Abstract
Free full text

AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: Implication in stepwise leukemogenesis and response to Gleevec
Abstract
To explore the genetic abnormalities that cooperate with AML1-ETO (AE) fusion gene to cause acute myeloid leukemia (AML) with t(8;21), we screened a number of candidate genes and identified 11 types of mutations in C-KIT gene (mC-KIT), including 6 previously undescribed ones among 26 of 54 (48.1%) cases with t(8;21). To address a possible chronological order between AE and mC-KIT, we showed that, among patients with AE and mC-KIT, most leukemic cells at disease presentation harbored both genetic alteration, whereas in three such cases investigated during complete remission, only AE, but not mC-KIT, could be detected by allele-specific PCR. Therefore, mC-KIT should be a subsequent event on the basis of t(8;21). Furthermore, induced expression of AE in U937-A/E cells significantly up-regulated mRNA and protein levels of C-KIT. This may lead to an alternative way of C-KIT activation and may explain the significantly higher C-KIT expression in 81.3% of patients with t(8;21) than in patients with other leukemias. These data strongly suggest that t(8;21) AML follows a stepwise model in leukemogenesis, i.e., AE represents the first, fundamental genetic hit to initiate the disease, whereas activation of the C-KIT pathway may be a second but also crucial hit for the development of a full-blown leukemia. Additionally, Gleevec suppressed the C-KIT activity and induced proliferation inhibition and apoptosis in cells bearing C-KIT N822K mutation or overexpression, but not in cells with D816 mC-KIT. Gleevec also exerted a synergic effect in apoptosis induction with cytarabine, thus providing a potential therapeutic for t(8;21) leukemia.
Abnormalities in genes that encode transcription factors (TFs) and tyrosine kinases (TKs) represent two classes of the most frequently detected genetic events in human leukemias (1–3). Disruptions of TFs, often as results of recurring chromosomal translocations where fusion genes are generated, may lead to inhibition of hematopoietic differentiation and subsequent apoptosis, whereas mutations or alterations of TKs may confer proliferative and/or survival advantage to hematopoietic stem/progenitor cells. Recent evidence suggests that alterations in TFs and TKs are required to cooperate in causing full-blown leukemia (3, 4). However, whether a causal relationship or a chronological order exists between the two genetic events remains largely unknown in a clinical setting.
The t(8;21)(q22;q22), where coding sequences of the AML1 gene on chromosome 21 are juxtaposed to coding sequences of the ETO gene on chromosome 8 generating an AML1-ETO (AE) fusion transcript, represents the most common chromosomal translocation in acute myeloid leukemia (AML) (5). The AE chimeric protein recruits the N-CoR-mSin3-HDAC complex (6) and represses wild-type AML1, which is a crucial TF for hematopoiesis, modifies intranuclear targets of AML1 (5, 7), and even represses genes that normally are not regulated by AML1 (8). At cellular level, the AE fusion protein transforms NIH 3T3 cells and activates TF AP-1 (9), maintains and enhances the self-renewal of human pluripotent stem cells (10, 11), and promotes extensive self-renewal of human primary erythroid cells (12). However, reports also showed that ectopic expression of AE induces growth arrest and subsequent apoptosis in U937 cells (13). Although Fenske et al. (14) reported that targeted expression of the AE in mice using the Ly6A (ScaI) locus induces a spontaneous myeloproliferative disorder, the AE knock-in and transgenic mice using MRP promoter, a tetracycline-inducible system, or Cre/LoxP system and lethally irradiated mice reconstituted by AE-expressing hematopoietic stem/progenitor cells do not develop any malignancies (15–18) or leukemia (11, 19). In the presence of additional mutations, e.g., mutagen N-ethyl-N-nitrosourea (ENU), introduced mutations, or coexpression of TEL-PDGFRβ fusion gene, exogenous AE-expressing mice develop an AML phenotype (4, 16, 18). However, the genetic abnormalities that cooperate with AE to induce human t(8;21) AML remain elusive.
Perturbation of receptor and nonreceptor TK signaling has been implicated in the pathogenesis of a broad spectrum of human neoplasia, including leukemias (20). Altered regulation of members such as FLT3 of type III receptor TK (RTK) family in AML has recently received emphasis. A number of observations also suggest a role for C-KIT, another member of type III RTK family that is important for the development of a range of cells including hematopoietic cells (2), in leukaemogenesis. High expression of C-KIT in AML (60–80%) has been reported (21, 22), and point mutations of C-KIT have been identified in 33.3–45% of AML with inv (16) and 12.8–46.8% of AML M2 with t(8;21) (23–25). However, many of these studies screened for C-KIT mutations only in a proportion of C-KIT coding sequence, and others were limited by the small case number studied. Thus, the prevalence and clinical significance of C-KIT mutations in the t(8;21) leukemia are still unclear. Another important issue to be addressed is the possible chronological order or causal relationship between t(8;21) and C-KIT mutation and/or overexpression, and a clarification of this may shed light on the multistep leukemogenesis scenario. In addition, although results showed that cells with certain C-KIT mutation types, such as substitutional mutation at residue D816, were resistant to TK inhibitor Gleevec/STI-571/imatinib mesylate (26), the Gleevec response of leukemic cells with other types of C-KIT mutations and/or overexpression remains unclear.
To address these questions, we analyzed the structure and expression of C-KIT gene in a large cohort of patients with distinct leukemia subtypes and monitor the expression of AE and C-KIT in disease course. Our results show that C-KIT abnormalities are closely correlated with t(8;21) AML, and the t(8;21) should be an event preceding the C-KIT mutation in leukemogenesis, whereas the expression of AE may cause the overexpression of C-KIT. Our results also showed a potential benefit of using TK inhibitor in the treatment of t(8;21) leukemia with C-KIT abnormalities other than D816 substitution.
Materials and Methods
Patients. Fifty-four patients with t(8;21) AML and 57 with other types of malignancies were studied. The diagnosis was based on clinical data and examination of peripheral blood and bone marrow according to French–American–British classification. Dual color FISH was performed as described (27) by using LSI AML1-ETO dual-color, dual-fusion translocation probes (Vysis). Molecular analysis of AE, PML-RARα (for acute promyelocytic leukemia), CBFβ-MYH11 [for AML M4Eo with inv(16)], and BCR-ABL (for chronic myeloid leukemia) transcripts was carried out by using RT-PCR. Patients with t(8;21) leukemia were treated with the local chemotherapy protocol [daunorubicin at a daily dose of 60 mg/m2·day for 3 days and cytarabine (Ara-C) at 150 mg/m2·day for 7 days for induction therapy, followed by daunorubicin or mitoxantrone and Ara-C for consolidation].
Sequencing of cDNA and Genomic DNA of C-KIT Gene. Bone marrow samples were obtained with informed consent. Total RNA and DNA were extracted. All coding regions were RT-PCR amplified with specific primers (see Table 2, which is published as supporting information on the PNAS web site), and the resultant products were sequenced directly. The genomic DNA regions containing mutations of C-KIT as well as the gene's promoter region were also sequenced. The expression of AE and mutant C-KIT (mC-KIT) in patients during CR were detected by using TrueSNP allele-specific PCR primers (Proligo, Paris) with a locked nucleic acid at the 3′ end of the primers (28) (see Table 2).
Immunohistochemical Analysis. Immunohistochemical staining was performed by using polyclonal p145 anti-human CD117 (C-KIT) antibody (Dako), and immunoreactivity on leukemic blasts was scored according to Kindler et al. (29). The immunoreactivity score (IRS) was calculated as IRS (0–12) = RP (0–4) × SI (0–3), where RP is the percentage of receptor-positive blasts and SI is staining intensity.
Quantitative Assessment of the Expression of AE and C-KIT and Assay of C-KIT Protein TK Activity. After reverse transcription, cDNA was used for real-time RT-PCR using a SYBR Green PCR Master mix (Applied Biosystems), and the results was presented as the corrected copy numbers (CNs) as described for PML-RARα from our lab (30). The β-actin was coamplified as an endogenous control to standardize the amount of the sample RNA added to the reaction. The primers used are presented in Table 2. For assay of C-KIT PTK activity, cells were lysed, and C-KIT protein was immunoprecipitated from cell extracts containing 300 μg of total protein by using 1 μg of monoclonal anti-C-KIT antibody (BD Pharmingen) (31). Immunoprecipitation was carried out overnight at 4°C, and immunocomplexes were harvested by addition of protein A/G agarose (Santa Cruz Biotechnology). After extensive washing, the C-KIT PTK activity was analyzed by using a TK activity assay kit (Chemicon) according to the manufacturer's instruction.
Cell Culture. The N822K-type mC-KIT-bearing Kasumi-1 cells (32) (provided by D. E. Zhang, The Scripps Institute, La Jolla, CA), and primary leukemic cells isolated from t(8;21) leukemia patients were cultured in RPMI medium 1640 (Gibco/BRL) supplemented with 10% FCS (PAA, Linz, Austria). The ponasterone A (PA; Invitrogen)-inducible AE-expressing U937 cells (U937-A/E cells; provided by M. Lübbert, University of Freiburg Medical Center, Freiburg, Germany) were cultured in RPMI medium 1640 supplemented with 10% FCS. Stem cell factor (SCF) was supplemented to Kasumi-1 cells after cells were sera deprived as indicated. The expression of AE, TGF-β1, and C-KIT before and after PA induction were assayed with quantitative real-time RT-PCR, and C-KIT PTK activity was detected.
Assessment of Effects of Gleevec on AML M2 Leukemic Cells. The Kasumi-1 cells and fresh leukemic cells were incubated at the presence or absence of Gleevec and/or Ara-C. Cell proliferation was assayed by using a Cell Counting kit (Dojin Laboratories, Kumamoto, Japan), and cell apoptosis was analyzed by using an ApoAlert Annexin V-FITC Apoptosis kit (BD Clontech), according to manufacturers' instructions.
Results
Association of AE and C-KIT Mutations in t(8;21) AML. Gain-of-function C-KIT mutations are closely correlated with the t(8;21) AML. Through sequencing the whole C-KIT gene and eliminating a number of SNPs by typing >100 controls, a mutation in one of the two alleles was detected in leukemic cells from 26 of 54 (48.1%) patients with t(8;21) AML (Table 1 and Fig. 5, which is published as supporting information on the PNAS web site), whereas no mutation was detected in AML M2 without t(8;21) (5 cases), acute promyelocytic leukemia (20 cases), AML M4Eo with inv (16) (7 cases), AML M4 without inv (16) (5 cases), chronic myeloid leukemia (6 cases), and other hematological malignancies (14 cases), indicating that the mC-KIT was correlated with t(8;21) leukemia (R = 0.94, P < 0.01). In total, seven types of point mutations, three internal tandem duplication (ITD)-type mutations, and one amino acid deletion (D419) were characterized. The most frequently detected mutation was N822K (10 of 26, 38.5%). Point mutations affecting residues L773, D816, N822, and V825 at enzymatic “pocket” of C-KIT occurred in 21 of 26 patients (80.8%), constituting the “hot spot” for activating mutations (Table 1). The ITD-type mutations occurred in jaxtamembrane (JM) domain (two cases) and extracellular domain (one case). Analysis of both cDNA and genomic DNA sequences in these three cases confirmed that the insertion of the amino acid stretches was caused by an insertion of sequence at genomic DNA, and did not result from aberrant processing of mRNA. A point mutation, I748T, was found in the kinase insert sequence. In agreement with previous reports (33), mC-KIT was a factor for poor prognosis in this series (for details, see Fig. 6, which is published as supporting information on the PNAS web site).
Table 1.
| Sex
| Average age, years (range)
| ||||||
|---|---|---|---|---|---|---|---|
| Classes | CN | M | F | Type | DOM† | ||
| mC-KIT | 10 | 5 | 5 | 17.8 (3-56) | N822K | TK2 | R |
| 4 | 3 | 1 | 18.5 (5-31) | D816Y | TK2 | R | |
| 3 | 2 | 1 | 33.7 (17-46) | D816V | TK2 | R | |
| 2 | 2 | 11.5 (7-16) | D816H | TK2 | R | ||
| 1 | 1 | 55 | V825I | TK2 | N | ||
| 1 | 1 | 47 | L773S | TK2 | N | ||
| 1 | 1 | 34 | I748T | KIS | N | ||
| 1 | 1 | 24 | Q575+17(EFCELPYDHKWEFPRNR) | JM | N | ||
| 1 | 1 | 65 | I571+14(ESVDPTQLPYDHKW) | JM | N | ||
| 1 | 1 | 17 | 502(AYFNF) repeat | EC | N | ||
| 1 | 1 | 31 | D419 deletion | EC | R | ||
| Summary | 26 | 15 | 11 | 25.0 (3-65) | |||
| wtC-KIT | 28 | 17 | 11 | 29.6 (4-68) | |||
| Total | 54 | 32 | 22 | 27.4 (3-68) | Mutation rate: 48.1% | Six N mutations | |
CN, case number; M, male; F, female; DOM, domain of mutations; R, reported previously; N, previously undescribed.
*Capital letters represent abbreviations of certain amino acids.
Coexistence of t(8;21) and mC-KITs in a sizable proportion of leukemic cells at the disease presentation. Whether the t(8;21) and mC-KITs are acquired by different fractions of leukemic cell population or can coexist in one population is an intriguing question. To address this, mononuclear cells were harvested, and dual-color FISH with AML1 and ETO probes was performed in two cases. A total of 1,000 cells including metaphase and interphase were analyzed for each case studied. Our results showed that 87.4–96.0% of the cells had fusion signals (Fig. 1 A and B). We then assessed the percentage of mC-KIT-carrying leukemic cells by analyzing the peak height ratio of wild-type/mutant alleles in heterozygous sequencing chromatogram according to Nurpeisov et al. (34) and Blazej et al. (35). In the two patients described above, the peak height ratio of wild-type/mutant alleles was 0.8/0.6 and 1.2/0.7, respectively; thus, the mC-KIT-carrying leukemic cells were ≈85% and ≈70%, respectively (Fig. 1 C and D). Taken together, it is obvious that t(8;21) and mC-KITs must coexist in a large proportion of leukemic cells in newly diagnosed patients.
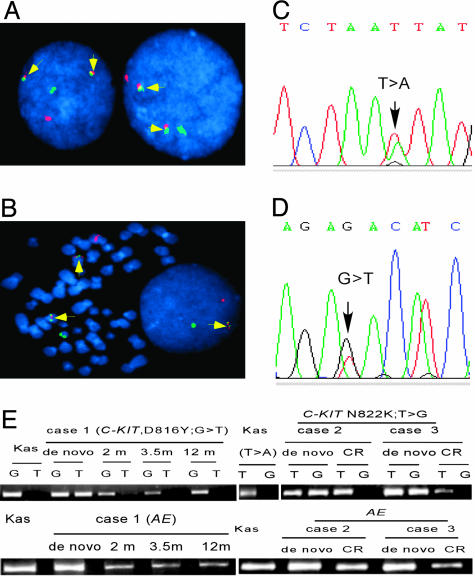
Coexistence of t(8;21) and mC-KITs in leukemic cells of de novo patients and persistence of AE but absence of mC-KIT in patients in CR. (A and B) Dual color FISH was performed, and 1,000 cells including interphase and metaphase were analyzed. The fusion signals (arrow) is seen in 96% in case 1 and 87.4% in case 2 of leukemic cells. Green signal, AML1 gene; orange signal, ETO gene. (C and D) Analysis of the peak height ratio of wild-type/mutant alleles in chromatogram of genomic DNA from leukemic cells of the two cases reveals that ≈85% in case 1 and 70% in case 2 of leukemic cells carried a mutant C-KIT allele. (E) The allele-specific RT-PCR is performed to detect the mutant alleles in three patients with t(8;21) leukemia at de novo and in remission. The primers used have an extra 2′-O,4′-C-methylene bridge at the ribose ring of nucleic acids at the 3′ end. The assays for Kasumi-1 (Kas) cells, which have a N822K (T → A) type mC-KIT, clearly show that these primers are sensitive and allele specific. At de novo, mC-KITs and AE are positive in the three patients, whereas the mC-KIT becomes undetectable and AE remains positive in CR.
Absence of mC-KIT, but persistence of AE during CR in patients with both abnormalities at disease presentation. The fact that, in this series, all patients with mC-KIT had t(8;21), whereas about half of patients with t(8;21) did not have detectable mC-KIT, seemed to be in favor of the idea that t(8;21) should be an earlier event in leukemogenesis. However, at individual patient level, we did not see any case with t(8;21) alone at disease onset subsequently evolving into a status with double genetic defects. A solution to address the interrelationship of the two genetic defects could be to look at the process in a reversed manner, because previous studies showed that AE remains positive in patients with t(8;21) AML over long-term remission and even after bone marrow transplantation (36, 37). Indeed, when samples from three patients in CR were assayed with real-time RT-PCR for AE, the fusion transcript was still detectable, although the level was extremely low as compared to the disease onset [a 167- to 986-fold (mean: 456-fold) reduction of copy number, Fig. 2A]. Next, AE and mC-KITs were further analyzed in three cases over 8–12 months of CR by using TrueSNP allele-specific PCR primers (28). For the results to be comparable, the sensitivity of the method for the transcripts of both AE and mC-KITs was carefully adjusted to similar level, 10–4. mC-KIT was also assayed at DNA level. Among the three patients, one had a D816Y-type and two had a N822K-type mC-KIT alleles, respectively (Fig. 1E). The results showed that AE transcripts remained positive 2–12 months after achieving CR, whereas D816Y-type mC-KIT gradually turned negative in patient 1, and N822K-type mC-KIT became undectable in patients 2 and 3 during CR (Fig. 1E). Thus, these data strongly suggested that mC-KITs might arise from a less aggressive “preleukemic” clone that harbors AE and acts as a “second” but crucial “hit” to provide the preleukemic cells with growth and survival advantages.
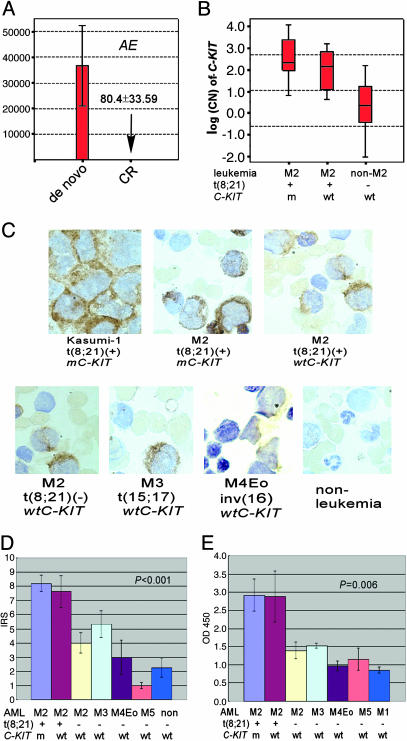
Reduction of AE in CR and expression of C-KIT in t(8;21) leukemia. (A) AE expression in patients in CR decreased significantly compared to that in disease presentation. (B) Median of C-KIT log(CN) in t(8;21) and non-t(8;21) leukemias. (C and D) Immunohistochemical analysis of CD117 expression on bone marrow specimens of leukemia patients. (C) CD117-positive cells and their staining intensity are higher in patients with t(8;21) and mC-KIT than in those with t(8;21) and wtC-KIT, whereas the percentage of CD117-positive cells and their staining intensity are higher in patients with t(8;21) and wtC-KIT than in patients without t(8;21). (D) Accordingly, the immunoreactivity score (IRS) and staining intensity is higher in t(8;21) leukemia than in other hemopathies. (E) The C-KIT PTK activity is higher in t(8;21) leukemia than in other subtypes of leukemias.
High Expression of C-KIT in t(8;21) Leukemia as an Alternative Way of C-KIT Activation. Overexpression of C-KIT in the majority of t(8;21) cases. The fact that mC-KIT was detected in only half of t(8;21) patient suggested that alternative mechanism of C-KIT activation, such as overexpression, might exist and cooperate with AE in leukemogenesis. Therefore, we quantified the C-KIT mRNA in t(8;21) patients negative for mC-KIT. The CN and log(CN) of C-KIT were calculated (Fig. 2B). In t(8;21) leukemia the log(CN) of C-KIT was significantly higher than that in leukemias negative for t(8;21) (P < 0.001), whereas no difference was detected between patients with either mC-KIT or wtC-KIT (P = 0.11). Because no difference was detected among log(CN) values for t(8;21)-negative leukemias, the C-KIT log(CN) values of these patients were pooled and the median was 0.38. We considered log(CN) values greater than (median + SD) of these cases (1.71) to be C-KIT high expression. Then, 16 of 18 (88.9%) and 10 of 14 (71.4%) t(8;21) leukemia patients with mC-KIT or wtC-KIT had a high expression of C-KIT mRNA, respectively. Thus, overexpression of C-KIT was found in 26 of 32 (81.3%) of patients with t(8;21) leukemia, whereas only 1/27 (3.7%) of leukemia patients without t(8;21) had a high expression of C-KIT. The standard immunohistochemical analysis showed that the C-KIT expression in patients with t(8;21) leukemia was higher than in other subtypes of leukemias (Fig. 2 C and D). Note that the C-KIT PTK activity was also increased in t(8;21) leukemia patients (Fig. 2E). Notably, the C-KIT expression was declined to a low, but still detectable, level after CR in all patients investigated (data not shown).
Significant up-regulation of C-KIT mRNA and protein expression by induction of AE in U937-A/E cells. Previous work showed that TGF-β1 was able to down-regulate expression of C-KIT in hematopoietic cells and inhibits stromal cell production of C-KIT ligand–SCF (38, 39), whereas a separate work suggested that the expression of TGF-β1 was repressed by AE (40). Therefore, we hypothesized that the expression of wtC-KIT might also be up-regulated in t(8;21) AML as a result of the presence of AE. We quantified C-KIT expression after AE was induced in U937-A/E cells. In these cells, the baseline expression of AE and C-KIT is very low and the transcription of AE can be specifically induced with PA. As shown in Fig. 3, upon PA treatment, both mRNA and protein levels of AE were increased and TGF-β1 decreased significantly. Intriguingly, the C-KIT expression was then enhanced substantially at 12–48 h at transcript, protein, and C-KIT PTK activity levels (Fig. 3 A–C), suggesting that AE could induce the C-KIT expression, probably by down-regulation of TGF-β1. We also examined whether the activation of C-KIT by SCF could alter AE expression in U937-A/E cells. Treatment of these cells with SCF did not affect the AE expression (data not shown). We then performed similar experiment in Kasumi-1 cells. Serum was withheld from these cells overnight before they were treated with SCF at a concentration of 200 μg/liter. At the presence of SCF, phosphorylated C-KIT (pC-KIT) was increased significantly (Fig. 3B Lower), indicating the activation of C-KIT signaling. However, the expression of AE and TGF-β1 at both mRNA (not shown) and protein (Fig. 3B Lower) levels was not altered, further supporting the hypothesis that the expression of AE is able to activate C-KIT kinase.
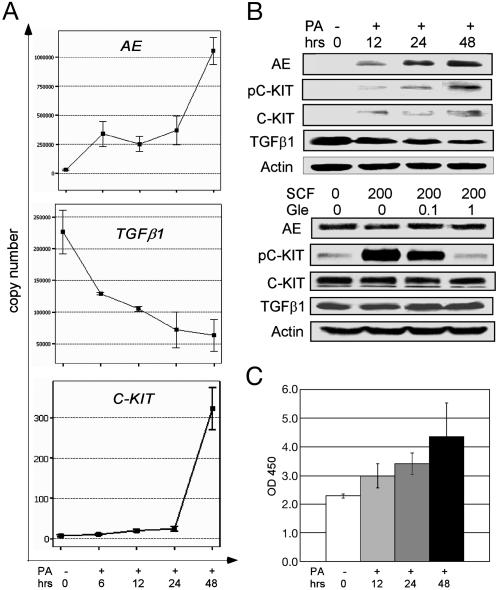
Induction of C-KIT expression by AE in vitro. (A) In U937-A/E cells treated with PA, the expression of AE at mRNA level increases gradually, whereas TGF-β1 is down-regulated at an early stage. The expression of C-KIT is up-regulated subtly at early stage (6–24 h) and dramatically at late stage (48 h). (B) At the protein level, the induced expression of AE down-regulates TGF-β1 and up-regulates phosphorylated (pC-KIT) and unphosphorylated C-KIT significantly (Upper). However, activation of C-KIT by SCF (μg/liter) do not modify AE and TGF-β1 expression (Lower). PA, ponasterone A (in μM); Gle, Gleevec (in μM). (C) The C-KIT PTK activity, which is reflected by absorbance at 450 nm on a spectrophotometer, is enhanced significantly 12 h after PA treatment.
Gleevec Response in t(8;21) Leukemic Cells with C-KIT Mutation/Overexpression. To explore the therapeutic potential of PTK inhibitor in the treatment of t(8;21) leukemia, AE-expressing cells were treated with Gleevec and the effects were evaluated. The IC50 of Gleevec for Kasumi-1 cells was 1.2 μM. After treatment with Gleevec at 0.5–5 μM for 48 h, the Annexin V positive/PI-negative cells were increased to up to 50% (Fig. 4A), and the C-KIT PTK activity was suppressed in a dose- and time-dependent manner (Fig. 4B). Gleevec also induced apoptosis and inhibited proliferation of primary leukemic cells from five patients with t(8;21), of which three had N822K-type mC-KIT and two had wild-type and overexpressed C-KIT (Fig. 4 C, D, and F), whereas cells from a patient with t(8;21) and D816H type mC-KIT were resistant to Gleevec but sensitive to Ara-C (Fig. 4E). A synergic effect was seen when Gleevec was combined with Ara-C (Fig. 4 D and F). However, cells from two AML M2 cases with neither t(8;21) nor C-KIT mutation/overexpression displayed no obvious response to Gleevec (data not shown).

Effects of Gleevec on t(8;21) leukemic cells. (A) Gleevec induces apoptosis of Kasumi-1 cells in a dose- and time-dependent manner. Gle, Gleevec, given in μM; time is given in hours. (B) Gleevec suppresses C-KIT PTK activity of the Kasumi-1 cell. (C–F) Effects of Gleevec and/or cytarabine (Ara-C, in μM) on primary leukemic cells from patients with t(8;21) leukemia. These effects are detected by annexin V assay (C–E) and analysis of morphological changes (F).
Discussion
In this work, we extended previous studies on C-KIT structural abnormality in AML by characterizing more mutation types and positions through whole-gene resequencing. However, is the C-KIT mutation an independent event, or is it associated with t(8;21) or other genetic defects in a multistep scenario of leukemogenesis? Here we showed that the C-KIT mutations were closely correlated with t(8;21) leukemia, with a mC-KIT rate of 48.1% in this AML subtype, whereas no C-KIT mutation was detected among 57 patients of other types of malignant hemopathies. Importantly, the coexistence of t(8;21) and mC-KIT demonstrated in most leukemic cells in newly diagnosed patients formed a base to further address the interrelationship between the two genetic abnormalities. A chronological order became clear when it was shown in three patients that, after CR, the AE remained positive and mC-KIT disappeared. Hence, AE should be a very early event in leukemogenesis, but clones with AE alone might have very limited proliferative potential in vivo. The addition of mC-KIT as a secondary oncogenic event based on t(8;21) may accelerate the disease process. The double hit leukemic cells with growth advantage could be more sensitive to the chemotherapy, whereas quiescent leukemic pluripotent progenitors with AE alone may escape the chemotherapy and persist in otherwise healthy, long-term disease-free survivors of t(8;21) leukemia. Our results have added more evidence to the concept that for a full-blown leukemia to occur in vivo, aberrations of both TFs and TKs are required. It may be worth pointing out that, in this work, no mC-KIT was detected among seven cases with inv (16) AML, a result that differs from previous reports (23–25), possibly because of the small number of cases in our series. More cases of this leukemia subtype should be investigated to clarify the issue.
Many studies have documented the complex and multifacial functions of AE in the last decade, including inhibiting differentiation and subsequent apoptosis (5, 6, 8, 41–43) and activating signals for cell proliferation (44–47). Here, we showed that the expression levels of C-KIT mRNA, protein, and PTK activity were much higher in t(8;21)AML with either wild-type or mutant C-KIT than those in leukemias negative for t(8;21) at the onset of disease, and were down-regulated simultaneously with decrease of AE during disease remission (data not shown). Furthermore, ectopic expression of AE in U937-A/E cells induces C-KIT expression at both mRNA and protein levels. These data unveil an important biological function of AE, and demonstrate that AE can activate PTK signal for cell proliferation by itself and may thus contribute to disease progression. Therefore, these data seem also to be in agreement with the ability of AE alone in inducing myeloproliferative disorder in mouse when appropriate promoter was used to drive the gene expression (14). In exploring the possible mechanism underlying AE-induced up-regulation of C-KIT, we found in U937-A/E cells that TGF-β1, a potent negative regulator of hematopoiesis, was down-regulated only 6 h after specific induction of AE. Previous studies showed that AE repressed basal promoter activity of Smad, the mediator of TGF-β signaling, and blocked the response to TGF-β1 (40). TGF-β1, on the other hand, could inhibit C-KIT expression by accelerated degradation of C-KIT mRNA (38, 39). Therefore, AE induced up-regulation of C-KIT may be at least partially mediated by TGF-β1. However, it is difficult to assess how essential the overexpression of C-KIT could be in leukemogenesis, particularly among t(8;21) leukemia patients without mC-KIT, in that other unknown secondary molecular abnormalities might also play a role in disease pathogenesis.
Finally, although reports suggested and our study confirmed that D816 mutations were resistant to Gleevec (26), our results clearly show that t(8;21) leukemic cells carrying N822K mC-KIT, the most frequently detected mC-KIT in t(8;21) leukemia, and C-KIT overexpression are sensitive, whereas cells from AML M2 without t(8;21) are resistant to Gleevec. Therefore, our results open therapeutic perspectives for PTK inhibitor in the treatment of leukemias with C-KIT mutation/overexpression. Further studies are required to evaluate the C-KIT targeting therapy as an additional treatment option for t(8;21) leukemia.
Acknowledgments
We thank Prof. L. J. Gu (Shanghai Children's Medical Center, Shanghai) for his kind collaboration and Dr. D. E. Zhang for her useful discussion on this work. We thank Prof M. Lübbert for providing the U937-A/E cells. This work was supported in part by the Chinese National High Tech Program (863, 2002BA711A04), the National Key Program for Basic Research (973, 2004CB518606), the National Natural Science Foundation of China, the Shanghai Municipal Commission for Science and Technology, the Shanghai Commission for Education, Shanghai Municipal Bureau for Public Health, the Shanghai Leading Academic Discipline Program, and the K. C. Wong Education Foundation, Hong Kong.
Notes
Author contributions: Z.C. and S.-J.C. designed research; Y.-Y.W., G.-B.Z., T.Y., B.C., J.-Y.S., W.-X.L., X.-L.J., G.Y., F.X., and Y.-W.L. performed research; G.-Q.C. contributed new reagents/analytic tools; Y.-Y.W., G.-B.Z., Z.C., and S.-J.C. analyzed data; G.-B.Z., Z.C., and S.-J.C. wrote the paper; J.-H.Y. performed clinical data analysis; and Z.-X.S., J.C., and S.-M.X. performed clinical work.
Abbreviations: TF, transcription factor; TK, tyrosine kinase; AML, acute myeloid leukemia; Ara-C, cytarabine; CN, copy number; PA, ponasterone A; SCF, stem cell factor.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0408831102
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc545849?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0408831102
Article citations
Risk-directed therapy based on genetics and MRD improves the outcomes of AML1-ETO-positive AML patients, a multi-center prospective cohort study.
Blood Cancer J, 13(1):168, 13 Nov 2023
Cited by: 3 articles | PMID: 37957175 | PMCID: PMC10643486
Protein-Nanocaged Selenium Induces t(8;21) Leukemia Cell Differentiation via Epigenetic Regulation.
Adv Sci (Weinh), 10(35):e2300698, 27 Oct 2023
Cited by: 1 article | PMID: 37888866 | PMCID: PMC10724402
[The study of clinical characteristics and prognosis of RUNX1-RUNX1T1 positive acute myeloid leukemia based on next-generation sequencing].
Zhonghua Xue Ye Xue Za Zhi, 44(10):851-854, 01 Oct 2023
Cited by: 0 articles | PMID: 38049338 | PMCID: PMC10694073
Targeting Stress Erythropoiesis Pathways in Cancer.
Front Physiol, 13:844042, 25 May 2022
Cited by: 8 articles | PMID: 35694408 | PMCID: PMC9174937
Review Free full text in Europe PMC
Prognostic Factors in Acute Myeloid Leukemia with t(8;21)/AML1-ETO: Strategies to Define High-Risk Patients.
Indian J Hematol Blood Transfus, 38(4):631-637, 01 Dec 2021
Cited by: 0 articles | PMID: 36258727 | PMCID: PMC9569252
Go to all (177) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The fusion protein AML1-ETO in acute myeloid leukemia with translocation t(8;21) induces c-jun protein expression via the proximal AP-1 site of the c-jun promoter in an indirect, JNK-dependent manner.
Oncogene, 22(36):5646-5657, 01 Aug 2003
Cited by: 26 articles | PMID: 12944913
AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) acute myeloid leukemia-M2.
Leukemia, 23(9):1598-1604, 21 May 2009
Cited by: 49 articles | PMID: 19458628
A new D816 c-KIT gene mutation in refractory AML1-ETO leukemia.
Haematologica, 91(9):1283-1284, 01 Sep 2006
Cited by: 18 articles | PMID: 16956837
AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML.
Oncogene, 20(40):5660-5679, 01 Sep 2001
Cited by: 112 articles | PMID: 11607817
Review





