for deposition in a subject-specific public repository (such as GenBank or ArrayExpress), one way to make that data available is to deposit it in the Dryad Digital Repository. As you may recall, we ask all authors to agree to make data available; this is one way to achieve that. A full list of recommended repositories can be found on our website.
The following link will take you to the Dryad record for your article, so you won't have to re‐enter its bibliographic information, and can upload your files directly:
http://datadryad.org/submit?journalID=pgenetics&manu=PGENETICS-D-22-01367R1
More information about depositing data in Dryad is available at http://www.datadryad.org/depositing. If you experience any difficulties in submitting your data, please contact gro.dayrdatad@pleh for support.
Additionally, please be aware that our data availability policy requires that all numerical data underlying display items are included with the submission, and you will need to provide this before we can formally accept your manuscript, if not already present.
----------------------------------------------------
Press Queries
If you or your institution will be preparing press materials for this manuscript, or if you need to know your paper's publication date for media purposes, please inform the journal staff as soon as possible so that your submission can be scheduled accordingly. Your manuscript will remain under a strict press embargo until the publication date and time. This means an early version of your manuscript will not be published ahead of your final version. PLOS Genetics may also choose to issue a press release for your article. If there's anything the journal should know or you'd like more information, please get in touch via gro.solp@scitenegsolp.
Acceptance letter
5 Jan 2023
PGENETICS-D-22-01367R1
Ubiquitination of stalled ribosomes enables mRNA decay via HBS-1 and NONU-1 in vivo
Dear Dr Arribere,
We are pleased to inform you that your manuscript entitled "Ubiquitination of stalled ribosomes enables mRNA decay via HBS-1 and NONU-1 in vivo" has been formally accepted for publication in PLOS Genetics! Your manuscript is now with our production department and you will be notified of the publication date in due course.
The corresponding author will soon be receiving a typeset proof for review, to ensure errors have not been introduced during production. Please review the PDF proof of your manuscript carefully, as this is the last chance to correct any errors. Please note that major changes, or those which affect the scientific understanding of the work, will likely cause delays to the publication date of your manuscript.
Soon after your final files are uploaded, unless you have opted out or your manuscript is a front-matter piece, the early version of your manuscript will be published online. The date of the early version will be your article's publication date. The final article will be published to the same URL, and all versions of the paper will be accessible to readers.
Thank you again for supporting PLOS Genetics and open-access publishing. We are looking forward to publishing your work!
With kind regards,
Zsuzsanna Gémesi
PLOS Genetics
On behalf of:
The PLOS Genetics Team
Carlyle House, Carlyle Road, Cambridge CB4 3DN | United Kingdom
gro.solp@scitenegsolp | +44 (0) 1223-442823
plosgenetics.org | Twitter: @PLOSGenetics
Articles from PLOS Genetics are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pgen.1010577
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosgenetics/article/file?id=10.1371/journal.pgen.1010577&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/141067731
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pgen.1010577
Article citations
High-resolution reconstruction of a <i>C. elegans</i> ribosome sheds light on evolutionary dynamics and tissue specificity.
RNA, 30(11):1513-1528, 16 Oct 2024
Cited by: 0 articles | PMID: 39209556
Molecular targets and strategies in the development of nucleic acid cancer vaccines: from shared to personalized antigens.
J Biomed Sci, 31(1):94, 09 Oct 2024
Cited by: 0 articles | PMID: 39379923 | PMCID: PMC11463125
Review Free full text in Europe PMC
Translation-coupled mRNA quality control mechanisms.
EMBO J, 42(19):e114378, 22 Aug 2023
Cited by: 14 articles | PMID: 37605642 | PMCID: PMC10548175
Review Free full text in Europe PMC
In vitro Analysis of Stalled Ribosomes Using Puromycin Incorporation.
Bio Protoc, 13(16):e4744, 20 Aug 2023
Cited by: 1 article | PMID: 37638299 | PMCID: PMC10450738
A ubiquitin language communicates ribosomal distress.
Semin Cell Dev Biol, 154(pt b):131-137, 22 Mar 2023
Cited by: 0 articles | PMID: 36963992 | PMCID: PMC10878831
Review Free full text in Europe PMC
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Protein structures in PDBe (3)
-
(1 citation)
PDBe - 3IZQView structure
-
(1 citation)
PDBe - 5M1JView structure
-
(1 citation)
PDBe - 1P3QView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay.
Elife, 8:e49117, 20 Jun 2019
Cited by: 103 articles | PMID: 31219035 | PMCID: PMC6598757
Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J, 38(5):e100276, 04 Jan 2019
Cited by: 154 articles | PMID: 30609991 | PMCID: PMC6396155
NONU-1 Encodes a Conserved Endonuclease Required for mRNA Translation Surveillance.
Cell Rep, 30(13):4321-4331.e4, 01 Mar 2020
Cited by: 47 articles | PMID: 32234470 | PMCID: PMC7184879
A ubiquitin language communicates ribosomal distress.
Semin Cell Dev Biol, 154(pt b):131-137, 22 Mar 2023
Cited by: 0 articles | PMID: 36963992 | PMCID: PMC10878831
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: T32 GM133391
Grant ID: R01 GM131012
National Institute of General Medical Sciences (2)
Grant ID: 5T32GM133391
Grant ID: 1R01GM131012

 *
*


