Abstract
Free full text

The compact Casπ (Cas12l) ‘bracelet’ provides a unique structural platform for DNA manipulation
Abstract
CRISPR-Cas modules serve as the adaptive nucleic acid immune systems for prokaryotes, and provide versatile tools for nucleic acid manipulation in various organisms. Here, we discovered a new miniature type V system, CRISPR-Casπ (Cas12l) (~860 aa), from the environmental metagenome. Complexed with a large guide RNA (~170 nt) comprising the tracrRNA and crRNA, Casπ (Cas12l) recognizes a unique 5′ C-rich PAM for DNA cleavage under a broad range of biochemical conditions, and generates gene editing in mammalian cells. Cryo-EM study reveals a ‘bracelet’ architecture of Casπ effector encircling the DNA target at 3.4 Å resolution, substantially different from the canonical ‘two-lobe’ architectures of Cas12 and Cas9 nucleases. The large guide RNA serves as a ‘two-arm’ scaffold for effector assembly. Our study expands the knowledge of DNA targeting mechanisms by CRISPR effectors, and offers an efficient but compact platform for DNA manipulation.
Å resolution, substantially different from the canonical ‘two-lobe’ architectures of Cas12 and Cas9 nucleases. The large guide RNA serves as a ‘two-arm’ scaffold for effector assembly. Our study expands the knowledge of DNA targeting mechanisms by CRISPR effectors, and offers an efficient but compact platform for DNA manipulation.
Introduction
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) genes function as the adaptive immune module for many prokaryotes and huge phages against invading nucleic acid.1,2 Generally, the CRISPR immune response comprises the DNA adaptation, effector biogenesis and nucleic acid interference stages.3 With excellent engineerable capacity, the CRISPR effectors that provide RNA-guided DNA targeting and cleaving activities are also effectively repurposed as genomic, epigenomic and transcriptional manipulation tools in many organisms.4,5
Though an increasing number of CRISPR-Cas effectors have confirmed DNA interference activity in vitro, only a few of them, like SpyCas9 and AsCas12a, substantially work and are widely used for efficient genome editing in vivo.6–8 Among these few effectors, the large molecular size of their Cas nucleases (1200–1400 amino acids (aa)) largely limits the options of delivering vehicles into the target cells. Furthermore, although several types of compact effectors with Cas nucleases < 1000 aa have recently been employed for genome editing (CasPhi (Cas12j) effector, 700–800 aa protein monomer with ~40 nt crRNA; Cas12f effector, 900–1000 aa protein dimer with ~190
nt crRNA; Cas12f effector, 900–1000 aa protein dimer with ~190 nt single guide RNA (sgRNA); CasX (Cas12e) effector, ~980 aa protein monomer with ~120
nt single guide RNA (sgRNA); CasX (Cas12e) effector, ~980 aa protein monomer with ~120 nt sgRNA), the initial versions of these compact systems all exhibit weak or moderate editing efficacy and require extensive and persisted optimization for further application,8–12 similar to how SpyCas9-based technology was developed in the last decade. Moreover, all these compact effectors recognize the T-rich protospacer adjacent motif (PAM), largely limiting the targeting scope during gene editing practice. Structural design and directed evolution have been performed to alter the PAM preference for Cas effectors, but the significant decrease of editing efficacy or fidelity has often been observed for those mutants.11,12 Therefore, compact but still efficient effectors which offer unique targeting scopes are essential to overcome the application limitations within the current gene editing toolbox.
nt sgRNA), the initial versions of these compact systems all exhibit weak or moderate editing efficacy and require extensive and persisted optimization for further application,8–12 similar to how SpyCas9-based technology was developed in the last decade. Moreover, all these compact effectors recognize the T-rich protospacer adjacent motif (PAM), largely limiting the targeting scope during gene editing practice. Structural design and directed evolution have been performed to alter the PAM preference for Cas effectors, but the significant decrease of editing efficacy or fidelity has often been observed for those mutants.11,12 Therefore, compact but still efficient effectors which offer unique targeting scopes are essential to overcome the application limitations within the current gene editing toolbox.
Here, via a home-developed bioinformatics pipeline using iterative Hidden Markov model (HMM), we identified a new and compact type V CRISPR-Cas family with four orthologous proteins from the environmental metagenome. We designated this new subtype as CRISPR-Casπ, or CRISPR-Cas12l referring to the recent version of complete classification for CRISPR.13 Different from the T-rich PAM preference within the reported type V effectors including those with compact sizes (750–1000 aa protein with 45–190 nt guide RNA (gRNA)),14,15 the Casπ (Cas12l) effectors (~860
nt guide RNA (gRNA)),14,15 the Casπ (Cas12l) effectors (~860 aa protein with ~170
aa protein with ~170 nt gRNA) recognize the 5′ C-rich PAM for DNA cleavage under various biochemical environments and exhibit efficient trans-activity promising for diagnosis application. Furthermore, even without optimization, the naive versions of Casπ (Cas12l) effectors behave effectively for DNA manipulation both in prokaryotic and eukaryotic cells. Cryo-EM study revealed that Casπ (Cas12l) protein presents a locked ‘bracelet’ architecture for DNA targeting, which is unique from the canonical ‘two-lobe’ Class 2 nucleases (Cas9 and Cas12). Notably, four non-reported structural domains are identified, including a 69
nt gRNA) recognize the 5′ C-rich PAM for DNA cleavage under various biochemical environments and exhibit efficient trans-activity promising for diagnosis application. Furthermore, even without optimization, the naive versions of Casπ (Cas12l) effectors behave effectively for DNA manipulation both in prokaryotic and eukaryotic cells. Cryo-EM study revealed that Casπ (Cas12l) protein presents a locked ‘bracelet’ architecture for DNA targeting, which is unique from the canonical ‘two-lobe’ Class 2 nucleases (Cas9 and Cas12). Notably, four non-reported structural domains are identified, including a 69 aa ‘proline-rich string’ loop and a ‘lock-catch’ domain which work together to tie up the Casπ (Cas12l) and lock it around the nucleic acid target. The large sgRNA composed of the tracrRNA and crRNA folds into a ‘two-arm’ scaffold to recruit and embrace the Casπ (Cas12l) nuclease, forming the stable DNA interference effector. Collectively, our results provide a novel and compact DNA manipulation platform to substantially expand the CRISPR toolbox and offer new aspects to further explore the CRISPR biology.
aa ‘proline-rich string’ loop and a ‘lock-catch’ domain which work together to tie up the Casπ (Cas12l) and lock it around the nucleic acid target. The large sgRNA composed of the tracrRNA and crRNA folds into a ‘two-arm’ scaffold to recruit and embrace the Casπ (Cas12l) nuclease, forming the stable DNA interference effector. Collectively, our results provide a novel and compact DNA manipulation platform to substantially expand the CRISPR toolbox and offer new aspects to further explore the CRISPR biology.
Results
Casπ (Cas12l) is a novel type of compact nuclease guided by a large tracr–crRNA hybrid
During the last decade, huge efforts have been made to explore the CRISPR systems in prokaryotic genome and revealed a large CRISPR kingdom with functional and structural diversities.1,13 Nowadays, it is challenging to identify novel systems to further expand the CRISPR biology. Therefore, we built an iterative bioinformatics pipeline and performed large-scale environmental sample screening over the land and ocean (Supplementary information, Fig. S1a). From the metagenome of sludge sample previously collected in Tianjin and Beijing for symbiotic bacteria research, we discovered a new Class 2 CRISPR family with three orthologous systems that bear significant phylogenetic distance from all reported subtypes (Fig. 1a; Supplementary information, Fig. S1b and Table S1).16,17 To reveal the entire CRISPR cassette, the metagenome was re-sequenced and updated (see Materials and methods; NCBI Accession ID: PRJNA857874).
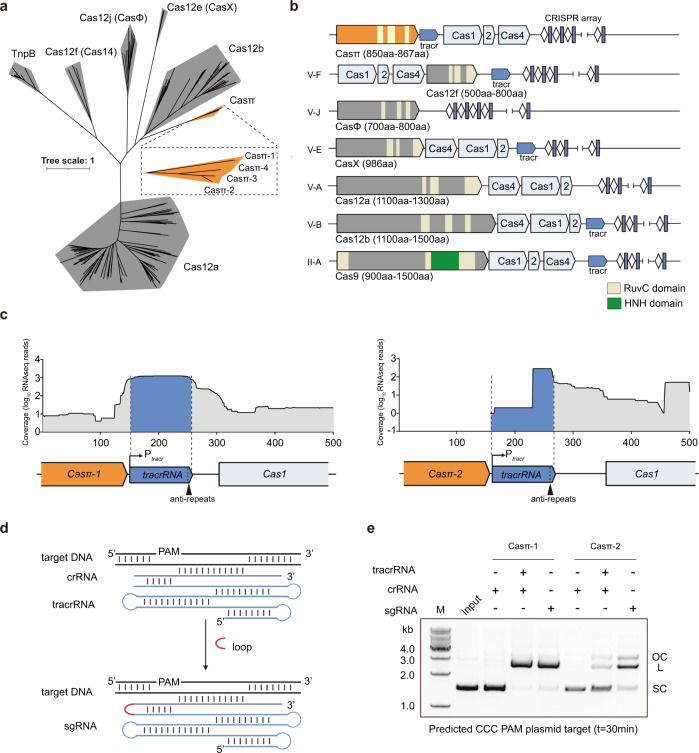
a Maximum likelihood phylogenic analysis of Casπ orthologs with reported type V Cas nucleases, which are employed in genome-editing application. Bootstrap =
= 1500, Casπ protein sequences are shown in Supplementary information, Table S1. b Illustrations of genomic loci of Class 2 CRISPR family members employed in genome-editing application. c Meta-transcriptome results mapped to the native genomic loci of Casπ tracrRNA (promoter regions were predicted by BDGP and labeled with Ptracr; anti-repeat region was labeled with arrow in black). d Schematic diagram of Casπ dual-guide RNA (crRNA in purple, tracrRNA in cyan). The RNA loop that connects the tracrRNA and crRNA is depicted in red. e In vitro cleavage of plasmids containing predicted CCC PAM by both Casπ proteins using crRNA, tracr–crRNA pair or sgRNA. SC supercoiled plasmids; L linearized plasmids; OC open-circle plasmids.
1500, Casπ protein sequences are shown in Supplementary information, Table S1. b Illustrations of genomic loci of Class 2 CRISPR family members employed in genome-editing application. c Meta-transcriptome results mapped to the native genomic loci of Casπ tracrRNA (promoter regions were predicted by BDGP and labeled with Ptracr; anti-repeat region was labeled with arrow in black). d Schematic diagram of Casπ dual-guide RNA (crRNA in purple, tracrRNA in cyan). The RNA loop that connects the tracrRNA and crRNA is depicted in red. e In vitro cleavage of plasmids containing predicted CCC PAM by both Casπ proteins using crRNA, tracr–crRNA pair or sgRNA. SC supercoiled plasmids; L linearized plasmids; OC open-circle plasmids.
Overall, this novel system includes the integration module with cas1, cas2 and cas4 genes, and an uncharacterized gene encoding an 867 aa protein that we designate as Casπ (or Cas12l referring to the recent version of complete classification for CRISPR, hereafter all mentioned as Casπ for convenient description) (Fig. 1b; Supplementary information, Fig. S1c). Via basic local alignment search (BLAST) in public database,18 we further discovered a fourth orthologous system, Casπ-4 (854
aa protein that we designate as Casπ (or Cas12l referring to the recent version of complete classification for CRISPR, hereafter all mentioned as Casπ for convenient description) (Fig. 1b; Supplementary information, Fig. S1c). Via basic local alignment search (BLAST) in public database,18 we further discovered a fourth orthologous system, Casπ-4 (854 aa), which shares ~45% protein sequence identity with Casπ-1 and ~62% identity with both Casπ-2 and Casπ-3 (Fig. 1a; Supplementary information, Fig. S1c, d).19 Of note, all four CRISPR-Casπ cassettes were validated to reside in the genomes of Armatimonadetes bacterium (Supplementary information, Fig. S1c). Remote homology detection, structural prediction and sequence alignment identified a RuvC nuclease domain near the Casπ C-terminus, with organization reminiscent of that found in type V CRISPR-Cas systems (Fig. 1b; Supplementary information, Fig. S1e and Data S1).20–22 The rest of the Casπ protein (~500 amino acids at the N-terminus) showed no detectable similarity to any annotated protein (probability
aa), which shares ~45% protein sequence identity with Casπ-1 and ~62% identity with both Casπ-2 and Casπ-3 (Fig. 1a; Supplementary information, Fig. S1c, d).19 Of note, all four CRISPR-Casπ cassettes were validated to reside in the genomes of Armatimonadetes bacterium (Supplementary information, Fig. S1c). Remote homology detection, structural prediction and sequence alignment identified a RuvC nuclease domain near the Casπ C-terminus, with organization reminiscent of that found in type V CRISPR-Cas systems (Fig. 1b; Supplementary information, Fig. S1e and Data S1).20–22 The rest of the Casπ protein (~500 amino acids at the N-terminus) showed no detectable similarity to any annotated protein (probability <
< 50% and E-value
50% and E-value >
> 200 by HH-suite),21 suggesting Casπ as a novel type V nuclease. Furthermore, the genomic organization of cas1–cas2–cas4 integration module in CRISPR-Casπ cassette is unique from the common cas4–cas1–cas2 pattern within type V systems (Fig. 1b). The 37
200 by HH-suite),21 suggesting Casπ as a novel type V nuclease. Furthermore, the genomic organization of cas1–cas2–cas4 integration module in CRISPR-Casπ cassette is unique from the common cas4–cas1–cas2 pattern within type V systems (Fig. 1b). The 37 bp CRISPR repeats within the four systems share ~68% DNA sequence identity, and the tracrRNA anti-repeat is well identified next to each casπ gene rather than proximal to CRISPR repeats as seen in other type V systems (Fig. 1c; Supplementary information, Fig. S1d and Table S1).
bp CRISPR repeats within the four systems share ~68% DNA sequence identity, and the tracrRNA anti-repeat is well identified next to each casπ gene rather than proximal to CRISPR repeats as seen in other type V systems (Fig. 1c; Supplementary information, Fig. S1d and Table S1).
Since the Casπ-1 and Casπ-2 nucleases bear the largest evolution distance within this new family (Fig. 1a; Supplementary information, Fig. S1d), we then chose these two orthologs for further experimental characterization. Via promoter prediction and meta-transcriptome mapping to the anti-repeat regions (see Materials and methods), the tracrRNA sequences for Casπ-1 and Casπ-2 systems were determined to be substantially long (> 100 nt) (Fig. 1c; Supplementary information, Fig. S1c, Tables S1 and S2). Further, the DNA cleavage activity of Casπ effectors guided by tracrRNA and crRNA was tested using predicated PAM by CRISPRTarget server (AGC PAM1 for Casπ-1 and CCC PAM2 for Casπ-2).23 While rarely recognizing PAM1, both Casπ nucleases robustly linearized the target plasmid containing PAM2 using the tracr–crRNA pair or a joint hybrid (sgRNA) (Fig. 1d, e; Supplementary information, Fig. S1f, g). Thus, Casπ (~860
nt) (Fig. 1c; Supplementary information, Fig. S1c, Tables S1 and S2). Further, the DNA cleavage activity of Casπ effectors guided by tracrRNA and crRNA was tested using predicated PAM by CRISPRTarget server (AGC PAM1 for Casπ-1 and CCC PAM2 for Casπ-2).23 While rarely recognizing PAM1, both Casπ nucleases robustly linearized the target plasmid containing PAM2 using the tracr–crRNA pair or a joint hybrid (sgRNA) (Fig. 1d, e; Supplementary information, Fig. S1f, g). Thus, Casπ (~860 aa) associated with a large tracr–crRNA hybrid (~170
aa) associated with a large tracr–crRNA hybrid (~170 nt) functions as a novel type of compact DNA interference effector.
nt) functions as a novel type of compact DNA interference effector.
Casπ cleaves DNA targets using 5′ C-rich PAM distinct from other Cas12 variants
To further determine the biochemical characteristics of Casπ, we started with identifying the PAM preference of both orthologs using a plasmid library containing five randomized DNA nucleotides upstream of the protospacer (Fig. 2a; Supplementary information, Fig. S2a). Deep sequencing analysis suggests that both Casπ effectors recognize the 5′-CCN-3′ PAM (Fig. 2a; Supplementary information, Fig. S2b, c and Table S3). Specifically, for Casπ-1 effector, the strictness of PAM requirement increases when increasing the salt concentration in the cleavage buffer (Supplementary information, Fig. S2b). Notably, this C-rich PAM preference for Casπ is different from the T-rich PAM preference for all reported type V nucleases (Supplementary information, Fig. S2d), which will help expand the targeting scope for type V-based technologies. Using the most favorable CCC PAM determined by plasmid screening assay, we observed efficient cleavage activity for both Casπ effectors on the double-stranded DNA (dsDNA) target even compared to the large Lachnospiraceae bacterium Cas12a (LbCas12a, 1228 aa) effector (Fig. 2b; Supplementary information, Table S3). A further screening showed that both Casπ effectors can only robustly cleave the dsDNA target with CCC or CCT (CCY) PAM, indicating a more stringent PAM requirement on dsDNA target (linearized substrate) compared to plasmid target (negative supercoiled substrate) (Supplementary information, Fig. S2e, f). Gel analysis of the cleavage products from the DNA non-target strand (NTS) and target strand (TS) showed that both effectors generate a staggered cut on the dsDNA (Fig. 2c). Consistent with the deep sequencing analysis result for plasmid cleavage (Supplementary information, Fig. S2a, g, h), the exact cleavage sites locate at 11–14 nt downstream of the PAM on the NTS and 2–4
nt downstream of the PAM on the NTS and 2–4 nt downstream of the protospacer on the TS, thus leaving a 5′ single strand overhang of 6–12
nt downstream of the protospacer on the TS, thus leaving a 5′ single strand overhang of 6–12 nt on the products (Fig. 2d, e). Moreover, we observed the single-stranded DNA (ssDNA) TS cleavage (cis-cleavage) by both effectors, and the cleavage efficacy and pattern are comparable to the TS cleavage within dsDNA (Supplementary information, Fig. S2i).
nt on the products (Fig. 2d, e). Moreover, we observed the single-stranded DNA (ssDNA) TS cleavage (cis-cleavage) by both effectors, and the cleavage efficacy and pattern are comparable to the TS cleavage within dsDNA (Supplementary information, Fig. S2i).
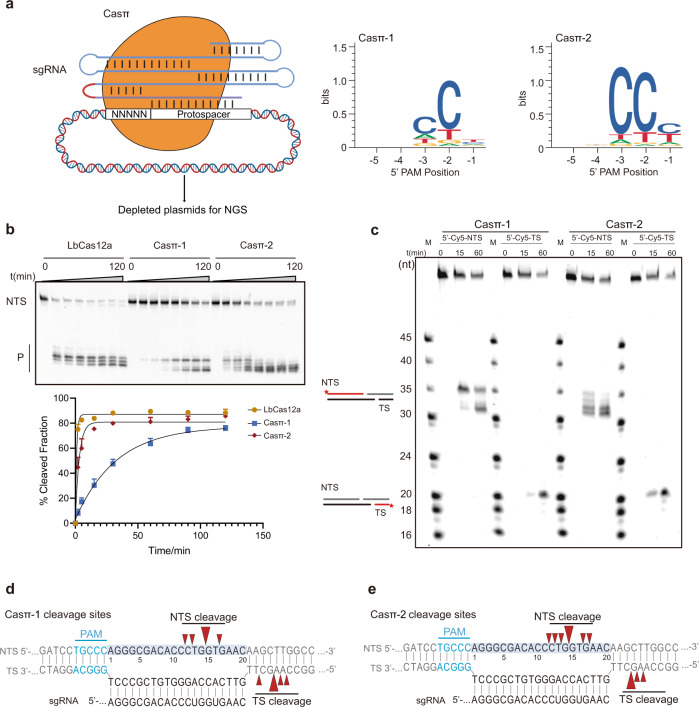
a Graphical representation of the in vitro PAM depletion assay and the resulting PAMs for both Casπ effectors. b Top, in vitro dsDNA cleavage comparison between LbCas12a, Casπ-1 and Casπ-2 revealed by denaturing PAGE. NTS denotes the non-target strand DNA which is cy5-labeled at 5′ end. P denotes the cleavage products. Bottom, the plot of NTS dsDNA cleavage efficiency by LbCas12a, Casπ-1 and Casπ-2 (n =
= 3 each; mean
3 each; mean ±
± SD). c Cleavage site mapping of Casπ-1 and Casπ-2 revealed by denaturing PAGE. Lane M shows cy5-labeled marker. d, e The cleavage sites for NTS and TS of Casπ-1 (d) and Casπ-2 (e) (marked in red arrows, large arrows indicate high probability of cleavage, PAM in blue) suggested by both gel analysis (n
SD). c Cleavage site mapping of Casπ-1 and Casπ-2 revealed by denaturing PAGE. Lane M shows cy5-labeled marker. d, e The cleavage sites for NTS and TS of Casπ-1 (d) and Casπ-2 (e) (marked in red arrows, large arrows indicate high probability of cleavage, PAM in blue) suggested by both gel analysis (n =
= 3 each) and NGS analysis.
3 each) and NGS analysis.
Casπ exhibits substantial tolerance of biochemical conditions with efficient trans-activity
To explore the application potential of Casπ, we performed a general screening for DNA cleavage by both effectors under various biochemical conditions in vitro. For RuvC-containing nucleases, divalent ions are typically important to coordinate the catalytic core for DNA hydrolysis. The ion screening suggested that either Mg2+ or Mn2+ can robustly activate the nuclease activity in Casπ (Fig. 3a; Supplementary information, Fig. S3a). Further experiments also showed that Casπ overcomes several disadvantages reported in other Cas nucleases. Normally, one common drawback of most compact CRISPR effectors (< 1000 aa) is their limited tolerance range of salt concentration in vitro. For example, the compact AsCas12f and CasPhi (Cas12j) prefer low salt concentration (< 150
aa) is their limited tolerance range of salt concentration in vitro. For example, the compact AsCas12f and CasPhi (Cas12j) prefer low salt concentration (< 150 mM NaCl) for detectable dsDNA cleavage, due to their limited dsDNA unwinding ability.12,15 Meanwhile, PlmCasX (Cas12e) robustly unwinds the dsDNA for cleavage in high salt concentration condition (300–450
mM NaCl) for detectable dsDNA cleavage, due to their limited dsDNA unwinding ability.12,15 Meanwhile, PlmCasX (Cas12e) robustly unwinds the dsDNA for cleavage in high salt concentration condition (300–450 mM NaCl), but gets denatured and precipitated in low-salt buffer (< 300
mM NaCl), but gets denatured and precipitated in low-salt buffer (< 300 mM NaCl) as seen.11 In contrast, the compact Casπ persists a stable effector status for dsDNA cleavage in a wide range of salt concentrations from 50
mM NaCl) as seen.11 In contrast, the compact Casπ persists a stable effector status for dsDNA cleavage in a wide range of salt concentrations from 50 mM to 300
mM to 300 mM NaCl (Fig. 3b; Supplementary information, Fig. S3b). Furthermore, unlike many Cas nucleases which get denatured and precipitated in solution when being concentrated to a high protein concentration (50–100
mM NaCl (Fig. 3b; Supplementary information, Fig. S3b). Furthermore, unlike many Cas nucleases which get denatured and precipitated in solution when being concentrated to a high protein concentration (50–100 μM), both Casπ nucleases behave robustly upon physical enrichment (30
μM), both Casπ nucleases behave robustly upon physical enrichment (30 kD molecular weight cut-off centrifugal filters; see Materials and methods).11 Therefore, we often stock the Casπ nucleases at the ultra-high protein concentration of 300
kD molecular weight cut-off centrifugal filters; see Materials and methods).11 Therefore, we often stock the Casπ nucleases at the ultra-high protein concentration of 300 μM for the following convenient use. Moreover, a huge limitation of employing biomolecular tools in different exogenous scenarios is that they only work efficiently in the temperatures that their source bacterial hosts prefer. To our surprise, although discovered in mesophilic environment, Casπ tolerates temperatures from 25
μM for the following convenient use. Moreover, a huge limitation of employing biomolecular tools in different exogenous scenarios is that they only work efficiently in the temperatures that their source bacterial hosts prefer. To our surprise, although discovered in mesophilic environment, Casπ tolerates temperatures from 25 °C even to 65
°C even to 65 °C (Fig. 3c; Supplementary information, Fig. S3c).
°C (Fig. 3c; Supplementary information, Fig. S3c).
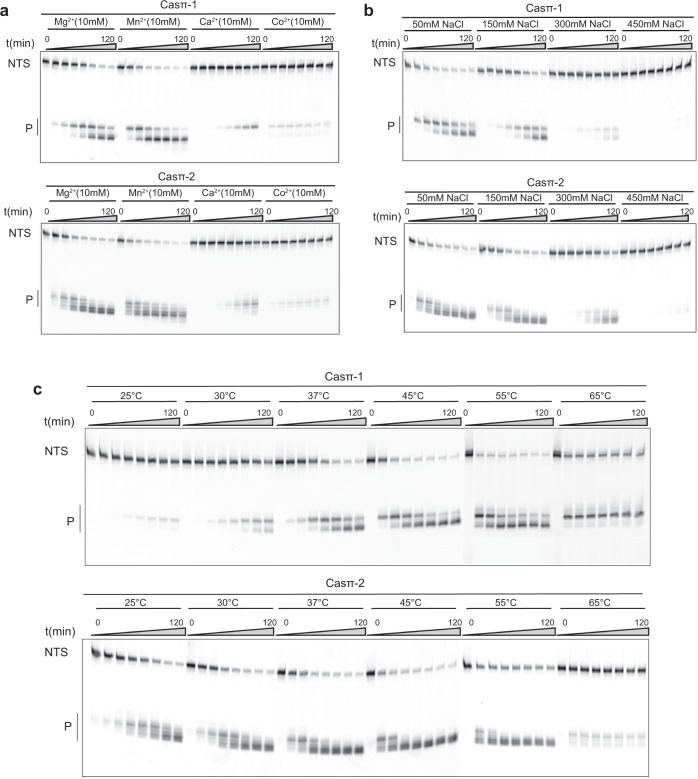
a In vitro dsDNA cleavage by Casπ effectors using different divalent ions revealed by denaturing PAGE. NTS denotes the non-target strand DNA which is cy5 labeled at 5′ end. Bottom, P means cleavage products (n =
= 3 each). b In vitro dsDNA cleavage by Casπ effectors in the buffers with different salt concentrations (n
3 each). b In vitro dsDNA cleavage by Casπ effectors in the buffers with different salt concentrations (n =
= 3 each). c In vitro dsDNA cleavage by Casπ effectors at different temperatures (n
3 each). c In vitro dsDNA cleavage by Casπ effectors at different temperatures (n =
= 3 each).
3 each).
To explore the cleavage specificity by Casπ effectors, we first performed the single mismatch screening on the DNA protospacer. The single mismatches between sgRNA and nucleotides 1–8 of the target DNA at the PAM-proximal region largely abolished the nuclease activity of Casπ, which suggests a ‘seed region’ located in the position of nucleotides 1–8 of the target DNA (Fig. 4a, b).24,25 Besides, single mismatches between nucleotides 13–16 at the PAM-distal region also significantly decreased the cleavage efficiency of Casπ (Fig. 4a, b). Additionally, many Cas12 nucleases cleave random ssDNA (trans-activity) when activated by ssDNA or dsDNA target (activator), which has been harnessed for nucleic acid diagnosis.10,26 Noteworthily, though compact in size, Casπ effectors show comparable trans-activity to the widely used LbCas12a with either ssDNA or dsDNA activator (Fig. 4c, d), indicating Casπ’s potential as a nucleic acid diagnosis tool. In summary, compared to many reported Cas effectors, Casπ presents a substantial advantage of flexibility and robustness for in vitro applications.
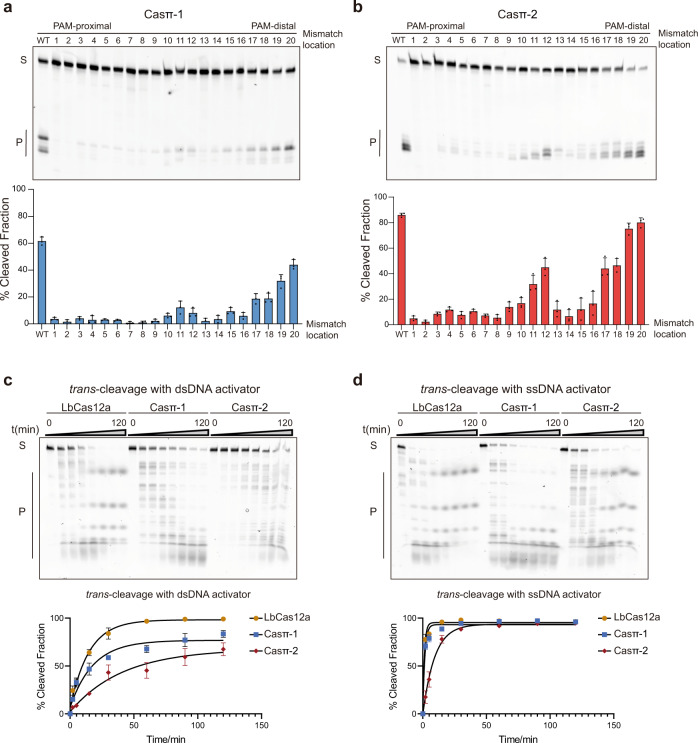
a Top, cleavage assay using single mismatched dsDNA targets by Casπ-1 effector at 1 h. Bottom, the bar plot of cleavage efficiency (WT indicates that sgRNA and dsDNA target are fully paired. Numbering means the position of the mismatches between sgRNA and dsDNA target used in this assay; n
h. Bottom, the bar plot of cleavage efficiency (WT indicates that sgRNA and dsDNA target are fully paired. Numbering means the position of the mismatches between sgRNA and dsDNA target used in this assay; n =
= 3 each; mean
3 each; mean ±
± SD). b Top, cleavage assay using single mismatched dsDNA targets by Casπ-2 effector at 1
SD). b Top, cleavage assay using single mismatched dsDNA targets by Casπ-2 effector at 1 h. Bottom, the bar plot of cleavage efficiency (n
h. Bottom, the bar plot of cleavage efficiency (n =
= 3 each; mean
3 each; mean ±
± SD). c Top, the trans-DNA cleavage by LbCas12a, Casπ-1 and Casπ-2 on ssDNA substrate with dsDNA activator (S means substrates and P means products). Bottom, the plot of trans-ssDNA substrate cleavage efficiency by LbCas12a, Casπ-1 and Casπ-2 with dsDNA activator (n
SD). c Top, the trans-DNA cleavage by LbCas12a, Casπ-1 and Casπ-2 on ssDNA substrate with dsDNA activator (S means substrates and P means products). Bottom, the plot of trans-ssDNA substrate cleavage efficiency by LbCas12a, Casπ-1 and Casπ-2 with dsDNA activator (n =
= 3 each; mean
3 each; mean ±
± SD). d Top, the trans-DNA cleavage by LbCas12a, Casπ-1 and Casπ-2 on ssDNA with ssDNA activator (S means substrates and P means products). Bottom, the plot of trans-ssDNA substrate cleavage efficiency by LbCas12a, Casπ-1 and Casπ-2 with ssDNA activator (n
SD). d Top, the trans-DNA cleavage by LbCas12a, Casπ-1 and Casπ-2 on ssDNA with ssDNA activator (S means substrates and P means products). Bottom, the plot of trans-ssDNA substrate cleavage efficiency by LbCas12a, Casπ-1 and Casπ-2 with ssDNA activator (n =
= 3 each; mean
3 each; mean ±
± SD).
SD).
Casπ orthologs are active for DNA manipulation both in prokaryotic and eukaryotic cells
To further explore whether the compact Casπ effectors can be employed for DNA cleavage in prokaryotes, we performed a plasmid interference assay using E. coli BW25141 strain carrying a ccdB toxin plasmid with arabinose-inducible promoter (Fig. 5a). While few survival clones were observed in the non-targeting control due to ccdB toxicity, expressing either Casπ-1 or -2 with the ccdB-targeting sgRNA led to significantly more survival clones (Fig. 5a, b; Supplementary information, Fig. S4a, b). This plasmid interference activity was further verified via PCR analysis (Supplementary information, Fig. S4c).
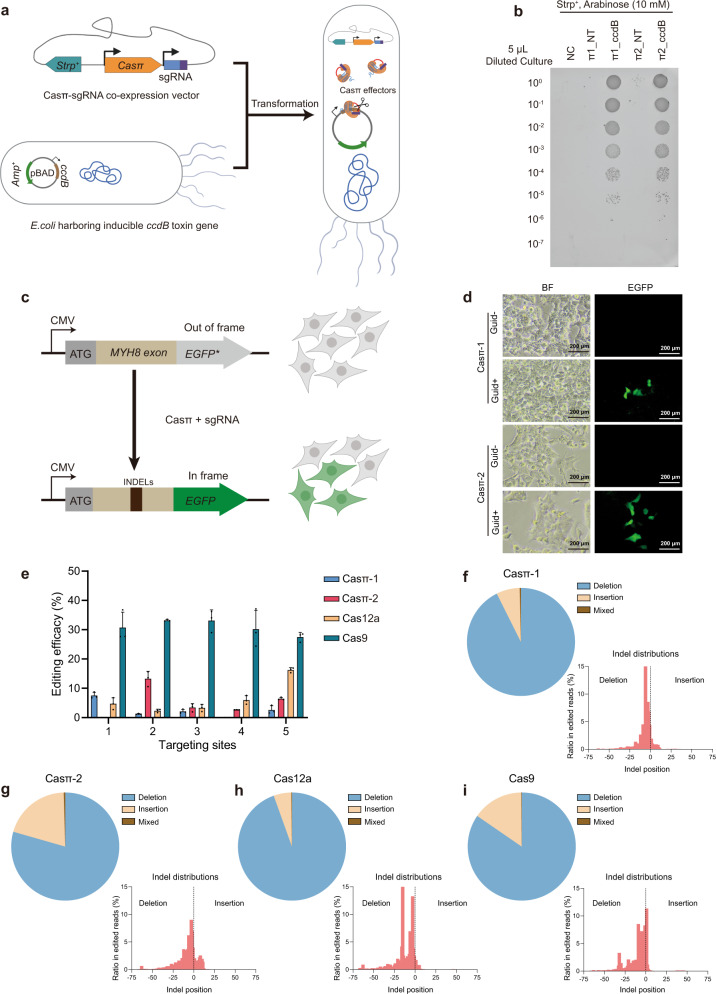
a Schematic illustration of the plasmid interference assay. b Bacteria survival assay on culture plates containing 10 mM arabinose. NT, plasmid with Casπ and non-target sgRNA; ccdB, plasmid with Casπ and sgRNA targeting ccdB gene. Dilution gradient is shown on the left. c Scheme of Casπ-mediated EGFP lighting up in HEK293A cells. d
EGFP lighting up results. Transfection of plasmids carrying Casπ and sgRNA activated EGFP (frame restored) with detectable green fluorescence signal. Both the bright field (BF) and fluorescent images of cultured cells are shown. e Editing efficacies determined by NGS from 5 targets mediated by Casπ-1, Casπ-2, Cas12a and Cas9 (n
mM arabinose. NT, plasmid with Casπ and non-target sgRNA; ccdB, plasmid with Casπ and sgRNA targeting ccdB gene. Dilution gradient is shown on the left. c Scheme of Casπ-mediated EGFP lighting up in HEK293A cells. d
EGFP lighting up results. Transfection of plasmids carrying Casπ and sgRNA activated EGFP (frame restored) with detectable green fluorescence signal. Both the bright field (BF) and fluorescent images of cultured cells are shown. e Editing efficacies determined by NGS from 5 targets mediated by Casπ-1, Casπ-2, Cas12a and Cas9 (n =
= 3 each, mean
3 each, mean ±
± SD). f Analysis of INDELs generated by Casπ-1 editing within all 15 editing experiments. Left, pie chart showing percentage of each INDEL within all 15 editing experiments analyzed by NGS (Mixed means mixed editing with both insertion and deletion). Right, INDEL size distributions within all 15 editing experiments. g Analysis of INDELs generated by Casπ-2 editing within all 15 editing experiments. h Analysis of INDELs generated by Cas12a editing within all 15 editing experiments. i Analysis of INDELs generated by Cas9 editing within all 15 editing experiments.
SD). f Analysis of INDELs generated by Casπ-1 editing within all 15 editing experiments. Left, pie chart showing percentage of each INDEL within all 15 editing experiments analyzed by NGS (Mixed means mixed editing with both insertion and deletion). Right, INDEL size distributions within all 15 editing experiments. g Analysis of INDELs generated by Casπ-2 editing within all 15 editing experiments. h Analysis of INDELs generated by Cas12a editing within all 15 editing experiments. i Analysis of INDELs generated by Cas9 editing within all 15 editing experiments.
Next, to investigate the genome-editing ability of Casπ in eukaryotic cells, we constructed a HEK293A cell line with the genome-integrated ORF containing the MYH8 exon and the out-of-frame EGFP (Fig. 5c; see Materials and methods). Expression of either Casπ-1 or -2 with sgRNA targeting the MYH8 exon efficiently lit up the cells with in-frame EGFP signal, which indicates that the DNA insertions or deletions (INDELs) were generated by Casπ editing (Fig. 5d). To compare the editing activity between Casπ effectors and the well-developed LbCas12a and SpyCas9 effectors, we designed five parallel targeting sites across the MYH8 exon (Supplementary information, Fig. S4d and Table S5). The edited genomes were PCR amplified, and the editing efficacies were validated by T7 endonuclease I (T7E1) assays and quantified by targeted sequencing (Supplementary information, Fig. S4e). Next-generation sequencing (NGS) revealed that both Casπ effectors introduced INDELs nearby the cleavage sites in TS as observed in vitro (Fig. 2d, e; Supplementary information, Fig. S4f, g). Overall, SpyCas9 presents an average editing efficacy of 30.9% across the five sites and a maximum efficacy of 37.1% at site 4 (Fig. 5e). LbCas12a shows an average editing efficacy of 6.7% and a maximum efficacy of 16.8% at site 5 (Fig. 5e). Casπ-1 shows an average editing efficacy of 2.7% and a maximum efficacy of 8.0% at site 1 (Fig. 5e). Casπ-2 shows an average editing efficacy of 5.4% and a maximum efficacy of 15.4% at site 2 (Fig. 5e). The combined INDEL analysis on the five targets shows that SpyCas9, LbCas12a and Casπ effectors mainly generate deletions on the targeted genome (Fig. 5f–i; Supplementary information, Fig. S4h). Of note, SpyCas9 may generate long deletions of ~40 nt, while Cas12a and Casπ editing dominantly contributes to shorter deletions of < 25
nt, while Cas12a and Casπ editing dominantly contributes to shorter deletions of < 25 nt (Fig. 5f–i). Further, three more endogenous targets on B2M and TP53 genes were edited by Casπ effectors and the editing efficacies were quantified by NGS (Supplementary information, Fig. S4i–k).
nt (Fig. 5f–i). Further, three more endogenous targets on B2M and TP53 genes were edited by Casπ effectors and the editing efficacies were quantified by NGS (Supplementary information, Fig. S4i–k).
Therefore, even without any optimizations, the naive version of compact Casπ effectors works comparably to LbCas12a and maximumly reaches over half of the editing ability of the well-developed SpyCas9, supporting Casπ’s potential to be a competitive and compact DNA manipulation platform with further engineering.
Unique structural domains in Casπ responsible for DNA interference
To understand the molecular details underlying the DNA targeting behavior by Casπ effector and provide structural information for editing optimization in future studies, we achieved the cryo-EM map of the R-loop complex containing the deactivated Casπ-1 (D537A, E643A), sgRNA and dsDNA at 3.4-Å resolution (Supplementary information, Figs. S5a–c, S6a–e). The EM density of Casπ R-loop complex is well resolved, which allows us to build the complete atomic model ab initio (Fig. 6a–c; Supplementary information, Fig. S6e, f and Video S1). Consistent with the primary sequence BLAST suggesting no significant similarity to reported proteins, Casπ also exhibits a unique 3D architecture compared to other CRISPR-Cas nucleases revealed by structural alignment with Dali server (Supplementary information, Fig. S7a, b).27 Only moderate similarity was observed between Casπ and Cas12 nucleases, mainly within the RuvC domain and oligonucleotide binding domain (OBD) (Supplementary information, Fig. S7c, d). Then, referring to CasX (Cas12e) which shares the top structural similarity with Casπ and also uses a large RNA guide (Supplementary information, Fig. S7e), we further located the conserved bridge helix (BH) element and four unique structural domains within Casπ, including the ‘lock-catch’ (LC) domain, proline-rich string (PRS), Helical-I domain and NTSB (non-target strand binding domain) chimera (HNC), and Casπ (Pi) C-terminal (PCT) domain (Fig. 6a–c; Supplementary information, Video S1).
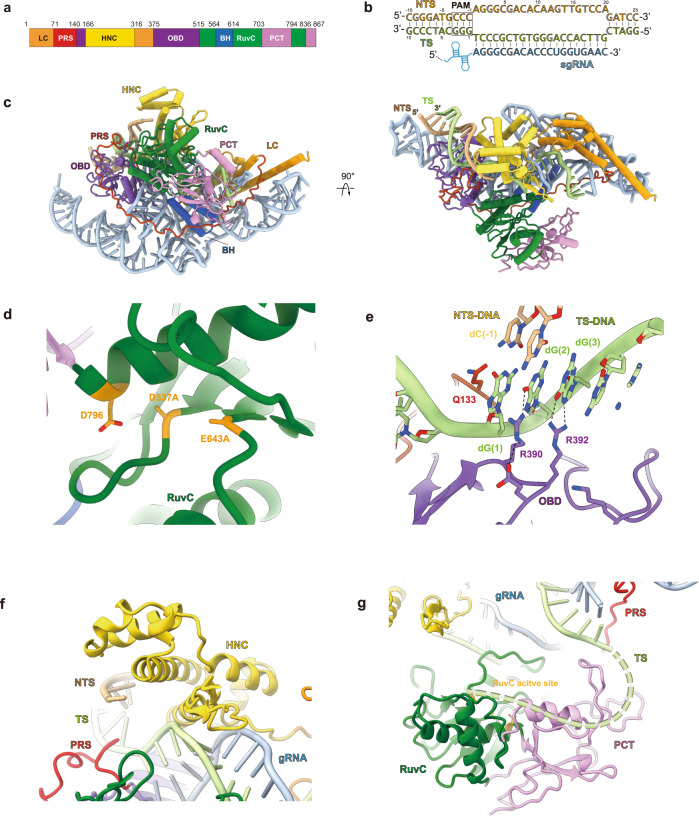
a The domain organization aligned with primary sequence. LC domain is colored in dark orange, PRS in red, HNC in yellow, OBD in purple, RuvC in dark green, BH in dark blue, and PCT in pink. b The base pairing details for the R-loop region. The sequences for NTS (light orange color), TS (light green color) and sgRNA spacer (cyan color) are presented. The PAM region is marked with rectangle. c The atomic model for Casπ R-loop complex. The protein domains, DNA and sgRNA are colored referring to a and b. The front and top views are presented. d The structural details within Casπ RuvC domain (dark green color). The three catalytic residues were highlighted with dark yellow color. In this complex, D537 and E643 were mutated to alanine. e The molecular details for PAM recognition. The amino acids involved in dG(2) and dG(3) recognition are labeled. The key hydrogen bonds are shown by dashed lines. f The structural details within Casπ HNC domain (yellow color). g The structure of Casπ PCT domain (pink color) and TS DNA loading model. The 5′ end of TS DNA is hypothetically modeled as dashed line (light green) and loaded into RuvC nuclease pocket by PCT domain.
The RuvC domain in Casπ displays a canonical DNA cleavage pocket with the conserved triplet of catalytic residues D537, E643 and D796 (Fig. 6d). D537 and E643 are mutated to alanine in this study for stabilizing the complex (Supplementary information, Fig. S5a–c). Different from other type V CRISPR nucleases which prefer T-rich PAM, two unique residues in Casπ OBD domain, Arg390 and Arg392, were observed to recognize the two guanine nucleotides (dG(2) and dG(3) in the TS) complementary to the CCN PAM (in the NTS) (Fig. 6b, e). Both the single mutations (R390A or R392A) and double mutation (R390A/R392A) totally abrogated the nuclease activity of Casπ (Fig. 6e; Supplementary information, Fig. S8a, b). In addition, the side chain of Gln133 inserts into the downstream site of PAM duplex, which may lead to local dsDNA melting for sgRNA–spacer invading as discussed in other type V nucleases (Fig. 6e).24
The HNC domain, which presents as a structural chimera of Helical-I domain and NTSB domain in CasX, interacts with both the ‘seed region’ of sgRNA–DNA heteroduplex at the PAM-proximal region and the backbone of DNA NTS to stabilize the R-loop conformation (Fig. 6f; Supplementary information, Fig. S8c). Meanwhile, neither primary sequence BLAST nor structural search for PCT domain (Trp703–Asp794 and Arg836–Ile867) reveals any suggestive similarity to annotated proteins, indicating that this unique feature is specific to Casπ nucleases (Supplementary information, Fig. S7b). Since the PCT domain sits at similar primary and spatial locations to the target-strand loading (TSL) domain of CasX (Supplementary information, Fig. S8d), we then hypothesize that the PCT domain may help with the target strand loading into RuvC nuclease domain (Fig. 6g), and this needs to be further explored in future studies.11
Casπ presents a ‘bracelet’ architecture encircling the nucleic acid target
Strikingly, a long ‘proline-rich string’ (PRS) loop composed of 69 aa (Pro72–Trp140) is largely resolved in the EM map (Fig. 7a; Supplementary information, Fig. S6f and Video S1). There are 14 prolines and 17 charged residues within this ‘string’ which makes it adopt high structural accessibility and electrostatic capacity to tie up the whole complex via multi-interactions with other protein domains, sgRNA and also the DNA target (Fig. 7a; Supplementary information, Fig. S9a). Directly next to the PRS N-terminus, Casπ folds into a two-helix structure (Met1–Asp71) which serves as a ‘lock’ and tightly interacts with a three-helix ‘catch’ module (Val317–Ala375) through multiple interactions, such as the hydrogen bonds (E28 and Y61 interact with R339 and E337, respectively) (Fig. 7b), the charged interactions and van der Waals interactions (not shown in the figure). Via this unique structure never observed in other Cas nucleases, the ‘lock-catch’ (LC) domain further locks the ‘tie-up’ conformation mediated by the PRS (Fig. 7a; Supplementary information, Fig. S9a and Video S1). Moreover, similar to the Helical II domain in CasX (Cas12e),11 the ‘lock’ part in LC domain also intensively interacts with the sgRNA stem to stabilize the assembly of R-loop complex (Fig. 7b; more details discussed in next section). Remarkably different from the canonical ‘two-lobe’ architecture for Class 2 Cas nucleases, the PRS and LC domains string all other protein domains together, and make the Casπ fold as a locked ‘bracelet’ encircling the nucleic acid target (Fig. 7c, d; Supplementary information, Fig. S9b, c).
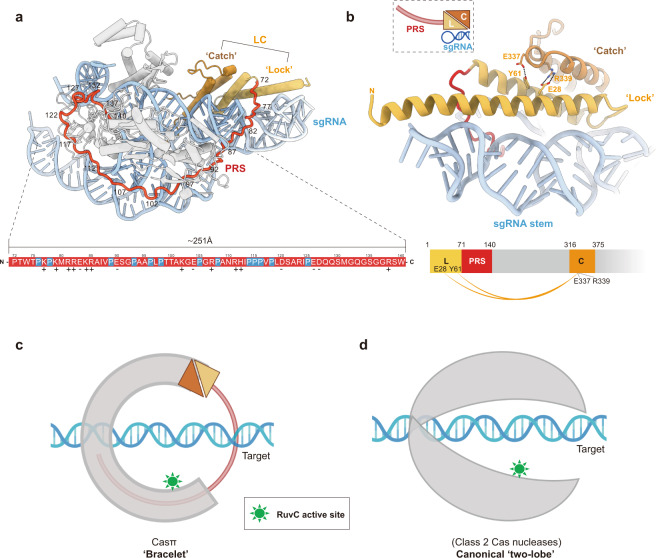
a The structural distribution of PRS domain across the Casπ R-loop complex. For clear presentation, the sgRNA and DNA are shown in cyan. ‘Lock’ part in LC domain is shown in light orange and the ‘catch’ part in deep orange. PRS is shown in red and all other protein domains in gray. The primary sequence for PRS loop is colored in red and shown at the bottom. The prolines in PRS are specially depicted in blue. The positively charged amino acids are labeled with ‘+’ and negatively charged ones with ‘–’. b The structural details of LC domain. The elements are colored referring to a. Side chains of the amino acids involved in the interactions between ‘lock’ and ‘catch’ are shown, and the formed hydrogen bonds are presented as dashed lines. The charged interactions and van der Waals interactions are not shown. The cartoon shapes for the ‘lock (L)’, ‘catch (C)’, PRS and sgRNA stem are outlined and presented at the top left. The domain organization of ‘lock (L)’, ‘catch (C)’ and PRS is presented at the bottom. The interactions between ‘lock (L)’ and ‘catch (C)’ are connected with orange curves. c A cartoon model for Casπ ‘bracelet’. The PRS is modeled as a half-ring colored in red. The LC is modeled referring to b, and all other domains are modeled as a half-ring colored in gray. The nucleic acid target is also modeled and labeled, accordingly. The RuvC active sites are indicated in green color. d The cartoon model of ‘two-lobe’ architecture for canonical Class 2 Cas nuclease. The protein part was colored in gray and nucleic acid part in cyan. The RuvC active sites are indicated in green color.
The large tracr–crRNA hybrid forms a ‘two-arm’ scaffold for effector assembly
The compact Casπ uses a large sgRNA (tracr–crRNA hybrid) for DNA interference. Well-resolved in the cryo-EM map (Supplementary information, Fig. S6), the sgRNA hybrid presents as a ‘two-arm’ architecture and embraces the Casπ monomer forming the ribonucleoprotein (RNP) effector (Fig. 8a). Referring both to the 2D and 3D structural details, we located four structural elements within this large sgRNA scaffold: arm-I (A-I), junction region (JR), arm-II (A-II) and pseudoknot region (PR) (Fig. 8a, b). Both A-I and A-II are built by the three-way junction, and these two three-way junctions are connected by JR. While A-I (previously labeled as ‘sgRNA stem’ in Fig. 7b) forms intensive interactions with Casπ protein (Fig. 8c, d), A-II largely stretches out from the effector complex (Fig. 8a, b). Noteworthily, both 12 nt and 24
nt and 24 nt truncations on the A-II increased the DNA cleavage activity by Casπ, suggesting a promising engineering site within the sgRNA for improving the genome-editing capability (Supplementary information, Fig. S10a, b). Likewise, this stretched A-II may provide a flexible engineering site for functional module integration without affecting the Casπ effector assembly. In addition, beyond the electrostatic interactions with RNA backbone (Fig. 8c), the binding between Casπ and sgRNA is also developed in a sequence-specific way. For example, the bases of nucleotides C48 and G49 in A-I was recognized by Arg23 and Arg26 residues in the LC domain, respectively (Fig. 8c, d). Moreover, the U148GAAAG153 in crRNA part pairs with the C100UUUCA105 loop from the tracrRNA part, forming a pseudoknot structure (corresponding to the PR) followed by the single-stranded spacer (Fig. 8a, b). This PR element tightly binds to Casπ PRS, BH, RuvC and OBD domains via backbone interactions and base-specific recognitions (Fig. 8c, e, f). Noteworthily, the sgRNA PR also gets shielded by the Casπ PRS domain (Fig. 8e). In summary, mainly mediated by the A-I and PR elements, the sgRNA hybrid provides a structurally continuous ‘two-arm’ scaffold to recruit the Casπ ‘bracelet’ via both backbone interactions and base-specific recognitions, forming a compact and ‘locked’ effector for DNA interference (Fig. 8; Supplementary information, Video S1).
nt truncations on the A-II increased the DNA cleavage activity by Casπ, suggesting a promising engineering site within the sgRNA for improving the genome-editing capability (Supplementary information, Fig. S10a, b). Likewise, this stretched A-II may provide a flexible engineering site for functional module integration without affecting the Casπ effector assembly. In addition, beyond the electrostatic interactions with RNA backbone (Fig. 8c), the binding between Casπ and sgRNA is also developed in a sequence-specific way. For example, the bases of nucleotides C48 and G49 in A-I was recognized by Arg23 and Arg26 residues in the LC domain, respectively (Fig. 8c, d). Moreover, the U148GAAAG153 in crRNA part pairs with the C100UUUCA105 loop from the tracrRNA part, forming a pseudoknot structure (corresponding to the PR) followed by the single-stranded spacer (Fig. 8a, b). This PR element tightly binds to Casπ PRS, BH, RuvC and OBD domains via backbone interactions and base-specific recognitions (Fig. 8c, e, f). Noteworthily, the sgRNA PR also gets shielded by the Casπ PRS domain (Fig. 8e). In summary, mainly mediated by the A-I and PR elements, the sgRNA hybrid provides a structurally continuous ‘two-arm’ scaffold to recruit the Casπ ‘bracelet’ via both backbone interactions and base-specific recognitions, forming a compact and ‘locked’ effector for DNA interference (Fig. 8; Supplementary information, Video S1).
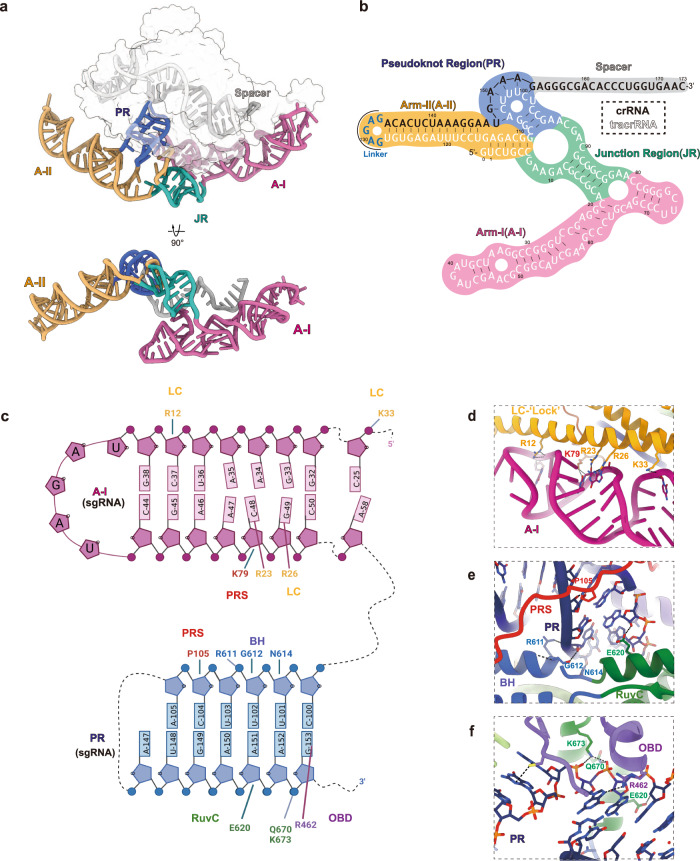
a The overall 3D structure of sgRNA. The A-I region is colored in plum, JR in green, A-II in orange, PR in blue and spacer in gray. Both front and bottom views are shown. The protein density is shown by transparent surface in the top panel. b The secondary structure details for the sgRNA. The background of different regions is colored according to a. The sequences for tracrRNA part, joint-loop and crRNA part are shown in white, blue and black, respectively. c The interaction details between Casπ protein and the sgRNA. Only the sgRNA A-I and PR regions are shown in this cartoon. The protein domains, associated amino acids and RNA nucleotides are labeled. The interaction pairs are linked with solid lines. d The structural details for the interaction interface between LC domain and A-I element. e, f The interaction details between PR element and PRS, BH (e), and RuvC, OBD (f) domains of Casπ. The protein domains are colored and labeled referring to Fig. 6a.
Discussion
Casπ provides a unique DNA targeting platform with a large potential given further engineering
In this study, via large-scale bioinformatics screening and manual annotation, we identified the CRISPR-Casπ as a novel type V system distinct from reported families which provides unique potentials for gene editing application, like the C-rich PAM preference, compact size, tolerance of various biochemical conditions and efficient trans-activity. Significantly, without any optimization, the naive version of Casπ effectors (~860 aa) shows substantial editing ability compared to SpyCas9 and LbCas12a benchmarks. This strongly suggests that Casπ has a huge potential to be largely improved via rational design or directed evolution, similar to how SpyCas9 or other effector-based technologies were developed in the last decade. Meanwhile, our cryo-EM study revealed the ‘bracelet’ architecture for Casπ which provides a brand-new structural platform for functional module integration and engineering. Furthermore, given the well-illustrated recognition details by Casπ protein, the ‘two-arm’ sgRNA also offers large engineering capacity, especially within the stretched-out A-II element.
aa) shows substantial editing ability compared to SpyCas9 and LbCas12a benchmarks. This strongly suggests that Casπ has a huge potential to be largely improved via rational design or directed evolution, similar to how SpyCas9 or other effector-based technologies were developed in the last decade. Meanwhile, our cryo-EM study revealed the ‘bracelet’ architecture for Casπ which provides a brand-new structural platform for functional module integration and engineering. Furthermore, given the well-illustrated recognition details by Casπ protein, the ‘two-arm’ sgRNA also offers large engineering capacity, especially within the stretched-out A-II element.
Strictness for PAM preference varies in different scenarios
PAM sequence is essential for dsDNA targeting by Class 2 Cas nucleases, and it is often determined by the cleavage of plasmid library containing randomized PAM either in vitro or in vivo. In our experience, Cas effectors usually show more robust cleavage on the plasmid target than linearized dsDNA,8 as plasmids contain melting bubbles in the supercoil conformation.28 Compared to the plasmid, a more stringent PAM requirement was observed on the linearized dsDNA target (Supplementary information, Fig. S2e, f). Moreover, we also found that the dC gradually dominated the third position of the PAM in the depletion analysis for Casπ-1 while increasing the salt concentration in the cleavage buffer, which indicates a more stringent PAM preference for Casπ-1 effectors in high-salt buffer (Supplementary information, Fig. S2b). Similar patterns were observed in CasX enzymes (unpublished data). Referring to previous biophysical studies, either linearizing the plasmid (relax the supercoil and re-anneal the bubbled strands in plasmids) or increasing the salt concentration (stabilize the dsDNA conformation) may contribute to ‘tougher’ targets for Cas effectors to unwind.28 Therefore, we would suggest that a stringent PAM sequence determined in the ‘tough’ condition (linearized dsDNA target in the buffer with the highest salt concentration that Cas effectors can tolerate) may be the prioritized choice for gene editing application.
A hypothetical evolution trend underlying Class 2 CRISPR effectors starting from the ‘RNA world’
The wet-lab validation and structural information allow us to accurately identify the functional size of each component in Cas effectors, especially for the tracrRNA whose exact length is usually challenging to determine bioinformatically. When arranging the structurally validated Class 2 effectors (using tracr–crRNA guide) together with our newly discovered Casπ effector, an interesting trend was observed: the size of tracr–crRNA hybrid (RNA part) gradually decreases as the Cas protein size increases within the RNP effectors (Supplementary information, Fig. S11a–d). Moreover, analysis of 383 bioinformatically identified Cas9 effectors also suggests a negative linear correlation (correlation coefficient of –0.439) between the sizes of the tracr–crRNAs and Cas proteins (Supplementary information, Fig. S11e). Considering that the linear correlation is sensitive to extreme values, we only selected the effectors with Cas9’s molecular weight of 100,000–200,000 Da and tracr–crRNA of 30,000–60,000
Da and tracr–crRNA of 30,000–60,000 Da for analysis. Notably, a recent structural study shows that the IscB effector (commonly-acknowledged ancestor for type II Cas9 effectors) comprises an IscB nuclease monomer smaller than reported Cas9s and an ωRNA significantly larger than reported tracr–crRNA hybrids (Supplementary information, Fig. S11a).29
Da for analysis. Notably, a recent structural study shows that the IscB effector (commonly-acknowledged ancestor for type II Cas9 effectors) comprises an IscB nuclease monomer smaller than reported Cas9s and an ωRNA significantly larger than reported tracr–crRNA hybrids (Supplementary information, Fig. S11a).29
Then starting from the IscB or other ancestors like TnpB for type V effectors,1,30,31 this trend may suggest an RNA-protein co-evolution path underlying the CRISPR effectors (Supplementary information, Fig. S11a, b).32,33 As proteins play more robust structural and enzymatic roles than RNAs, during the molecular evolution, the functional and structural domains of the RNA part are gradually replaced by Cas protein for efficient DNA interference (Supplementary information, Fig. S11a, b). This has actually often been the case that the CRISPR effectors with large Cas proteins and small gRNAs work better for DNA editing than the effector with small Cas protein and large gRNA.11,12,32
Further, even ancestral to the IscB or TnpB ‘intermediate’ ancestors, it is also reasonable to hypothesize the RNA and RNA-dominated ancestors for CRISPR effectors, in which the RNA part (ribozymes) but not the protein may play the enzymatic role for nucleic acid interference (Fig. 9).33–38 Though probably not existing in the current protein-dominated world, reconstruction of those RNA and RNA-dominated ancestors originated from the ‘RNA world’ will provide brand-new insights for molecular tool development, as well as the evolutionary evidence of enzymatic function transition from RNA to protein. While due to the lack of available knowledge, our current discussion is only focused on the molecular size of a limited number of CRISPR effectors. Thereby, a large-scale identification of new CRISPR effectors in the current protein-dominated world and a comprehensive understanding of the functional and structural replacement events between the RNA and protein may help understand the ‘co-evolutionary principle’ starting from the ‘RNA world’ (Fig. 9). Using this ‘co-evolutionary principle’, it is promising to reconstitute those RNA and RNA-dominated ancestors in silico.
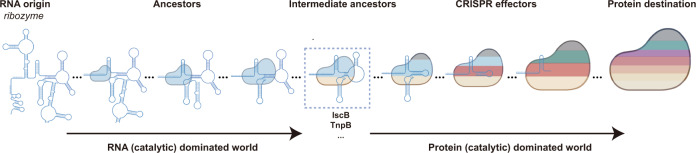
The RNA part is depicted by secondary structure model. The protein part is modeled with irregular circle. The molecular size of the RNA and protein is positively correlated with the cartoon size. Color codes in the protein cartoon indicated the abundance of functional or structural domains. The RNA origin (ribozyme), three RNA-dominated ancestors, intermediate ancestors (IscB, TnpB, etc.), three CRISPR effectors, and the protein destination (protein-only system) are arranged according to our hypothetical evolution trend.
Materials and methods
Metagenomics
The genetic materials were purified from bioreactor sludge sample as previously described, and sequenced on the Illumina NovaSeq 6000 platform using the PE150 sequencing strategy.17 All raw datasets were trimmed by Trim Galore v0.6.5 using default parameters, which generated data containing clean reads that were subsequently assembled using SPAdes v3.15.4 for detection of CRISPR-Cas system.39
Casπ detection and phylogenic analysis of type V CRISPR systems
The assembled contigs were scanned for Cas nucleases using HMM profiles, which were built using the HMMER,40 based on Cas nuclease sequence alignments from Clustal Omega (1.2.4).41 CRISPR arrays were identified using local version of the CRISPRCasFinder (4.2.20) and CRISPRidentify (v1.1.0).42,43 Loci that contained both cas1 and the CRISPR array were further analyzed to identify the proteins located within the range from 20,000 nt upstream to 20,000
nt upstream to 20,000 nt downstream of the CRISPR array. Potential functions of these proteins were annotated by HMMs and the local version of eggNOG mapper (2.1.6, eggNOG DB version: 5.0.2, MMseqs2 version: 13.45111).44,45 Proteins larger than 600
nt downstream of the CRISPR array. Potential functions of these proteins were annotated by HMMs and the local version of eggNOG mapper (2.1.6, eggNOG DB version: 5.0.2, MMseqs2 version: 13.45111).44,45 Proteins larger than 600 aa were selected as potential Class 2 Cas nucleases with nucleic-acid interference activity, and were further clustered by phylogenetic analysis.
aa were selected as potential Class 2 Cas nucleases with nucleic-acid interference activity, and were further clustered by phylogenetic analysis.
For phylogenetic analysis, sequences of reported Cas nucleases were collected from UniProt database by searching keywords of each nucleases, like Cas9 and Cas12a.10,12,13,46,47 Sequence alignment of Casπ with the selected type V Cas nucleases was generated using Clustal Omega (1.2.4).41 Phylogenic reconstruction was performed using IQ-TREE2 (2.0.7) with VT +
+ F
F +
+ R7 as the substitution model and 1500 bootstrap sampling.48 Reconstruction result was visualized and edited using iTOL v6.5.8.49
R7 as the substitution model and 1500 bootstrap sampling.48 Reconstruction result was visualized and edited using iTOL v6.5.8.49
Protein sequence and CRISPR repeat analysis
The protein and CRISPR repeat sequences of four Casπ orthologs were analyzed by Clustal Omega server with default parameters,41 and the two heatmaps illustrating the sequence similarity were built using the similarity score matrix (Sequences shown in Supplementary information, Table S1). For protein alignment with other type V CRISPR, the protein sequences of four Casπ orthologs were aligned with LbCas12a, AsCas12a, AaCas12b and DpbCas12e proteins using NCBI COBALT program,22 and the key amino acids in RuvC domains of Casπ were inferred from the alignment results.7,11,22,50
tracrRNA identification and PAM prediction
For CRISPR-Casπ system, tracrRNA 3′-region was determined by anti-repeat identification, transcriptome mapping and promoter prediction. Anti-repeats were searched against a 5 kb window upstream of the CRISPR locus using blastn with (E-value
kb window upstream of the CRISPR locus using blastn with (E-value <
< 0.2).18 Subsequently, the meta-transcriptomic reads of the sludge sample were extracted and mapped to their native genome locus around the anti-repeat region to analyze the tracrRNA expression. The transcript coverage was calculated by log10 formula. Finally, the 5′-boundary of tracrRNA was determined by promoter prediction using BDGP-Promoter Prediction program.51 All tracrRNAs were determined in this manner as shown in Fig. 1c and the sequences were shown in Supplementary information, Table S1.
0.2).18 Subsequently, the meta-transcriptomic reads of the sludge sample were extracted and mapped to their native genome locus around the anti-repeat region to analyze the tracrRNA expression. The transcript coverage was calculated by log10 formula. Finally, the 5′-boundary of tracrRNA was determined by promoter prediction using BDGP-Promoter Prediction program.51 All tracrRNAs were determined in this manner as shown in Fig. 1c and the sequences were shown in Supplementary information, Table S1.
To predict the PAM sequence for Casπ-1 and Casπ-2, all the spacers present in both CRISPR arrays were manually extracted and aligned against the default databases using CRISPRTarget to search the potential protospacer sequences.23 Sequences 3 bp upstream of the identified protospacers were extracted and aligned to predict the PAM sequences. The PAMs ranking at the top for both Casπs were further used for plasmid cleavage in vitro.
bp upstream of the identified protospacers were extracted and aligned to predict the PAM sequences. The PAMs ranking at the top for both Casπs were further used for plasmid cleavage in vitro.
Plasmid construction
Bacterial and human codon-optimized casπ-1 and casπ-2 genes were ordered from Sangon Biotech. For Casπ protein expression in E. coli, casπ genes were cloned into pET28a-based vector with an N-terminal hexa-histidine tag and a SUMO tag by homologous recombination (One Step Seamless Cloning Mix, CWBIO). For the D537A and E643A mutations in RuvC domain, R390A and R392A mutations in OBD domain of Casπ-1, mutated fragments were PCR amplified via mutagenetic PCR primers containing mutated sequences and inserted into pET28a-based vector by homologous recombination. For PAM depletion assay, the plasmid library containing five randomized nucleotides upstream of the target sequence was constructed as previously described.52 For in vitro plasmid cleavage, pUC19-based plasmids containing target sequence with different PAMs were constructed via homologous recombination. For bacterial plasmid interference, pBAD-driven arabinose inducible ccdB toxin plasmid (p11-LacY-wtx1) was requested from Prof. Wei Li group in the Institute of Zoology, Chinese Academy of Sciences.53 casπ genes were cloned into MCSI of pCDFDuet vector by Gibson assembly with a sgRNA region, containing 2 SapI sites for target spacer exchange by Golden Gate, inserting into MCSII of pCDFDuet (sgRNA spacer sequences were listed in Supplementary information, Table S5).
For constructing the EGFP report cell line, the CMV-driven fusion fragment of MYH8 (270 bp), a flanking sequence (32
bp), a flanking sequence (32 bp) and EGFP (1436
bp) and EGFP (1436 bp) was cloned into psi-LVRU6MP vector by Gibson assembly. For cell editing assay, plasmid vector was obtained from circular PCR amplification of pBLO62.5 (Addgene plasmid# 123124) with two primers respectively pairing to N-terminal and C-terminal NLS sequence.8 Subsequently, Casπ (SpyCas9 or LbCas12a) genes were inserted into the region downstream of the CMV promoter and N-terminal NLS by homologous recombination. Then, sgRNAs (containing 2 SapI sites for spacer insertion) were inserted into the circular PCR-amplified vector containing Casπ (SpyCas9 or LbCas12a) genes with a U6 promoter and a poly-T terminal signal by homologous recombination. Primers containing the target spacer sequences were annealed and phosphorylated prior to Golden Gate assembly (SapI restriction sites) for stuffer–spacer exchange insertion (target protospacer sequences were listed in Supplementary information, Table S5).
bp) was cloned into psi-LVRU6MP vector by Gibson assembly. For cell editing assay, plasmid vector was obtained from circular PCR amplification of pBLO62.5 (Addgene plasmid# 123124) with two primers respectively pairing to N-terminal and C-terminal NLS sequence.8 Subsequently, Casπ (SpyCas9 or LbCas12a) genes were inserted into the region downstream of the CMV promoter and N-terminal NLS by homologous recombination. Then, sgRNAs (containing 2 SapI sites for spacer insertion) were inserted into the circular PCR-amplified vector containing Casπ (SpyCas9 or LbCas12a) genes with a U6 promoter and a poly-T terminal signal by homologous recombination. Primers containing the target spacer sequences were annealed and phosphorylated prior to Golden Gate assembly (SapI restriction sites) for stuffer–spacer exchange insertion (target protospacer sequences were listed in Supplementary information, Table S5).
A list of plasmids and a brief description are summarized in Supplementary information, Table S4.
Protein expression and purification
Casπ expression plasmids were transformed into E. coli BL21(DE3) (TIANGEN) and incubated overnight at 37 °C on LB-Kan+ agar plates (50
°C on LB-Kan+ agar plates (50 μg/mL Kanamycin). Single colony was overnight cultured as seed in LB-Kan+ medium (50
μg/mL Kanamycin). Single colony was overnight cultured as seed in LB-Kan+ medium (50 μg/mL Kanamycin) at 37
μg/mL Kanamycin) at 37 °C. Each 1
°C. Each 1 L of LB-Kan+ medium (50
L of LB-Kan+ medium (50 μg/mL Kanamycin) was then inoculated with 100
μg/mL Kanamycin) was then inoculated with 100 mL seed culture and incubated at 37
mL seed culture and incubated at 37 °C. As the culture OD reached 1.0, the protein expression was induced with 0.2
°C. As the culture OD reached 1.0, the protein expression was induced with 0.2 mM IPTG for 20
mM IPTG for 20 h at 16
h at 16 °C. Bacterial cells were collected and resuspended in lysis buffer (800
°C. Bacterial cells were collected and resuspended in lysis buffer (800 mM NaCl, 20
mM NaCl, 20 mM HEPES-Na, pH 7.5, 10% glycerol, 40
mM HEPES-Na, pH 7.5, 10% glycerol, 40 mM imidazole, 1
mM imidazole, 1 mM TCEP and 1
mM TCEP and 1 mM PMSF) and lysed by sonication. The lysate was centrifuged at 15,000×
mM PMSF) and lysed by sonication. The lysate was centrifuged at 15,000× g for 80
g for 80 min at 4
min at 4 °C and applied to Ni-NTA gravity column. The resin was then washed with 20 column volumes (CVs) of wash buffer (500
°C and applied to Ni-NTA gravity column. The resin was then washed with 20 column volumes (CVs) of wash buffer (500 mM NaCl, 20
mM NaCl, 20 mM HEPES-Na, pH 7.5, 10% glycerol, 40
mM HEPES-Na, pH 7.5, 10% glycerol, 40 mM imidazole, 1
mM imidazole, 1 mM TCEP), and resuspended in 5 CVs of tag-removal buffer (500
mM TCEP), and resuspended in 5 CVs of tag-removal buffer (500 mM NaCl, 20
mM NaCl, 20 mM HEPES-Na, pH 7.5, 10% glycerol, 40
mM HEPES-Na, pH 7.5, 10% glycerol, 40 mM imidazole, 1
mM imidazole, 1 mM TCEP and 0.6
mM TCEP and 0.6 μg/mL ulp1 protease) for 1
μg/mL ulp1 protease) for 1 h incubation at 4
h incubation at 4 °C. Next, the supernatant was loaded into 5
°C. Next, the supernatant was loaded into 5 mL HiTrap Heparin HP column (GE Healthcare) and eluted with a linear gradient of heparin elution buffer (buffer A: 20
mL HiTrap Heparin HP column (GE Healthcare) and eluted with a linear gradient of heparin elution buffer (buffer A: 20 mM HEPES-Na, pH 7.5, 10% glycerol, 1
mM HEPES-Na, pH 7.5, 10% glycerol, 1 mM TCEP; buffer B: 2
mM TCEP; buffer B: 2 M NaCl, 20
M NaCl, 20 mM HEPES-Na, pH 7.5, 10% glycerol, 1
mM HEPES-Na, pH 7.5, 10% glycerol, 1 mM TCEP). Elution fractions with Casπ were pooled together and concentrated using 30 kD molecular weight cut-off centrifugal filters (Merck Millipore), and further purified by size exclusion chromatography (SEC) column (Superdex 200 Increase 10/300, GE Healthcare) with S200 buffer (400
mM TCEP). Elution fractions with Casπ were pooled together and concentrated using 30 kD molecular weight cut-off centrifugal filters (Merck Millipore), and further purified by size exclusion chromatography (SEC) column (Superdex 200 Increase 10/300, GE Healthcare) with S200 buffer (400 mM NaCl, 20
mM NaCl, 20 mM HEPES-Na, pH 7.5, 10% glycerol, 1
mM HEPES-Na, pH 7.5, 10% glycerol, 1 mM TCEP). Protein concentrations were measured by NanoDrop One (Thermo Scientific) and protein samples were stocked at –80
mM TCEP). Protein concentrations were measured by NanoDrop One (Thermo Scientific) and protein samples were stocked at –80 °C after flash-frozen in liquid nitrogen. The Casπ protein samples are usually stocked at the concentration of 300
°C after flash-frozen in liquid nitrogen. The Casπ protein samples are usually stocked at the concentration of 300 μM. LbCas12a was expressed as previously described.26
μM. LbCas12a was expressed as previously described.26
In vitro transcription of CRISPR RNA
DNA sequences containing T7 RNA polymerase promoter upstream of the Casπ tracrRNA, crRNA and sgRNA were assembled by overlap PCR and validated by Sanger sequencing. The validated sequences were then PCR amplified as the template for in vitro transcription (IVT). All reactions were performed in IVT buffer (30 mM Tris, pH 8.1, 25
mM Tris, pH 8.1, 25 mM MgCl2, 0.01% Triton, 2
mM MgCl2, 0.01% Triton, 2 mM spermidine) with 4
mM spermidine) with 4 mM NTP mix and 0.4
mM NTP mix and 0.4 mg/mL T7 RNA polymerase. The transcribed product was loaded into 10% Urea-PAGE with 2× formamide loading buffer (95% formamide, 0.02% SDS, 0.02% BPB, 0.01% xylene cyanole FF, 1
mg/mL T7 RNA polymerase. The transcribed product was loaded into 10% Urea-PAGE with 2× formamide loading buffer (95% formamide, 0.02% SDS, 0.02% BPB, 0.01% xylene cyanole FF, 1 mM EDTA) for electrophoresis. The gel region containing the target RNA band was extracted, smashed and soaked in soaking buffer (0.38
mM EDTA) for electrophoresis. The gel region containing the target RNA band was extracted, smashed and soaked in soaking buffer (0.38 M NaAc, pH 5.2, 0.8
M NaAc, pH 5.2, 0.8 mM EDTA, 0.8% SDS) for 8
mM EDTA, 0.8% SDS) for 8 h at 4
h at 4 °C. The dissolved RNA was then concentrated using 3 kD molecular weight cut-off centrifugal filters (Merck Millipore) and stocked at –80
°C. The dissolved RNA was then concentrated using 3 kD molecular weight cut-off centrifugal filters (Merck Millipore) and stocked at –80 °C. The RNA samples are usually stocked at the concentration of 50
°C. The RNA samples are usually stocked at the concentration of 50 μM. The RNA sequences and related description are listed in Supplementary information, Table S5.
μM. The RNA sequences and related description are listed in Supplementary information, Table S5.
PAM depletion assay and analysis
PAM depletion assay was performed as previously described with modifications (Supplementary information, Fig. S2a).52 Plasmids containing a PAM library were transformed into E. coli DH5α (TIANGEN) and incubated overnight at 37 °C on LB-Amp+ agar plates (100
°C on LB-Amp+ agar plates (100 μg/mL Ampicillin), and then all colonies were harvested to extract the plasmids using HighPure Maxi Plasmid Kit (TIANGEN). For cleavage reaction, sgRNA was diluted to the concentration of 30
μg/mL Ampicillin), and then all colonies were harvested to extract the plasmids using HighPure Maxi Plasmid Kit (TIANGEN). For cleavage reaction, sgRNA was diluted to the concentration of 30 μM in refolding buffer (50
μM in refolding buffer (50 mM KCl, 5
mM KCl, 5 mM MgCl2) and refolded at 72
mM MgCl2) and refolded at 72 °C for 5
°C for 5 min, and then slowly cooled down to room temperature (RT). Subsequently, active RNP complexes were assembled by incubating 1
min, and then slowly cooled down to room temperature (RT). Subsequently, active RNP complexes were assembled by incubating 1 μM Casπ protein with 1.2
μM Casπ protein with 1.2 μM sgRNA in assembly buffer (100
μM sgRNA in assembly buffer (100 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 5
mM TCEP, 5 mM MgCl2) at RT for 30
mM MgCl2) at RT for 30 min. The reaction was initiated by adding 20
min. The reaction was initiated by adding 20 nM plasmid and performed as three individual replicates in cleavage buffers (50–300
nM plasmid and performed as three individual replicates in cleavage buffers (50–300 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 10
mM TCEP, 10 mM MgCl2) at 37
mM MgCl2) at 37 °C for 1
°C for 1 h, and then quenched with loading buffer (Gel Loading Dye Purple 6×, NEB) supplemented with 20
h, and then quenched with loading buffer (Gel Loading Dye Purple 6×, NEB) supplemented with 20 mM EDTA and 25
mM EDTA and 25 μg/mL heparin. The cleaved products were analyzed and purified by electrophoresis on the 1.2% agarose gel with GelRed staining (Vazyme). Then, the end of linearized products was repaired by T4 DNA polymerase (Thermo Fisher Scientific) with 1
μg/mL heparin. The cleaved products were analyzed and purified by electrophoresis on the 1.2% agarose gel with GelRed staining (Vazyme). Then, the end of linearized products was repaired by T4 DNA polymerase (Thermo Fisher Scientific) with 1 mM dNTP (Sangon Biotech). dA oligo was further added to the 3′ end of the products by Dreamtaq polymerase (Thermo Fisher Scientific) with 1
mM dNTP (Sangon Biotech). dA oligo was further added to the 3′ end of the products by Dreamtaq polymerase (Thermo Fisher Scientific) with 1 mM dATP (Sangon Biotech). Adapters with 3′ dT overhang were ligated with the products containing 3′ dA overhang by fast T4 DNA ligase (Beyotime). The DNA fragments containing the recognized PAM sequence were PCR amplified using a primer pairing to the adapter and the other primer pairing to the 120
mM dATP (Sangon Biotech). Adapters with 3′ dT overhang were ligated with the products containing 3′ dA overhang by fast T4 DNA ligase (Beyotime). The DNA fragments containing the recognized PAM sequence were PCR amplified using a primer pairing to the adapter and the other primer pairing to the 120 bp upstream region of the PAM. Next, the PCR-amplified PAM-containing products were purified by VAHTS DNA Clean Beads (Vazyme) and further amplified by TIANSeq Fast DNA Library Prep Kit (TIANGEN) for Illumina Novaseq PE150 sequencing. In control groups, the plasmids were treated with blank buffer instead of Casπ effectors, and DNA fragments containing PAM library were directly amplified by two primers covering the PAM region for the following process as described above. The depletion fold-change for each PAM was analyzed using the number of matched reads in Casπ and control groups normalized with total reads.
bp upstream region of the PAM. Next, the PCR-amplified PAM-containing products were purified by VAHTS DNA Clean Beads (Vazyme) and further amplified by TIANSeq Fast DNA Library Prep Kit (TIANGEN) for Illumina Novaseq PE150 sequencing. In control groups, the plasmids were treated with blank buffer instead of Casπ effectors, and DNA fragments containing PAM library were directly amplified by two primers covering the PAM region for the following process as described above. The depletion fold-change for each PAM was analyzed using the number of matched reads in Casπ and control groups normalized with total reads.
A list of depleted PAMs and related fold-change values are summarized in Supplementary information, Table S3.
In vitro cleavage assays
For cleavage assays with labeled NTS, the dsDNA substrate was prepared by PCR extension using a 65 nt ssDNA template and a 5′-cy5-labeled 16
nt ssDNA template and a 5′-cy5-labeled 16 nt primer (ordered from Sangon Biotech). Then the extended dsDNA was purified by DNA Clean & Concentrator-25 (Zymo Research) and diluted to 1
nt primer (ordered from Sangon Biotech). Then the extended dsDNA was purified by DNA Clean & Concentrator-25 (Zymo Research) and diluted to 1 μM in nuclease-free water (Invitrogen). The sgRNA was diluted to the concentration of 30
μM in nuclease-free water (Invitrogen). The sgRNA was diluted to the concentration of 30 μM in refolding buffer (50
μM in refolding buffer (50 mM KCl, 5
mM KCl, 5 mM MgCl2) and refolded as described above. Subsequently, Casπ effectors were assembled in a 1:1.2 protein to sgRNA ratio (1
mM MgCl2) and refolded as described above. Subsequently, Casπ effectors were assembled in a 1:1.2 protein to sgRNA ratio (1 μM Casπ protein and 1.2
μM Casπ protein and 1.2 μM refolded sgRNA) in assembly buffer (100
μM refolded sgRNA) in assembly buffer (100 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 5
mM TCEP, 5 mM MgCl2) at RT for 30
mM MgCl2) at RT for 30 min. The reaction was started by mixing 1
min. The reaction was started by mixing 1 μM RNP with 20
μM RNP with 20 nM dsDNA substrate in cleavage buffer (150
nM dsDNA substrate in cleavage buffer (150 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 10
mM TCEP, 10 mM MgCl2) at 37
mM MgCl2) at 37 °C and aliquots were collected at the following time points: 0 mim, 2
°C and aliquots were collected at the following time points: 0 mim, 2 min, 5
min, 5 min, 15
min, 15 min, 30
min, 30 min, 60
min, 60 min, 90
min, 90 min and 120
min and 120 min. For biochemical screenings, only the reaction buffers were modified accordingly, such as the salt concentration (50
min. For biochemical screenings, only the reaction buffers were modified accordingly, such as the salt concentration (50 mM, 150
mM, 150 mM, 300
mM, 300 mM or 450
mM or 450 mM NaCl with 10
mM NaCl with 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 10
mM TCEP, 10 mM MgCl2), type of divalent ions (10
mM MgCl2), type of divalent ions (10 mM Mg2+, Mn2+, Ca2+ or Co2+ with 150
mM Mg2+, Mn2+, Ca2+ or Co2+ with 150 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP) and temperatures (25
mM TCEP) and temperatures (25 °C, 30
°C, 30 °C, 37
°C, 37 °C, 45
°C, 45 °C, 55
°C, 55 °C or 65
°C or 65 °C with 150
°C with 150 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 10
mM TCEP, 10 mM MgCl2). The products were analyzed as described above.
mM MgCl2). The products were analyzed as described above.
For cleavage assays with labeled TS, the 5′-cy5-labeled TS ssDNA was synthesized by Sangon Biotech and diluted to 10 μM in nuclease-free water (Invitrogen). dsDNA was prepared by mixing 5′-cy5-labeled TS and unlabeled complementary oligo at the molar ratio of 1:1.2 in annealing buffer (10
μM in nuclease-free water (Invitrogen). dsDNA was prepared by mixing 5′-cy5-labeled TS and unlabeled complementary oligo at the molar ratio of 1:1.2 in annealing buffer (10 mM HEPES-Na, pH 7.5, 150
mM HEPES-Na, pH 7.5, 150 mM KCl), followed by heating for 5
mM KCl), followed by heating for 5 min at 95
min at 95 °C and slow cooling down to RT. Cleavage reactions were initiated by mixing 1
°C and slow cooling down to RT. Cleavage reactions were initiated by mixing 1 μM RNP with 20
μM RNP with 20 nM ssDNA or dsDNA substrate in cleavage buffer (150
nM ssDNA or dsDNA substrate in cleavage buffer (150 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 10
mM TCEP, 10 mM MgCl2) at 37
mM MgCl2) at 37 °C and the product aliquots were collected at the following time points: 0
°C and the product aliquots were collected at the following time points: 0 min, 2
min, 2 min, 5
min, 5 min, 15
min, 15 min, and 60
min, and 60 min.
min.
For mismatched cleavage assay, the dsDNA substrates with single mismatches were prepared by PCR extension using a 65 nt ssDNA template with single mismatch and a 5′-cy5-labeled 16-nt primer (ordered from Sangon Biotech). Then the extended dsDNA was purified by DNA Clean & Concentrator-25 (Zymo Research) and diluted to 1
nt ssDNA template with single mismatch and a 5′-cy5-labeled 16-nt primer (ordered from Sangon Biotech). Then the extended dsDNA was purified by DNA Clean & Concentrator-25 (Zymo Research) and diluted to 1 μM in nuclease-free water (Invitrogen). Cleavage reactions were initiated by mixing 1
μM in nuclease-free water (Invitrogen). Cleavage reactions were initiated by mixing 1 μM RNP with 20
μM RNP with 20 nM dsDNA substrate in cleavage buffer (150
nM dsDNA substrate in cleavage buffer (150 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 10
mM TCEP, 10 mM MgCl2) at 37
mM MgCl2) at 37 °C and the product aliquots were collected at 1
°C and the product aliquots were collected at 1 h.
h.
For trans-cleavage assay, 1 μM Casπ or LbCas12a RNP was first incubated with 1.5
μM Casπ or LbCas12a RNP was first incubated with 1.5 μM dsDNA or ssDNA activator at 37
μM dsDNA or ssDNA activator at 37 °C for 30
°C for 30 min. Then 20
min. Then 20 nM 5′-cy5-labeled random 60
nM 5′-cy5-labeled random 60 nt ssDNA was mixed into the reaction. The product aliquots were collected at the following time points: 0
nt ssDNA was mixed into the reaction. The product aliquots were collected at the following time points: 0 min, 2
min, 2 min, 5
min, 5 min, 15
min, 15 min, 30
min, 30 min, 60
min, 60 min, 90
min, 90 min and 120
min and 120 min.
min.
All cleavage products collected above were quenched with 2× Urea-loading buffer (8 M urea and 2
M urea and 2 mM Tris-Cl, pH 7.5) supplemented with 20
mM Tris-Cl, pH 7.5) supplemented with 20 mM EDTA and 25
mM EDTA and 25 μg/mL heparin, and then analyzed in 15% urea-PAGE and visualized using Amersham Typhoon 5 (GE Healthcare). Product bands were quantified using ImageJ and cleaved fraction was calculated using the intensity of product bands divided by input intensity.54 Curves of cleavage efficiency were plotted using a One-Phase-Decay model in Prism 8 (GraphPad).
μg/mL heparin, and then analyzed in 15% urea-PAGE and visualized using Amersham Typhoon 5 (GE Healthcare). Product bands were quantified using ImageJ and cleaved fraction was calculated using the intensity of product bands divided by input intensity.54 Curves of cleavage efficiency were plotted using a One-Phase-Decay model in Prism 8 (GraphPad).
For plasmid cleavage assay, 1 μM Casπ RNP effectors were incubated with 20
μM Casπ RNP effectors were incubated with 20 nM target plasmids at 37
nM target plasmids at 37 °C for 30
°C for 30 min and then quenched with loading buffer (Gel Loading Dye Purple 6×, NEB) supplemented with 20
min and then quenched with loading buffer (Gel Loading Dye Purple 6×, NEB) supplemented with 20 mM EDTA and 25
mM EDTA and 25 μg/mL heparin. The samples were analyzed by electrophoresis on a 1.2% agarose gel with GelRed staining (Vazyme). For non-labeled dsDNA cleavage assay, the dsDNA target was PCR amplified from the plasmid containing the protospacer and purified by DNA Clean & Concentrator-25 (Zymo Research). The reaction was initiated by incubating 1
μg/mL heparin. The samples were analyzed by electrophoresis on a 1.2% agarose gel with GelRed staining (Vazyme). For non-labeled dsDNA cleavage assay, the dsDNA target was PCR amplified from the plasmid containing the protospacer and purified by DNA Clean & Concentrator-25 (Zymo Research). The reaction was initiated by incubating 1 μM Casπ RNP effectors with 20
μM Casπ RNP effectors with 20 nM dsDNA target at 37
nM dsDNA target at 37 °C for 30
°C for 30 min and then quenched with loading buffer (Gel Loading Dye Purple 6×, NEB) supplemented with 20
min and then quenched with loading buffer (Gel Loading Dye Purple 6×, NEB) supplemented with 20 mM EDTA and 25
mM EDTA and 25 μg/mL heparin. The samples were analyzed by electrophoresis on the 1.2% agarose gel with GelRed staining.
μg/mL heparin. The samples were analyzed by electrophoresis on the 1.2% agarose gel with GelRed staining.
All experiments were performed at least three times for replicability. A list of oligonucleotides used in this study and related description are summarized in Supplementary information, Table S5.
Determination of cleavage sites
The cleavage products and sites on dsDNA were analyzed by electrophoresis using 15% urea-PAGE as described above. To determine the cleavage sites on plasmids, linearized plasmids were purified and subjected to NGS library construction for Illumina Novaseq PE150 sequencing as described in PAM depletion assay. Paired-end reads were mapped to the target sequence using BWA and 3′-ends were selected to determine the cleavage sites. The abundance of each site was normalized to the total reads and plotted using Prism 8 (GraphPad).
Plasmid interference in bacteria
E. coli BW25141 cells were requested from Prof. Guangdong Shang group in College of Life Sciences, Nanjing Normal University. E. coli BW25141 competent cells carrying the ccdB toxin plasmid (p11-LacY-wtx1) was prepared following the protocol previously described.53 For each group, 200 ng plasmid expressing Casπ and sgRNA (ccdB-targeting or non-targeting) was electroporated into 50
ng plasmid expressing Casπ and sgRNA (ccdB-targeting or non-targeting) was electroporated into 50 μL competent cells with 0.2
μL competent cells with 0.2 cm cuvette (BIO-RAD) under 2.5
cm cuvette (BIO-RAD) under 2.5 kV using Eppendorf eporator. After 1.5
kV using Eppendorf eporator. After 1.5 h of recovering in 5
h of recovering in 5 mL SOC medium (Sangon Biotech) under 37
mL SOC medium (Sangon Biotech) under 37 °C, the bacterial cells were enriched by centrifugation and resuspended in 5
°C, the bacterial cells were enriched by centrifugation and resuspended in 5 mL liquid LB-Strep+ medium (50
mL liquid LB-Strep+ medium (50 μg/mL streptomycin), and cultured for an extra 8
μg/mL streptomycin), and cultured for an extra 8 h. Subsequently, to investigate the effects on bacterial survival by Casπ editing, 5
h. Subsequently, to investigate the effects on bacterial survival by Casπ editing, 5 µL of culture with gradient dilutions from 100 to 10–7 was spotted onto the LB-Amp+ agar plates (100
µL of culture with gradient dilutions from 100 to 10–7 was spotted onto the LB-Amp+ agar plates (100 μg/mL ampicillin) or LB-Strep+-Ara+ agar plates (50
μg/mL ampicillin) or LB-Strep+-Ara+ agar plates (50 μg/mL streptomycin, 10
μg/mL streptomycin, 10 mM arabinose), respectively, and incubated overnight at 37
mM arabinose), respectively, and incubated overnight at 37 °C. In the meantime, to validate the transformation efficiency of Casπ–sgRNA expression plasmids, 10
°C. In the meantime, to validate the transformation efficiency of Casπ–sgRNA expression plasmids, 10 μL of culture was spreaded on LB-Strep+ agar plates (50
μL of culture was spreaded on LB-Strep+ agar plates (50 μg/mL streptomycin) for overnight incubation at 37
μg/mL streptomycin) for overnight incubation at 37 °C, and colony number on each plate was manually counted. 5
°C, and colony number on each plate was manually counted. 5 μL of edited bacterial cells was used for PCR validation of the plasmid interference with Phanta Max Super-Fidelity DNA Polymerase Mastermix (Vazyme).
μL of edited bacterial cells was used for PCR validation of the plasmid interference with Phanta Max Super-Fidelity DNA Polymerase Mastermix (Vazyme).
Construction of EGFP report cell line
To obtain a natural target sequence with diverse targeting windows (different GC contents and PAMs), a sequence survey was performed in mouse genome. Via screening by 20 nt window, we allocated a 270
nt window, we allocated a 270 bp fragment within the Mus musculus myosin heavy polypeptide 8 (MYH8) exon (NCBI accession: NM_177369.3 (3650-3919)) which presents a well distribution of targeting windows with various GC contents (30%–85%) and PAMs (Supplementary information, Table S3). This region shows low sequence similarity to human genome. Frameshifting EGFP (3n
bp fragment within the Mus musculus myosin heavy polypeptide 8 (MYH8) exon (NCBI accession: NM_177369.3 (3650-3919)) which presents a well distribution of targeting windows with various GC contents (30%–85%) and PAMs (Supplementary information, Table S3). This region shows low sequence similarity to human genome. Frameshifting EGFP (3n +
+ 2) was created by fusing the MYH8 fragment, a 32
2) was created by fusing the MYH8 fragment, a 32 bp random flanking sequence and EGFP ORF (1436
bp random flanking sequence and EGFP ORF (1436 bp). The MYH8-EGFP was further inserted into lentiviral packaging plasmid. The LV-MAX lentiviral production system (Thermo Fisher Scientific) was used to produce the lentivirus for inserting the MYH8-EGFP (3n
bp). The MYH8-EGFP was further inserted into lentiviral packaging plasmid. The LV-MAX lentiviral production system (Thermo Fisher Scientific) was used to produce the lentivirus for inserting the MYH8-EGFP (3n +
+ 2) fragment into HEK293A cell genome via infection. The selection and enrichment of genome-modified cells were performed according to the manufacturer’s protocol (Thermo Fisher Scientific).
2) fragment into HEK293A cell genome via infection. The selection and enrichment of genome-modified cells were performed according to the manufacturer’s protocol (Thermo Fisher Scientific).
Gene editing assay in human cells
For EGFP activation editing assay in human cells, the EGFP HEK293A reporter cells were cultured in DMEM (Gibco) supplemented with 10% (v/v) FBS (Gemini) and 1% (v/v) penicillin streptomycin (Gibco) at 37 °C in 5% CO2. About 8.0
°C in 5% CO2. About 8.0 ×
× 104 cells were seeded onto the each well of 48-well plate for ~16
104 cells were seeded onto the each well of 48-well plate for ~16 h incubation. When the cell confluency reached 60%–70%, 300
h incubation. When the cell confluency reached 60%–70%, 300 ng plasmid expressing NLS-Casπ- or Cas9-P2A-PuroR-NLS with sgRNA (MYH8-targeting and non-targeting) was transfected into the cells within each well using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s protocols. One day after transfection, the old medium was replaced by fresh DMEM-Puro+ medium (1.5
ng plasmid expressing NLS-Casπ- or Cas9-P2A-PuroR-NLS with sgRNA (MYH8-targeting and non-targeting) was transfected into the cells within each well using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s protocols. One day after transfection, the old medium was replaced by fresh DMEM-Puro+ medium (1.5 μg/mL puromycin, Sigma) for 3-day culturing. Then the enriched cells were further cultured for another 3 days using fresh DMEM medium without puromycin for gene editing analysis. The EGFP signal was observed with fluorescent microscopy (Nikon Eclipse TS2FL fluorescence microscope). Edited cells were also collected and stored at –80
μg/mL puromycin, Sigma) for 3-day culturing. Then the enriched cells were further cultured for another 3 days using fresh DMEM medium without puromycin for gene editing analysis. The EGFP signal was observed with fluorescent microscopy (Nikon Eclipse TS2FL fluorescence microscope). Edited cells were also collected and stored at –80 °C. For more endogenous gene editing assay, the HEK293T cells were treated the same as mentioned above, but transfected with NLS-Casπ-P2A-PuroR-NLS with sgRNA targeting other endogenous genes.
°C. For more endogenous gene editing assay, the HEK293T cells were treated the same as mentioned above, but transfected with NLS-Casπ-P2A-PuroR-NLS with sgRNA targeting other endogenous genes.
A list of targeting sequences is summarized in Supplementary information, Table S5.
Evaluation of gene editing efficacy
For T7E1 assay, the genome of edited cells was extracted using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech). The edited genome was used as the template for PCR amplification of target region using Phanta Max Super-Fidelity DNA Polymerase Mastermix (Vazyme) (primers listed in Supplementary information, Table S4). The PCR product was gel-purified, and ~200 ng purified DNA was re-annealed for T7E1 cleavage assay according to the manufacturer’s protocol (Vazyme). Cleavage products were analyzed by electrophoresis using 2% agarose gel with GelRed staining (Vazyme).
ng purified DNA was re-annealed for T7E1 cleavage assay according to the manufacturer’s protocol (Vazyme). Cleavage products were analyzed by electrophoresis using 2% agarose gel with GelRed staining (Vazyme).
For NGS, ~210 bp regions nearby the target protospacers were amplified via PCR with Q5 polymerase (NEB) and primers containing Illumina adaptor sequences. Amplicons were verified by electrophoresis using 2% agarose gel with GelRed staining (Vazyme), purified by VAHTS DNA Clean Beads according to the manufacturer’s protocol (Vazyme) and further loaded onto Illumina Novaseq PE150 sequencing by Tianjin Novogene Bioinformatic Technology Co., Ltd. Sequencing reads were analyzed by CRISPResso2 with the following parameters: quantification window centered at 3
bp regions nearby the target protospacers were amplified via PCR with Q5 polymerase (NEB) and primers containing Illumina adaptor sequences. Amplicons were verified by electrophoresis using 2% agarose gel with GelRed staining (Vazyme), purified by VAHTS DNA Clean Beads according to the manufacturer’s protocol (Vazyme) and further loaded onto Illumina Novaseq PE150 sequencing by Tianjin Novogene Bioinformatic Technology Co., Ltd. Sequencing reads were analyzed by CRISPResso2 with the following parameters: quantification window centered at 3 bp for Casπ-1 (2
bp for Casπ-1 (2 bp for Casπ-2, 1
bp for Casπ-2, 1 bp for Cas12a and –3
bp for Cas12a and –3 bp for Cas9) according to cleavage sites of both Casπs (Supplementary information, Fig. S2g, h), quantification window size of 14
bp for Cas9) according to cleavage sites of both Casπs (Supplementary information, Fig. S2g, h), quantification window size of 14 bp for both Casπs (8
bp for both Casπs (8 bp for Cas9), and plot window size of 40
bp for Cas9), and plot window size of 40 bp (to visualize large indels).55 Cells treated with plasmids carrying codon-optimized Cas genes with a non-targeting sgRNA were evaluated at every spacer sequence within every read as a negative control. Percentage of each indel plotted (regardless of substitution) was based on the results of modified reads from the CRISPResso2 output. For the indel size distribution plots, unmodified reads (indel length of 0
bp (to visualize large indels).55 Cells treated with plasmids carrying codon-optimized Cas genes with a non-targeting sgRNA were evaluated at every spacer sequence within every read as a negative control. Percentage of each indel plotted (regardless of substitution) was based on the results of modified reads from the CRISPResso2 output. For the indel size distribution plots, unmodified reads (indel length of 0 bp) were plotted as 0% of the total reads for clarify and the remaining reads were grouped and plotted based on the modified results.
bp) were plotted as 0% of the total reads for clarify and the remaining reads were grouped and plotted based on the modified results.
Reconstitution of Casπ R-loop complex
Deactivated Casπ-1 (dCasπ-1, D537A, E643A) was purified as described above. The sgRNA was diluted to 40 μM in refolding buffer (50
μM in refolding buffer (50 mM KCl, 5
mM KCl, 5 mM MgCl2) and refolded as described above. The dCasπ-1–sgRNA binary was reconstituted by incubating 20
mM MgCl2) and refolded as described above. The dCasπ-1–sgRNA binary was reconstituted by incubating 20 μM dCasπ-1 and 25
μM dCasπ-1 and 25 μM sgRNA for 30
μM sgRNA for 30 min at RT in a total volume of 150
min at RT in a total volume of 150 μL assembly buffer (100
μL assembly buffer (100 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 5
mM TCEP, 5 mM MgCl2). To facilitate the R-loop formation, the bubbled dsDNA substrate with 10
mM MgCl2). To facilitate the R-loop formation, the bubbled dsDNA substrate with 10 nt mismatch in the protospacer was used for R-loop ternary complex assembly. The bubbled dsDNA was diluted to 30
nt mismatch in the protospacer was used for R-loop ternary complex assembly. The bubbled dsDNA was diluted to 30 μM in 150
μM in 150 μL assembly buffer, and mixed with 150
μL assembly buffer, and mixed with 150 μL binary complex at RT for 30
μL binary complex at RT for 30 min incubation. Subsequently, the assembled sample was purified by size exclusion column (Superdex 200 Increase 10/300, GE Healthcare) in SEC buffer (150
min incubation. Subsequently, the assembled sample was purified by size exclusion column (Superdex 200 Increase 10/300, GE Healthcare) in SEC buffer (150 mM NaCl, 10
mM NaCl, 10 mM HEPES-Na, pH 7.5, 1
mM HEPES-Na, pH 7.5, 1 mM TCEP, 0.1% glycerol, 5
mM TCEP, 0.1% glycerol, 5 mM MgCl2) at 4
mM MgCl2) at 4 °C. After flash freezing by liquid nitrogen, the aliquots of purified sample were stocked at –80
°C. After flash freezing by liquid nitrogen, the aliquots of purified sample were stocked at –80 °C. The reconstituted complex was usually stocked at the concentration of 3
°C. The reconstituted complex was usually stocked at the concentration of 3 μM. A list of DNA oligonucleotides and sgRNA sequences with brief descriptions are presented in Supplementary information, Table S5.
μM. A list of DNA oligonucleotides and sgRNA sequences with brief descriptions are presented in Supplementary information, Table S5.
Cryo-EM sample preparation and data collection
4 μL of purified Casπ R-loop complex (~1.5
μL of purified Casπ R-loop complex (~1.5 μM) was crosslinked by BS3 (Sigma-Aldrich) and applied to the graphene oxide grid from Shuimu Biosciences Ltd. (Quantifoil Au 1.2/1.3, 300 mesh), which was glow-discharged (in a HARRICK PLASMA) for 10
μM) was crosslinked by BS3 (Sigma-Aldrich) and applied to the graphene oxide grid from Shuimu Biosciences Ltd. (Quantifoil Au 1.2/1.3, 300 mesh), which was glow-discharged (in a HARRICK PLASMA) for 10 s at middle level after 2
s at middle level after 2 min evacuation. The grid was then blotted by a pair of 55
min evacuation. The grid was then blotted by a pair of 55 mm filter papers (Ted Pella) for 0.5
mm filter papers (Ted Pella) for 0.5 s at 22
s at 22 °C with 100% humidity, and flash-frozen in liquid ethane using FEI Vitrobot Marke IV. Cryo-EM data were collected on a Titan Krios electron microscope operated at 300
°C with 100% humidity, and flash-frozen in liquid ethane using FEI Vitrobot Marke IV. Cryo-EM data were collected on a Titan Krios electron microscope operated at 300 kV equipped with a Cs-corrector and Gatan K3 direct electron detector with Gatan Quantum energy filter using EPU. Micrographs were recorded in counting mode at a nominal magnification of 105,000×, resulting in a physical pixel size of 0.856
kV equipped with a Cs-corrector and Gatan K3 direct electron detector with Gatan Quantum energy filter using EPU. Micrographs were recorded in counting mode at a nominal magnification of 105,000×, resulting in a physical pixel size of 0.856 Å per pixel. The defocus was set between –1.5
Å per pixel. The defocus was set between –1.5 μm and –2.5
μm and –2.5 μm. The total exposure time of each movie stack led to a total accumulated dose of 50 electrons per Å2 which fractionated into 32 frames. More parameters for data collection are shown in Supplementary information, Table S6.
μm. The total exposure time of each movie stack led to a total accumulated dose of 50 electrons per Å2 which fractionated into 32 frames. More parameters for data collection are shown in Supplementary information, Table S6.
Image processing and 3D reconstruction
The raw dose-fractionated image stacks were 2× Fourier binned, aligned, dose-weighted, and summed using MotionCor2.56 CTF-estimation, blob particle picking, 2D reference-free classification, initial model generation, final 3D refinement and local resolution estimation were performed in cryoSPARC.57 Two rounds of 3D reference-based classification were performed in RELION.58 The details of data processing were summarized in Supplementary information, Fig. S5 and Table S6.
Model building and refinement
The initial protein model was generated using AlphaFold2 and manually revised in UCSF-Chimera and Coot.20,59,60 The DNA substrates and sgRNA were manually built in Coot based on the cryo-EM density. The complete model was refined against the EM map by PHENIX in real space with secondary structure and geometry restraints.61 The final model was validated in PHENIX software package. The structural validation details for the final model are summarized in Supplementary information, Table S6.
Quantification and statistical analysis
Statistical details for each experiment can be found in the figure legends and the details of corresponding methods. Graphs show the average of replicates with individual points overlaid, unless stated otherwise.
Supplementary information
Acknowledgements
EM data were collected at the Tsinghua Cryo-EM facility and Shuimu Bioscience. The data were analyzed using the Bio-Computation platform at the Tsinghua University Branch of the Chinese National Center for Protein Sciences (Beijing). We thank the supports from the Tsinghua University Technology Center for Protein Research, Genome Sequencing and Analysis. We thank J.L. Lei, X.M. Li, and X.D. Li for expert electron microscopy assistance. We thank T. Yang, Y.K. Wang, A.B. Jia for computational support. We thank D. Chia, Y. Lin, and N. Liu for their kind advice on the manuscript. The work was supported by the National Key R&D Program of China (2022YFF1002801 to J.J.G.L.), the Ministry of Agriculture and Rural Affairs of China (J.J.G.L.), the National Natural Science Foundation of China (32150018 to J.J.G.L and 32101195 to S.Z.), and start-up funds from Tsinghua University, Beijing (J.J.G.L.).
Author contributions
J.J.G.L. supervised the project. J.J.G.L., J.W., A.S., C.P.L., Z.C., S.Z., and S.L. designed the experiments. C.P.L., S.Z., S.L., and J.L. collected and analyzed the environmental metagenome. C.P.L. and S.Z. built the bioinformatics pipeline and discovered the new system. A.S. purified the Casπ proteins and performed the biochemical assays and analyses. J.W. and C.P.L. did the structural analysis and built the atomic model. Z.C., A.S., D.Y.L., Y.Y., L.Q.L., Y.Z., K.W., and Z.L. did the gene editing experiments in bacterial and mammalian cells. J.J.G.L., J.W., A.S., C.P.L., and Z.C. wrote the manuscript with help from all authors.
Data availability
The electron density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession number of EMD-33983 which are publicly available as of the date of publication. The atomic coordinates and structure factors have been deposited to the Protein Data Bank (PDB) under the accession number of 7YOJ which are publicly available as of the date of publication. The raw cryo-EM micrographs and movies used in this study will be shared by corresponding author upon request. The raw sequencing result of metagenome is uploaded to NCBI database with the accession ID of PRJNA857874. Any additional information required to re-analyze the data reported in this paper is available from the corresponding author upon request.
Material availability
Plasmids generated in this study will be deposited to Addgene or are available upon request. Requests for materials should be addressed to the lead contact J.J.G.L. ([email protected]).
Competing interests
Tsinghua University has filed a patent that includes work described in this paper.
Footnotes
These authors contributed equally: Ao Sun, Cheng-Ping Li, Zhihang Chen, Shouyue Zhang.
Contributor Information
Jia Wang, Email: nc.ude.auhgnist@6102aijgnaw.
Jun-Jie Gogo Liu, Email: nc.ude.auhgnist@uilogogeijnuj.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00771-2.
References
Articles from Cell Research are provided here courtesy of Nature Publishing Group
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/141456440
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/s41422-022-00771-2
Article citations
Recent Advances in the CRISPR/Cas-Based Nucleic Acid Biosensor for Food Analysis: A Review.
Foods, 13(20):3222, 10 Oct 2024
Cited by: 0 articles | PMID: 39456285 | PMCID: PMC11507162
Review Free full text in Europe PMC
TracrRNA reprogramming enables direct PAM-independent detection of RNA with diverse DNA-targeting Cas12 nucleases.
Nat Commun, 15(1):5909, 13 Jul 2024
Cited by: 0 articles | PMID: 39003282 | PMCID: PMC11246509
Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630(8016):484-492, 29 May 2024
Cited by: 0 articles | PMID: 38811729
Flexible TAM requirement of TnpB enables efficient single-nucleotide editing with expanded targeting scope.
Nat Commun, 15(1):3464, 24 Apr 2024
Cited by: 0 articles | PMID: 38658536 | PMCID: PMC11043419
Utilization of CRISPR-Cas genome editing technology in filamentous fungi: function and advancement potentiality.
Front Microbiol, 15:1375120, 28 Mar 2024
Cited by: 0 articles | PMID: 38605715 | PMCID: PMC11007153
Review Free full text in Europe PMC
Go to all (12) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioProject
- (1 citation) BioProject - PRJNA857874
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NM_177369.3
Electron Microscopy Data Bank (EMDB) at PDBe
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structure of the type V-C CRISPR-Cas effector enzyme.
Mol Cell, 82(10):1865-1877.e4, 01 Apr 2022
Cited by: 15 articles | PMID: 35366394 | PMCID: PMC9522604
Identification of RNA Binding Partners of CRISPR-Cas Proteins in Prokaryotes Using RIP-Seq.
Methods Mol Biol, 2404:111-133, 01 Jan 2022
Cited by: 1 article | PMID: 34694606
Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing.
Nucleic Acids Res, 52(6):3234-3248, 01 Apr 2024
Cited by: 1 article | PMID: 38261981 | PMCID: PMC11013384
Discovery of Diverse CRISPR-Cas Systems and Expansion of the Genome Engineering Toolbox.
Biochemistry, 62(24):3465-3487, 16 May 2023
Cited by: 14 articles | PMID: 37192099 | PMCID: PMC10734277
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Ministry of Agriculture and Rural Affairs of the People’s Republic of China
Ministry of Science and Technology of the People’s Republic of China (1)
Grant ID: 2022YFF1002801
National Natural Science Foundation of China (2)
Grant ID: 32150018
Grant ID: 32101195
 1,2 and
1,2 and 


