Abstract
Free full text

Modification of human β-globin locus PAC clones by homologous recombination in Escherichia coli
Abstract
We report here modifications of human β-globin PAC clones by homologous recombination in Escherichia coli DH10B, utilising a plasmid temperature sensitive for replication, the recA gene and a wild-type copy of the rpsL gene which allows for an efficient selection for plasmid loss in this host. High frequencies of recombination are observed even with very small lengths of homology and the method has general utility for introducing insertions, deletions and point mutations. No rearrangements were detected with the exception of one highly repetitive genomic sequence when either the E.coli RecA- or the lambdoid phage encoded RecT and RecE-dependent recombination systems were used.
INTRODUCTION
The ability to obtain high level, tissue-specific and integration site-independent expression of genes requires the use of large genomic fragments. This often precludes the use of plasmids, λ phage vectors and cosmids. The use of YAC (yeast artificial chromosome) vectors has largely overcome this problem as they can propagate large DNA fragments. YACs also allow the manipulation of cloned sequences by the use of homologous recombination in yeast (1). However, the system has limitations. YACs have a high degree of chimerism and clonal instability (2), and the isolation and preparation of YAC clone DNAs is slow with low yields. An alternative to cloning large sequences in YACs would be the use of Escherichia coli-based cloning vectors. Both BAC (bacterial artificial chromosome) and PAC (P1 derived artificial chromosome) vectors are being increasingly used as cloning vehicles for genomic libraries (3,4). These vectors can be stably maintained and propagated with DNA insert sizes of up to 300 kb in length as one or two copy plasmids in bacteria. Their advantages over YAC vectors lies in the ease of library construction and the higher yield of supercoiled plasmid DNA that is more resistant to breakage. Sequences cloned in BAC and PAC vectors also show a high level of stability and low chimerism. No rearrangements have been observed in sequences cloned in these vectors following several generations of growth (4).
Two methods for the genetic modification and manipulation of BAC and PAC clones are available. Yang et al. (5) reported the modification of a BAC clone by homologous recombination in E.coli. Subsequently, Zhang et al. (6) reported a method that involves the use of RecE and RecT, encoded by a lambdoid phage, to modify cloned sequences in E.coli by homologous recombination.
In this paper we describe a general, highly efficient two-step method that has enabled the precise modifications of a PAC clone of the human β-globin locus by E.coli RecA-dependent homologous recombination and the use of a simple and powerful selection procedure in the E.coli DH10B host. The method employs positive and counter selection markers that independently ensure the monitoring of each step. Following integration of the recombination plasmid by a single reciprocal homologous exchange, the E.coli DH10B host not only acquires antibiotic resistance provided by the plasmid-derived positive selection marker, but also loses the host genome-borne resistance to another antibiotic. The stringent counter selection step ensures loss of the excised plasmid following a second homologous recombination event, thereby generating the desired modification of PAC clones in a proportion of the excisants.
MATERIALS AND METHODS
Bacterial host and plasmids
DH10B was grown in l-broth supplemented with the appropriate antibiotics. This strain is streptomycin resistant as a consequence of a mutation in the rpsL gene. The recombination vector pDF25 was constructed by cloning a 3.3 kb BamHI fragment, containing the E.coli recA gene, into the BglII site of the vector pLN135 (7) derived from the plasmid pSC101, and carrying a mutation in the replication protein RepA that renders the plasmid temperature-sensitive for DNA replication. The human β-globin PAC clones, PAC 100 (insert size 175 kb) and PAC 148 (insert size 185 kb) were a gift from Dr P. Ioannou (Institute of Neurology, Nicosia, Cyprus).
Media
l-broth and l-agar plates were prepared as described in Sambrook et al. (8). Chloramphenicol (Cm) was used at 34 µg/ml, kanamycin (Km) at 25 µg/ml, ampicillin (Ap) at 20 µg/ml and streptomycin (Str) at either 20 or 200 µg/ml.
Sensitivity to high streptomycin concentration and preparation of E.coli DH10B electrocompetent cells
Since our method requires a selection step which relies on the DH10B host acquiring sensitivity to Str after transformation with the recombination vector pDF25, we first tested the sensitivity of the untransformed host to low (20 µg/ml) and high (200 µg/ml) concentrations of Str. The host was resistant to both concentrations of the antibiotic as a consequence of its mutation in the rpsL gene. However, on transformation by the recombination vector, which carries the rpsL+ allele, the host became sensitive to the higher concentration of Str (7,9). Electrocompetent E.coli DH10B bacteria were prepared as described (10) but the water was supplemented with 10% glycerol throughout the procedure.
Construction of the plasmid pD3γlox
A loxP sequence was cloned at position –1659 of a human Gγ-globin gene plasmid in the BglII site using the oligonucleotide: 5′-ATCGGGGCCATGGATAACCTTCGTATAGCATACATTATACGAAGTTATGGATC-3′. This sequence includes a diagnostic NcoI site. A 2 kb DraI fragment (–2431 to –393), which harbours the loxP sequence, was isolated from the plasmid and cloned into the unique NaeI site of the recombination vector pDF25. The resulting construct, pD3γlox, was used to target loxP into the corresponding sequence of the Gγ-globin gene in the PAC 100 and PAC 148 clones.
Preparation of PAC clone DNAs, plasmid DNAs and Southern blot hybridisations
PAC DNA was prepared using the modified alkaline lysis method (11). All other plasmid DNAs were prepared by the standard alkaline lysis procedure (8). DNA fragments used for hybridisations were labelled using the DNA oligo labelling method of Feinberg and Vogelstein (12). Southern blots were as described earlier (8).
Induction of recombinase RecE from pBAD-ETγ and mapping gel
The vector pBAD-ETγ (6), with cloned copies of recT and recE genes, was transformed into E.coli DH10B with either PAC 100 or PAC 148 and the cells grown under Ap and Km selection. A single colony was grown overnight at 37°C in 5 ml l-broth cultures supplemented with the same antibiotics. An aliquot was grown to OD600 0.7 and further supplemented with 0.1% l-arabinose for 1 h before harvesting, resulting in the induction of RecE gene from the ara promoter. The expression of RecE recombinase together with RecT facilitates RecA-independent homologous recombination in E.coli (6). Cultures were then plated on l-agar plates with Km alone and grown at 37°C. Absence of Ap selection leads to a loss of the plasmid pBAD-ETγ. Km-resistant but Ap-sensitive clones were used for PAC DNA analysis on a 0.4% agarose gel run in TBE buffer (8).
RESULTS
To perform modifications of PAC clone insert DNAs we took advantage of RecA-mediated homologous recombination in E.coli using the recombination vector pDF25 (Fig. (Fig.1a).1a). This plasmid has a Cm-resistance marker (CmR) for positive selection of the vector, an rpsL+ allele for counter selection (gain of the plasmid-borne wild-type rpsL+ allele in a strain with the a chromosomal mutant rpsL allele, which specifies strR in the haploid state, converts the strain to strS from strR), a polylinker cloning site, a temperature-sensitive replication initiation protein (RepAts), an origin of replication and the recA gene of E.coli. Target sequences were cloned either in the NaeI site or one of the other unique cloning sites of the vector and used for transformation of the bacterial host carrying PAC recombinant clones. Transformants were grown overnight at 30°C on l-agar plates supplemented with Km and Cm. Between 30 and 50 single colonies were picked and transferred to a fresh plate with the same antibiotics and grown overnight at the non-permissive temperature of 43°C. Under these conditions the recombination plasmid cannot replicate and resistance to Cm is conferred by the plasmid integrating either into the host genome or the region of homology contained within the PAC plasmid. Recombination can take place in the region of homology giving a type I or a type II integration upstream of the Gγ-globin gene (Fig. (Fig.1b).1b). The predicted sizes of bands from an NcoI digestion of co-integrant DNAs probed with the 2 kb DraI fragment cloned in the plasmid pD3γlox is shown (Fig. 1b). Blotting analysis of a targeted integration of the plasmid pD3γlox in PAC 148 gave the expected result. Both type I and type II integrations were observed (Fig. (Fig.2a,2a, lanes 4 and 5). The frequency of integration in experiments varied from 30 to 90% (Table (Table1).1). Single colonies of a mixed genotype were also observed. Figure Figure2a2a shows a clone comprising both type I and type II recombination (lane 7). The 1.3 kb band (Fig. 2a, lanes 2–7) is due to cross-hybridisation with the probe and is observed depending on hybridisation conditions and exposure times (Fig. (Fig.2b).2b). Clones giving the correct bands were grown overnight at 43°C on l-agar with Km alone allowing excision and loss of the integrated vector. In this recombination either the original copy of the targeted sequence or the modified copy is left behind. Excised vector clones were selected overnight at 43°C on a fresh plate with Km and high Str. Only clones that have lost the excised recombination vector (ie the rpsL+ allele) grow in high concentrations of Str (7,9). Blotting of these clones showed the desired modification resulting in the generation of 1.5 and 1 kb NcoI bands due to the introduction of the NcoI site present in the loxP oligo instead of the original, unmodified 2.5 kb NcoI band (Fig. (Fig.2b,2b, lanes 4–7 and 9). The frequency of modification was 40% following excision of the plasmid. Loss of the excised plasmid was at a frequency of 100%. Occasionally, both the modified and the unmodified copies of the PAC were seen in the same clone (Fig. (Fig.2b,2b, lane 9 and Table Table1).1). These were separated by replating. Mapping the modified PAC 148 clones (PAC 148γlox), showed that the region corresponding to the β-globin locus was unaltered and showed only the required change (Fig. 2c).
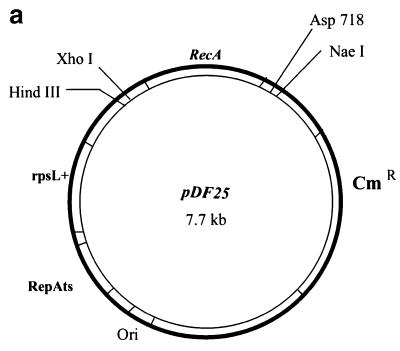
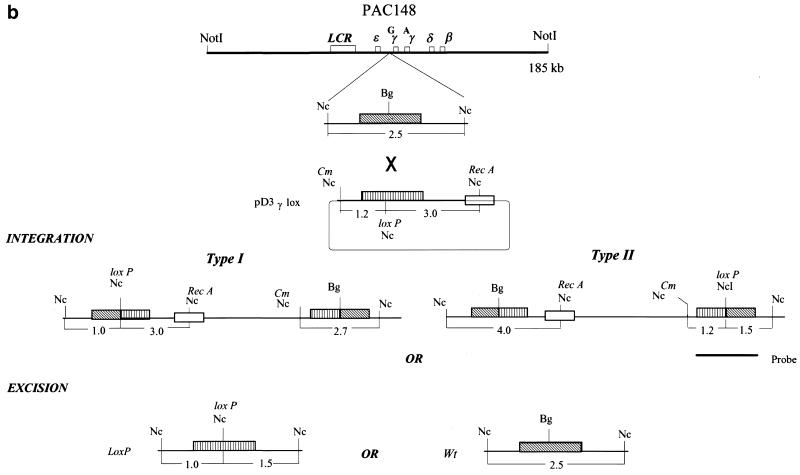
(a) A map of the recombination vector pDF25. A 3.3 kb BamHI fragment of the E.coli recA gene was cloned into the polylinker BglII site of the plasmid pLN135 (9). The plasmid carries an origin of replication (Ori), a temperature-sensitive mutation in the replication initiation protein RepA (RepAts), the gene for resistance to chloramphenicol (CmR) and the rpsL+ gene (the wild-type allele of the rpsL gene). Some of the unique cloning sites are indicated. (b) A schematic representation of the sizes of NcoI fragments hybridising to the 2 kb DraI insert following integration of the plasmid pD3γ-lox into PAC 148 insert and its subsequent excision. The original 2.5 kb NcoI fragment is now interrupted by the new NcoI site giving fragments of 1.5 and 1.0 kb. The abbreviations used in the figure are: Bg, BglII; Nc, NcoI; RecA Nc, NcoI site in the recA gene; Cm Nc, NcoI site in the Cm gene; loxP Nc, NcoI site in the loxP oligo; Wt, unmodified PAC; and LoxP, modified PAC with a targeted insertion of the loxP oligo.
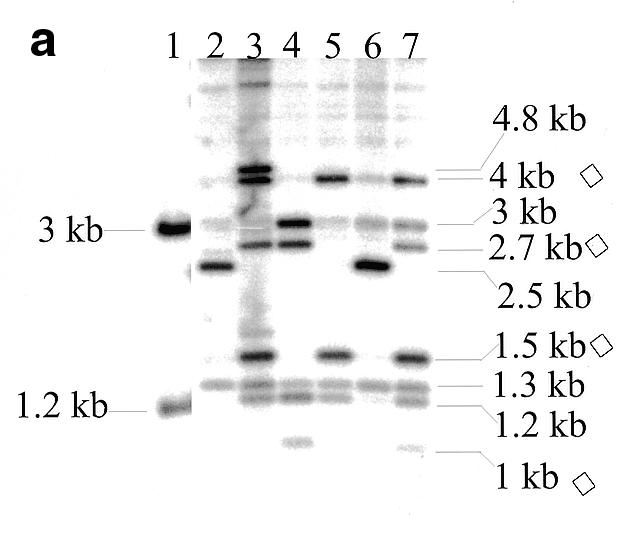

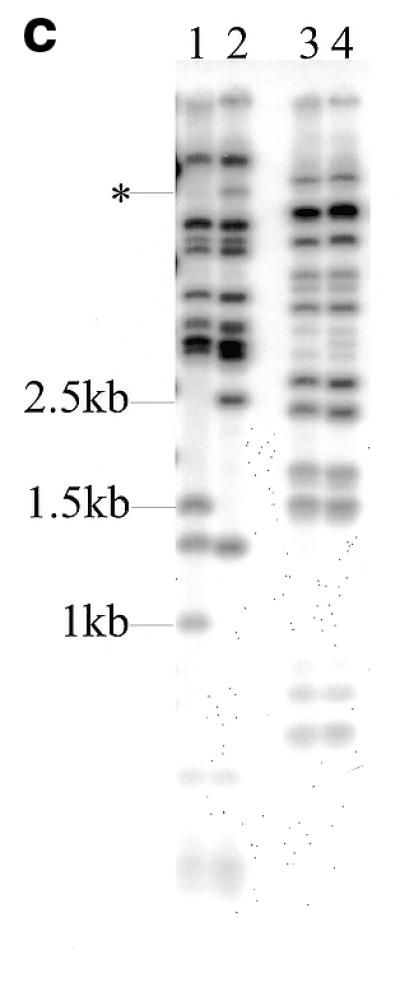
(a) Southern blot hybridisation of NcoI-digested miniprep DNAs of PAC clones following integration of pD3γlox into PAC 148 insert and probing with the fragment indicated in Figure 1b. Unintegrated pD3γlox DNA digested with NcoI (lane 1), PAC 148 DNA (lane 2) and DNAs of clones selected for the integration of pD3γlox into PAC 148 insert (lanes 3–7). Bands marked with a diamond are diagnostic for the integration. Both the 3 and the 1.2 kb bands are also seen in the unintegrated plasmid (lane 1), consequently they are not marked as diagnostic. The 1.3 kb band is due to cross-hybridisation with the probe. (b) Introduction of the loxP sequence and a diagnostic NcoI site into PAC 148 insert after excision of the plasmid pD3γlox. All DNAs were digested with NcoI and probed as in (a). Unintegrated pD3γlox digested with NcoI (lane 1), PAC 148 (lane 2), DNAs from clones selected following excision of the recombination vector construct pD3γlox (lanes 3–9). The 1.5 and 1.0 kb bands are diagnostic for a successful homologous recombination. The 1.3 kb cross-hybridising band seen in (a) is observed only on longer exposures. (c) Mapping of PAC 148 (lanes 2 and 4) and PAC 148γlox (lanes 1 and 3). DNAs were digested with either NcoI (lanes 1 and 2) or with EcoRI (lanes 3 and 4) and the blot probed with the cosmid clone Cosγγδβ (22). The band marked with an asterisk is a partial digest. The 1.5 and 1.0 kb bands are the result of homologous recombination.
Table 1.
| Recombination plasmid | Length of homology | Recombination frequency | Targeted PAC clone | ||
|---|---|---|---|---|---|
| 5′ | 3′ | % Co-integrants | % Mixed genotype | ||
| pD3γlox | 700 bp | 1.2 kb | 96 | 47 | PAC 100 |
| 30 | 10 | PAC 148 | |||
| pDA1 | 620 bp | 600 bp | 60 | 40 | PAC 148 |
| pDA2 | 157 bp | 136 bp | 10 | n/a | PAC 148 |
| pDA3 | 2 kb | 1.5 kb | 80 | 20 | PAC 148 |
| pDA4 | 630 bp | 950 bp | 60 | n/a | PAC 148 |
| pDA5 | 1.2 kb | 1.6 kb | 85 | 30 | PAC 148 |
| pDA6 | 1 kb | 475 bp | 20 | n/a | PAC 148 |
| pDA7 | 600 bp | 1 kb | 85 | n/a | PAC 148 |
| pDmcp3f | 1.6 kb | 1.3 kb | 97 | n/a | PAC-MCP |
Plasmid pD3γlox and plasmids pDA1–7 were used for modifying different sequences along the human β-globin locus. Plasmid pDmcp3f was used to modify the PAC clone of the MCP locus (PAC-MCP). A total of 50–100 colonies were analysed for each experiment. The percentage of co-integrants shown in the table represents the sum total of all the clones which are either of a pure or of a mixed genotype, comprising clones which have either the type I and type II co-integrants or have either of these together with the unrecombined PAC molecule.
n/a, Data not available.
This method was used for modifying several positions in PAC 148γlox and also employed to modify a 120 kb PAC clone of the MCP (membrane cofactor protein CD46) locus (13). We observed frequencies of recombination between 10 and 97% (Table (Table1).1). Lengths of homology at the 5′ and 3′ ends of the desired changes varied respectively from 157 and 136 bp (pDA2) to 1.6 and 1.3 kb (pDmcp3f). Although the shorter homology regions (pDA2), resulted in lower recombination frequencies (10%) they were high enough to efficiently modify the PAC. Longer regions of homology gave higher frequencies, ranging from 30 and 96% (pD3γlox) to 83% (pDA5) and 97% (pDmcp3f) although there are some unexplained variations in the observed frequencies e.g. pD3γlox of 30% (PAC 148) and 96% (PAC 100).
We were also interested in examining the stability of highly repetitive sequences when propagated either in the presence of RecA or the recombinases RecE and RecT. Both PAC 100, which contains a large region of highly repetitive sequence 3′ to the human β-globin locus, and PAC 148 which lacks this region were propagated in E.coli DH10B which is normally without these recombinases. This did not result in any rearrangements following several generations of growth (Fig. 3, lanes 1 and 7). But on introducing either the plasmid pDF25 (Fig. (Fig.3,3, lanes 2, 5, 8 and 11), which harbours the recA gene, or the plasmid pBAD-ETγ (Fig. (Fig.3,3, lanes 3, 6, 9 and 12), which has cloned copies of the RecE and RecT recombinases, we noticed a rearrangement in PAC 100. Following digestion with XhoI or ClaI, a 13 or 10 kb band (Fig. (Fig.3,3, lanes 1 and 4), containing highly repetitive DNA sequences (14), rearranges to a 7.5 or a 5.2 kb band (Fig. (Fig.3,3, lanes 2 and 3 and also lanes 5 and 6). PAC 148 did not show any changes (Fig. (Fig.3,3, lanes 7–12).
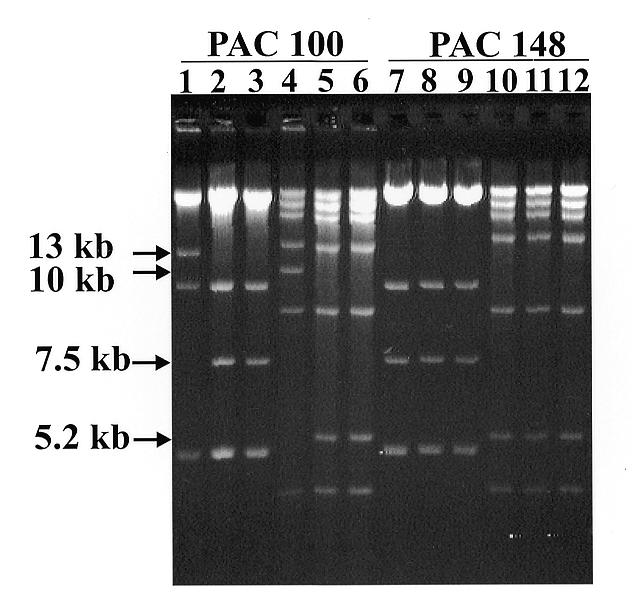
PAC 100 (lane 1) and PAC 148 (lane 7) grown in the absence of RecA and the recombinases RecE and RecT. The effect of either RecA (lanes 2, 5, 8 and 11) or the recombinases RecE and RecT (lanes 3, 6, 9 and 12) on PAC 100 and 148 inserts. DNAs were digested with either XhoI (lanes 1–3 and 7–9) or ClaI (lanes 4–6 and 10–12), run on a 0.4% agarose gel in TBE buffer (8) and visualised by staining with 0.5 µg/ml ethidium bromide.
DISCUSSION
The means to modify large cloned sequences has long been possible in YACs, thus enabling the introduction of point mutations, deletions, inversions etc. and a method describing the modification of cosmid clones by homologous recombination in E.coli has been reported (15). This facility is now being extended to the E.coli BAC and PAC vectors, which have a bigger cloning capacity than cosmids and are easier and faster to prepare than YACs. We have used the method described here to introduce both a copy of the loxP site upstream of the Gγ-globin gene and to modify the human β-globin locus further at several other positions. The use of RecA-mediated homologous recombination to modify both the E.coli genome and episomes has been reported earlier (5,7,16–18). Yang et al. (5) earlier reported the modification of a BAC clone using a plasmid temperature-sensitive for replication, a cloned copy of the recA gene and the TetR gene for both positive and counter selection. However, counter selections using the TetR gene and fusaric acid have been reported to be inefficient (19). Yang and co-workers observed an integration frequency of 10% and a modification frequency of 4% using homology regions of 1 and 1.6 kb. In our experiments using similar lengths of homology (pD3γlox, pDA5 and pDA8) we consistently observed higher frequencies of integration (83–97%) and modification (40–80%). Only when very short 5′ and 3′ lengths of homology are used do we observe a decrease in the integration frequency (10%) and a concomitant drop in the frequency of modification (5%). The use of the wild-type rpsL+ allele in our recombination vector pDF25 allows for a very efficient counter selection procedure in E.coli DH10B, a host normally used to propagate PAC and BAC clones, as it carries a mutated genomic copy of the rpsL gene that confers resistance to Str. The presence of two alleles of the gene conferring sensitivity to Str selection in E.coli is well established (20) and has been used earlier to modify the E.coli genome (7,9). This counter selection also obviates the need to use specialised reagents. An alternative is the use of the sacB gene of Bacillus subtilis for counter selection (6,21). However, this inevitably results in deletions in the sacB gene following growth in sucrose resulting in a high background (6).
The presence of mixed genotypes was observed particularly when very high frequencies of integrations are obtained (Table 1 and Fig. Fig.2a,2a, lane 7). Given that there is more than one copy of the PAC molecule per cell and that multiple copies of the recombination vector pDF25 are present in the same cell, both type I and type II recombinations could proceed in that cell giving this result. We have also observed clones with either type I or type II co-integrants together with the unrecombined PAC (data not shown).
In the literature there is no reference to the fate of highly repetitive sequences propagated in YAC, BAC and PAC clones in the presence of recombinases. Mapping of the human β-globin PAC 100 clone showed a reproducible rearrangement when either RecA or the recombinases RecT and RecE were expressed in the host. Previously, a region ~50 kb downstream from the human β-globin gene, present in PAC 100, has been found to contain highly repetitive sequences (14). PAC 148 lacks this region and remains unaltered under the same conditions. The existence of repeat sequences per se, such as Alu and L1, mapped within the 70 kb region of the β-globin locus do not result in any instability. The possibility of highly repetitive sequences in cloned regions of the eukaryote genome undergoing changes, such as those seen here in PAC 100, would seem to limit the use of this method, but the use of alternative clones and careful mapping following the desired recombination event can overcome this limitation.
The use of bacterial homologous recombination procedures, such as the method described in this paper, allow for efficient introduction of insertions, point mutations and deletions in large DNA fragments cloned in the E.coli PAC and BAC vectors. This technology will facilitate the analysis and organisation of complex genomes and the re-introduction of defined mutations into the eukaryotic genome.
ACKNOWLEDGEMENTS
This work is supported by NWO and the EC. G.P.P. is a recipient of an EC fellowship (contract # HPMFCT199900175). E.K. is a recipient of a short term EMBO fellowship.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/28.12.e65
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/28/12/e65/9904595/2800e65.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Erythropoietic defect associated with reduced cell proliferation in mice lacking the 26S proteasome shuttling factor Rad23b.
Mol Cell Biol, 33(19):3879-3892, 29 Jul 2013
Cited by: 3 articles | PMID: 23897431 | PMCID: PMC3811862
Improved seamless mutagenesis by recombineering using ccdB for counterselection.
Nucleic Acids Res, 42(5):e37, 24 Dec 2013
Cited by: 67 articles | PMID: 24369425 | PMCID: PMC3950717
A dual reporter mouse model of the human β-globin locus: applications and limitations.
PLoS One, 7(12):e51272, 14 Dec 2012
Cited by: 10 articles | PMID: 23272095 | PMCID: PMC3522686
Back to BAC: the use of infectious clone technologies for viral mutagenesis.
Viruses, 4(2):211-235, 03 Feb 2012
Cited by: 16 articles | PMID: 22470833 | PMCID: PMC3315213
Review Free full text in Europe PMC
Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex.
J Cell Biol, 196(6):681-688, 01 Mar 2012
Cited by: 46 articles | PMID: 22431748 | PMCID: PMC3308700
Go to all (42) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Insertion of modifications in the beta-globin locus using GET recombination with single-stranded oligonucleotides and denatured PCR fragments.
Mol Biotechnol, 23(1):29-36, 01 Jan 2003
Cited by: 7 articles | PMID: 12611267
Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system.
Gene Ther, 6(3):442-447, 01 Mar 1999
Cited by: 103 articles | PMID: 10435094
Interplay of SOS induction, recombinant gene expression, and multimerization of plasmid vectors in Escherichia coli.
Biotechnol Bioeng, 80(1):84-92, 01 Oct 2002
Cited by: 7 articles | PMID: 12209789
Homologous pairing proteins encoded by the Escherichia coli recE and recT genes.
Mol Microbiol, 11(1):23-30, 01 Jan 1994
Cited by: 63 articles | PMID: 8145642
Review




