Abstract
Free full text

Human RNPS1 and Its Associated Factors: a Versatile Alternative Pre-mRNA Splicing Regulator In Vivo
Abstract
Human RNPS1 was originally purified and characterized as a pre-mRNA splicing activator, and its role in the postsplicing process has also been proposed recently. To search for factors that functionally interact with RNPS1, we performed a yeast two-hybrid screen with a human cDNA library. Four factors were identified: p54 (also called SRp54; a member of the SR protein family), human transformer 2β (hTra2β; an exonic splicing enhancer-binding protein), hLucA (a potential component of U1 snRNP), and pinin (also called DRS and MemA; a protein localized in nuclear speckles). The N-terminal region containing the serine-rich (S) domain, the central RNA recognition motif (RRM), and the C-terminal arginine/serine/proline-rich (RS/P) domain of RNPS1 interact with p54, pinin, and hTra2β, respectively. Protein-protein binding between RNPS1 and these factors was verified in vitro and in vivo. Overexpression of RNPS1 in HeLa cells induced exon skipping in a model β-globin pre-mRNA and a human tra-2β pre-mRNA. Coexpression of RNPS1 with p54 cooperatively stimulated exon inclusion in an ATP synthase γ-subunit pre-mRNA. The RS/P domain and RRM are necessary for the exon-skipping activity, whereas the S domain is important for the cooperative effect with p54. RNPS1 appears to be a versatile factor that regulates alternative splicing of a variety of pre-mRNAs.
Most pre-mRNAs in higher eukaryotes complete accurate splicing in the nucleus, as a prerequisite for carrying the correct genetic information to the cytoplasm for translation. Constitutive splicing is highly precise and is sensitive to mutations in critical signal sequences of the pre-mRNA. Indeed, human genetic diseases are often caused by point mutations that lie in 5′ or 3′ splice site elements or by creation of new ones at inappropriate locations, which result in splicing defects (reviewed in references 33 and 50). On the other hand, a subset of pre-mRNAs shows sufficient flexibility for alternative potential splice sites to be used, often in a regulated way in response to tissue-specific or developmentally regulated states (reviewed in references 24, 55, and 82). This process, called alternative splicing, is a basic strategy for the regulation of eukaryotic gene expression. An unexpectedly small set of protein-coding genes, at most 30,000 has recently been estimated in the human genome (51, 72, 79). Indeed, a higher prevalence of alternative splicing than earlier estimates is likely responsible for a larger number of, and more complex, protein products (51, 64).
Pre-mRNA splicing takes place within a large complex, or spliceosome, which includes the small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U4/U6, and U5, together with a large number of non-snRNP protein factors. Biochemical characterization of the spliceosome, together with genetic studies in fission and budding yeast, predicted that more than 50 proteins are essential for constitutive splicing (reviewed in references 15 and 73). The members of the serine/arginine-rich protein (SR protein) family are well-studied non-snRNP protein factors that have diverse functions in both constitutive and alternative pre-mRNA splicing (reviewed in references 18, 36, and 77). SR proteins have a modular structure and contain one or two N-terminal RNA-recognition motifs (RRM) that interacts with RNA, and a C-terminal arginine/serine-rich domain (RS domain) that functions in protein-protein interaction. RS domain-containing proteins with or without a RRM, which are distinct from SR proteins, are often splicing-related factors, e.g., the splicing coactivators SRm160/300, the alternative splicing regulator human Tra2α/β (hTra2α/β), U2 auxiliary factors U2AF35/U2AF65, the SR protein kinase Clk/Sty, the U1 snRNP components U1-70K, hLuc7p, and so on (reviewed in references 6 and 35).
In the course of purification of an activity that stimulates splicing via a distal alternative 3′ splice site, an ~50-kDa protein (on sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) was purified from HeLa cell nuclear extracts and identified as human RNPS1 (59). The human RNPS1 cDNA encodes a typical RNA-binding protein containing a single canonical RRM, a characteristic serine-rich domain (S domain) in the N-terminal region, and an arginine/serine/proline-rich domain (RS/P domain) in the C-terminal region (2, 59). Purified native or recombinant RNPS1 with a limited amount of SF2/ASF synergistically stimulates both constitutive and alternative splicing in vitro (59). Therefore, RNPS1 was characterized as a general splicing activator, and it was suggested that RNPS1 itself is not sufficient for the alternative 3′ splice site switch from proximal to distal that was observed with partially purified fractions.
Human RNPS1 colocalizes with SR proteins to the nucleus in an intense speckled pattern (56, 59), which is characteristic of many other splicing factors (reviewed in reference 63). The major structural component of the nuclear speckles is the interchromatin granule cluster, which is thought to represent storage or assembly locations for splicing components (reviewed in reference 63). Immunoprecipitation by anti-RNPS1 antibody demonstrated that RNPS1, like SR proteins, is stably associated with active spliceosomes including both pre-mRNA and spliced mRNA (59). Recently, RNPS1 was identified as a component of a postsplicing mRNP complex, termed the exon-exon junction complex (EJC), together with other factors involved in splicing, mRNA export, and nonsense-mediated mRNA decay (NMD) (52). The EJC, therefore, was suggested to mark spliced mRNA specifically to promote subsequent splicing-dependent processes. RNPS1 as well as any of three human Upf proteins (hUpf1, hUpf2, and hUpf3) was demonstrated to be capable of triggering NMD when tethered downstream of a premature termination codon (reference 57 and references therein). Recently, it has been suggested that RNPS1 facilitates 3′ end processing of mRNA and enhances the translational activity but does not affect nuclear mRNA export in human cells (83; M. J. Moore, personal communication). RNPS1 was also identified in the complex termed the apoptosis- and splicing-associated protein (ASAP) together with SAP18 and different isoforms of Acinus protein, suggesting a possible role for the ASAP complex in linking RNA processing and apoptosis (75).
Interactions of RNPS1 with other proteins may underlie the multiple functions of RNPS1, such as splicing activation of constitutive pre-mRNAs, regulation of alternatively splicing, and a potential role in postsplicing processes. To search systematically for factors that functionally interact with human RNPS1, we performed a yeast two-hybrid screen of a human cDNA library with human RNPS1 as bait. We identified several known splicing-related factors and confirmed their interaction with RNPS1 in vitro. We performed in vivo splicing assays to analyze the function of these protein interactions. Significantly, we found functional synergy between RNPS1 and its associated factors in the modulation of alternative splicing in vivo.
MATERIALS AND METHODS
Construction of plasmids.
All the variant RNPS1 constructs are summarized with their names and schematic protein structures in Table Table11.
TABLE 1.
Summary of RNPS1 variant proteins used in this study
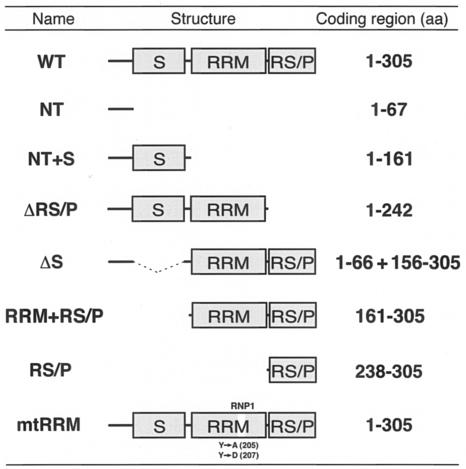 |
Full-length and a series of variant RNPS1 fragments were amplified from RNPS1 (E5.1) cDNA (2) by PCR with Deep Vent DNA polymerase (New England Biolabs) and the following specific pairs of DNA primers: S1-WT-F (5′-CCGAATTCATGGATTTATCAGGAGTG-3′) and S1-WT-R (5′-AACTCGAGGTTTATCGGGAGGAGTTG-3′) for wild-type (WT) RNPS1 (Table (Table1);1); S1-WT-F (see above) and 5′-TACTCGAGTTAGGTGGGCTTAGGAGATG-3′ for RNPS1-NT+S (Table (Table1);1); S1-WT-F (see above) and 5′-AACTCGAGTTACCAGGGGGCCAGCAC-3′ for RNPS1-ΔRS/P (Table (Table1);1); 5′-AAGAATTCACCAAAGTGCACATTGGG-3′ and S1-WT-R (see above) for RNPS1-RRM+RS/P (Table (Table1);1); and S1-CT-F (5′-TTGAATTCGTGCTGGCCCCCTGGCCT-3′) and S1-WT-R (see above) for RNPS1-RS/P (Table (Table1).1). These PCR-amplified fragments were digested with EcoRI and XhoI and subcloned into the corresponding sites of the LexA DNA-binding vector pLexA (Clontech) to generate plasmids, pLexA-RNPS1, pLexA-RNPS1-NT+S, pLexA-RNPS1-ΔRS/P, pLexA-RNPS1-RRM+RS/P, and pLexA-RNPS1-RS/P.
The two coiled-coil domains of pinin cDNA (pinin-C23; amino acids 134 to 347) were amplified from MH22 DRS cDNA plasmid (gift from W.W. Franke) (11) by PCR with the primers 5′-TTGGATCCATGGATGAAAAGGGAAAGC-3′ and 5′-GGGAATTCTTAACTATCCTGCTGCTTCTC-3′. The fragment was subcloned between the BamHI and EcoRI sites of pGEX-2T vector (Amersham) and the expression plasmid pGEX-pinin-C23 was obtained. The excised BamHI fragment of pcDNA3-p54 (a gift from J. Y. Wu) (86) was subcloned into the corresponding site of pFLAG-CMV-2 (Sigma) to generate the expression plasmid pFLAG-p54. The excised NdeI-BamHI fragment of pET14b-htra2β (a gift from J. L. Manley) (78) was filled in by Klenow fragment (New England Biolabs) and subcloned into the EcoRV site of pFLAG-CMV-2 to create the expression plasmid pFLAG-hTra2β.
To construct pFLAG-GFP, the HindIII-EcoRV fragment excised from pCMX-hGFP-S65A/Y145F (a gift from H. Sakamoto) (69) was subcloned into the corresponding sites of pFLAG-CMV-2. Two synthetic DNAs (5′-GCCTGCAGAATTCATTAAAGAGGAGTGCAGCCACCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGACTACAAGGATGACGATGACAAGGTCGACAGATCTACTAGTGGATCCGC-3′ and its complementary fragment) were annealed, cleaved with PstI and BamHI, and the DNA fragment was subcloned into the corresponding sites of pCMV-SPORT (Invitrogen) to generate pCMV-3×FLAG. The RNPS1-coding region was excised with BamHI from pVL-RNPS1 (59) and subcloned into the BglII site of pCMV-3×FLAG to generate N-terminal FLAG-tagged plasmid pCMV-3×FLAG-RNPS1. The RNPS1-coding region was excised with SalI and BamHI from pCMV-3×FLAG-RNPS1 and subcloned into the corresponding sites of pDsRed2-C1 (Clontech) to generate pDsRed-RNPS1.
To construct RNPS1-ΔS plasmids (Table (Table1),1), three steps of PCR were performed with Platinum Pfx DNA polymerase (Invitrogen). First, two short DNA fragments of RNPS1 were amplified from pCMV-3×FLAG-RNPS1 with two sets of primers, SP6-PRO (5′-GATTTAGGTGACACTATAG-3′)/5′-TAGGAGATGGGCGCCTCTTTCGGGTTTTGTC-3′ and 5′-AAGAGGCGCCCATCTCCTAAGCCCACCAA-3′/S1-C-R (5′-CACACAGATCTTATCGGGAGGAGTTGG-3′). These two amplified fragments, which are capable of annealing to each other (see italic sequences), were purified from a 2% agarose gel and mixed together for the second PCR (five cycles in 20 μl). The primers SP6-PRO and S1-C-R (see above) were added directly to this reaction, and the last PCR was performed (25 cycles in 100 μl). The amplified fragment was cleaved with EcoRI and BglII and subcloned into the corresponding sites of pCMV-3×FLAG to obtain plasmid pCMV-3×FLAG-RNPS1-ΔS. The SalI-BamHI fragment from pCMV-3×FLAG-RNPS1-ΔS was subcloned into the corresponding sites of pDsRed2-C1 to obtain pDsRed-RNPS1-ΔS. The pCMV-3×FLAG-RNPS1-mtRRM plasmid (Table (Table1)1) was constructed in a similar way, except that the following sets of primers were used for the first PCR: SP6-PRO (see above)/5′-CTACGTCCGCAGCGCCTTTGGACAGATGG-3′ and 5′-GGCGCTGCGGACGTAGAGTTTGAGAATCC-3′/S1-C-R (see above) (italic sequences indicate annealing site).
To construct RNPS1-ΔRS/P (Table (Table1)1) plasmids, the DNA fragment was PCR-amplified from pCMV-3×FLAG-RNPS1 with two primers SP6-PRO (see above) and 5′-TTTAGATCTTACCAGGGGGCCAGCAC-3′. The amplified fragment was digested with EcoRI and BglII, purified from a 2% agarose gel, and subcloned into the corresponding sites of pCMV-3×FLAG to obtain pCMV-3×FLAG-RNPS1-ΔRS/P. The SalI-BamHI fragment of this plasmid was subcloned into the corresponding sites of pDsRed2-C1 to obtain pDsRed-RNPS1-ΔRS/P. The pCMV-3×FLAG-RNPS1-RS/P (Table (Table1)1) plasmid was constructed in a similar way, with the following primers: S1-CT-F2 (5′-CCTGGATCCGTGCTGGCCCCCTGGCCTAG-3′) and RNPS1-C (59). The amplified fragment was digested with BamHI, purified from a 3% agarose gel, and subcloned into the BglII sites of pCMV-3×FLAG.
pCMV-1×FLAG was generated in a similar way as pCMV-3×FLAG (see above), with the following synthetic DNAs: 5′-GCCTGCAGAATTCATTAAAGAGGAGTGCAGCCACCATGGACTACAAGGATGACGATGACAAGGTCGACAGATCTACTAGTGGATCCGC-3′ and its complementary fragment. The green fluorescent protein (GFP)-coding region was PCR amplified from plasmid pMG2 (46) with two primers (5′-CGACTAGTATCGATAGTAAAGGAGAAGGACTTTTC-3′ and 5′-AACTCGAGGGATCCTCTAGATTAGTGGTGGTGGTGGTGGTGGGCATAGTCCACGTCATAGGGATATTTGTATAGTTCATCCATGCC-3′) and subcloned into the SpeI and XhoI sites of pCMV-1×FLAG to generate pCMV-1×FLAG-GFP. The fragments of NT and RS/P domains (Table (Table1)1) were PCR-amplified from RNPS1 cDNA (see above) with the following primer combinations: RNPS1-N2 (59)/5′-CCGATCGATGCTGCGCCTCTTTCGGGTTT-3′ for the NT fragment, and S1-CT-F2 (see above)/5′-CACATCGATTCGGGAGGAGTTGGAGCTGG-3′ for RS/P fragment. The amplified fragments were digested with BamHI and ClaI and subcloned into the BglII and ClaI sites of pCMV-1×FLAG-GFP to obtain plasmids pCMV-1×FLAG-RNPS1-NT-GFP and pCMV-1×FLAG-RNPS1-RS/P-GFP, respectively.
The enhanced green fluorescent protein (EGFP)-encoding DNA fragment (Clontech) was PCR amplified with flanking KpnI and SalI tags and subcloned in the pBluescript vector (Stratagene) to obtain plasmid pBlue-EGFP. The mouse RNPS1 encoding sequence, which has ~99% identity with human RNPS1 (59), was PCR amplified with flanking SalI and HindIII tags and subcloned in the corresponding sites of pBlue-EGFP. The in-frame fused EGFP-RNPS1 fragments were excised from the pBlue-EGFP with KpnI and NotI and subcloned into the corresponding sites of the eukaryotic expression vector pcDNA3 (Invitrogen) to obtain pcDNA3-EGFP-RNPS1. For the control plasmid (pcDNA3-EGFP), the KpnI- and NotI-tagged EGFP fragment was subcloned in the pcDNA3 vector as well.
Yeast two-hybrid assays.
The cDNA fragments for full-length RNPS1 and RNPS1-ΔRS/P (Table (Table1)1) were fused in-frame to the LexA DNA-binding domain of the pLexA vector to obtain pLexA-RNPS1 and pLexA-RNPS1-ΔRS/P, respectively. A human HeLa cell cDNA library subcloned into the B42 activation vector pB42AD (Clontech) was screened as described in the manufacturer's protocol. Briefly, Saccharomyces cerevisiae EGY48 harboring the β-galactosidase reporter plasmid p8op-lacZ (EGY48[p8op-lacZ]) was cotransformed with the pB42AD-human HeLa cDNA library and either pLexA-RNPS1 or pLexA-RNPS1-ΔRS/P. Positive clones were identified by galactose-dependent growth in medium lacking leucine and confirmed by β-galactosidase activity of the colonies. Duplicate clones were eliminated by comparing the lengths of their cDNA inserts amplified by PCR with two primers, pB42AD-F (5′-CCAGCCTCTTGCTGAGTGGAGATG-3′) and pB42AD-R (5′-GGCAAGGTAGACAAGCCGACAACC-3′), and by their Sau3AI-digested fragments.
To eliminate false-positive clones, plasmids from putative positive clones were transformed and tested again for galactose-dependent growth in medium lacking leucine and for β-galactosidase activity. pLexA-Lam (which encodes human lamin C66-230 in pLexA; Clontech) and the pLexA vector were used as negative controls. The cDNA inserts in the selected positive plasmids were sequenced with the pB42AD-F primer. Independent transformants (~4 × 106) were screened by their ability to grow in the absence of leucine as well as their β-galactosidase activity. A total of 157 colonies were isolated from the initial screening. After false-positive and duplicate clones were eliminated, 20 clones were shown to activate the reporters in combination with LexA-RNPS1 or LexA-RNPS1-ΔRS, but not with LexA alone or LexA-lamin. The sequences of each clone (~200 bp) were used to search sequence databases.
Preparation of recombinant proteins.
Expression and purification of glutathione S-transferase (GST) and GST fusion proteins were performed with Escherichia coli strain BL21-CodonPlus (DE3)-RP (Stratagene) following the manufacturer's instructions (Amersham). The E. coli strain JM101 containing pGEX-hTra2β was a gift from J. L. Manley, and expression and purification of the protein were performed as described (78).
Cell culture and transient transfections.
Adherent HeLa cells were cultured as described (38). For transient transfection, cells were plated at density of ~3.0 × 105 cells per 3.5-cm dish or ~2.0 × 106 cells per 10-cm dish the day before transfection. Each expression plasmid was transfected with Lipofectamine 2000 (Invitrogen) in serum-free Opti-MEM (Invitrogen) as described in the manufacturer's protocol. After 4 h incubation, Opti-MEM with 20% fetal bovine serum (FBS) was added to the medium to a final 10% FBS concentration. Approximately 85% of HeLa cells were transfected, as determined by expression of transfected pFLAG-GFP under a fluorescent microscope. Ehrlich ascites tumor cells (suspension culture) were transiently transfected by electroporation as described (74).
Preparation of whole HeLa cell extracts.
HeLa cells were transiently transfected with 15 μg of either pFLAG-p54, pFLAG-hTra2β, or pFLAG-GFP plasmids in 10-cm dishes. At 24 h after transfection, cells were lysed in 150 μl of lysis buffer [20 mM HEPES-NaOH (pH 7.3), 3.2 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride] by freezing and thawing. The lysate was centrifuged at 20,000 × g, and the supernatant was used for immunoprecipitation assays.
In vitro-coupled transcription-translation.
In vitro translation was performed with the TNT T7 coupled wheat germ extract (see Fig. Fig.1A),1A), and reticulocyte lysate (see Fig. Fig.1C)1C) systems (Promega). Briefly, 0.5 to 1 μg of the E. coli expression plasmid pSBETc-RNPS1 (59) and the control luciferase T7 plasmid were incubated (in 50 μl) with TNT master mixture with 20 μCi of [35S]methionine (Amersham) for 2 h at 30°C. The translation efficiency was checked by analyzing 2 μl of lysate on a 10% SDS-PAGE gel.
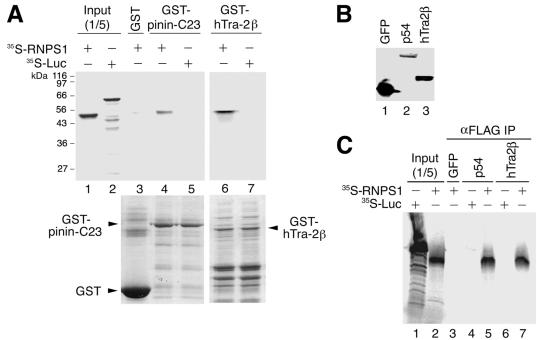
In vitro binding assays of RNPS1 to pinin, hTra2β, and p54 proteins. (A) The indicated recombinant GST-tagged fusion proteins coupled to glutathione Sepharose beads were incubated with in vitro-translated 35S-labeled RNPS1 or luciferase (Luc) as a control, and the pelleted beads were washed and analyzed by 10% SDS-PAGE (GST pull-down assays). The upper panel shows the autoradiograph. A portion (one fifth) of each 35S-labeled protein used in the pull-down reactions was loaded as the input control (Input). Marker protein sizes are indicated on the left. The lower panel shows a 10% SDS-PAGE gel (stained with Coomassie blue) that was used for autoradiography. The corresponding proteins are indicated with arrowheads. (B) Expression of FLAG-tagged constructs (GFP, p54, and hTra2β) in HeLa whole-cell extracts. The extracts (5 μl each) were separated by 10% SDS-PAGE and detected by immunoblotting with anti-FLAG antibody. (C) Immunoprecipitation analyses of 35S-labeled RNPS1 and luciferase (Luc) as a control with the indicated expressed FLAG-fused proteins (GFP as a control). Anti-FLAG antibody immobilized on protein G-Sepharose was used for the immunoprecipitation. Immunoprecipitated 35S-labeled RNPS1 protein was detected by autoradiography. A portion (one fifth) of each 35S-labeled protein used in the immunoprecipitation was loaded as control (Input).
GST pull-down and immunoprecipitation assays.
GST-tagged fusion proteins (pinin-C23, hTra2β, or GST alone for a control) coupled to glutathione Sepharose beads (Amersham) were incubated for 120 min at 4°C with 1 μl of in vitro-translated RNPS1 (or luciferase as a control) in a total volume of 300 μl with binding buffer A (20 mM HEPES-KOH [pH 7.3], 3.2 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 33.3 U of DNase I [Promega] per ml, 10.7 μg of RNase A [Worthington] per ml, and 0.5 mg of bovine serum albumin [Sigma] per ml). After incubation, the beads were washed four to five times with binding buffer A. The pelleted beads were heated at 92°C for 5 min with 15 μl of SDS sample buffer. After removing insoluble material by microcentrifugation, the proteins were separated on a 10% SDS-PAGE gel.
FLAG-tagged cDNA constructs (p54, hTra2β, or GFP as a control) were transiently transfected into HeLa cells and their expression was checked by immunoblotting. Immunoblotting was performed as described (59) with anti-FLAG M2 monoclonal antibody (Sigma) and anti-mouse IgG horseradish peroxidase-linked whole antibody (Amersham). The immunoreactive proteins were detected by chemiluminescence according to the manufacturer's protocol (ECL detection reagents; Amersham). A washed protein G-Sepharose slurry (20 μl = 10 μl of beads; Amersham) was first added to the FLAG-tagged expressed-protein extracts and rocked at 4°C for 1 h to preadsorb nonspecific protein G binding proteins. After brief microcentrifugation, the supernatants were recovered and 3 μg of anti-FLAG M2 monoclonal antibody, 3 μl of in vitro-translated RNPS1 (or luciferase as a control), and another 20 μl of fresh protein G-Sepharose beads mixture were added in a total volume of 300 μl with binding buffer A. Incubation was continued at 4°C with rocking for 2 h. The protein G-Sepharose beads were washed four times in 500 μl of binding buffer A, and the bound proteins were analyzed on a 10% SDS-PAGE gel as described above.
The 35S-labeled RNPS1 in the SDS-polyacrylamide gel was analyzed with a phosphorimager (Storm 840; Molecular Dynamics) or a Bio-imaging analyzer (BAS2000; Fujifilm).
In vivo splicing and RT-PCR analyses.
The reporter plasmids expressing pre-mRNA substrates were as follows; pHD1012-DUP51 (a gift from R. Kole; 30) for duplicated human β-globin pre-mRNA (DUP51), pCR3.1-hTra2-β1 (a gift from J. L. Manley) (66) for human transformer-2-β1 minigene (hTra2β), and pCMV-F1γEx8-10 (38) for human mitochondrial ATP synthase γ-subunit minigene (F1γ). HeLa cells were transiently transfected in a 3.5-cm dish with 2 μg of the reporter plasmids and various amounts of protein expression plasmids; pFLAG-p54 (0 or 2 μg), pFLAG-hTra2β (0 or 2 μg), pCMV-3×FLAG-RNPS1 (0, 0.5, or 2 μg), pCMV-3×FLAG-RNPS1-ΔS (0, 0.5, or 2 μg), pCMV-3×FLAG-RNPS1-ΔRS/P (0, 0.5, or 2 μg), pCMV-3×FLAG-RNPS1-mtRRM (0, 0.5, or 2 μg), and pCMV-3×FLAG-RNPS1-RS/P (0, 0.5, or 2 μg). The final amount of transfected DNA was kept constant at 6 μg by addition of control pFLAG-CMV-2 vector. At 24 h after transfection, cells were harvested and total RNA was isolated with Trizol reagent (Invitrogen) as described in the manufacturer's protocol. The isolated total RNA was treated with DNase I for 1 h, denatured at 65°C for 10 min, and immediately chilled on ice prior to reverse transcription (RT).
RT-PCR was used to detect in vivo splicing products. First strand cDNA was synthesized (in 10 μl) by Superscript II (50 units; Invitrogen) for 50 min at 42°C with 400 ng of total RNA in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM each of each deoxyribonucleoside-5′-triphosphate (dNTP), and either 0.1 mM oligo(dT)16 primer or 50 nM specific RT primer. The RT primers used were Glo-E2 (5′-CAGGTGAGCCAGGCCATCACTAAAG-3′) for DUP51 substrate, pCR3.1RT (5′-GCCCTCTAGACTCGAGCTCGA-3′) for hTra2β, and oligo(dT)16 primer for F1γ substrate. PCR amplification was performed (in 25 μl) with 1 μl of first-strand cDNA mixture, and 1 unit of Taq DNA polymerase (Takara) in 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each of each dNTP, and 0.2 μM each of the forward and reverse PCR primers. The PCR primers used were DUP-F (5′-CACTAGCAACCTCAAACAGACACCATGC-3′) and DUP-R (5′-ATGAGCCTTCACCTTAGGGTTGCCC-3′) for DUP51 substrate, MGTraBam (5′-GGGGATCCGACCGGCGCGTCGTGCGGGGCT-3′) and MGTraXho (5′-GGGCTCGAGTACCCGATTCCCAACATGACG-3′) for the hTra2β substrate, and the sense and antisense primers (in SP6 promoter and exon 10, respectively) for the F1γ substrate were previously described (38). The PCR conditions for each cycle were 94°C for 20 s, 62°C for 30 s, and 72°C for 1 min, and a total of 30 cycles were performed. The amplified PCR products were analyzed by electrophoresis on a 3% agarose gel and visualized by ethidium bromide staining. The essential RT-PCR experiments were repeated at least three times, and all these data gave consistent results.
Fluorescence and immunofluorescence microscopic analyses.
HeLa cells were transfected in a 3.5-cm glass-bottomed dish (Matsunami) with expression constructs (2 μg each plasmid); pDsRed-RNPS1, pDsRed-RNPS1-ΔS and pDsRed-RNPS1-ΔRS/P, in combination with pFLAG-p54 (2 μg) or pFLAG-hTra2β (2 μg). For the expression of GFP fusion proteins, pCMV-1×FLAG-RNPS1-NT-GFP (2 μg) and pCMV-1×FLAG-RNPS1-RS/P-GFP (2 μg) were transfected. Since overexpression of RNPS1 fusion protein beyond 48 h resulted in lethality (data not shown) (56), cells were examined 12 to 18 h after transfection. Cells were rinsed twice with phosphate buffered saline (PBS), fixed and permeabilized with PBS containing 4% paraformaldehyde and 0.4% Triton X-100 for 15 min at room temperature, and rinsed three times with PBS. The GFP fusion proteins were visualized under microscopy at this point (see below).
For fluorescent staining, fixed cells were incubated overnight at 4°C with anti-FLAG antibody [Sigma; 1:1,000 dilution in PBS containing 2% FBS and 0.05% Tween 20 (same in the following dilutions)]. After washing four times with PBS, cells were incubated at room temperature for 1 h with diluted secondary antibodies, Alexa Fluor 488 goat anti-mouse IgG conjugate (1:1,000; Molecular Probes), and washed with PBS extensively. Images were obtained with a Micro-Radiance confocal laser scanning microscope system (MR/AG-2; Bio-Rad). Dual-wavelength channels (at 488 and 568 nm) were used to excite Alexa Fluor 488 and DsRed, respectively. Digital pictures were processed by Lasersharp (Bio-Rad) and iProps (Nihon Visual Science).
RESULTS
RNPS1-interacting factors identified by yeast two-hybrid screen.
To search for proteins that interact specifically with RNPS1, we performed a yeast two-hybrid screen. For the bait, full-length RNPS1 or a C-terminal RS/P domain-deleted mutant (RNPS1-ΔRS/P) were fused to the LexA DNA-binding domain. The LexA-RNPS1 bait was used to screen a HeLa cell cDNA library. Out of ~4 × 106 independent transformants, 20 clones (7 clones from the screen with full-length RNPS1, 13 clones from RNPS1-ΔRS/P) were isolated, sequenced, and identified (summarized in Table Table2).2). The following proteins are intriguing candidates that may be functionally associated with RNPS1.
TABLE 2.
Proteins that interact with RNPS1, as determined by two-hybrid screening
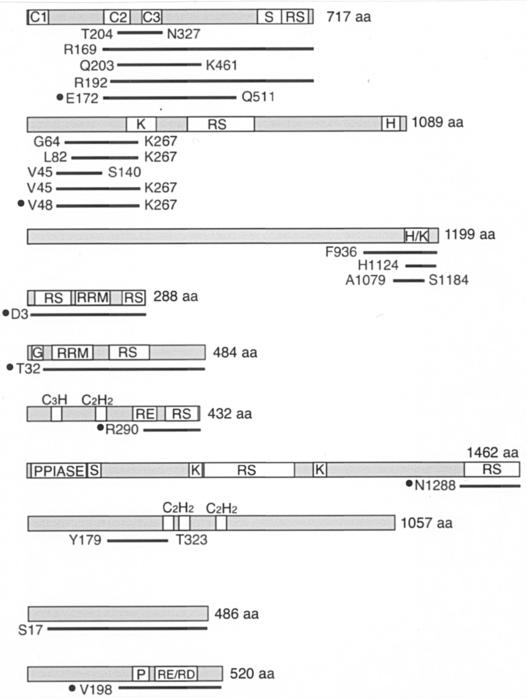 |
Pinin (also known as DRS and MemA) was originally identified as a desmosome-associated protein (71), but it was also shown to localize in nuclear speckles (12, 70, 81). This protein contains a coiled-coil domain at its N terminus and an S domain at its C terminus, but lacks a RRM (12, 27). Recently, it was shown that overexpression of pinin is prone to decrease splicing via distal 5′ and 3′ splice sites in adenovirus E1A pre-mRNA and a chimeric calcitonin/dihydrofolate reductase (dhfr) pre-mRNA, respectively (81).
CIF150 is a human homologue of Drosophila melanogaster TAFII150, a TATA-binding protein-associated factor (TAF). CIF150 is associated with the human transcription factor IID (TFIID) and may play an important role in the regulation of cell growth and cell cycle progression (58). Some TAFs are present not only in TFIID but also in TATA-binding protein-free TAFII-containing complex (TFTC), which plays additional roles in chromatin remodeling (10). Interestingly, one of the components of TFTC (9) is the splicing factor SAP130, which is a subunit of a U2 snRNP-associated protein SF3b (26).
Tra2 is a well-documented alternative splicing regulator of sex determination in D. melanogaster (reviewed in reference 16). Two human homologues, hTra2α and hTra2β, were characterized as RNA binding proteins that bind efficiently and specifically to purine-rich exonic splicing enhancers (ESE) (78). It was demonstrated that both hTra2α and hTra2β activate ESE-dependent splicing in vitro. hTra2β was also reported to promote exon 7 (which contains purine-rich (ESE) inclusion of survival motor neuron 2 (SMN2) transcript in vivo (41).
p54 is a member of the SR protein family and thus is also termed SRp54 (reviewed in reference 36). It was first identified by its structural and antigenic characteristics (21). HeLa cell cytosolic S100 extract contains many splicing factors but by itself is not competent for splicing because it has limiting amount of SR proteins that are required for efficient splicing (reference 60 and references therein). Like other SR proteins, p54 can complement splicing-deficient S100 extracts and thus functions as a constitutive splicing factor (86). However, p54 shows less conservation in its domain structure and has properties distinct from those of other SR protein family members. Authentic SR proteins, such as SF2/ASF and SC35, interact with either the U1 small nuclear ribonucleoprotein U1-70K or the 35-kDa subunit of U2AF (U2AF35), but not with its 65-kDa subunit (U2AF65) (84). In contrast, p54 directly interacts with U2AF65 but interacts neither with U1-70K nor with U2AF35 (86). As for its effect on alternative splicing, p54 promotes the use of the distal 5′ splice site in E1A pre-mRNA (86), whereas the use of the proximal 5′ splice site is promoted by SF2/ASF and SC35 (20, 40, 80). A D. melanogaster orthologue of human p54, dSRp54, was characterized as a C-rich intronic element binding factor, which is necessary for the splicing of short introns (45).
hLuc7A/CROP is one of three human counterparts homologous to yeast Luc7p, which is an essential component of U1 snRNP, interacts with the nuclear cap binding complex, and influences 5′ splice site selection (34, 68). hLuc7A contains N-terminal zinc fingers and multiple RE and RS repeats in the C-terminal region.
RNPS1 binds to hTra2β, pinin, and p54 in vitro.
To confirm physical binding of RNPS1 to pinin and hTra2β, in vitro translated 35S-labeled RNPS1 was subjected to pull-down assays with recombinant GST-pinin and GST-hTra2β fusion proteins in the presence of DNase and RNase. Since full-length recombinant GST-pinin could not be expressed in E. coli, we used the second and third coiled-coil domains of pinin (pinin-C23), which include the minimal region shared by all of five selected pinin cDNA clones (Table (Table2).2). We found that GST-pinin-C23 and GST-hTra2β coprecipitated with RNPS1 (Fig. (Fig.1A,1A, lanes 4 and 6), but not with the control luciferase (lanes 5 and 7), or GST alone (lane 3). Since recombinant GST-p54 could not be expressed in E. coli, we performed immunoprecipitations with extracts from HeLa cells in which FLAG-tagged p54 was overexpressed (Fig. (Fig.1B).1B). After addition of 35S-labeled RNPS1 to the HeLa cell extract, p54 was immunoprecipitated with an anti-FLAG antibody in the presence of DNase and RNase. The coprecipitated RNPS1 was analyzed by SDS-PAGE followed by autoradiography. RNPS1, but not control luciferase, coimmunoprecipitated with p54, but not with GFP as a negative control (Fig. (Fig.1C,1C, lanes 3 to 5). Consistent results, as in the GST pull-down assays, were obtained with HeLa cell extracts expressing FLAG-tagged hTra2β (lanes 6 and 7). We conclude that RNPS1 can bind to p54, hTra2β and the coiled-coil domain of pinin via protein-protein interactions in vitro.
p54, hTra2β, and pinin interact with different domains of RNPS1.
To map the domains of RNPS1 that are responsible for its interaction with p54, hTra2β, and pinin, respectively, deletion mutants of RNPS1 were used in the yeast two-hybrid assays (Fig. (Fig.2A).2A). RNPS1-NT+S (Table (Table1)1) and RNPS1-ΔRS/P (Table (Table1),1), both of which possess the S domain, interacted with p54 (Fig. (Fig.2A).2A). In contrast, RNPS1-RRM+RS/P (Table (Table1)1) and RNPS1-RS/P (Table (Table1),1), which share the RS/P domain, interacted with hTra2β. RNPS1-ΔRS/P and RNPS1-RRM+RS/P, which share the RRM, interacted with pinin. These data indicate that distinct domains of RNPS1 are responsible for the interactions with the different factors (Fig. (Fig.2B).2B). Previously, the N-terminal region of RNPS1, which includes the S domain, was shown to be necessary for the interaction with PITSLRE p110 kinase and a tumor-rejection antigen, SART3 (37, 56).
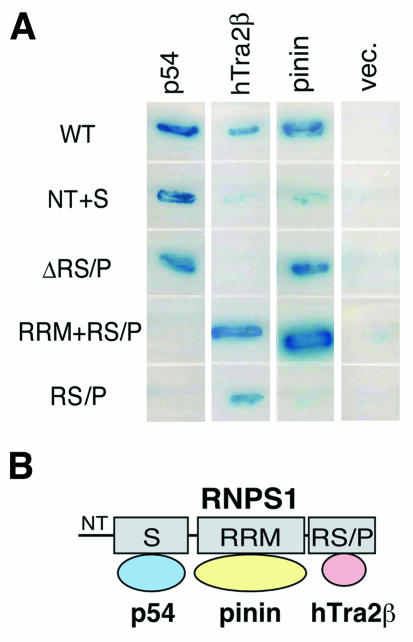
Protein interaction domains of RNPS1 analyzed by yeast two-hybrid assays. (A) Yeast host strain EGY48 was cotransformed with the indicated combinations of RNPS1 deletion constructs (in pLexA; see Table Table1)1) and selected clones (p54, hTra2β, and pinin in pB42AD). As a negative interaction control, the empty vector pB42AD (vec.) was used. Protein interactions were detected by β-galactosidase activity and auxotrophy for leucine. (B) A model of protein interactions of RNPS1 with p54, pinin, and hTra2β is shown.
RNPS1 localizes to nuclear speckles in living cells.
It was previously shown that human RNPS1 often localizes to enlarged (or mega) nuclear speckles when transiently overexpressed, and its distribution partially overlaps that of SC35 (an SR protein) in a typical nuclear speckle pattern (56). Here, we examined the expression pattern of mouse RNPS1 (99% identical to human RNPS1) in living Ehrlich ascites cells (Fig. (Fig.3A).3A). EGFP-RNPS1 exhibited exclusive nuclear localization with a canonical speckled pattern, while the control EGFP distributed throughout the whole cell (data not shown). In agreement with this observation, mouse RNPS1 was shown to be enriched in biochemically purified interchromatin granule clusters (part of the structural component of the nuclear speckles), which also contain many known splicing factors, including snRNPs and SR proteins (62). Continued observation of the transfected living cells showed dynamic intranuclear movements and redistribution of the speckled particles (data not shown).
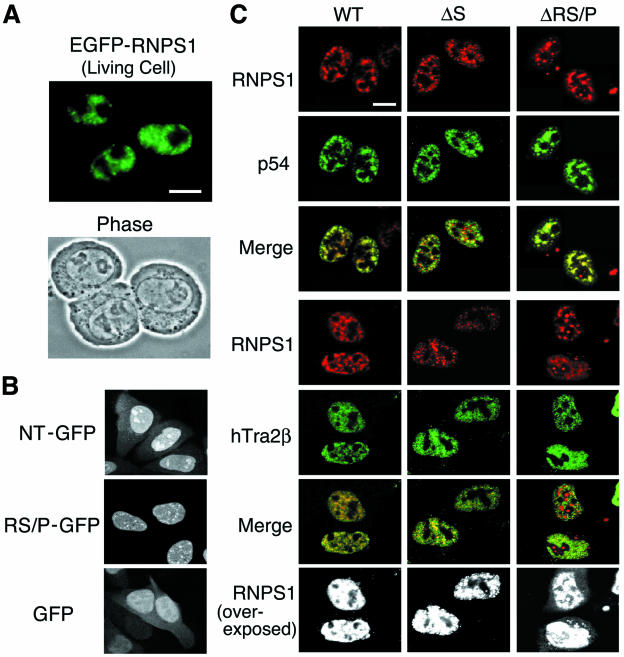
Subcellular localization of WT RNPS1 and deletion mutants, p54, and hTra2β. (A) EGFP-fused mouse RNPS1 was transiently transfected into living Ehrlich ascites tumor cells and analyzed by fluorescence microscopy (24 h after transfection). A phase contrast micrograph taken at the same time is shown below. (B) The GFP fusion constructs with either NT or RS/P domain (GFP alone as a control) were transiently transfected into HeLa cells. The cells were fixed and permeabilized at 12 h after transfection and analyzed by fluorescence microscopy. (C) The DsRed fusion proteins of human RNPS1 (WT) and two deletion mutants, RNPS1-ΔS and RNPS1-ΔRS/P, were transiently cotransfected into HeLa cells with FLAG-tagged p54 and FLAG-tagged hTra2β. The cells were fixed and permeabilized at 15 h after transfection, treated with an anti-FLAG antibody, and analyzed by confocal microscopy. Both images were also superimposed, and the yellow color indicates colocalization of the two proteins in nuclear speckles (Merge). Overexposed images (black and white) are also shown to highlight the cytoplasmic localization that is specifically observed in RNPS1-ΔRS/P protein. Bar, 10 μm.
C-terminal but not N-terminal NLS of RNPS1 may be functional.
Previously, a putative simian virus 40 T antigen-type nuclear localization signal (NLS) (amino acids 278 to 287) was reported in RNPS1 (2), and if it is extended to amino acid 296 it matches the consensus bipartite-type NLS (reviewed in references 28 and 29). We also found a bipartite type-NLS in the N-terminal region (amino acids 19 to 47) of RNPS1, which is predicted by PROSITE motif search (32).
To test the function of these NLS candidates, two RNPS1 domain-GFP fusion constructs were transiently expressed in HeLa cells and their subcellular localization was examined (Fig. (Fig.3B).3B). The NT-GFP fusion protein (Table (Table1),1), which has the N-terminal putative NLS, showed both nuclear and cytoplasmic localization, as observed with the control GFP protein. In contrast, RS/P-GFP protein (Table (Table1),1), which includes the C-terminal putative NLS, reveals exclusive nuclear localization. These data suggest that the C-terminal, but not N-terminal, NLS (amino acids 278 to 287 or 296) functions as an active nuclear localization signal.
p54 and hTra2β colocalize with RNPS1 in nuclear speckles.
To investigate the colocalization of RNPS1-associated factors with RNPS1, p54 and hTra2β cDNA expression plasmids were transiently transfected into HeLa cells (we did not detect expression of the full-length pinin cDNA). The localization of DsRed-RNPS1 and epitope-tagged proteins FLAG-p54 and FLAG-hTra2β, was analyzed by confocal laser microscopy with a red fluorescent protein and an antibody against the FLAG tag. All three fusion proteins localized in nuclear speckles (Fig. (Fig.3C,3C, column WT) and these distribution patterns are consistent with previous immunofluorescence studies (21, 59, 66). While RNPS1 and p54 showed relatively little signal in the nucleoplasm, hTra2β was found somewhat more prominently in the nucleoplasm. The merged images revealed that RNPS1 colocalizes with both p54 and hTra2β in nuclear speckles.
We examined the role of two specific RNPS1-domains in the subcellular localization of the protein. To this end, we constructed two deletion mutants; lacking either the S domain (RNPS1-ΔS; Table Table1)1) or the RS/P domain (RNPS1-ΔRS/P; Table Table1),1), which were fused to the C terminus of DsRed, and transiently coexpressed together with either FLAG-p54 or FLAG-hTra2β in HeLa cells. The RNPS1-ΔS protein localized mostly in the nuclear speckles. However, additional numerous small foci that did not necessarily colocalize with p54 (see red spots in the merged image) were found dispersed throughout the nucleoplasm (Fig. (Fig.3C,3C, column ΔS). In contrast, cells coexpressing hTra2β displayed colocalization of this protein with RNPS1-ΔS in both speckles and small nucleoplasmic foci (see the merged image). Overexpressed RNPS1-ΔRS/P was predominantly found in enlarged nuclear speckles (column ΔRS/P), however, some of the large spots were also detected in nucleoli (see the merged image). Consistent with the presence of C-terminal functional NLS in the RS/P domain, a small amount of the ΔRS/P protein was often observed in the cytoplasm, which was never observed with the wild-type RNPS1 and RNPS1-ΔS proteins (see overexposed). RNPS1-ΔRS/P did not colocalize well with hTra2β (see red spots in the merged image). These observations are generally consistent with our yeast two-hybrid results obtained with deletion mutants of RNPS1 (Fig. (Fig.2B2B).
RNPS1 promotes exon skipping in vivo.
RNPS1 and SF2/ASF synergistically stimulate both constitutive and alternative splicing in vitro, and thus RNPS1 was characterized as a general splicing activator (59). As RNPS1 was purified based on the activation of distal alternative 3′ splice sites (SF7 activity), we assumed that factor(s) associated with RNPS1 may be necessary for the SF7 activity that was observed in partially purified fractions. Here, we examined the activity of RNPS1 with its associated factors in the regulation of alternative splicing in vivo.
cDNAs encoding these protein factors were transiently overexpressed in HeLa cells, and the splicing patterns of three reporter minigenes containing alternatively spliced exons were examined by RT-PCR (Fig. (Fig.4).4). We first tested the exon/inclusion-skipping model substrate DUP51, which consists of three exons and two identical introns of the human β-globin gene (30). Besides a majority of middle-exon inclusion product (271 nucleotides), we also observed a middle-exon skipping product (220 nucleotides) specifically upon overexpression of either RNPS1 or p54 (Fig. (Fig.4A,4A, lanes 1 to 3, 12). This p54 effect is consistent with the previous in vivo splicing results, i.e., p54 stimulates the use of the distal 5′ splice site in E1A pre-mRNA (86). The activity of RNPS1 in promoting exon skipping was verified with a natural, alternatively spliced substrate, human Tra2-β1 (hTra2β) pre-mRNA (1, 66). Overexpression of RNPS1 induced double-exon skipping in hTra2β pre-mRNA (Fig. (Fig.4B,4B, lanes 1 to 3; beta 3 product). In the human mitochondrial ATP synthase γ-subunit minigene (F1γ) pre-mRNA (38), however, RNPS1 could not induce exon skipping (muscle type). Rather, we observed decreasing products of both exon skipping and exon inclusion (Fig. (Fig.4C,4C, lanes 1 to 3). In these transfections, we observed increasing unspliced pre-mRNA by RT-PCR with two primers between the 5′ end of exon 8 and the downstream intron (data not shown), and therefore the decreasing RT-PCR signal of spliced products indicate the actual repression of splicing. We also took internal control of the RT-PCR (under the same condition) by detecting endogenous β-actin mRNA, and we did not observe any changes of RT-PCR products (data not shown). We conclude that RNPS1 promotes exon skipping in a substrate-specific manner in vivo.
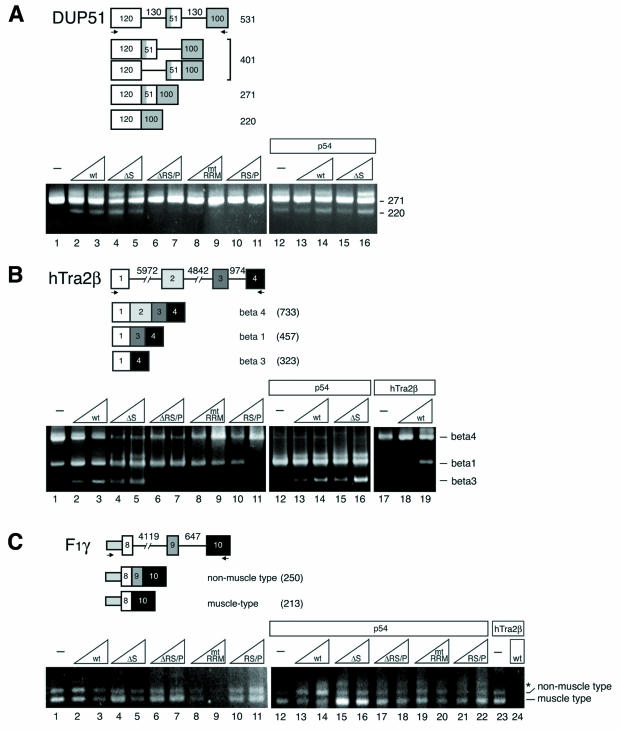
Effect of transiently overexpressed RNPS1, p54, and hTra2β on three kinds of alternatively spliced pre-mRNAs. (A) In vivo splicing of a model β-globin DUP51 pre-mRNA. (B) In vivo splicing of an hTra2β minigene pre-mRNA (C) In vivo splicing of a human F1γ minigene pre-mRNA. The structures of the pre-mRNAs and the spliced products are schematically illustrated on the top of each panel (numbers denote length in nucleotides). HeLa cells were transfected with a constant amount of individual pre-mRNA reporter plasmid along with a constant (indicated by rectangles) or increasing (indicated by triangles) amounts of protein expression plasmids pCMV-3×FLAG-RNPS1 (wild type), pCMV-3×FLAG-RNPS1-ΔS (ΔS), pCMV-3×FLAG-RNPS1-ΔRS/P (ΔRS/P), pCMV-3×FLAG-RNPS1-mtRRM (mtRRM), pCMV-3×FLAG-RNPS1-RS/P (RS/P), pFLAG-p54 (p54), and pFLAG-hTra2β (hTra2β). An empty vector, pFLAG-CMV-2, was used to keep the total amount of transfected DNA constant. Spliced products were analyzed by RT-PCR with specific DNA primers (indicated by arrows), followed by agarose gel electrophoresis. The length (in nucleotides) of splicing products (A) or specific alternatively spliced isoforms (B and C) are indicated on the right of the panels. The bands above the beta 1 (lanes 4, 5, and 12 to 16 in B) and non-muscle type (* in C) were found as nonspecific amplified products unrelated to splicing (data not shown) (42).
RRM and RS/P domains of RNPS1 are responsible for exon-skipping activity.
To determine which of the RNPS1 domains mediate its exon-skipping activity, we analyzed in vivo splicing upon overexpression of various RNPS1 mutants (Fig. 4A and B, lanes 4 to 11). Previously, it was shown that mutation of two solvent-exposed aromatic residues in the RRM of SF2/ASF and hnRNP A1 completely abolishes their functions in constitutive splicing and alternative splicing, respectively (17, 61). Therefore, we introduced the corresponding mutations in the RRM of RNPS1 to inactivate it (Table (Table1).1). Deletion of the RS/P domain (ΔRS/P) and mutations in the RRM (mtRRM) completely abolished exon skipping in the DUP51 pre-mRNA (220 nucleotides; lanes 6 to 9) and double-exon skipping in the hTra2β pre-mRNA (beta 3; lanes 6 to 9), whereas deletion of the S domain (ΔS) had no effect on exon-skipping induction in both substrates (220-nucleotide and beta 3; lanes 4 and 5). However, the RS/P domain alone had no exon-skipping activity in both substrates (lanes 10 and 11). The domain requirement for the exon-skipping activity is essentially consistent with these two different substrates, DUP51 and hTra2β pre-mRNAs. We conclude that both the RRM and RS/P domains are necessary, while the S domain is dispensable, for the exon-skipping activity of RNPS1.
RNPS1 and p54 cooperatively modulate alternative splicing in vivo.
To test whether the identified RNPS1-associated factors, p54 and hTra2β, cooperate with RNPS1 in the regulation of alternative splicing in vivo, the splicing patterns of all three substrates were examined upon coexpression of these factors in HeLa cells (Fig. (Fig.4,4, lanes 12 and above). Unfortunately, transfected full-length pinin was not detectably expressed in HeLa cells.
With the DUP51 pre-mRNA, coexpression of both p54 and RNPS1 stimulated exon skipping (220-nucleotide; lanes 12 to 14, compare lanes 1 to 3). This stimulation was also observed with RNPS1-ΔS (lanes 15 and 16), which lacks p54-interacting S domain (Fig. (Fig.2B).2B). Thus, the stimulation of exon skipping is independent from p54 interaction to S domain.
With the hTra2β pre-mRNA, coexpression of these factors induced totally distinctive splicing patterns. Overexpression of p54 produced beta 1 (exon 2-skipping) as a major spliced product (lane 12), whereas hTra2β overexpression generated beta 4 (exon inclusion) almost exclusively (lane 17). Coexpression of either p54 or hTra2β together with RNPS1 induced the exon-skipping products beta 3 and beta 1, respectively (lanes 14 and 19). As in the DUP51 pre-mRNA, stimulated exon skipping, even stronger, was also observed with RNPS1-ΔS, which lacks a p54 interacting S domain (beta 3; lanes 15 and 16). These data, taken together, indicate that cotransfection of either p54 or hTradβ does not interfere with the exon-skipping activity of RNPS1, which was observed with both DUP51 and hTra2β substrates (Fig. 4A and B, lanes 1 to 3).
In contrast, we observed synergistic effects upon the coexpression with the F1γ mini-gene pre-mRNA. Coexpression of RNPS1 with p54 promoted a shift from exon skipping (muscle type) to exon inclusion (non-muscle type; Fig. Fig.4C,4C, lanes 12 to 14), whereas the same level of RNPS1 overexpression alone repressed both splicing (lanes 1 to 3). Coexpression of RNPS1 with hTra2β also repressed both splicing (lanes 23 and 24). With the DUP51 and hTra2β pre-mRNAs, equivalent coexpression of RNPS1 and p54 rather stimulated exon skipping (Fig. 4A and B, lanes 12 to 14). Therefore, this modulation of alternative splicing under the p54 coexpression is F1γ substrate specific. Observed negative activity of RNPS1 (Fig. (Fig.4C,4C, lanes 1 to 3) is not due to nonspecific inhibition, but rather reveals substrate-specific negative regulation, since the same level of RNPS1 overexpression did not cause comparable inhibition with other substrates (Fig. 4A and B, lanes 1 to 3). Moreover, splicing assays with RNPS1 mutants suggested that RS/P domain is necessary for the splicing repression effect of RNPS1, while either S domain or RRM is dispensable (Fig. (Fig.4C,4C, lanes 4 to 9). However, RS/P domain itself has no repression activity (lanes 10 and 11). Interestingly, this domain requirement is different from that for the exon-skipping activity (see above). Therefore, the negative effect appears to be a domain-specific intrinsic activity of RNPS1 to the particular substrate.
S domain of RNPS1 is responsible for the cooperative effect with p54.
To examine whether the observed synergistic effect is due to protein-protein interaction between RNPS1 and p54, we used various RNPS1 mutant proteins to coexpress with p54 in F1γ pre-mRNA (Fig. (Fig.4C,4C, lanes 15 to 22). A drastic change was observed only upon deletion of the S domain (ΔS), which is the p54-binding site (Fig. (Fig.2B),2B), i.e., the exon inclusion (non-muscle type) product was not induced, and rather the exon skipping (muscle type) product was generated (Fig. (Fig.4C,4C, lanes 15 and 16) as observed upon expression of p54 alone (lane 12). However, coexpression of all the other RNPS1 mutants showed a similar pattern of products (lanes 17 to 22) as observed with the wild-type RNPS1 (lanes 13 and 14), albeit with somewhat less induction of exon inclusion (non-muscle type) but clearly distinct from p54 expression alone (lane 12). These data suggest that the observed induction of exon inclusion (non-muscle type) is due to the interaction of p54 with the S domain of RNPS1, which is dispensable for the exon-skipping activity (in the DUP51 and hTra2β pre-mRNAs) and for splicing repression (in the F1γ pre-mRNA).
DISCUSSION
RNPS1-interacting factors.
We have searched for factors that interact with RNPS1 with a yeast two-hybrid screen of a HeLa cDNA library. Intriguingly, four known factors that are directly or indirectly involved in pre-mRNA splicing were isolated: p54, a member of the SR protein family; hTra2β, a purine-rich ESE-binding factor; hLuc7, a human homologue of a yeast U1 snRNP component; and pinin, which localizes in the nuclear speckles and has been suggested to associate with U2 snRNP proteins (see below). RNPS1 may be capable of interacting with all these factors directly and simultaneously, since different domains of RNPS1 are required for the interaction with p54, hTra2β and pinin (Fig. (Fig.2B2B).
Previously, RNPS1 was identified by two-hybrid screens with the following proteins as bait: the protein kinases, Clk/Sty and PITSLRE p110 (23, 56); the tumor-rejection RNA-binding antigen SART3 (37); and pinin (53; P. Ouyang and W. Y. Tarn, unpublished data). However, of these proteins we only detected pinin using RNPS1 as bait. Although we observed functional synergy between RNPS1 and SF2/ASF (or SC35) for the stimulation of splicing in vitro (59), p54 is the only member of the SR protein family isolated by a yeast two-hybrid screen. The failure to identify these proteins in our yeast two-hybrid screen might be due to technical problems. For instance, hybrid formation with particular proteins (preys) may be affected by the structure or folding of the LexA-RNPS1 fusion protein used as bait, or some of the proteins may not be well represented in the library.
Recently, protein interactions between the components of the EJC, of which RNPS1 is a component, have also been reported. Y14, another component of the EJC, binds to 35S-labeled RNPS1 in vitro (43). Coimmunoprecipitation with tagged-RNPS1 has identified interactions of RNPS1 with TAP (a nuclear export receptor) and hUpf protein complex (central components of NMD) (57). However, using a yeast two-hybrid screen, we did not isolate these proteins. It is possible that these mammalian factors individually cannot form stable hybrids to activate transcription in yeast, since the interactions among EJC components are specific to postsplicing process in higher eukaryotes. For instance, it is conceivable that posttranslational modifications of RNPS1, which are probably critical for interaction with some human proteins, are lacking in yeast. Indeed, potential posttranslational modification of human RNPS1 was suggested to be important for its function (59).
Nuclear speckle colocalization of RNPS1 with hTra2β and p54.
A two-step subcellular localization mechanism was suggested for the RS domain containing D. melanogaster splicing regulator Tra protein: the first step is transport across the nuclear membrane via the NLS, while the second step is localization in the speckles via the RS domain (39). A similar mechanism is conceivable for the nuclear speckle localization of RNPS1.
We found that the RS/P domain of RNPS1, which contains several RS or SR dipeptide sequences (though too short for a typical RS domain) and a putative simian virus 40 T antigen-type or bipartite-type NLS (amino acids 278 to 287 or 296), directs localization exclusively to the nucleus. Both the RS/P and the S domains of RNPS1 are required for proper nuclear speckle localization, since the mutant proteins deleted for these domains (ΔS and ΔRS/P) showed almost exclusively nuclear localization, but did not distribute in a typical speckled pattern. The RS domain of some, but not all, SR and SR-related proteins can serve as a targeting signal to nuclear speckles (19, 39, 54). We showed that the SR protein p54 interacts with the S domain of RNPS1, and that this protein-protein interaction may direct RNPS1 to the proper nuclear speckles. A similar mechanism has been proposed from the study of the D. melanogaster Tra protein. Tra mutant protein lacking an RS domain, which shows diffuse nucleoplasmic localization, can be directed to the nuclear speckles via protein-protein interaction with Tra2 (39).
Possible mechanisms of splicing activation by RNPS1.
We previously reported that RNPS1 and SR proteins cooperatively activate general splicing in vitro (59), however the underlying mechanism remained to be elucidated. Our discovery of associated factors provides insights into the mechanisms of splicing activation.
The splicing coactivator complex SRm160/300, which also possesses an S domain and an RS/P domain, promotes splicing in a purine-rich ESE-dependent manner (7, 31). The association of SRm160/300 and U2 snRNP with pre-mRNA requires both U1 snRNP and hTra2β, the latter of which binds to ESEs, suggesting that SRm160/300 may promote critical interactions between hTra2β and the snRNP machinery in the spliceosome (31). Since we showed that possible interactions of RNPS1 with both hTra2β and the putative U1 snRNP component hLuc7A, an analogous mechanism of splicing stimulation, involving RNPS1, hTra2β, and hLuc7A, is conceivable in pre-mRNAs that have a purine-rich ESE. In contrast with the case of SRm160/300, however, splicing activation by RNPS1 is not highly dependent upon the pre-mRNA substrates (59), and thus it is unlikely that the corresponding purine-rich ESEs exist in all the interactive substrates.
RNPS1 may activate ESE-independent splicing by a different mechanism that involves pinin and U2 snRNP and/or p54 and U2AF65. A potential interaction between RNPS1 and pinin has recently been suggested by a yeast two-hybrid screen (53; P. Ouyang and W. Y. Tarn, unpublished data), and here we demonstrate that the RRM of RNPS1 can directly bind to the second and third coiled-coil region of pinin. Recently, the interaction of RNPS1 with the coiled-coil region of pinin has also been reported (53). Pinin is enriched in nuclear fractions containing the U2 snRNP proteins SF3a and SF3b (12), which are required for assembly of the splicing complex A and for stable association of U2 snRNP with the branch site region (5, 13, 14, 49). On the other hand, we found that RNPS1 can bind to p54 through its S domain, and p54 can directly interact with U2AF65 (86). Based on all these observations, we propose that RNPS1 stimulates recruitment of U2 snRNP and U2AF65 via interaction with pinin and p54, respectively, to activate purine-rich ESE-independent pre-mRNA splicing. In agreement with this hypothesis, our splicing complex analysis revealed that RNPS1 stimulated formation of the ATP-dependent splicing complex A in vitro (J. H. Trembley et al., submitted for publication).
RNPS1 as a versatile regulator of alternative splicing.
Constitutive splicing of several substrates was stimulated by adding purified RNPS1 or recombinant RNPS1 into splicing reactions in vitro, however, we failed to observe RNPS1 activity in modulating alternative splicing with several pre-mRNA tested in vitro (59). In vivo splicing assays, however, uncovered a significant substrate-specific effect of RNPS1 on alternative splicing. We found that RNPS1 can promote alternative exon skipping of model β-globin (DUP51) pre-mRNA and natural hTra2β minigene pre-mRNA in vivo. The observed exon-skipping activity is consistent with in vitro splicing results, i.e., the activation of distal alternative 3′ splice site observed with partially purified RNPS1 (59). However, RNPS1 was not able to induce exon skipping (muscle-type splicing) in F1γ pre-mRNA, rather overall splicing was repressed. In adenovirus E1A pre-mRNA, which has three alternative 5′ splice sites, overexpression of RNPS1 represses the distal 5′ splice site (9S) and stimulates the proximal 5′ splice site (13S) in vivo (81). Therefore, RNPS1 shows distinctive modulation of alternative splicing in a substrate-dependent manner. The fact that RNPS1 is unable to cause exon skipping indiscriminately, but rather selectively or depending upon specific substrates, is likely to be biologically significant.
The observed splicing repression of RNPS1 with particular substrates in vivo is not necessarily an artifact of overexpression (since the same level of overexpression did not cause repression in other substrates), but rather RNPS1 might be involved in the negative regulation of alternative splicing. Indeed, we could identify a responsible domain (RS/P domain) of RNPS1 for splicing repression in the F1γ pre-mRNA, and the domain requirement is distinct from that for the exon skipping and for the cooperative function with p54. It has been reported that several mammalian SR protein-like factors, SRrp86, SRrp35, and SRrp40/SRp38 (also known as NSSR-1 and TASR-2), have significant roles in the repression of splicing (3, 25, 48, 76, 85).
It is of considerable interest that the effects of cotransfection of RNPS1-associated factors were not only additive but also synergistic with regards to the regulation of alternative splicing. In a calcitonin/dhfr chimeric pre-mRNA, it was reported that the coexpression of both RNPS1 and SART3 represses the relative use of a distal 3′ splice site, however, overexpression of either protein alone did not cause significant changes in the alternative 3′ splice site choice (37). In the hTra2β pre-mRNA, overexpression of RNPS1 alone induces alternative double-exon skipping (beta 3 splicing) and this effect is additive upon coexpression of either hTra2β or p54, which cause single-exon (beta 1) or double-exon (beta 3) skipping, respectively. In contrast, overexpression of RNPS1 alone suppresses overall splicing of F1γ pre-mRNA, while it promotes alternative exon 9 inclusion (non-muscle type splicing) when it was coexpressed with p54. Intriguingly, deletion of the S domain does not affect on the exon-skipping activity in the DUP51 and hTra2β pre-mRNAs, and on the splicing repression in the F1γ pre-mRNA, but it abolishes the cooperative effect with p54 to induce exon inclusion (non-muscle type) in F1γ pre-mRNA. These data indicate that the different domains of RNPS1 are responsible for these distinctive activities of RNPS1.
Taken together, our results indicate that RNPS1 regulates alternative splicing both negatively and positively through interaction with associated factors in vivo. Since RNPS1 has the ability to promote a variety of alternative splicing events in a substrate-specific manner, it appears to be a versatile splicing regulator for a wide variety of alternatively spliced genes.
Acknowledgments
We thank J. L. Manley for the pET14b-hTra2β and pGEX-hTra2β plasmids; W. W. Franke for the DRSP-MH22 plasmid; J. Y. Wu for the pcDNA3-p54 plasmid; S. Stamm for the hTra2-β1 minigene plasmid; R. Kole for the pHD1012-DUP51 plasmid; H. Sakamoto for the pCMX-hGFP-S65A/Y145F plasmid; and J. Brandner for anti-DRSP-M2 and M3 antibodies. We are grateful to M. P. Deutscher, A. R. Krainer, T. Misteli, J. F. Cáceres, C. Jain, and members of the Mayeda laboratory for helpful discussions and critical reading of the manuscript and to M. J. Moore and W. Y. Tarn for communicating results prior to publication.
This work was supported by funds awarded by the L. P. Markey Trust, an institutional research grant (IRG-98-277-04) from A.C.S., Florida Biomedical Research Program grant (BM031) from F.D.H. to A.M., and by grants-in-aid from the Ministry of Education, Science and Culture of Japan to H.E. E.S. was supported by a postdoctoral fellowship from the Uehara Memorial Biological Science Foundation and a grant from the Jichi Medical School Young Investigator Award in Japan. A.M. is a research member of the Sylvester Comprehensive Cancer Center.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.24.3.1174-1187.2004
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc321435?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Metastasis-Associated Lung Adenocarcinoma Transcript 1 (<i>MALAT1</i>) lncRNA Conformational Dynamics in Complex with RNA-Binding Protein with Serine-Rich Domain 1 (RNPS1) in the Pan-cancer Splicing and Gene Expression.
ACS Omega, 9(41):42212-42226, 03 Oct 2024
Cited by: 0 articles | PMID: 39431102 | PMCID: PMC11483381
Nonsense-Mediated mRNA Decay in Human Health and Diseases: Current Understanding, Regulatory Mechanisms and Future Perspectives.
Mol Biotechnol, 12 Sep 2024
Cited by: 0 articles | PMID: 39264527
Review
Messenger RNA Surveillance: Current Understanding, Regulatory Mechanisms, and Future Implications.
Mol Biotechnol, 27 Feb 2024
Cited by: 1 article | PMID: 38411790
Review
A molecular brake that modulates spliceosome pausing at detained introns contributes to neurodegeneration.
Protein Cell, 14(5):318-336, 01 May 2023
Cited by: 4 articles | PMID: 37027487 | PMCID: PMC10166177
The rice blast fungus SR protein 1 regulates alternative splicing with unique mechanisms.
PLoS Pathog, 18(12):e1011036, 08 Dec 2022
Cited by: 4 articles | PMID: 36480554 | PMCID: PMC9767378
Go to all (74) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation.
Mol Cell Biol, 25(4):1446-1457, 01 Feb 2005
Cited by: 34 articles | PMID: 15684395 | PMCID: PMC547998
Modulation of alternative pre-mRNA splicing in vivo by pinin.
Biochem Biophys Res Commun, 294(2):448-455, 01 Jun 2002
Cited by: 45 articles | PMID: 12051732
Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing.
EMBO J, 18(16):4560-4570, 01 Aug 1999
Cited by: 98 articles | PMID: 10449421 | PMCID: PMC1171530
Nuclear mRNA binding proteins couple pre-mRNA splicing and post-splicing events.
Mol Cells, 12(1):1-10, 01 Aug 2001
Cited by: 25 articles | PMID: 11561715
Review





