Abstract
Free full text

Comparison of Identification Systems for Classification of Bacteria Isolated from Water and Endolithic Habitats within the Deep Subsurface
Abstract
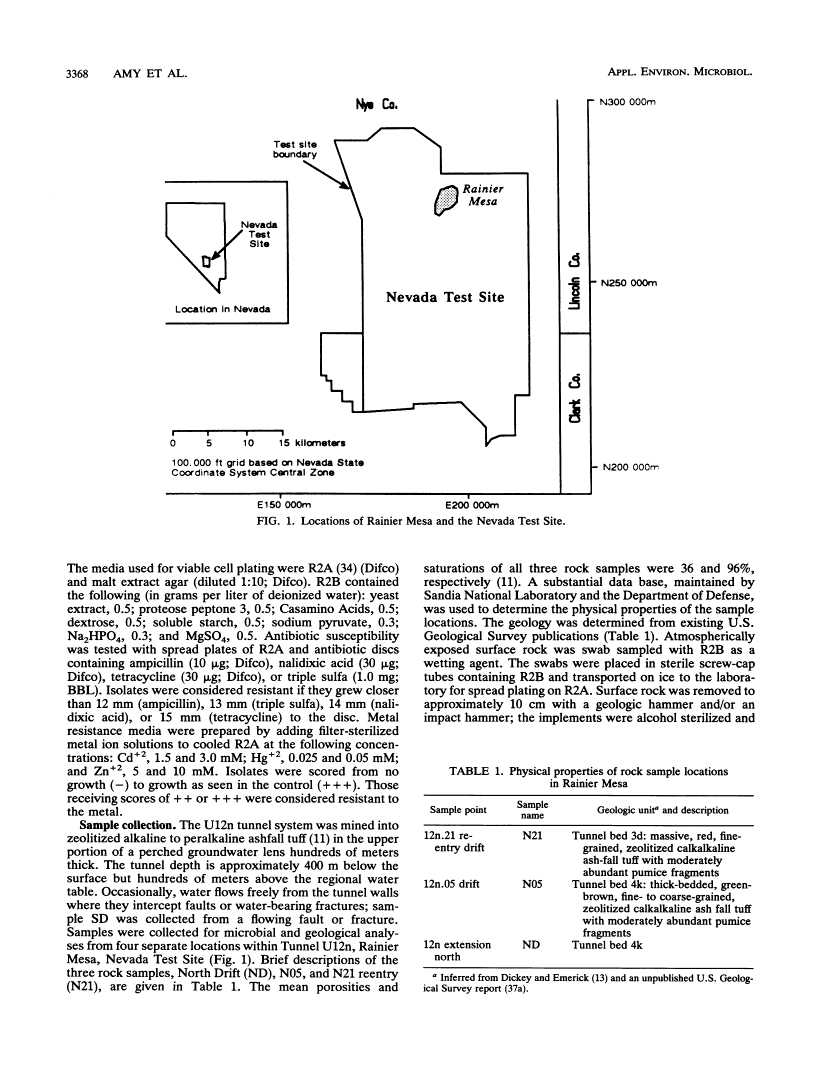
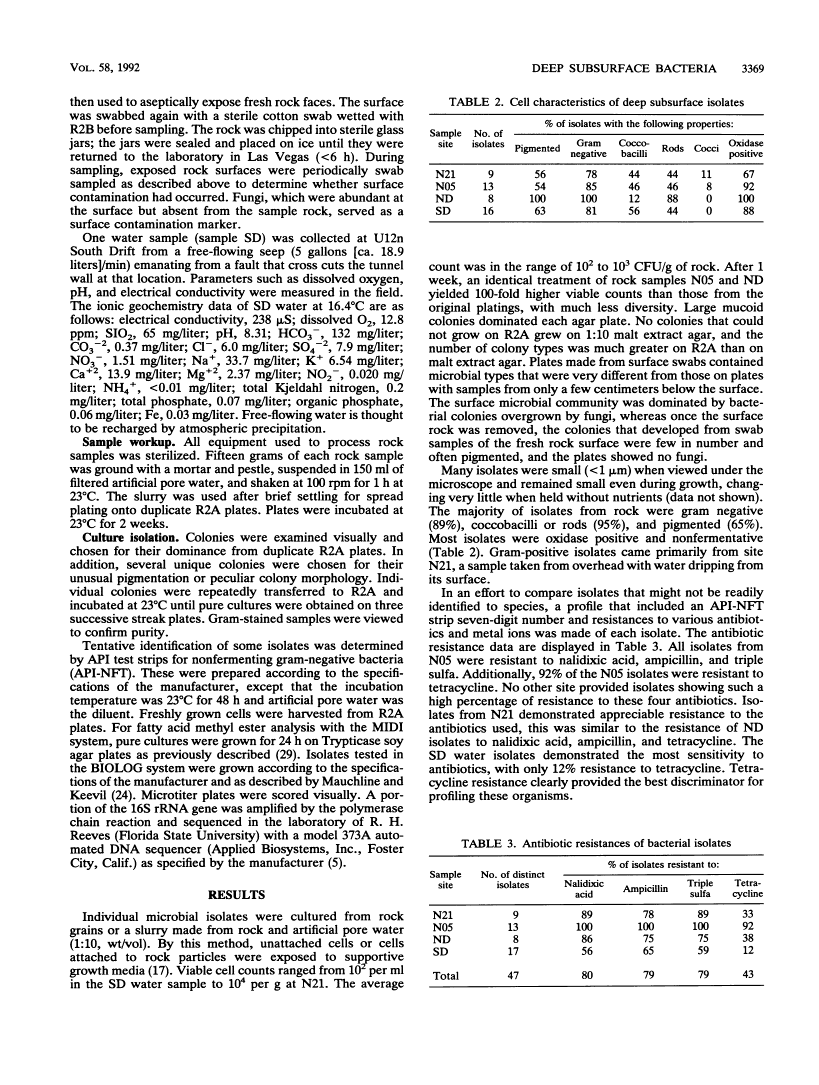
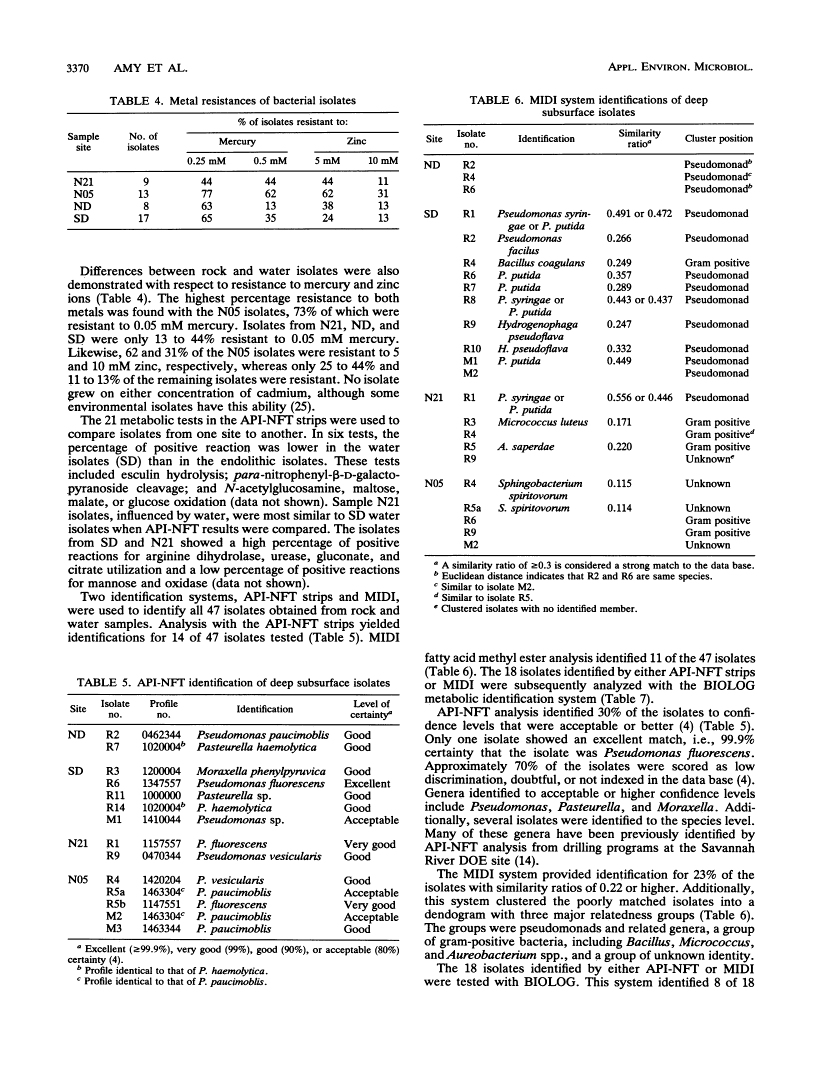
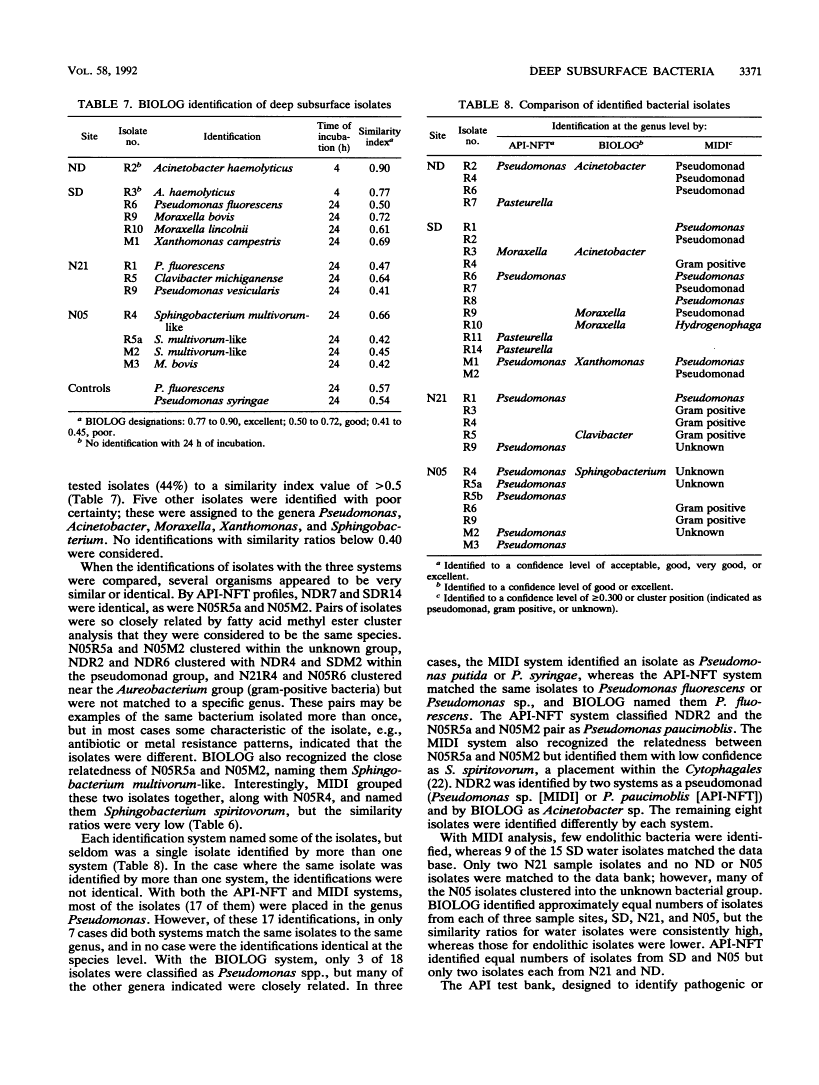
One water and three rock samples were taken from a mined tunnel system, U12n, in Rainier Mesa at the Nevada Test Site. Endolithic microorganisms were cultured from ashfall tuff, which was crushed and made into slurries with a formulation of artificial pore water, on R2A agar plates. Microbial counts ranged from 102 to 104 viable cells per g (dry weight) of rock sampled. The cultured water sample yielded 102 viable cells per ml. Many of the isolates were very small (<1 μm) when viewed in the rock matrix and remained small even when cultured. Most were gram-negative rods. Individual isolates were profiled by API-NFT strip number, antibiotic and metal resistance patterns, and colony and cellular morphologies. Three identification systems, API-NFT strips, BIOLOG, and MIDI, were compared. Each system identified only a small percentage of the total isolates, and in only seven cases were the isolates identified the same way by more than one system. The same genus was identified in three of these cases, but different species were indicated. The genus Pseudomonas was the most commonly identified. The isolate profiles and the three identification systems demonstrated that water isolates were considerably different from endolithic isolates.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.3M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy PS, Morita RY. Starvation-survival patterns of sixteen freshly isolated open-ocean bacteria. Appl Environ Microbiol. 1983 Mar;45(3):1109–1115. [Europe PMC free article] [Abstract] [Google Scholar]
- Amy PS, Pauling C, Morita RY. Starvation-survival processes of a marine Vibrio. Appl Environ Microbiol. 1983 Mar;45(3):1041–1048. [Europe PMC free article] [Abstract] [Google Scholar]
- Amy PS, Pauling C, Morita RY. Recovery from nutrient starvation by a marine Vibrio sp. Appl Environ Microbiol. 1983 May;45(5):1685–1690. [Europe PMC free article] [Abstract] [Google Scholar]
- Balkwill DL, Fredrickson JK, Thomas JM. Vertical and horizontal variations in the physiological diversity of the aerobic chemoheterotrophic bacterial microflora in deep southeast coastal plain subsurface sediments. Appl Environ Microbiol. 1989 May;55(5):1058–1065. [Europe PMC free article] [Abstract] [Google Scholar]
- Balkwill DL, Ghiorse WC. Characterization of subsurface bacteria associated with two shallow aquifers in oklahoma. Appl Environ Microbiol. 1985 Sep;50(3):580–588. [Europe PMC free article] [Abstract] [Google Scholar]
- Fredrickson JK, Balkwill DL, Zachara JM, Li SM, Brockman FJ, Simmons MA. Physiological diversity and distributions of heterotrophic bacteria in deep cretaceous sediments of the atlantic coastal plain. Appl Environ Microbiol. 1991 Feb;57(2):402–411. [Europe PMC free article] [Abstract] [Google Scholar]
- Geesey GG, Morita RY. Capture of arginine at low concentrations by a marine psychrophilic bacterium. Appl Environ Microbiol. 1979 Dec;38(6):1092–1097. [Europe PMC free article] [Abstract] [Google Scholar]
- Harvey RW, Smith RL, George L. Effect of organic contamination upon microbial distributions and heterotrophic uptake in a Cape Cod, Mass., aquifer. Appl Environ Microbiol. 1984 Dec;48(6):1197–1202. [Europe PMC free article] [Abstract] [Google Scholar]
- Iacobellis NS, Devay JE. Long-term storage of plant-pathogenic bacteria in sterile distilled water. Appl Environ Microbiol. 1986 Aug;52(2):388–389. [Europe PMC free article] [Abstract] [Google Scholar]
- Jiménez L. Molecular analysis of deep-subsurface bacteria. Appl Environ Microbiol. 1990 Jul;56(7):2108–2113. [Europe PMC free article] [Abstract] [Google Scholar]
- Kurath G, Morita RY. Starvation-Survival Physiological Studies of a Marine Pseudomonas sp. Appl Environ Microbiol. 1983 Apr;45(4):1206–1211. [Europe PMC free article] [Abstract] [Google Scholar]
- Macdonell MT, Hood MA. Isolation and characterization of ultramicrobacteria from a gulf coast estuary. Appl Environ Microbiol. 1982 Mar;43(3):566–571. [Europe PMC free article] [Abstract] [Google Scholar]
- Mauchline WS, Keevil CW. Development of the BIOLOG substrate utilization system for identification of Legionella spp. Appl Environ Microbiol. 1991 Nov;57(11):3345–3349. [Europe PMC free article] [Abstract] [Google Scholar]
- Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. [Europe PMC free article] [Abstract] [Google Scholar]
- Osterhout GJ, Shull VH, Dick JD. Identification of clinical isolates of gram-negative nonfermentative bacteria by an automated cellular fatty acid identification system. J Clin Microbiol. 1991 Sep;29(9):1822–1830. [Europe PMC free article] [Abstract] [Google Scholar]
- Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985 Jan;49(1):1–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Torrella F, Morita RY. Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl Environ Microbiol. 1981 Feb;41(2):518–527. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.58.10.3367-3373.1992
Read article for free, from open access legal sources, via Unpaywall:
https://aem.asm.org/content/aem/58/10/3367.full.pdf
Free after 4 months at aem.asm.org
http://aem.asm.org/cgi/reprint/58/10/3367
Free to read at aem.asm.org
http://aem.asm.org/cgi/content/abstract/58/10/3367
Citations & impact
Impact metrics
Citations of article over time
Article citations
A glimpse of the paleome in endolithic microbial communities.
Microbiome, 11(1):210, 25 Sep 2023
Cited by: 0 articles | PMID: 37749660 | PMCID: PMC10518947
From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada).
Microorganisms, 10(1):129, 08 Jan 2022
Cited by: 4 articles | PMID: 35056578 | PMCID: PMC8781179
Thermoanaerosceptrum fracticalcis gen. nov. sp. nov., a Novel Fumarate-Fermenting Microorganism From a Deep Fractured Carbonate Aquifer of the US Great Basin.
Front Microbiol, 10:2224, 27 Sep 2019
Cited by: 2 articles | PMID: 31611860 | PMCID: PMC6776889
Microbial distribution and turnover in Antarctic microbial mats highlight the relevance of heterotrophic bacteria in low-nutrient environments.
FEMS Microbiol Ecol, 94(9):fiy129, 01 Sep 2018
Cited by: 9 articles | PMID: 29982398
Microbial mobilization of plutonium and other actinides from contaminated soil.
J Environ Radioact, 150:277-285, 25 Sep 2015
Cited by: 5 articles | PMID: 26406590
Go to all (35) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Bacterial heterogeneity in deep subsurface tunnels at Rainier Mesa, Nevada test site.
Microb Ecol, 25(2):183-194, 01 Mar 1993
Cited by: 14 articles | PMID: 24189814
Phenotypic characterization and 16S rDNA identification of culturable non-obligate halophilic bacterial communities from a hypersaline lake, La Sal del Rey, in extreme South Texas (USA).
Aquat Biosyst, 8(1):5, 02 Feb 2012
Cited by: 10 articles | PMID: 22480362 | PMCID: PMC3310331
Changes in Bacteria Recoverable from Subsurface Volcanic Rock Samples during Storage at 4 degrees C.
Appl Environ Microbiol, 60(8):2697-2703, 01 Aug 1994
Cited by: 8 articles | PMID: 16349343 | PMCID: PMC201711
Resuscitation of microorganisms after gamma irradiation.
Radiat Res, 152(1):71-75, 01 Jul 1999
Cited by: 4 articles | PMID: 10381843