Abstract
Free full text

A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation
Abstract
The NF-κB family members p65 (RelA) and c-Rel recognize similar DNA sequences, yet the phenotypes of mutant mice suggest that these proteins regulate distinct sets of genes. Here we demonstrate that 46 unique residues within an 86-residue segment of the Rel homology region (RHR) of c-Rel are responsible for the c-Rel requirement for Il12b gene induction by lipopolysaccharide in bone marrow-derived macrophages. These same residues were responsible for the c-Rel requirement for Il12a induction in dendritic cells, and in both instances, no evidence of c-Rel-specific coactivator interactions was found. Although the residues of c-Rel and p65 that contact specific bases and the DNA backbone within nuclear factor-κB (NF-κB) recognition sequences are identical, homodimers of c-Rel and of a chimeric p65 protein containing the critical c-Rel residues bound with high affinity to a broader range of NF-κB recognition sequences than did wild-type p65 homodimers. These results demonstrate that the unique functions of closely related transcription factor family members can be dictated by differences in the range of DNA sequences recognized at high affinity, despite having similar binding site consensus sequences and DNA contact residues.
Combinatorial gene regulation strategies required for the development of a multicellular eukaryote depend on a limited number of transcription factor families whose members are characterized by a conserved DNA-binding domain. In some families, such as the nuclear hormone receptor family, individual proteins recognize DNA sequences that are readily distinguishable from those recognized by other family members, providing an explanation for their functional differences (Rastinejad 2001). However, in many other families, multiple family members recognize identical DNA sequences and are expressed in the same cell types, raising the question of how they regulate distinct sets of target genes (Carlsson and Mahlapuu 2002; Hollenhorst et al. 2004).
Members of the Rel/nuclear factor-κB (NF-κB) family of transcription factors are key regulators of innate and adaptive immune responses and of several cellular processes, including survival, growth, and proliferation (Zhang and Ghosh 2001; Bonizzi and Karin 2004). In mammals, the Rel family consists of five members—c-Rel, p65 (RelA), RelB, NF-κB1 (p50/p105), and NF-κB2 (p52/p100)—that bind DNA as homodimers and heterodimers through a conserved N-terminal Rel homology region (RHR) (Ghosh et al. 1998). The RHR contains two immunoglobulin (Ig) domains, one of which is responsible for DNA binding and the other, dimerization (Ghosh et al. 1995, 1998). Rel dimers are often maintained in an inactive state by cytoplasmic association with IκB proteins (Karin and Ben-Neriah 2000). In the classical pathway, cell activation leads to signal transduction cascades that result in the phosphorylation and degradation of the IκBs, allowing rapid nuclear translocation of Rel dimers (Bonizzi and Karin 2004; Hayden and Ghosh 2004).
Mice lacking individual Rel family members display distinct phenotypes, suggesting that different Rel dimers regulate different target genes (Attar et al. 1997; Gerondakis et al. 1999). Indeed, studies of mutant mice have led to the identification of genes that require specific family members for expression, although considerable redundancy and compensation have been documented (Attar et al. 1997; Gerondakis et al. 1999; Hoffmann et al. 2003). Furthermore, recent studies have revealed the existence of two distinct signaling pathways leading to the activation of Rel dimers; the type 1 pathway activates most Rel dimers via phosphorylation of IkBα and IkBβ, whereas the type 2 pathway activates p52/RelB heterodimers by proteolytic processing of a p100 precursor (Pomerantz and Baltimore 2002; Bonizzi and Karin 2004).
Within the Rel family, the distinctions between c-Rel and p65 are of particular interest because they possess different biological functions, despite similar biochemical properties. p65-deficient mice die during mid-gestation due to massive TNF-α-induced liver apoptosis, whereas mice deficient in c-Rel develop normally (Kontgen et al. 1995; Beg and Baltimore 1996; Gerondakis et al. 1996). Neither c-Rel nor p65 is essential for the maturation of hematopoietic cells. However, the loss of either factor leads to unique activation defects in mature B, T, and myeloid cells (Kontgen et al. 1995; Grigoriadis et al. 1996; Gerondakis et al. 1999).
Despite major functional differences, c-Rel and p65 complexes coexist in many cell types and are activated by the same type 1 pathway (Pomerantz and Baltimore 2002; Bonizzi and Karin 2004). In addition, both c-Rel and p65 contain transactivation domains at their C termini, whereas p50 and p52 lack transactivation domains (Siebenlist et al. 1994; Li and Verma 2002). The c-Rel and p65 RHRs are more closely related to each other than to the other Rel family members (Li and Verma 2002), and identical c-Rel and p65 residues contact the DNA backbone and specific bases within NF-κB recognition sequences (Chen and Ghosh 1999; Huang et al. 2001). Consistent with the identical DNA contact residues, binding site selection studies failed to reveal clear differences in DNA consensus sequences for c-Rel and p65, although c-Rel homodimers selected a slightly broader range of DNA sequences than p65 homodimers (Kunsch et al. 1992). Electrophoretic mobility shift assays (EMSAs) performed with NF-κB sequences from a few c-Rel-dependent genes have suggested preferential binding of c-Rel homodimers or c-Rel/p50 heterodimers (e.g., Himes et al. 1996; Shapiro et al. 1997; Chen et al. 2000). However, no quantitative information regarding preferential binding has been reported. Sequences that appear to preferentially bind p65 dimers have also been reported (Paxton et al. 1997; Saccani et al. 2001). Most recently, NF-κB recognition sequences from a number of p65-dependent genes were compared, with no distinguishing characteristics to explain the p65 dependence (Hoffmann et al. 2003).
Another mechanism that may allow p65 and c-Rel to regulate distinct sets of target genes is through interactions with specific coactivators. Several coactivators interact with p65 (Paal et al. 1997; Sheppard et al. 1999; Zhong et al. 2002), but little is known about coactivator interactions with c-Rel. c-Rel can be modified by phosphorylation (Druker et al. 1994; Fognani et al. 2000; Martin and Fresno 2000), raising the possibility that c-Rel-specific phosphorylation events support interactions with unique coactivators at c-Rel-dependent genes. Interestingly, a recent study revealed that a requirement for p50/p65 heterodimers versus p65/p65 homodimers can be dictated by the differential recruitment of coactivator complexes, with the DNA recognition sequence responsible for determining which complex is recruited (Leung et al. 2004).
Il12b, which encodes the IL-12 p40 protein, is one example of a gene that can be activated by c-Rel, but not by p65 (Sanjabi et al. 2000). Biologically active IL-12 p70 is composed of two subunits, p40 and p35, which are induced in macrophages and dendritic cells at the level of transcription (Trinchieri 2003). IL-12 serves as a key inducer of T helper 1 cell development and therefore provides a bridge between innate and adaptive immunity (Trinchieri 2003). Il12b expression was greatly reduced following lipopolysaccharide (LPS) stimulation of peritoneal and fetal liver-derived macrophages from c-Rel-/- mice, whereas expression was only slightly reduced in p65-/- macrophages (Sanjabi et al. 2000). Interestingly, the c-Rel requirement for endogenous Il12b expression could not be recapitulated in transfection assays, as an Il12b promoter-reporter plasmid was activated to a similar extent by overexpressed c-Rel or p65 (Sanjabi et al. 2000). Furthermore, p65/p50 and c-Rel/p50 heterodimers bound the known NF-κB recognition sequence in the Il12b promoter with comparable affinities (Sanjabi et al. 2000).
In this study, we pursued the mechanism underlying the c-Rel requirement for Il12b transcription. The results reveal that 46 residues derived from the RHR of c-Rel can support Il12b transcription when substituted for the corresponding residues of p65 in the context of the full-length p65 protein. These same residues selectively increase the affinity of c-Rel for NF-κB recognition sequences that diverge from the consensus, with relatively little effect on binding to canonical sites. These data provide strong evidence that the c-Rel requirement is due largely or exclusively to the ability of c-Rel homodimers to recognize a broader range of NF-κB sequences than p65 homodimers.
Results
Restoration of Il12b expression in c-Rel-/- macrophages by retroviral transduction
To determine why c-Rel is required for Il12b induction in LPS-stimulated macrophages, we set out to identify regions of c-Rel that support Il12b induction when substituted for the corresponding regions of p65. Because overexpressed p65 and c-Rel transactivated Il12b promoter-reporter plasmids comparably in transient transfection assays (Sanjabi et al. 2000), we relied on the use of retroviral vectors to express Rel proteins at approximately physiological concentrations in c-Rel-/- macrophages differentiated in the presence of M-CSF from bone marrow precursors. Like peritoneal and fetal liver-derived c-Rel-/- macrophages (Sanjabi et al. 2000), bone marrow-derived macrophages from c-Rel-/- mice expressed little Il12b mRNA and IL-12 p40 protein following stimulation with LPS + IFN-γ (Fig. 1A [lanes 1-8], C [mock]). mRNA and protein were monitored by RT-PCR and ELISA, respectively, with the ELISAs performed with an antibody that detects total secreted p40 protein in the form of both p40 homodimers and p40/p35 (p70) heterodimers.
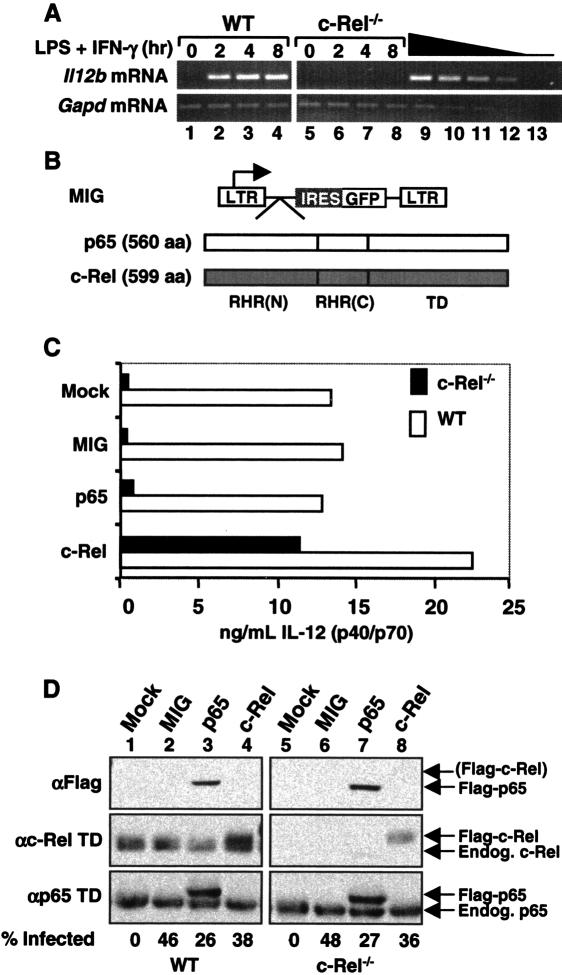
Retroviral expression of c-Rel in c-Rel-/- macrophages rescues Il12b expression. (A) RT-PCR was performed using RNA from wild-type and c-Rel-/- bone marrow-derived macrophages at the indicated time points after activation with LPS (10 μg/mL) and IFN-γ (10 U/mL). Twofold titrations of wild-type RNA (lanes 9-12) and a negative control (lane 13) were included. (B) Diagrams of the MIG vector and murine p65 and c-Rel cDNAs are shown. The N-terminal [RHR(N)] and C-terminal [RHR(C)] portions of the RHR and transactivation domain (TD) are depicted. (C) Secreted IL-12 (p40/p70) protein levels observed in LPS/IFN-γ-activated wild-type and c-Rel-/- macrophages are shown. Cells were left untransduced (mock) or were transduced with the MIG virus or with viruses expressing p65 or c-Rel. (D) Protein levels in untransduced and transduced macrophages were monitored by Western blot, using antibodies directed against the Flag epitope or transactivation domains of c-Rel or p65. The percentage of cells transduced by each retroviral preparation was determined by flow cytometry analysis of GFP fluorescence.
Murine c-Rel and p65 containing N-terminal Flag tags were expressed from MIG, a murine stem cell virus (MSCV)-derived vector that contains an internal ribosome entry site (IRES) and green fluorescent protein (GFP) coding sequences following the cDNA cloning site (Fig. 1B). After retroviral transduction and differentiation, the wild-type and c-Rel-/- bone marrow macrophages were activated with LPS + IFN-γ for 24 h. IL-12 p40 protein was undetectable in unactivated wild-type and c-Rel-/- macrophages (data not shown). In activated c-Rel-/- macrophages, flag-c-Rel greatly enhanced p40 expression, whereas flag-p65 and the empty MIG vector had little effect relative to mock-infected cells (Fig. 1C). In wild-type cells, p40 expression was also selectively increased by flag-c-Rel (Fig. 1C), suggesting that endogenous c-Rel is limiting for Il12b expression. The results in Figure 1C are representative of several independent experiments but are shown alone to allow a direct comparison with the Western blot and GFP expression data in Figure 1D, which were derived from the same experiment.
In Western blots performed with cell extracts from the transduced cells, flag-p65 was readily detected when using Flag antibodies, but flag-c-Rel was usually undetectable (Fig. 1D, top panel). Although undetectable with Flag antibodies, flag-c-Rel was detected when using a c-Rel antibody that recognizes the transactivation domain (αc-Rel TD) (Fig. 1D, middle panel). The concentration of flag-c-Rel present in extracts from wild-type and c-Rel-/- macrophages was comparable to that of endogenous c-Rel in wild-type extracts (Fig. 1D, middle panel). However, flow cytometry analysis of GFP revealed that only 38% and 36% of the wild-type and c-Rel-/- macrophages, respectively, were transduced by the flag-c-Rel retrovirus (Fig. 1D, bottom). Thus, in the transduced cells, the average concentration of flag-c-Rel was approximately three times higher than that of endogenous c-Rel. A similar evaluation of flag-p65 suggested that it was overexpressed by approximately four-fold relative to endogenous p65 (Fig. 1D, bottom panel). To summarize, flag-c-Rel restored endogenous p40 expression in c-Rel-/- macrophages much more efficiently than did flag-p65. Thus, the retroviral transduction assay was suitable for identifying regions of c-Rel that are critical for Il12b induction.
c-Rel selectivity is dictated by 46 residues within an 86-residue segment of the RHR
To identify the region of c-Rel that is required for Il12b transcription, p65/c-Rel chimeric proteins were analyzed using the retroviral transduction assay. Interestingly, a chimeric protein containing the c-Rel RHR and p65 transactivation domain (p65/N,C) restored p40 expression as efficiently as wild-type c-Rel, but a protein containing the p65 RHR and c-Rel transactivation domain (p65/TD) was largely inactive (Fig. 2B, rows 4-6). These results were surprising because they suggest that selectivity is dictated by residues within the c-Rel RHR, which is highly homologous to the p65 RHR (Fig. 2A), rather than by residues within c-Rel's unique transactivation domain. Although the p65/N,C chimera was far more active than the p65/TD chimera, it was expressed at a lower concentration (Fig. 2C, lanes 5,6). It is noteworthy that, although the c-Rel and p65 transactivation domains were interchangeable in this assay, mutant c-Rel proteins lacking a transactivation domain were inactive (Supplementary Fig. 1).
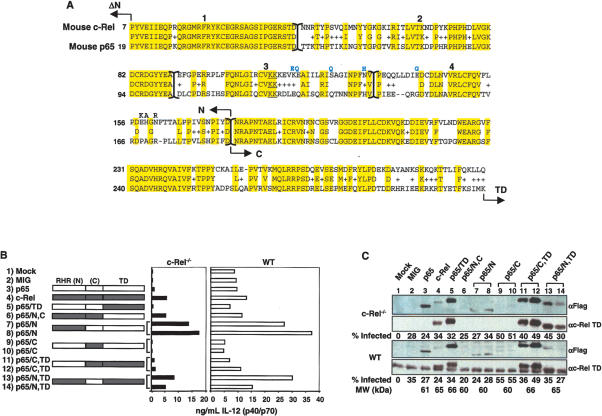
The N-terminal portion of the c-Rel RHR is necessary for Il12b induction in macrophages. (A) Primary amino acid sequences of mouse c-Rel and p65 RHRs were compared. Arrows indicate the boundaries between the N-terminal RHR (N), the C-terminal RHR (C), the transactivation domain (TD), and the first nonhomologous six and 18 residues of c-Rel and p65, respectively (ΔN; residues not shown). The N-terminal portion of the RHR was further divided into four regions labeled 1-4 (separated by brackets). The conserved DNA-contacting lysines in region 3 are underlined. The eight residues that were introduced in the modified proteins described in Supplementary Figure 5 are shown above the main sequence. (B) IL-12 (p40/p70) protein levels were measured after transduction of c-Rel-/- or wild-type macrophages with viruses expressing the indicated chimeras, and 24 h after stimulation with LPS + IFN-γ. Regions in white are from p65, and shaded regions are from c-Rel. The chimeras were named according to the following code: p65/domain X (N, C, and/or TD) from c-Rel. (C) Protein expression and the percentage of transduced cells in each sample were monitored as described in the legend to Figure 1D. The molecular weight of each protein is also shown.
Additional chimeras were analyzed to determine whether the c-Rel selectivity region resides within the N-terminal portion of the RHR, which is primarily responsible for sequence-specific DNA binding, or the C-terminal portion, which mediates dimerization (Chen and Ghosh 1999) (for chimera fusion sites, see N-terminal and C-terminal in Fig. 2A). Efficient Il12b induction was observed only with chimeras containing the N-terminal portion of the c-Rel RHR (Fig. 2B, rows 7, 8, 13, 14 vs. rows 9-13). The protein expression data in Figure 2C revealed that these chimeric proteins were expressed at variable concentrations, but expression level did not correlate with the capacity of each chimera to restore Il12b expression in c-Rel-/- macrophages.
To localize the c-Rel selectivity region more precisely, the N-terminal DNA-binding region of the RHR was divided into four segments, which are numbered 1-4 in Figure 2A. Chimeric proteins were generated with variable numbers of segments from c-Rel or p65 fused to the remaining segments from the alternate protein. The results (see Supplementary Fig. 2) provided strong evidence that segments 3 and 4 within the N-terminal portion of the c-Rel RHR are responsible for the selective activation of Il12b transcription in macrophages. Segments 3 and 4 span 86 amino acids, 46 of which differ between p65 and c-Rel (Fig. 2A).
To test more rigorously the significance of segments 3 and 4, a new chimeric protein was analyzed. In this new chimera, p65/N3,4, segments 3 and 4 alone from c-Rel were substituted for the corresponding segments of p65 in the context of full-length p65 (Fig. 3A,D). Thus, this chimeric protein differs from p65 by only 46 residues within segments 3 and 4.
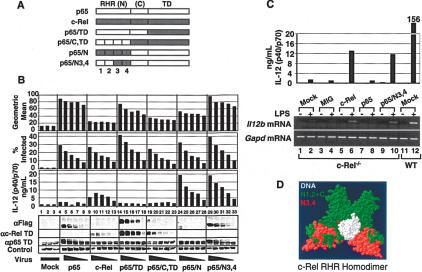
A p65/c-Rel chimera containing 46 residues from the c-Rel RHR activates Il12b expression in c-Rel-/- macrophages. (A) The wild-type and p65/c-Rel chimeras analyzed in this experiment are shown. (B) c-Rel-/- macrophages were transduced with various concentrations (twofold titrations) of titered viruses expressing each of the proteins depicted in A. (Rows 1, 2) Flow cytometry monitoring GFP fluorescence was used to determine the geometric mean fluorescence within each population, as well as the percentage of infected cells. (Row 3) IL-12 p40/p70 protein was monitored by ELISA. Western blots with antibodies against the Flag epitope, c-Rel TD, and p65 TD were used to monitor protein expression. An unknown protein recognized nonspecifically by the Flag antibody served as a control. (C) IL-12 p40/p70 protein (top) and Il12b mRNA (bottom) were analyzed by ELISA and RT-PCR, respectively, after transduction of c-Rel-/- macrophages and activation with LPS/IFN-γ for 8 h. Gapd mRNA was monitored as a control. (D) A structural model of the c-Rel RHR homodimer bound to the CD28 response element from the IL-2 promoter is shown (Huang et al. 2001). The DNA (white), region N3,4 of the c-Rel RHR (red), and regions N1,2 and C (green) are highlighted.
Bone marrow macrophages from c-Rel-/- mice were transduced with retroviruses expressing wild-type c-Rel, wild-type p65, the p65/N3,4 chimera, and three of the chimeras analyzed previously (Fig. 3A). In this experiment, the cells were transduced with five different concentrations of each viral stock (twofold serial dilutions), after determining relative titers in NIH 3T3 cells. Using flow cytometry, we first monitored the percentage of GFP-expressing cells in each transduced sample, as well as the geometric mean fluorescence in the GFP-expressing subset. Increasing concentrations of virus resulted in a linear increase in the percentage of infected cells, with comparable percentages infected with the different viral stocks (Fig. 3B, row 2). In contrast, the geometric mean fluorescence increased by less than twofold in any given virus titration (Fig. 3B, row 1). Thus, increasing the virus amount increases the percentage of cells infected, with little effect on the number of “hits” per cell. Interestingly, the wild-type c-Rel virus yielded the lowest geometric mean fluorescence, suggesting that this viral mRNA is transcribed or translated relatively poorly, consistent with the low c-Rel expression observed in Figures Figures11 and and22.
Next, IL-12 p40 expression was monitored by ELISA, revealing strong, viral dose-dependent expression in the presence of c-Rel, p65/N, and p65/N3,4, but not in the presence of p65, p65/TD, or p65/C,TD (Fig. 3B, row 3). Most importantly, the viruses expressing wild-type p65 and p65/N3,4 yielded similar geometric means and similar percentages of infected cells, but exhibited very different abilities to restore p40 expression (Fig. 3B, lanes 4-8,29-33). Interestingly, although wild-type p65 was capable of restoring low levels of p40 expression in comparison to mock-infected cells, p40 expression did not increase with increasing concentrations of the p65 virus (Fig. 3B, row 3, lanes 1-8). It is also noteworthy that, in this and other experiments, a decrease in p40 expression was observed with the highest concentration of c-Rel virus (Fig. 3B, row 3, lane 9). The reason for this decrease is not known, but may be due to toxicity caused by high concentrations of c-Rel. Western blots performed with Flag, p65, and c-Rel antibodies were used to monitor relative concentrations of the wild-type and mutant proteins (Fig. 3B, rows 4-6). Most importantly, the transduced p65 and p65/N3,4 proteins were expressed at similar concentrations, as determined by using the Flag antibody (Fig. 3B, lanes 4-8,29-33). As above, the wild-type c-Rel and p65/N proteins were poorly expressed, despite their abilities to restore p40 expression. In a separate experiment, IL12b mRNA levels were monitored by RT-PCR (Fig. 3C, top and bottom, respectively). The results confirmed that wild-type c-Rel and p65/N3,4 can restore both p40 protein and Il12b mRNA, with no restoration observed with wild-type p65.
Together, these results demonstrate that 46 residues within the c-Rel RHR make a major contribution to the selective activation of Il12b by c-Rel and appear to be solely responsible for selectivity. These 46 residues span β-strands C′ through G′, α-helices 1 and 2, and loop 2 of the N-terminal DNA-binding Ig domain of the c-Rel RHR (see regions highlighted in red in Fig. 3D; Huang et al. 2001).
Activation of other c-Rel target genes by the selectivity region
To determine whether the 46 residues responsible for c-Rel selectivity at Il12b are responsible for selective activation of other c-Rel target genes, Affymetrix microarrays were used to identify additional c-Rel-dependent genes. RNA was prepared from wild-type and c-Rel-/- macrophages before and after stimulation with LPS + IFN-γ for 8 h. Although 293 genes were identified that appeared to be stimulated by LPS + IFN-γ by more than twofold in wild-type macrophages (data not shown), only four were found to be dependent (fourfold or more) on c-Rel (Fig. 4A). Interestingly, Il12b exhibited the strongest c-Rel dependence (65-fold difference between wild-type and c-Rel-/- macrophages), with less c-Rel dependence observed at the other three genes, Lox (20-fold), Il4i1 (ninefold), and Gfi1 (fourfold). Semi-quantitative RT-PCR experiments confirmed that these three genes are induced by LPS + IFN-γ and that their induction is reduced only modestly in c-Rel-/- macrophages (Fig. 4B, lanes 1-6).
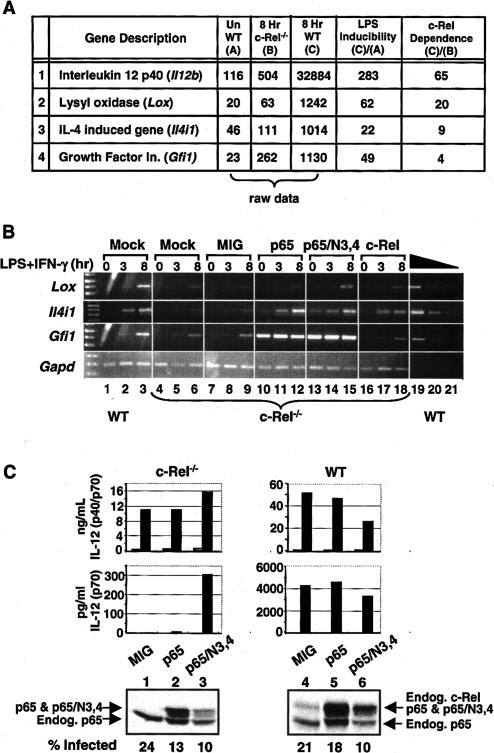
Activation of other c-Rel-dependent genes by retroviral transduction. (A) Affymetrix microarrays were used to monitor gene expression in unstimulated wild-type macrophages (bone marrow-derived) and in stimulated wild-type and c-Rel-/- macrophages (10 μg/mL LPS + 10 U/mL IFN-γ for 8 h). Of 4849 expressed genes, 293 were up-regulated twofold or more by LPS + IFN-γ in wild-type cells, and 65 exhibited reduced expression (twofold or more) in stimulated c-Rel-/- cells versus stimulated wild-type cells. However, only four LPS/IFN-γ-stimulated genes exhibited c-Rel dependence. The raw data represent the normalized expression level from the Affymetrix arrays. (B) Wild-type and c-Rel-/- bone marrow-derived macrophages were mock-treated or transduced with the indicated viruses. After LPS/IFN-γ activation for 0, 3, or 8 h, total RNA was isolated and analyzed by semiquantitative RT-PCR. A two-fold titration of the wild-type 8-h-activated sample was used to monitor linearity of the RT-PCR assay. (C) Bone marrow precursors were differentiated into dendritic cells and were transduced with the MIG, p65, and p65/N3,4 viruses, followed by stimulation with LPS (1 μg/mL). Total IL-12 p40 concentrations (p40/p70, top graph) were compared with IL-12 p70 concentrations (bottom graph) as a strategy for monitoring the c-Rel requirement for IL-12 p35 expression (Grumont et al. 2001). Results from unstimulated and stimulated cells are shown (open and filled bars, respectively). Protein expression was monitored by Western blot, using a mixture of antibodies directed against the p65 TD and c-Rel TD.
To determine whether the selectivity region defined above could explain the c-Rel dependence of these genes, their expression was monitored in c-Rel-/- macrophages transduced with the c-Rel, p65, and p65/N3,4 retroviruses. Only Lox yielded results resembling those obtained with Il12b, with expression restored to a slightly greater extent with transduced c-Rel and p65/N3,4 than with p65 (Fig. 4B, lanes 7-18). However, the Lox results were more difficult to evaluate than were the Il12b results presented above, perhaps because Lox exhibited only modest dependence on c-Rel (Fig. 4B, cf. lanes 3 and 6). Analysis of the other two c-Rel-dependent genes, Il4ia and Gfi1, yielded different results: Il4i1 expression was restored by all three proteins in an LPS/IFN-γ-dependent manner, whereas Gfi1 expression was restored by p65 and p65/N3,4 in the absence of cell stimulation (Fig. 4B). Thus, modest overexpression of p65 was sufficient to overcome the c-Rel preference at these genes.
Another gene that has been reported to exhibit a strong requirement for c-Rel is Il12a, which encodes the p35 subunit of IL-12 (Grumont et al. 2001). Interestingly, Il12a requires c-Rel for expression in LPS-stimulated dendritic cells but not in LPS-stimulated macrophages, whereas Il12b requires c-Rel in LPS-stimulated macrophages but not in LPS-stimulated dendritic cells (Sanjabi et al. 2000; Grumont et al. 2001). To determine whether the c-Rel selectivity region is responsible for the c-Rel requirement at Il12a, bone marrow cells were differentiated into dendritic cells (Ouaaz et al. 2002). IL-12 p35 expression was then monitored by comparing the concentration of the IL-12 p70 heterodimer to the total concentration of the IL-12 p40 subunit. Consistent with the results of Grumont et al. (2001), the IL-12 p40 concentration was reduced by only fourfold in c-Rel-/- dendritic cells in comparison to wild-type cells (Fig. 4C, top, MIG results; please note Y-axis scale differences), with this modest decrease likely due to decreased numbers of viable cells in the c-Rel-/- cultures. In contrast, IL-12 p70 concentrations were reduced by ~1000-fold (Fig. 4C, bottom graph, MIG results), reflecting the decreased expression of Il12a that was reported previously (Grumont et al. 2001). When c-Rel-/- dendritic cells were reconstituted with p65/N3,4, the concentration of IL-12 p70, but not of IL-12 p40, was greatly enhanced. In contrast, wild-type p65 had little effect. These results demonstrate that the same 46 residues are critical for the c-Rel-dependent expression of both Il12a and Il12b.
The selectivity region modulates the DNA-binding properties of c-Rel
Two possible functions for the c-Rel selectivity region must be considered. First, it could interact with another molecule (e.g., a coactivator) that is essential for Il12a and Il12b transcription. Alternatively, the 46 unique residues could influence the DNA-binding properties of c-Rel. To distinguish between these models, we first attempted to define more precisely the residues that are responsible for c-Rel selectivity. However, chimeric proteins in which segments 3 and 4 were divided into six independent regions and replaced with the corresponding regions from p65 failed to disrupt Il12b activation by c-Rel (Supplementary Fig. 3A,B). Further efforts to test the effect of various combinations of these six segments in the context of c-Rel or p65 were also unsuccessful, as they had no significant effect on activity or destabilized the protein (Supplementary Fig. 3C). Mutations in potential phosphorylation sites were also examined, again with little or no effect on gene activation (Supplementary Fig. 4).
To reduce the potential for structural abnormalities and instability, modified versions of c-Rel and p65/N3,4 were analyzed in which eight solvent-exposed residues from among the 46 unique residues were simultaneously changed to the corresponding p65 residues (above c-Rel sequence in Fig. 2A); although these eight substitutions substantially altered the surface of the protein, the mutant proteins retained the ability to restore p40 expression (Supplementary Fig. 5). We also created modified versions of c-Rel and p65/N3,4 in which 11 solvent-exposed residues were altered, but these mutations dramatically destabilized the protein (data not shown).
To summarize, no single residues or small clusters of residues within the selectivity region were essential for selectivity, and we found that a p65 variant containing only 38 residues from c-Rel was sufficient for Il12b induction. Because the most solvent-exposed residues in the selectivity region can be altered without any functional loss, these results argue against a protein-protein interaction model.
We initially considered it unlikely that the c-Rel selectivity region would influence DNA binding because the RHR residues that contact DNA bases and the sugarphosphodiester backbone are conserved between c-Rel and p65 (Chen and Ghosh 1999; Huang et al. 2001). In addition, only two of these residues (K100 and K111) are located within the selectivity region (Fig. 2A). Furthermore, our previous studies suggested that p50/p65 and p50/c-Rel heterodimers bind the Il12b NF-κB site with comparable relative affinities (Sanjabi et al. 2000). However, an analysis of the DNA-binding properties of c-Rel, p65, and p65/N3,4, after overexpression in HEK 293T cells with small amounts of p50, revealed a surprising difference: The homodimeric complexes containing c-Rel or p65/N3,4 were enhanced relative to the p65 homodimeric complex (Fig. 5B, lanes 1-12). This difference was most readily apparent when comparing p65 to p65/N3,4, which yielded very similar heterodimeric complexes with p50. Flag and p50 antibodies were used in supershift experiments to confirm that the upper complexes consisted of homodimers (Fig. 5B, lanes 13-18). Results matching those shown here were obtained when a probe containing only the proximal NF-κB recognition sequence (and lacking the distal recognition sequence described below) was tested (data not shown).
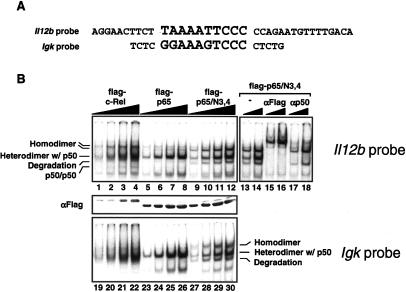
Enhanced binding of c-Rel and p65/N3,4 homodimers to NF-κB recognition sequences. (A) The sequences of EMSA probes containing the Il12b and Igk NF-κB recognition sequences are shown. (B) HEK 293T cells were transfected with plasmids expressing flag-tagged versions of c-Rel, p65, and p65/N3,4, with a small amount of a plasmid expressing murine p50. After extract preparation, EMSA experiments were performed using the Il12b (lanes 1-12) and Igk (lanes 19-30) probes, with different concentrations (twofold titrations) of each extract. (Lanes 13-18) The composition of each complex was determined by supershift analysis using Flag and p50 antibodies. (Middle panel) A Western blot performed with Flag antibodies was used to monitor protein expression.
Interestingly, the increased abundance of the p65/N3,4 homodimeric complex was also observed in EMSA experiments when using a probe containing a consensus NF-κB recognition site from the Igκ enhancer (Fig. 5B, lanes 23-30). The surface-modified versions of c-Rel and p65/N3,4 that retained their ability to restore p40 expression also retained the ability to bind DNA with a relatively high affinity (Supplemental Fig. 5C). These results suggest that the selectivity region allows c-Rel and p65/N3,4 homodimers to bind NF-κB sites with higher affinities than p65 homodimers.
The Il12b promoter contains a functionally important NF-κB homodimer-binding site
The above results suggest that the c-Rel selectivity region enhances the affinity of homodimers for DNA, but not heterodimers. This finding was unexpected because the known NF-κB site identified in the Il12b promoter binds much more tightly to p50/p65 and p50/c-Rel heterodimers (Sanjabi et al. 2000). However, a closer inspection of the Il12b promoter revealed a sequence immediately adjacent to the previously described NF-κB site (Plevy et al. 1997) that matches at eight out of nine positions the CD28 response element in the IL-2 promoter, which may preferentially bind homodimers (CD28 RE, Fig. 6A). In our previous mutant analysis, mutations at this site had no effect on Il12b promoter activity in a transient transfection assay (Plevy et al. 1997). We therefore considered the possibility that this site may be important only in a more physiological setting. To examine this possibility, wild-type and mutant Il12b promoters were analyzed in a stable transfection assay in the J774 macrophage line. An Il12b promoter fragment extending from -350 to +55 was inserted upstream of an EGFP reporter gene in a vector containing flanking insulator elements from the chicken β-globin locus (Chung et al. 1997; Muljo and Schlissel 2002). A newly discovered Il12b enhancer (L. Zhao and S.T. Smale, unpubl.) was placed upstream of the promoter because the isolated promoter was largely inactive in stable transfection assays when flanked by insulator elements (Fig. 6A, see Il12bP vs. Il12bEP; L. Zhao and S.T. Smale, unpubl.). The vector also contains a puromycin resistance gene on the opposite side of the insulator elements. After transfection into J774 cells, monoclonal/oligoclonal cell populations that had integrated each reporter plasmid were expanded in 96-well plates in the presence of puromycin. After stimulation, the wild-type Il12b promoter supported GFP expression in a fraction of the cell populations (Fig. 6A). Importantly, expression was reduced by mutations in either the proximal or distal NF-κB-binding site, with the most dramatic reduction observed with a promoter containing mutations at both sites (Fig. 6A). These results suggest that both sites contribute to Il12b promoter activity. Both sites are highly conserved in the mouse and human genomes (Fig. 6A).
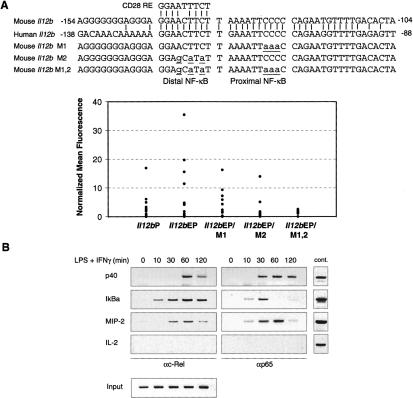
Analysis of NF-κB recognition site mutants in a stable transfection assay. (A) At the top, the mouse Il12b promoter sequence from -154 to -104 was compared with the human Il12b promoter sequence from -138 to -88. The similarity between the distal NF-κB recognition sequence and the CD28 response element is shown, as are the sequences of the M1, M2, and M1,2 mutants analyzed. J774 cells were transfected with GFP reporter plasmids containing the Il12b promoter alone (Il12bP), Il12b promoter with an upstream Il12b enhancer (Il12bEP), or mutant Il12b promoters with the upstream enhancer (M1, M2, and M1,2). Cell clones were stimulated with LPS and GFP reporter activity monitored by flow cytometry. Each dot represents the mean fluorescence observed with a different cell clone, after subtraction of the mean fluorescence observed with the unstimulated cells of the same clone. (B) Binding of c-Rel and p65 to the endogenous Il12b promoter in bone marrow-derived macrophages stimulated with LPS + IFN-γ was monitored by ChIP. Primers that amplify the IκBα and MIP-2 promoters, which are also induced by LPS, were included as positive controls. Primers that amplify the IL-2 promoter were included as a negative control. Control genomic DNA samples at the right demonstrate the relative amplification efficiencies of the different primer pairs, and input samples at the bottom demonstrate that similar amounts of chromatin from each time point were analyzed.
To determine whether c-Rel and p65 can interact with the Il12b promoter, a chromatin immunoprecipitation (ChIP) assay was employed. The results revealed that both proteins can associate with the promoter in stimulated cells (Fig. 6B). At face value, the ChIP results suggest that p65 may associate more rapidly with the promoter following stimulation. However, the significance of the different kinetics observed in this assay remains unknown.
The stable transfection and ChIP results are consistent with a model in which the Il12b promoter contains two adjacent NF-κB sites. The proximal site may interact with p50/p65 and/or p50/c-Rel heterodimers, with the distal site interacting specifically with c-Rel homodimers (see Discussion).
The c-Rel selectivity region allows c-Rel homodimers to bind NF-κB sites with higher affinity than p65 homodimers
The results in Figure 5B suggest that c-Rel and p65/N3,4 homodimers bind the original Il12b NF-κB site and a consensus NF-κB site from the Igκ enhancer with a higher affinity than p65 homodimers. To examine binding affinities in greater depth, the c-Rel, p65, and p65/N3,4 RHRs were expressed as homodimers in Escherichia coli and purified. Binding affinities for several NF-κB sites were then determined by EMSA, after normalization for the percentage of active protein in each purified sample (Carey and Smale 2000; Cowley and Graves 2000). Under the binding conditions used, the equilibrium dissociation constant (Kd) for c-Rel RHR homodimers bound to the proximal NF-κB site within the Il12b promoter (1.2 nM) (Fig. 7A,B) was 42.8 times lower than that for p65 RHR homodimers bound to this site (51.4 nM). Importantly, p65/N3,4 homodimers bound this site with a 11.7-fold higher affinity than p65 homodimers. This result confirms that the 46 residues responsible for selectivity increase the affinity of the RHR for this recognition site. Similar results were obtained when the distal NF-κB site from the Il12b promoter or the IL-2 CD28 RE was examined.
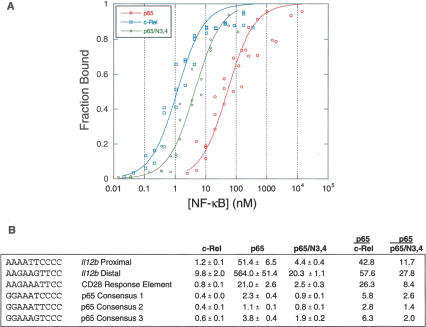
Relative affinities of the c-Rel, p65, and p65/N3,4 RHRs for NF-κB recognition sequences. (A) Compiled results are shown from three independent EMSA titrations comparing the binding of p65, p65/N3,4, and c-Rel to the Il12b proximal recognition site. Results were normalized for the percentage of active protein within each purified protein sample. The Y-axis shows the fraction of probe molecules bound by protein, and the X-axis shows the concentration of NF-κB homodimer added to the reaction. The plot was created by the Kaleidograph program, using a nonlinear least squares analysis as described by Cowley and Graves (2000). (B) Kd values (nanomolar, with standard deviations) are shown for recombinant NF-κB RHR homodimers bound to probes containing the indicated recognition sequences. The results were derived from three independent EMSA titrations performed with each probe and each RHR homodimer. Kd values were determined as described (Cowley and Graves 2000). At the right, Kd ratios for p65 versus c-Rel and for p65 versus p65/N3,4 are shown.
To determine whether NF-κB sites could be identified that bound p65 with a higher affinity than c-Rel or p65/N3,4, we examined three different sequences identified in a binding site selection analysis performed with p65 homodimers (Kunsch et al. 1992). Although the p65 homodimer affinities for these sites were indeed much higher than for the other sites examined, the c-Rel and p65/N3,4 homodimer affinities remained slightly higher. It is important to note that the sites recognized by c-Rel and p65/N3,4 with a substantially higher affinity than p65 (Il12b proximal and distal and CD28 RE) diverge from the generic NF-κB consensus site at one half-site (i.e., at least one of the two key GC base pairs is missing), whereas the sites bound by all three proteins with comparable affinities (p65 consensus 1, 2, and 3) closely match the consensus. Together, these results suggest that c-Rel homodimers possess an intrinsic capacity to bind divergent NF-κB recognition sites with much higher affinities than p65 homodimers. The 46 unique residues that comprise the c-Rel selectivity region are largely responsible for this enhanced binding affinity.
Discussion
The results of this study provide strong evidence that the primary reason c-Rel is required for Il12b transcription is because c-Rel homodimers can bind a broader range of NF-κB recognition sequences with high affinity than can p65 homodimers. In this analysis, the only region of c-Rel that was not functionally redundant with p65 corresponded to an N-terminal portion of the RHR containing 46 unique residues. Because these residues greatly increased the affinity of c-Rel homodimers for some NF-κB recognition sequences, the increased affinity is likely to be responsible for the functional differences between c-Rel and p65 at the Il12b and Il12a genes. Interestingly, c-Rel homodimers and p65 homodimers bound NF-κB recognition sequences that resemble the previously defined consensus (Kunsch et al. 1992) with similar affinities and with the highest affinities, consistent with previous evidence that the DNA-contact residues of c-Rel and p65 are identical (Chen and Ghosh 1999; Huang et al. 2001). Thus, unlike other transcription factor families that use DNA recognition differences to regulate distinct sets of genes, the preferred binding sites for c-Rel and p65 homodimers do not differ. Instead, the functionally important difference appears to be the range of sequences than can be recognized with high affinity.
The first evidence that c-Rel homodimers could recognize a broader range of sequences than do p65 homodimers was provided by the binding site selection studies of Kunsch et al. (1992). Although a few sequences were identified in that study that bound c-Rel homodimers but not p65 homodimers, the resulting consensus sequences for c-Rel and p65 homodimers were very similar, and the functional relevance of the differences observed was difficult to determine. The results shown here demonstrate that c-Rel's ability to recognize a broader range of sequences is due to a maximum of 46 residues within the RHR and is responsible for the c-Rel-dependent expression of Il12b and Il12a.
It is noteworthy that our attempts to further localize the residues responsible for c-Rel selectivity were unsuccessful. This suggests that multiple residues contribute to the unique three-dimensional structure of the c-Rel RHR that allows it to recognize a broad range of sequences with high affinity. Although a structural explanation for the affinity differences remains to be determined, it is interesting to note that p65 homodimers (and presumably c-Rel homodimers) interact strongly with two G/C base pairs at each end of a consensus NF-κB recognition sequence (Chen and Ghosh 1999). c-Rel homodimers can also bind with high affinity to the CD28 response element, which lacks one G/C base pair at one end, and to the Il12b proximal element, which contains no G/C base pairs within one of its half-sites. The affinity of p65 homodimers for this Il12b site is ~40-fold lower. These observations lead us to propose that the 46 key residues of c-Rel lead to greater flexibility within the DNA-binding domain, allowing the DNA-binding domain of one c-Rel subunit to adapt more effectively to the nonconsensus half-site after the other subunit provides an anchor at the consensus half-site. The selectivity region threads through the entire N-terminal portion of the RHR and includes the extended L2 loop, which contains two α-helices (Huang et al. 2001). This extended loop approaches the DNA from the minor groove and contains two residues that contact the phosphate backbone to clamp the DNA.
The affinity differences observed in this analysis were apparent only when p65, c-Rel, and p65/N3,4 homodimers were compared. In contrast, the relative affinities of the p50/p65, p50/c-Rel, and p50/p65/N3,4 heterodimers for the proximal Il12b site appeared to be indistinguishable. These data support the hypothesis that c-Rel selectivity is due to differences between c-Rel homodimers and p65 homodimers. Additional evidence that c-Rel homodimers are important for Il12b activation is provided by studies of p50-/-p52-/- mice, which express Il12b mRNA at wild-type levels (Franzoso et al. 1997). The reason p50/c-Rel heterodimers do not appear to bind DNA with a higher affinity than p50/p65 heterodimers is not known. However, an attractive hypothesis is that the p50 subunit of these heterodimers binds its half-site much more tightly than the c-Rel or p65 subunit at the other half-site, thereby masking the differences between p65 and c-Rel.
Although our results provide strong evidence that the c-Rel requirement is due to preferential binding of c-Rel homodimers, the DNA sequence that is responsible for the c-Rel requirement at the Il12b locus remains unknown. One possibility is that a c-Rel homodimer binds to the proximal NF-κB site in the promoter. This model is attractive because c-Rel homodimers bind this site with a much higher affinity than p65 homodimers. Furthermore, this site is important for promoter activity in both transient and stable transfection assays (Murphy et al. 1995; Plevy et al. 1997). However, EMSA experiments previously suggested that p50/p65 and p50/c-Rel heterodimers bind this site more tightly than either c-Rel homodimers or p65 homodimers (Sanjabi et al. 2000). One possibility is that p50/p65 or p50/c-Rel heterodimers are recruited (perhaps redundantly) to the promoter and then exchange with c-Rel homodimers prior to transcription initiation. This model is consistent with the evidence that both p65 and c-Rel can bind the promoter in ChIP experiments, although a ChIP assay cannot address functional relevance.
A second possibility is that c-Rel homodimers bind the distal NF-κB site in the Il12b promoter. This hypothesis is attractive because the distal site is homologous to the CD28 response element that requires c-Rel for function (Kontgen et al. 1995). In addition, this site is likely to bind p50/p65 and p50/c-Rel heterodimers with relatively low affinities, due to the absence of the third G/C base pair in the distal half-site (Kunsch et al. 1992; Ghosh et al. 1995). However, c-Rel homodimers bound this site with a lower affinity than the Il12b proximal site. Furthermore, it seems unlikely that c-Rel homodimers and p50/p65 or p50/c-Rel heterodimers can simultaneously occupy the distal and proximal sites because of their close proximity. Thus, regardless of whether the c-Rel homodimer binds the proximal or distal site, the possibility of sequential rather than simultaneous binding of heterodimers and the c-Rel homodimer must be considered. We must also consider the possibility that the critical c-Rel homodimer binds elsewhere in the promoter, in a distant enhancer, or acts indirectly to regulate Il12b transcription.
One surprising finding in our analysis was that the transactivation domains of p65 and c-Rel were interchangeable. Given that several coactivators have been reported to interact with p65 and c-Rel (Paal et al. 1997; Sheppard et al. 1999; Zhong et al. 2002), critical c-Rel-specific coactivator interactions were expected. These results suggest that c-Rel and p65 interact with the same coactivators or that the p65 and c-Rel coactivators are functionally interchangeable. We must also consider the possibility that nonphysiological features of the retroviral reconstitution assay may have caused us to overlook additional contributions to selectivity. According to our calculations (Fig. 1), retrovirus-derived c-Rel was, on average, overexpressed by approximately threefold relative to endogenous c-Rel in wild-type cells. Perhaps, this threefold overexpression masked requirements for specific coactivators. In addition, it is important to emphasize that the threefold overexpression represents an average value within a transduced population. Some cells were likely to have overexpressed c-Rel to a much greater extent, which may have masked the requirement for specific coactivators but not the requirement for high-affinity DNA binding.
The DNA affinity results suggest that p65 homodimers may not bind preferentially to any DNA sequences. We propose that p65 homodimers will preferentially bind DNA only when c-Rel is not present in a cell or when p65 homodimer binding is preferentially stabilized by interactions with adjacent DNA-binding proteins. Preferential transactivation by p65 complexes could be due to selective interactions with coactivators.
Although Il12b transcription depends strongly on c-Rel in LPS-stimulated macrophages, it does not depend on c-Rel in LPS-stimulated dendritic cells (Grumont et al. 2001) or in macrophages infected with Toxoplasma gondi (Mason et al. 2002). The reason for this differential requirement is unknown, but it can be hypothesized that other transcription factors expressed in LPS-stimulated dendritic cells and T. gondi-infected macrophages allow Il12b activation in the absence of c-Rel homodimers.
Finally, we were surprised to find that the c-Rel requirement for Il12b expression was quite unusual in LPS-stimulated macrophages. Although we anticipate that other c-Rel-dependent genes were missed by the microarrays, the actual number of c-Rel-dependent, LPS-induced genes is likely to be small. On the basis of these results, we anticipate that the c-Rel requirement for Il12a expression in LPS-stimulated dendritic cells is also likely to be unusual. Therefore, it may be appropriate to speculate that the strong and unusual requirement for c-Rel for production of biologically active IL-12 p70 in macrophages and dendritic cells is biologically meaningful.
Materials and methods
Primary bone marrow-derived macrophages
Bone marrow was isolated from wild-type and c-Rel-/- mice. Red blood cells were lysed in ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM EDTA at pH 7.3), and the remaining cells were washed twice with PBS. Cells (106) were plated on 10-cm plates in 5 mL of BM-medium (DMEM [GIBCO] supplemented with 20% low-endotoxin fetal bovine serum [Omega Scientific], 30% L929-cell conditioned medium, 1% L-glutamine, 1% Pen/Strep, 0.5% Na Pyruvate, 0.1% β-mercaptoethanol). The cells were fed with 2.5 mL of fresh medium every 2 d for up to 7 d. After the cells reached 70%-90% confluence, the nonadherent cells were washed away, and the adherent bone marrow-derived macrophages were used.
Retroviral constructs and production of retroviruses
All expression constructs were prepared in the MSCV-derived MIG vector, which contains a cDNA insertion site followed by an IRES and GFP coding sequences. Murine c-Rel and p65 cDNAs with 5′ sequences encoding a Flag epitope were amplified by PCR with BclII and EcoRI sites at the 5′ and 3′ ends, respectively. The PCR products were inserted into MIG cleaved with BclII and EcoRI. Retroviral plasmids for expression of c-Rel/p65 chimeras were generated by a two-step PCR sewing protocol. All constructs were verified by DNA sequencing.
HEK 293T cells were transfected with the retroviral plasmids and the helper plasmid pθeco (4 μg of each DNA) on 10-cm plates using a calcium phosphate method. Supernatants (5 mL) containing virions were collected 36, 48, and 60 h post-transfection. Viral titers were calculated by infecting NIH 3T3 cells with serial dilutions of the virus and determining the number of GFP positive cells by flow cytometry.
Retroviral transductions
On day 0, bone marrow cells were isolated and 106 cells/well were seeded on six-well plates in BM-medium for differentiation into macrophages. Spin infections of the cells was performed for three consecutive days. On days 1-3, the cells were first spun at 2500 rpm for 5 min, the medium was removed, and the retroviral supernatants were added with 8 μg/mL polybrene and 10 μL/mL of 1M HEPES (pH 7.55). Spin-infections continued for 1.5 h at 2500 rpm and 30°C, followed by gentle removal of the virus-containing medium and addition of fresh BM-medium. On day 5, the medium and the nonadherent cells were removed, and either fresh medium (unactivated) or medium containing 100 ng/mL LPS (Sigma) and 10 U/mL of recombinant murine IFN-γ (PharMingen) was added. On day 6, 20-24 h after activation, the supernatant was removed and secreted IL-12 p40/p70 levels measured by sandwich ELISA (BD-Pharmingen) as described (Sanjabi et al. 2000).
For differentiation into dendritic cells and retroviral transduction, 106 bone marrow cells/well were seeded on 24-well plates in DC-medium (RPMI-1640 [GIBCO-BRL] supplemented with 5% fetal bovine serum, 5% Pen/Strep, 0.1% β-mercaptoethanol, and 10 ng/mL of recombinant purified mouse GM-CSF and IL-4 [R&D Systems]). Spin infections were carried out for three consecutive days as described above. Dendritic cell clusters were harvested after 6 d of culture and transferred to new culture plates prior to activation with LPS (1 μg/mL).
Western blots
Cells were harvested by removing medium and adding ice-cold TEN buffer (0.04 M Tris-HCl at pH 7.4, 1 mM EDTA, 0.15 M NaCl). After incubating on ice for 5-10 min, macrophages were detached from the plates by gentle pipetting and were then washed in PBS. One-fifth of the cells were analyzed by flow cytometry to determine the percentage of GFP-positive cells. The remaining cells were resuspended in 100 μL sample buffer (2.86% SDS, 285 mM Tris at pH 6.8). After heating at 80°C-90°C for 5 min, protein were separated on a 10% SDS-polyacrylamide gel, followed by Western blot analysis using anti-Flag (Sigma M2 Monoclonal Ab, 1:1000 dilution), anti-c-Rel (Santa Cruz c-Rel[C], 1:10,000), and anti-p65 (Santa Cruz p65[C-20], 1:20,000) antibodies.
RT-PCR
RNA was extracted from macrophages using TRI-reagent (MRC). RNA was treated with RNase-free DNase I and purified by phenol-chloroform extraction, followed by ethanol precipitation. Reverse transcription was performed with random hexamers or the following specific primers: Il12b, 5′-CTTGAT GTTGAACTTCAAGTCC-3′; Gapd, 5′-ATTTCTCGTGGTTC ACAC-3′. cDNAs were analyzed by PCR using the following primers: Il12b, 5′-TTTCTTTGCACCAGCCATGAGC-3′; Gapd, 5′-TCCAGTATGACTCCACTC-3′ coupled with the primers used for reverse transcription. Primers for analysis of other c-Rel-dependent genes were as follows: Lox forward, 5′-TCTCAAG TAAGGCATCTCCTC-3′; Lox reverse, 5′-CAGTCTGGCTCA GTATTCATA-3′; Gfi1 forward, 5′-ACAGCTATCACCAGCC GCGTT-3′; Gfi1 reverse, 5′-GCTGCACTCCCGATAGCTCTG-3′; Il4i1 forward, 5′-CTTGGTCTGGCAGGCTCTCTG-3′; Il4i1 reverse 5′-CATCATACTGTGTGAACTGAG-3′.
Nuclear extracts, gel shift assays, and ChIP
HEK 293T cells were transfected with Rel expression plasmids in the pcDNA3 (Invitrogen) vector. Flag-p65 or Flag-c-Rel expression plasmids (1-10 μg) were cotransfected with a limiting amount (20 ng) of a murine p50 expression plasmid. After 48 h, cells were washed with PBS and incubated for 15 min on ice in 400 μL cold buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 4 μg/mL Leupeptin, 1 μM Pepstatin A, 1 μg/mL Aprotinin). Ten percent NP-40 (25 μL) was then added, and samples were mixed vigorously for 10 sec. Nuclei were pelleted by centrifugation, and supernatant (cytoplasmic extract) was saved. The nuclei were suspended in 200 μL cold buffer C (20 mM HEPES at pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 1 mM DTT, 1 mM PMSF, 4 μg/mL Leupeptin, 1 μM Pepstatin A, 1 μg/mL Aprotinin), subjected to periodic agitation for 20 min, and then pelleted by centrifugation for 5 min at 4°C. Extracts with equal amounts of Flag-tagged proteins were used for gel shift analysis as described previously (Plevy et al. 1997; Sanjabi et al. 2000). Supershift analyses were performed using anti-Flag (Sigma M2, 2 μL) and anti-p50 (Santa Cruz p50[D-17], 2 μL) antibodies. ChIP experiments were performed with bone marrow macrophages stimulated with LPS + IFN-γ as previously described (Saccani et al. 2001).
Microarray analysis
Wild-type and c-Rel-/- bone marrow-derived macrophages were left unactivated or were activated for 8 h with 10 μg/mL LPS plus 10 U/mL recombinant murine IFN-γ. Total RNA was isolated using TRI-reagent (MRC) and was further purified using an RNeasy-Qiagen Maxi kit. Quality analysis of total RNA (Agilent), cRNA synthesis, testing of labeling, hybridization, scanning, and data extraction were performed at the University of California Los Angeles Microarray Core Facility. MG-U74Av2 expression probe arrays (Affymetrix, Inc.) were used for gene expression screening. Results were captured and analyzed using GeneChip analysis software (Affymetrix, Inc.).
Stable transfection experiments
The plasmid used for the analysis of Il12b promoter activity by stable transfection will be described elsewhere (L. Zhou, A. Nazarian, and S.T. Smale, in prep.) and was derived from a vector provided by M. Schlissel (University of California, Berkeley). In brief, this reporter plasmid contains an Il12b enhancer upstream of a 400-bp Il12b promoter fragment, which drives expression of an enhanced GFP reporter gene. The reporter cassette is flanked by two pairs of insulator elements from the chicken β-globin locus and a neo resistance gene external to the insulators. Murine J774 macrophages were transfected with linearized reporter plasmids containing the wild-type and mutant Il12b promoters. After electroporation, cells were cultured in 96-well plates in the presence of G418 (GIBCO-BRL, 364 μg/mL). Drug resistant clones were expanded, stimulated with LPS (10 μg/mL) for 24 h, and GFP expression analyzed by flow cytometry.
Purification of recombinant NF-κB proteins and affinity measurements
The murine c-Rel (residues 1-282), p65 (residues 1-314), and p65/N3,4 (residues 1-314) RHRs were inserted into the pET11a expression vector (Novagen). Transformed E. coli cells (BL21[DE3]) were grown to an OD600 of 0.2, induced with 0.1 mM IPTG, and grown for 18 h at 30°C. Recombinant proteins were purified as described (Chen et al. 1999). The Q-Sepharose and SP-Sepharose purifications steps were uncoupled, and purified proteins were concentrated after each step using a 50 K MWCO Macrosep Centrifugal Device (Pall Life Sciences).
The dissociation constants and amounts of active protein in each sample were determined as described previously (Carey and Smale 2000). Binding reactions were performed in the presence of 50 mM Tris-HCL (pH 7.5), 200 mM NaCl, 5 mM DTT, 20% glycerol, 80 ng/μL dI-dC, and 200 ng/μL BSA, with limiting amounts of double-stranded 32P-labeled probes (~10-11 M). Each reaction was incubated at 4°C for 45 min before gel electrophoresis. Data from three independent EMSA titrations were subjected to nonlinear least squares analysis using the Kaleidograph program as described (Cowley and Graves 2000).
Acknowledgments
We thank Barbara Graves for advice on the calculation of dissociation constants; Mark Schlissel and Luk Van Parijs for providing the GFP-insulator and MIG vectors, respectively; Gary Kleiger for help with structural analysis; the UCLA Microarray Core Facility for help with microarray data analysis; and Amer Beg, Sankar Ghosh, Thomas Leung, Einhard Schmidt, and Ranjan Sen for valuable discussions. This work was supported by U.S. P.H.S. grant T32-AI07323, a Warsaw Family Fellowship, a Dorothy Radcliffe Lee Fellowship, and a UCLA Dissertation Year Fellowship to S.S, and U.S. P.H.S. grant T32-CA09120 to K.J.W. S.T.S. is an Investigator with the Howard Hughes Medical Institute.
Notes
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1329805.
References
- Attar R.M., Caamaño, J., Carrasco, D., Iotsova, V., Ishikawa, H., Ryseck, R.-P., Weih, F., and Bravo, R. 1997. Genetic approaches to study Rel/NF-κB/IκB function in mice. Semin. Cancer Biol. 8: 93-101. [Abstract] [Google Scholar]
- Beg A.A. and Baltimore, D. 1996. An essential role for NF-κBin preventing TNF-α-induced cell death. Science 274: 782-784. [Abstract] [Google Scholar]
- Bonizzi G. and Karin, M. 2004. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25: 280-288. [Abstract] [Google Scholar]
- Carey M. and Smale, S.T. 2000. Transcriptional regulation in eukaryotes: Concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Carlsson P. and Mahlapuu, M. 2002. Forkhead transcription factors: Key players in development and metabolism. Dev. Biol. 250: 1-23. [Abstract] [Google Scholar]
- Chen F.E. and Ghosh, G. 1999. Regulation of DNA binding by Rel/NF-κB transcription factors: Structural views. Oncogene 18: 6845-6852. [Abstract] [Google Scholar]
- Chen F.E., Kempiak, S., Huang, D.-B., Phelps, C., and Ghosh, G. 1999. Construction, expression, purification and functional analysis of recombinant NFκB p50/p65 heterodimer. Protein Eng. 12: 423-428. [Abstract] [Google Scholar]
- Chen C., Edelstein, L.C., and Gelinas, C. 2000. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-x(L). Mol. Cell. Biol. 20: 2687-2695. [Europe PMC free article] [Abstract] [Google Scholar]
- Chung J.H., Bell, A.C., and Felsenfeld, G. 1997. Characterization of the chicken β-globin insulator. Proc. Natl. Acad. Sci. 21: 575-580. [Europe PMC free article] [Abstract] [Google Scholar]
- Cowley D.O. and Graves, B.J. 2000. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes & Dev. 14: 366-376. [Europe PMC free article] [Abstract] [Google Scholar]
- Druker B.J., Neumann, M., Okuda, K., Franza Jr., B.R., and Griffin, J.D. 1994. Rel is rapidly tyrosine-phosphorylated following granulocyte-colony stimulating factor treatment of human neutrophils. J. Biol. Chem. 18: 5387-5390. [Abstract] [Google Scholar]
- Fognani C., Rondi, R., Romano, A., and Blasi, F. 2000. cRel-TD kinase: A serine/threonine kinase binding in vivo and in vitro c-Rel and phosphorylating its transactivation domain. Oncogene 27: 2224-2232. [Abstract] [Google Scholar]
- Franzoso G., Carlson, L., Xing, L., Poljak, L., Shores, E.W., Brown, K.D., Leonardi, A., Tran, T., Boyce, B.F., and Siebenlist, U. 1997. Requirement for NF-κB in osteoclast and B-cell development. Genes & Dev. 11: 3482-3496. [Europe PMC free article] [Abstract] [Google Scholar]
- Gerondakis S., Strasser, A., Metcalf, D., Grigoriadis, G., Scheerlinck, J.-P.Y., and Grumont, R.J. 1996. Rel-deficient T cells exhibit defects in production of interleukin 3 and granuloctye-macrophage colony stimulating factor. Proc. Natl. Acad. Sci. 93: 3405-3409. [Europe PMC free article] [Abstract] [Google Scholar]
- Gerondakis S., Grossmann, M., Nakamura, Y., Pohl, T., and Grumont, R. 1999. Genetic approaches in mice to understand Rel/NF-κB and IκB function: Transgenics and knockouts. Oncogene 18: 6888-6895. [Abstract] [Google Scholar]
- Ghosh G., van Duyne, G., Ghosh, S., and Sigler, P.B. 1995. Structure of NF-κB p50 homodimer bound to a κB site. Nature 373: 303-310. [Abstract] [Google Scholar]
- Ghosh S., May, M.J., and Kopp, E.B. 1998. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16: 225-260. [Abstract] [Google Scholar]
- Grigoriadis G., Zhan, Y., Grumont, R.J., Metcalf, D., Handman, E., Cheers, C., and Gerondakis, S. 1996. The Rel subunit of NF-κB-like transcription factors is a positive and negative regulator of macrophage gene expression: Distinct roles for Rel in different macrophage populations. EMBO J. 15: 7099-7107. [Europe PMC free article] [Abstract] [Google Scholar]
- Grumont R., Hochrein, H., O'Keeffe, M., Gugasyan, R., White, C., Caminschi, I., Cook, W., and Gerondakis, S. 2001. c-Rel regulates interleukin 12 p70 expression in CD8(+) dendritic cells by specifically inducing p35 gene transcription. J. Exp. Med. 194: 1021-1032. [Europe PMC free article] [Abstract] [Google Scholar]
- Hayden M.S. and Ghosh, S. 2004. Signaling to NF-κB. Genes & Dev. 18: 2195-2224. [Abstract] [Google Scholar]
- Himes S.R., Coles, L.S., Reeves, R., and Shannon, M.F. 1996. High mobility group I(Y) is required for function and for c-Rel binding to CD28 response elements within the GM-CSF and IL-2 promoters. Immunity 5: 479-489. [Abstract] [Google Scholar]
- Hoffmann A., Leung, T.H., and Baltimore, D. 2003. Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. EMBO J. 22: 5530-5539. [Europe PMC free article] [Abstract] [Google Scholar]
- Hollenhorst P.C., Jones, D.A., and Graves, B.J. 2004. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 32: 5693-5702. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang D.B., Chen, Y.Q., Ruetsche, M., Phelps, C.B., and Ghosh, G. 2001. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure 9: 669-678. [Abstract] [Google Scholar]
- Karin M. and Ben-Neriah, Y. 2000. Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol. 18: 621-663. [Abstract] [Google Scholar]
- Kontgen F., Grumont, R.J., Strasser, A., Metcalf, D., Li, R., Tarlinton, D., and Gerondakis, S. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes & Dev. 9: 1965-1977. [Abstract] [Google Scholar]
- Kunsch C., Ruben, S.M., and Rosen, C.A. 1992. Selection of optimal κ B/Rel DNA-binding motifs: Interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol. 12: 4412-4421. [Europe PMC free article] [Abstract] [Google Scholar]
- Leung T.H., Hoffmann, A., and Baltimore, D. 2004. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 20: 453-464. [Abstract] [Google Scholar]
- Li Q. and Verma, I.M. 2002. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2: 725-734. [Abstract] [Google Scholar]
- Martin A.G. and Fresno, M. 2000. Tumor necrosis factor-α activation of NF-κB requires the phosphorylation of Ser-471 in the transactivation domain of c-Rel. J. Biol. Chem. 275: 24383-24391. [Abstract] [Google Scholar]
- Mason N., Aliberti, J., Caamano, J.C., Liou, H.C., and Hunter, C.A. 2002. Cutting edge: Identification of c-Rel-dependent and -independent pathways of IL-12 production during infectious and inflammatory stimuli. J. Immunol. 15: 2590-2594. [Abstract] [Google Scholar]
- Muljo S.A. and Schlissel, M.S. 2002. The variable, C(H)1, C(H)2, and C(H)3 domains of Ig heavy chain are dispensable for pre-BCR function in transgenic mice. Int. Immunol. 14: 577-584. [Abstract] [Google Scholar]
- Murphy T.L., Cleveland, M.G., Kulesza, P., Magram, J., and Murphy, K.M. 1995. Regulation of interleukin 12 p40 expression through an NF-κB half-site. Mol. Cell. Biol. 15: 5258-5267. [Europe PMC free article] [Abstract] [Google Scholar]
- Ouaaz F., Arron, J., Zheng, Y., Choi, Y., and Beg, A.A. 2002. Dendritic cell development and survival require distinct NF-κB subunits. Immunity 16: 257-270. [Abstract] [Google Scholar]
- Paal K., Baeuerle, P.A., and Schmitz, M.L. 1997. Basal transcription factors TBP and TFIIB and the viral coactivator E1A 13S bind with distinct affinities and kinetics to the transactivation domain of NF-κB p65. Nucleic Acids Res. 25: 1050-1055. [Europe PMC free article] [Abstract] [Google Scholar]
- Paxton L.L., Li, L.J., Secor, V., Duff, J.L., Naik, S.M., Shibagaki, N., and Caughman, S.W. 1997. Flanking sequences for the human intercellular adhesion molecule-1 NF-κB response element are necessary for tumor necrosis factor α-induced gene expression. J. Biol. Chem. 272: 15928-15935. [Abstract] [Google Scholar]
- Plevy S.E., Gemberling, J.H., Hsu, S., Dorner, A.J., and Smale, S.T. 1997. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: Evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 17: 4572-4588. [Europe PMC free article] [Abstract] [Google Scholar]
- Pomerantz J.L. and Baltimore, D. 2002. Two pathways to NF-κB. Mol. Cell 10: 693-695. [Abstract] [Google Scholar]
- Rastinejad F. 2001. Retinoid X receptor and its partners in the nuclear receptor family. Curr. Opin. Struct. Biol. 11: 33-38. [Abstract] [Google Scholar]
- Saccani S., Pantano, S., and Natoli, G. 2001. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 193: 1351-1359. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanjabi S., Hoffmann, A., Liou, H.C., Baltimore, D., and Smale, S.T. 2000. Selective requirement for c-Rel during IL-12 p40 gene induction in macrophages. Proc. Natl. Acad. Sci. 97: 12705-12710. [Europe PMC free article] [Abstract] [Google Scholar]
- Shapiro V.S., Truitt, K.E., Imboden, J.B., and Weiss, A. 1997. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol. Cell. Biol. 17: 4051-4058. [Europe PMC free article] [Abstract] [Google Scholar]
- Sheppard K.A., Rose, D.W., Haque, Z.K., Kurokawa, R., McInerney, E., Westin, S., Thanos, D., Rosenfeld, M.G., Glass, C.K., and Collins, T. 1999. Transcriptional activation by NF-κB requires multiple coactivators. Mol. Cell. Biol. 19: 6367-6378. [Europe PMC free article] [Abstract] [Google Scholar]
- Siebenlist U., Franzoso, G., and Brown, K. 1994. Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 10: 405-455. [Abstract] [Google Scholar]
- Trinchieri G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3: 133-146. [Abstract] [Google Scholar]
- Zhang G. and Ghosh, S. 2001. Toll-like receptor-mediated NF-κB activation: A phylogenetically conserved paradigm in innate immunity. J. Clin. Invest. 107: 13-19. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong H., May, M.J., Jimi, E., and Ghosh, S. 2002. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9: 625-636. [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.1329805
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/19/18/2138.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101916314
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gad.1329805
Article citations
Transcriptional repression by HDAC3 mediates T cell exclusion from <i>Kras</i> mutant lung tumors.
Proc Natl Acad Sci U S A, 121(42):e2317694121, 10 Oct 2024
Cited by: 1 article | PMID: 39388266
Selective regulation of a defined subset of inflammatory and immunoregulatory genes by an NF-κB p50-IκBζ pathway.
Genes Dev, 38(11-12):536-553, 19 Jul 2024
Cited by: 4 articles | PMID: 38918046 | PMCID: PMC11293394
Molecular definition of the endogenous Toll-like receptor signalling pathways.
Nature, 631(8021):635-644, 03 Jul 2024
Cited by: 3 articles | PMID: 38961291
Transient interactions modulate the affinity of NF-κB transcription factors for DNA.
Proc Natl Acad Sci U S A, 121(23):e2405555121, 28 May 2024
Cited by: 0 articles | PMID: 38805268
Non-canonical IKB kinases regulate YAP/TAZ and pathological vascular remodeling behaviors in pulmonary artery smooth muscle cells.
Physiol Rep, 12(7):e15999, 01 Apr 2024
Cited by: 1 article | PMID: 38610069 | PMCID: PMC11014870
Go to all (93) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Nature, 391(6665):410-413, 01 Jan 1998
Cited by: 376 articles | PMID: 9450761
Divergent C-terminal transactivation domains of Rel/NF-kappa B proteins are critical determinants of their oncogenic potential in lymphocytes.
Oncogene, 23(5):1030-1042, 01 Feb 2004
Cited by: 20 articles | PMID: 14647412
X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2.
Structure, 9(8):669-678, 01 Aug 2001
Cited by: 63 articles | PMID: 11587641
The structure of the NF-kappa B p50:DNA-complex: a starting point for analyzing the Rel family.
FEBS Lett, 369(1):113-117, 01 Aug 1995
Cited by: 22 articles | PMID: 7641872
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: T32 CA009120
Grant ID: T32-CA09120
NIAID NIH HHS (2)
Grant ID: T32-AI07323
Grant ID: T32 AI007323




