Abstract
Free full text

SRp54 (SFRS11), a Regulator for tau Exon 10 Alternative Splicing Identified by an Expression Cloning Strategy
Abstract
The tau gene encodes a microtubule-associated protein that is critical for neuronal survival and function. Splicing defects in the human tau gene lead to frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17), an autosomal dominant neurodegenerative disorder. Genetic mutations associated with FTDP-17 often affect tau exon 10 alternative splicing. To investigate mechanisms regulating tau exon 10 alternative splicing, we have developed a green fluorescent protein reporter for tau exon 10 skipping and an expression cloning strategy to identify splicing regulators. A role for SRp54 (also named SFRS11) as a tau exon 10 splicing repressor has been uncovered using this strategy. The overexpression of SRp54 suppresses tau exon 10 inclusion. RNA interference-mediated knock-down of SRp54 increases exon 10 inclusion. SRp54 interacts with a purine-rich element in exon 10 and antagonizes Tra2β, an SR-domain-containing protein that enhances exon 10 inclusion. Deletion of this exonic element eliminates the activity of SRp54 in suppressing exon 10 inclusion. Our data support a role of SRp54 in regulating tau exon 10 splicing. These experiments also establish a generally useful approach for identifying trans-acting regulators of alternative splicing by expression cloning.
Tauopathy is characterized by the formation of filamentous tau protein-containing inclusions in the affected brain regions. This is a group of genetically and phenotypically heterogeneous diseases, including Alzheimer's disease, Down's syndrome, several variants of prion diseases, progressive supranuclear palsy, amyotrophic lateral sclerosis, Pick's disease, corticobasal degeneration, and frontotemporal dementia (FTD) (reviewed in references 14 and 29). Mutations in the tau gene cause frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17), an autosomal dominant neurodegenerative disorder. FTDP-17 is characterized by behavioral and personality changes as well as muscular rigidity similar to that found in Parkinson's disease.
Located on chromosome 17, the human tau gene encodes the microtubule-associated protein tau (33). The tau protein is critical for microtubule assembly and stabilization, playing an important role in axonal growth and neuronal function (reviewed in references 1, 10, 14, 20, 25, 29, 31, and 43). The expression of the tau gene is under complex regulation at multiple levels. In addition to the extensive posttranslational modifications of the tau protein, tau gene expression is regulated at the level of pre-mRNA splicing, the most upstream step of posttranscriptional RNA processing. In the adult human brain, six isoforms of tau proteins are produced as a result of alternative splicing of exons 2 and 3 and exon 10. Alternative pre-mRNA splicing of exon 10, with either exclusion or inclusion of exon 10, leads to the formation of tau proteins containing either three or four microtubule binding repeats (tau3R or tau4R). In normal human adult brain, the ratio of tau4R to tau3R is approximately 1. This delicate balance between tau4R and tau3R isoforms is crucial for neuronal function in maintaining learning and memory.
Two types of tau mutations have been identified in patients with frontotemporal dementia, missense mutations that affect tau protein function and splicing mutations that change the balance of different tau splicing isoforms (reviewed in references 1, 10, 14, 20, 25, 29, 31, and 43). Most missense mutations occur inside or near the microtubule-binding domain and may reduce the ability of tau protein to bind to microtubules. Splicing mutations, including most intronic mutations and some exonic mutations, are associated with changes in the tau4R/tau3R ratio. The identification of these tau splicing mutations in FTDP-17 patients demonstrates the importance of controlling the balance of different tau isoforms by alternative splicing in neuronal function.
Molecular mechanisms underlying tau exon 10 splicing regulation remain to be elucidated. Only a few trans-acting splicing factors have been identified that regulate tau exon 10 splicing in the context of its native splice sites, although several known splicing factors have been tested using tau chimeric splicing cassettes (reviewed in references 1, 10, and 25). Our previous study established a tau minigene system containing tau exons 9, 10, and 11 in their native context. Using this minigene with biochemical approaches, we identified Tra2β, a splicing regulator that binds to the purine-rich exonic splicing enhancer element in exon 10 and stimulates the inclusion of exon 10 (22). In this paper, we report the development of an expression cloning approach that can be generally applicable to identifying trans-acting alternative splicing regulators. Using the expression cloning method, we have identified trans-acting factors that promote tau exon 10 skipping. One of these factors is a protein of the SR family, SRp54 (also named SFRS11). Using biochemical assays and transfection experiments, we have validated the role of SRp54 in suppressing exon 10 inclusion. Overexpression of SRp54 reversed the aberrant splicing of tau pre-mRNA containing an intronic mutation. Furthermore, UV cross-linking and splicing assays indicate that SRp54 suppresses exon 10 inclusion by antagonizing Tra2β.
MATERIALS AND METHODS
Construction of tau minigene and GFP reporter genes.
The tau exon 9-10-11 minigene, containing a single-nucleotide mutation in intron 10 and named the tau exon 9-10-11 DDPAC minigene, was described previously (21). One base pair was inserted by site-directed mutagenesis into exon 10, and the BamHI-EcoRI fragment containing the 94-bp exon 10 and flanking intronic sequences was used to replace the same region in the original tau exon 9-10-11 DDPAC minigene. The cDNA fragment encoding the enhanced green fluorescent protein (GFP) was inserted into the XhoI site to fuse in frame with exon 11. This resulted in the tau3RDDPAC-GFP reporter gene, and its sequence was confirmed by sequence analysis.
Construction of the cDNA library and expression cloning screening.
A human fetal brain cDNA library was prepared using mRNA purchased from Clontech. A mammalian expression ZAP Express cDNA library system (Stratagene) was used to construct the human fetal brain cDNA library. The quality of this cDNA library was tested using reverse transcription-PCR (RT-PCR) and small-scale screenings. The library, representing 1.2 million individual cDNA clones, was subdivided into 400 primary cDNA pools (A1, A2,…A20; B1, B2,..B20;…T1, T2, …T20). Each pool contained approximately 3,000 cDNA clones. Each of these 400 primary pools was amplified for 6 to 8 h at 37°C after being carefully plated on 20- by 20-cm plates. cDNAs were prepared from each pool using the medium-scale plasmid preparation method, combining polyethylene glycol precipitation with an endotoxin-free DNA preparation kit (QIAGEN) to ensure a high quality of DNA preparations. For the first round of transfection, 40 combined primary pools were used for transfecting cells stably expressing the tau-GFP reporter genes. Each combined primary pool contained 20 individual primary pools that were combined in a grid fashion (A1-A20, B1-B20, …T1-T20 in 20 rows; A1-T1, A2-T2,…A20-T20 in 20 columns) (see Fig. Fig.2,2, part I, below). Using this grid design, 400 cDNA pools were tested in 40 transfections in 15-cm tissue culture dishes. Two methods were tested for identifying positive clones: fluorescence-activated cell sorting and visual inspection under an inverted fluorescence microscope. The latter method proved to be more efficient and was used in our screening. Eight positive pools were identified in the primary round of screening, and each pool was subdivided into 20 subpools. DNA was prepared from each pool and rescreened following transfection into reporter-expressing cells in 10-cm dishes or six-well-dishes. This process of redividing and rescreening was repeated for six rounds, at which time single cDNA clones were identified. Out of eight primary positive pools, six individual cDNA clones were isolated after six rounds of subdivision rescreening of cDNA pools.
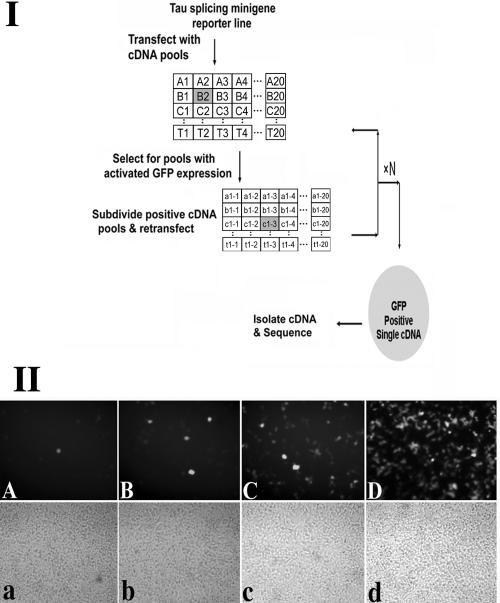
Part I. Flow chart for the cDNA library screening. The detailed procedure is described in Materials and Methods. Part II. Activation of GFP expression from the tau3RDDPAC-GFP reporter during cDNA library screening. The stable HEK cell line expressing the tau3RDDPAC-GFP reporter gene was used for screening the cDNA library. A progressive subdividing-retransfecting method was used to identify cDNAs encoding potential regulators of tau alternative splicing. GFP expression was monitored by fluorescence microscopy following the transfection of pools of cDNAs prepared from the cDNA library. (A to D) GFP expression in cells transfected with the identified positive pools following the first, third, fifth, and sixth rounds of cDNA library subdivision and transfection, demonstrating the progressive enrichment of the positive cDNA(s) that increases GFP expression. (a to d) Corresponding phase-contrast images. After six rounds of retransfection, we were able to isolate single cDNA clones that had significant effects on tau exon 10 splicing.
Transfection, splicing assay, and RT-PCR.
HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and were seeded at 2 × 105 cells/well in a six-well dish 24 h prior to transfection. A modified calcium phosphate precipitation procedure was used for transfection with 1 to 5 μg of DNA as described elsewhere (23). Cells were harvested 48 h after transfection, and the RNA was extracted using the TRIzol reagent (Invitrogen). Splicing products derived from the expressed tau minigenes were detected using RT-PCR as previously described (23). To detect the endogenous tau and SRp54 expression in the human brain, poly(A)+ RNA samples of human fetal and adult brains were purchased from Clontech. RT-PCR was used to detect tau, SRp54, and actin RNAs using the corresponding specific primers.
UV cross-linking, immunoprecipitation assay, and RNA-protein interaction assays.
Uniformly or site-specifically labeled tau pre-mRNA transcripts were prepared by in vitro transcription as described in our previous study (22). Equal amounts of 32P-labeled RNA transcripts, either uniformly labeled or site-specifically labeled, corresponding to different regions of tauEx10-11wt, were incubated for 30 min on ice with in vitro-translated SRp54-Flag or Tra2β-myc proteins. Following UV-cross-linking and then RNase treatment, corresponding proteins were immunoprecipitated using specific antibodies. After immunoprecipitation using a monoclonal anti-Flag antibody or anti-myc antibody, the beads were washed three times in cold phosphate-buffered saline with 20 mM HEPES (pH 7.4), 175 mM NaCl, and 0.1% NP-40. The precipitated proteins were dissolved in sample buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and detected by autoradiography.
RNA interference assay.
An RNA interference assay was carried out in HEK293 cells as described previously (22). The small interfering RNA oligonucleotides (Dharmacon, Lafayette, CO) were transfected at 0.2 μM using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The target site sequence for SRp54 corresponds to the coding region of nucleotides 361 to 380 (GenBank accession number NM_004768).
RESULTS
Construction of a tau-splicing GFP-reporter gene for cloning trans-acting splicing factors regulating tau exon 10 splicing.
To construct a reporter gene for identifying trans-acting splicing factors regulating tau exon 10 splicing, we made use of the human tau minigene model that we previously established (21). In this system, alternative inclusion or exclusion of the 93-bp exon 10 leads to the formation of tau4R and tau3R, respectively, similar to the endogenous human tau gene (21). To develop an expression cloning strategy that allows us to systematically screen for factors repressing tau exon 10 inclusion in an unbiased manner, we utilized the tau minigene containing a single-nucleotide DDPAC mutation in intron 10 (19, 21). The predominant splicing product of this DDPAC minigene is the exon 10 inclusion isoform (exon 10+), tau4R. A tau exon 10 reporter gene, named tau3RDDPAC-GFP, was constructed using this tau minigene and a cDNA fragment encoding GFP. In the tau3RDDPAC-GFP reporter gene, a translation start codon ATG was added upstream of exon 9 so that the tau minigene produced a fusion protein containing the tau peptide fragment fused to GFP (Fig. (Fig.1A).1A). One base pair was inserted by site-directed mutagenesis in exon 10 such that exon 10 and exon 11 were not in the same reading frame when exon 10 was included. This tau3RDDPAC-GFP reporter contained the GFP gene in frame with exon 11. Thus, the GFP gene was expressed as a functional protein product only when exon 10 was skipped (Fig. (Fig.1A).1A). This allowed us to use GFP expression as an indicator of repression of exon 10 splicing.
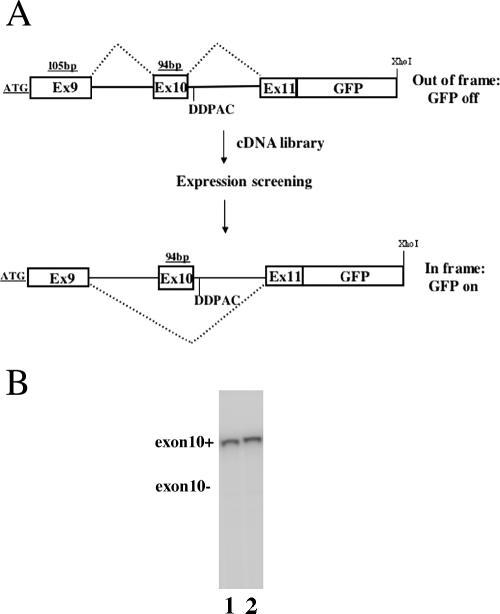
A. A tau exon 10 alternative splicing-dependent GFP reporter gene, tau3RDDPAC-GFP. B. The tau exon 10 splicing patterns of the tau3RDDPAC-GFP reporter gene and the original tau exon 9-10-11 (DDPAC) minigene. Specific RT-PCR was used to detect the tau exon 10 splicing isoforms using cDNAs isolated from cells expressing either the original tau exon 9-10-11 (DDPAC) minigene (lane 1) or tau3RDDPAC-GFP (lane 2). Both constructs produced the same splicing pattern with an almost exclusively tau exon 10+ isoform.
We carefully compared the splicing patterns of the original DDPAC tau minigene and of the tau3RDDPAC-GFP reporter gene containing the 1-bp insertion. The insertion of the 1 bp and GFP-ORF did not affect the minigene splicing or RNA stability. Aberrant splicing product was not detected, such as that produced by the activation of cryptic splice sites (data not shown) (Fig. (Fig.1B).1B). This tau3RDDPAC-GFP reporter gene was transfected into HEK293 cells. By a sensitive radioactive RT-PCR assay, we demonstrated that tau reporter gene splicing produced exclusively the tau exon 10 inclusion isoform, and the tau exon 10-skipping isoform was not detectable, similar to the original tau minigene. This was the same pattern as with the DDPAC mutant tau gene found in the brain tissue of patients with FTDP-17 (21). Consistent with this splicing pattern, the tau-GFP reporter showed a very low background of GFP reporter expression in HEK293 cells, and this facilitated the expression cloning screening described below.
Identification of trans-acting factors repressing tau exon 10 inclusion using expression cloning strategy.
tau3R, the tau isoform lacking exon 10, is the predominant tau splicing isoform produced in the human fetal brain (reviewed in references 1, 10, and 25) (see Fig. Fig.5,5, below), suggesting that factors suppressing the tau exon 10 inclusion are expressed in the fetal brain. Therefore, we used human fetal brain cDNA to prepare a cDNA library for this expression cloning screening. The cDNA library was prepared as described above in Materials and Methods. The library was subdivided into 400 primary cDNA pools. Following limited amplification, pooled cDNAs were prepared from each of these 400 primary pools and used for transfection, as described in Materials and Methods. For the first round of transfection, 40 combined pools of cDNAs were transfected in cells stably expressing the tau3RDDPAC-GFP reporter gene. Four hundred primary cDNA pools were combined in 40 group pools: A1-A20, B1-B20, …T1-T20 in 20 row-pools and A1-T1, A2-T2, …A20-T20 in 20 column-pools (Fig. (Fig.2,2, part I). For example, if GFP expression activation was detected in both row-pool B1-B20 and column-pool A2-T2, it would indicate the presence of positive cDNA(s) in the B2 primary pool, and this pool would be further tested. This grid design allowed us to test 400 primary cDNA pools in 40 transfections (see Materials and Methods). Positive pools were subdivided in a similar fashion and tested in another round of transfection.
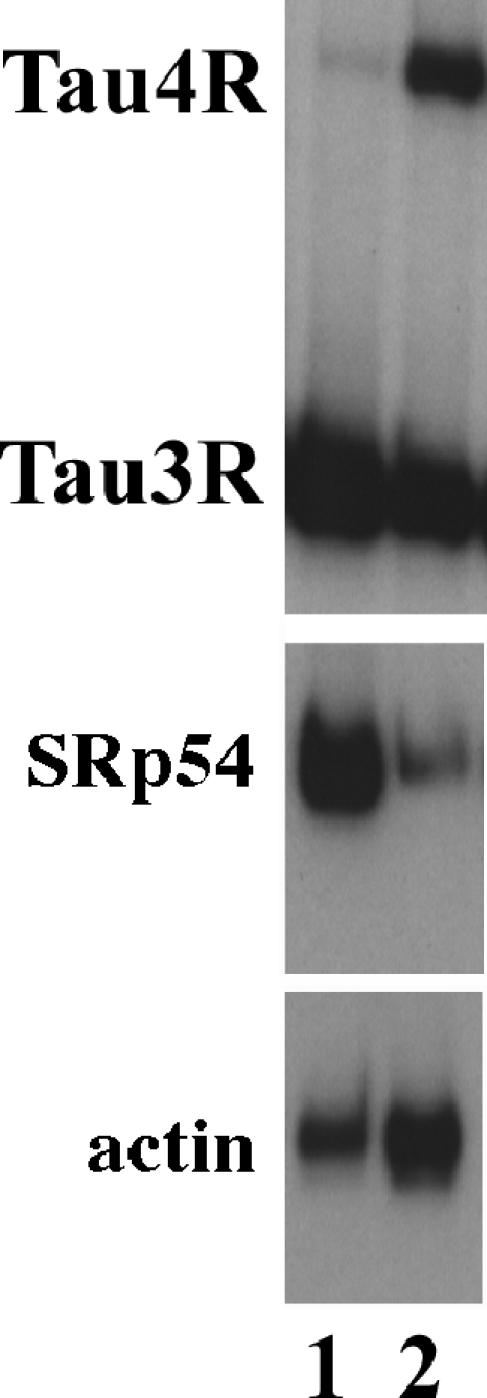
SRp54 expression is high in the fetal brain and reduced in the adult brain, correlating with its activity in repressing exon 10 splicing. The mRNA samples prepared from the frontal cortex regions of fetal (lane 1) and adult (lane 2) brains were used in the RT-PCR assay with specific primers to examine tau exon 10 splicing (top panel) and the expression of SRp54 (middle panel). Actin was used as a control for the RNA input (lower panel).
Because HEK293 cells had a high efficiency of transfection and a low background of GFP expression, we chose HEK293 cells to prepare a cell line stably expressing the tau reporter gene. A stable HEK293 cell line expressing the tau3RDDPAC-GFP reporter was established following selection with G418. The stable cell line showed the same pattern of tau exon 10 splicing as the original tau Ex9-10-11 DDPAC minigene, producing almost exclusively the exon 10+ tau isoform (Fig. (Fig.1B).1B). This stable cell line was used for screening the fetal brain cDNA library. Following each round of transfection, positive pools were identified by examining individual culture plates under an inverted fluorescence microscope for GFP expression (Fig. (Fig.2,2, part II). The primary positive pools that showed activated GFP expression were identified, and the cDNAs in these pools were further subdivided. For each primary positive pool, 20 subpools were prepared and transfected into stable cells expressing the tau3RDDPAC-GFP reporter gene. The positive subpools were identified again based on GFP expression using inverted fluorescence microscopy and subdivided for another round of transfection (Fig. (Fig.2).2). Six individual cDNA clones that activated GFP expression from the tau3RDDPAC-GFP reporter gene were isolated following six rounds of resubdivision and retransfection. Sequencing analyses of the cDNAs and databank searches showed that three clones encode novel proteins without detectable homology with known splicing factors, and these cDNAs are still under characterization. The fourth cDNA encodes the SRp75 protein and will be further pursued in a separate study. The remaining two cDNA clones encode a known SR protein, SRp54, and this protein is the focus of this report.
Confirmation of activity of SRp54 in regulating tau exon 10 splicing.
To eliminate the possibility of exon 10 splicing-independent GFP activation and to confirm the activity of SRp54 in tau exon 10 splicing, we cotransfected HEK293 cells using the original tau Ex9-10-11 DDPAC minigene without the GFP insert together with a cDNA plasmid expressing SRp54. As shown in Fig. Fig.3,3, transfection using the control vector plasmid (lane 1) or expression plasmid for another SR protein, SRp40 (lane 2), did not have any detectable effect on exon 10 splicing. Transfection of SRp54 (lane 3) led to a significant increase in exon 10 skipping. This demonstrates that the effect of SRp54 detected during expression cloning is a result of repression of exon 10 inclusion, rather than other indirect effects on GFP expression.
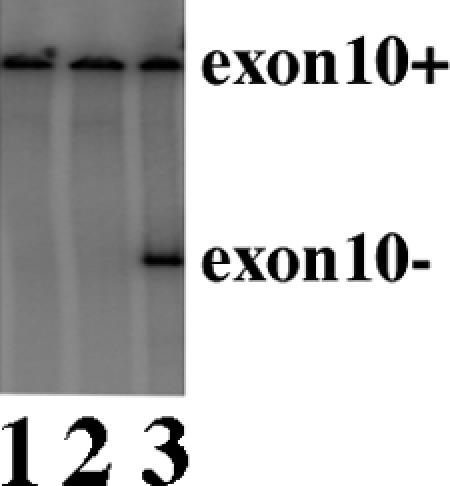
SRp54 stimulates exon 10 skipping when cotransfected with the tau minigene. The tau exon 9-10-11 (DDPAC) minigene was cotransfected into HEK cells together with the vector control, SRp40, or SRp54 (lanes 1, 2, and 3, respectively). tau exon 10 alternative splicing products were detected using RT-PCR.
To test the role of endogenously expressed SRp54 in regulating tau exon 10 splicing, we used RNA interference (RNAi) to reduce the expression of SRp54 and examine its effect on tau exon 10 splicing. Because the tau Ex9-10-11 DDPAC minigene gave rise to almost exclusively exon 10+ splicing product, it would be difficult to detect any further increase in the exon 10+ splicing isoform. Therefore, we used the wild-type tau exon 9-10-11 minigene to cotransfect cells in the RNAi experiments. The SRp54-specific RNAi was effective in reducing SRp54 expression at both RNA and protein levels (Fig. 4A and B, lane 3), whereas the control RNAi for either luciferase or SRp40 did not affect SRp54 expression (Fig. 4A and B, lanes 1 and 2). When the control or SRp40 RNAi was used, no changes were observed in the ratio of exon 10+/exon 10− tau-splicing isoforms (Fig. (Fig.4C,4C, lanes 1 and 2, respectively). However, when SRp54 expression was reduced by RNAi, a significant reduction in exon 10-skipping tau splicing isoform with a concomitant increase in exon 10 inclusion isoform was detected (Fig. (Fig.4C,4C, lane 3). The effect on SRp54 RNAi was specific for tau exon 10 splicing, because no effects were observed on the alternative splicing of other genes tested, including Bcl-x and caspase-2 (Fig. 4E and F, respectively).

Down-regulation of endogenous SRp54 by RNAi increases tau exon 10 inclusion. HEK cells were cotransfected using the wild-type tau exon 9-10-11 minigene together with control RNAi (lane 1), SRp40 RNAi (lane 2), or SRp54 RNAi (lane 3) in all panels. Following transfection, the expression of SRp54 was examined by RT-PCR (A) or Western blotting (B). tau exon 10 splicing was measured using RT-PCR with specific exon 9 and exon 11 primers (C). Actin was included as a control for the total RNA input (D). Alternative splicing of Bcl-x and caspase-2 (E and F, respectively) was examined using RT-PCR with the corresponding specific primers.
The expression level of the SRp54 gene in the human brain correlates with tau exon 10 alternative splicing changes.
The alternative splicing pattern of human tau exon 10 undergoes developmental changes, switching from almost exclusively tau exon 10-skipping isoform during the fetal stage to approximately a 1:1 ratio of exon 10+/exon 10- during the adult stage (reviewed in references 1, 10, and 25 and references therein). To begin to explore the potential involvement of SRp54 in regulating tau gene splicing during development, we examined the expression of the SRp54 gene in the fetal and adult human brain. Poly(A)+ RNA preparations of fetal and adult brains from Clontech were used in quantitative RT-PCR to examine tau exon 10 splicing and SRp54 expression. As shown in Fig. Fig.5,5, tau exon 10 splicing produced almost exclusively exon 10-skipping tau3R isoform in the fetal brain (Fig. (Fig.5,5, top panel, lane 1). However, this tau splicing pattern was switched to produce approximately an equal ratio of exon 10+ and exon 10- isoforms in the adult brain (Fig. (Fig.5,5, top panel, lane 2). SRp54 expression was detected at a high level in the fetal brain and significantly reduced in the adult brain (Fig. (Fig.5,5, middle panel, compare lanes 1 and 2). Actin was used as a control for the RNA input (Fig. (Fig.5,5, lower panel). The change in SRp54 expression correlates well with its activity in suppressing exon 10 inclusion, suggesting that SRp54 may play a role in regulating tau exon 10 splicing during development.
SRp54 represses tau exon 10 splicing by antagonizing Tra2β, suggesting a functional interaction between SRp54 and Tra2β.
Our previous study indicates that Tra2β enhances exon 10 inclusion (22). This prompted us to test whether SRp54 could antagonize the activity of Tra2β. HEK293 cells were cotransfected using the wild-type tau exon 9-10-11 minigene together with various amounts of SRp54 and Tra2β plasmids. As shown in Fig. Fig.6,6, when 1 μg Tra2β plasmid was cotransfected with increasing amounts of SRp54 plasmid (0, 2, and 4 μg, respectively, in lanes 1, 2, and 3), SRp54 antagonized the activity of Tra2β and switched the tau exon 10 splicing pattern from predominantly exon 10+ splicing to significantly more exon 10- splicing. Quantification of data from six independent experiments indicated that an increase in SRp54 expression changed the exon 10+/exon 10- ratio from 3.5 to 0.9 to 0.15, respectively (Fig. (Fig.6,6, lanes 1, 2, and 3). On the other hand, when SRp54 was held constant, increasing amounts of Tra2β expression stimulated tau exon 10 inclusion, switching the exon 10+/exon 10- ratio from 0.3 to 0.59 to 1.6 (Fig. (Fig.6,6, lanes 4, 5, and 6, respectively). This experiment demonstrates that SRp54 functionally antagonizes Tra2β in tau exon 10 splicing in transfected cells.
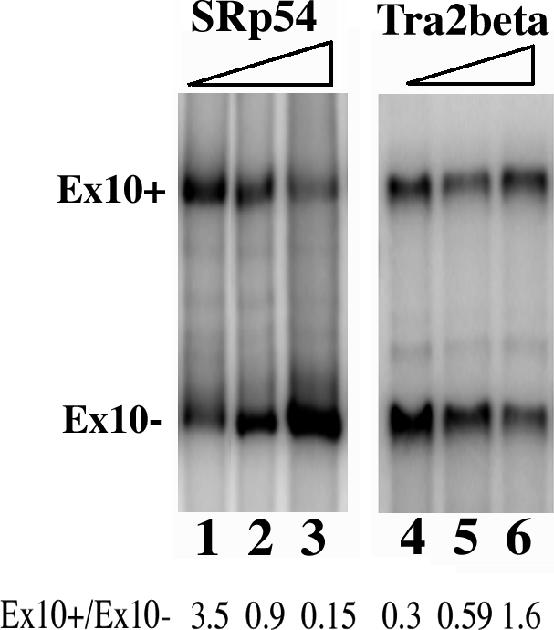
Functional interaction between SRp54 and Tra2β. HEK cells were cotransfected using the tau exon 9-10-11 (wt) minigene together with different amounts of SRp54 and Tra2β plasmids. In lanes 1, 2, and 3, 1 μg of Tra2β was cotransfected with 0, 2, and 4 μg of SRp54 plasmid. In lanes 4, 5, and 6, 1 μg of SRp54 was cotransfected with 0, 2, and 4 μg of Tra2β plasmid. tau exon 10 splicing was measured using RT-PCR with specific exon 9 and exon 11 primers. Increasing amounts of SRp54 counteracted the effect of Tra2β in stimulating exon 10 inclusion. The exon 10+/exon 10- ratio (Ex10+/Ex10-) was determined using a phosphorimager and is shown at the bottom of the corresponding lanes.
Competitive binding of SRp54 and Tra2β to the tau exon 10 splicing enhancer.
To explore the mechanisms of SRp54 function in regulating tau exon 10 splicing, we examined the interaction between SRp54 with different regions of tau pre-mRNA (Fig. (Fig.7A)7A) using UV cross-linking followed by immunoprecipitation. SRp54-Flag protein was prepared using in vitro translation and incubated with 32P-labeled tau RNA transcripts under splicing conditions. A UV cross-linking-coupled immunoprecipitation experiment was performed as described in Materials and Methods. In the absence of SRp54-Flag protein, no cross-linking signal was detected (Fig. (Fig.7B,7B, lane 1 and lane 5). In the presence of nonspecific RNA, there was no specific cross-linking signal detectable with SRp54-Flag protein (Fig. (Fig.7B,7B, lane 9). When tauEx10-11 pre-mRNA was used, SRp54 binding was detected (Fig. (Fig.7B,7B, lane 2), and such a cross-linking signal was stronger when A2 RNA (tauEx10) was used (Fig. (Fig.7B,7B, lane 3). When exon 10 was deleted from tauEx10-11 pre-mRNA, the cross-linking signal was significantly reduced (Fig. (Fig.7B,7B, lane 4). Because SRp54 functionally antagonizes Tra2β (Fig. (Fig.6)6) and Tra2β binds to exon 10 in the purine-rich enhancer region (22), we tested whether SRp54 interacted with the purine-rich region using tauEx10 transcript that was labeled with 32P at the AAGAAG region. Indeed, UV cross-linking and immunoprecipitation indicate that SRp54 interacts with this region inside exon 10 (Fig. (Fig.7B,7B, lane 6). This cross-linking signal was significantly reduced when an excess amount of unlabeled tau exon 10 RNA was added in the cross-linking reaction (Fig. (Fig.7B,7B, lane 8). This signal was not reduced in the presence of an excess amount of unlabeled nonspecific control RNA (Fig. (Fig.7B,7B, lane 7). These results demonstrate that SRp54 specifically interacts with the AAGAAG purine-rich enhancer region inside tau exon 10.
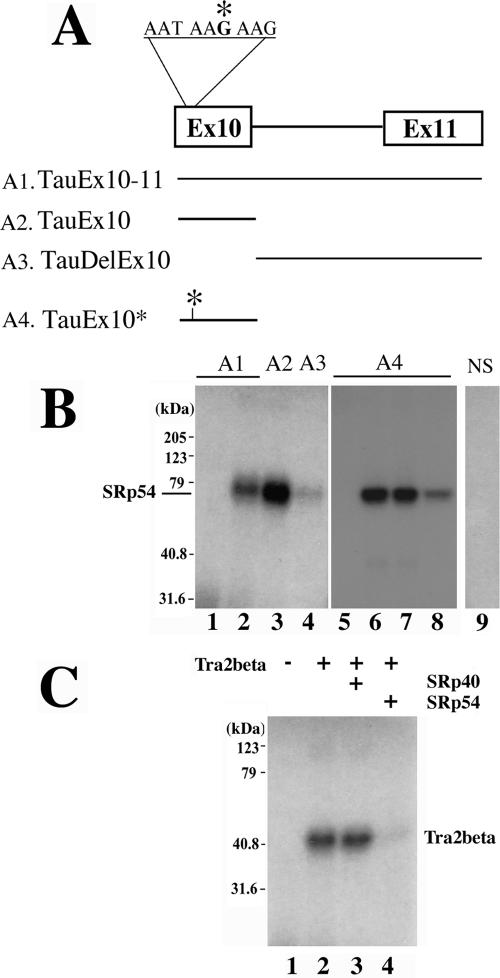
SRp54 interacts with tau pre-mRNA and competes with Tra2β in binding to the purine-rich element. A. Schematic illustration of RNA transcripts used for UV cross-linking experiments. RNA transcripts corresponding to either TauEx10-11 (A1), tau exon 10 (A2, TauEx10), or a downstream fragment (A3, TauDelEx10) or site-specific labeled tau exon 10 (A4, TauEx10*) were prepared and labeled with 32P. B. UV cross-linking assay demonstrating that SRp54 interacts with tau pre-mRNA preferentially in the exonic region. Either uniformly labeled (lanes 1 to 4) or site-specifically labeled (lanes 5 to 8) forms were used, as follows: TauEx10-11 (lanes 1 and 2), TauEx10 (lane 3), TauDelEx10 (lane 4), TauEx10* (lanes 5 to 8). Lane 9 shows the reaction containing a uniformly labeled nonspecific control RNA in the presence of SRp54 protein. UV cross-linking was performed using in vitro-translated cell lysates containing the vector control (lane 1) or SRp54-Flag (lanes 2 to 4) with 32P-labeled tau RNA transcripts, Tau Ex10-11 (lane 1 and 2), TauEx10 (lane 3), or TauDelEx10 (lane 4). The cross-linked species were immunoprecipitated using monoclonal antibody against the Flag epitope tag and detected by autoradiography. In lanes 5 to 8, similar UV cross-linking experiments were performed using site-specific-labeled TauEx10* RNA in the presence of cell ly sates containing the vector control (lane 5) or SRp54-Flag (lanes 6 to 8). Excess amounts of unlabeled competitor RNAs, either nonspecific control (lane 7) or tau exon 10 RNA (lane 8), were added, demonstrating the specificity of the interaction between SRp54 and tau exon 10. C. Binding of Tra2β to exon 10 was decreased by addition of the SRp54 protein. UV cross-linking was performed using in vitro translation cell lysates containing the vector control (lane 1) or Tra2β-myc (lanes 2 to 4) in the presence of an excess amount of mock preparation, SRp40, or SRp54 protein (lanes 2 to 4, respectively).
We further tested whether SRp54 competes with Tra2β, because they both bind to the same region in tau exon 10. Tra2β-myc protein was in vitro translated and incubated with the site-specifically labeled tau exon 10 RNA. Following UV cross-linking and immunoprecipitation with a monoclonal anti-myc antibody, the interaction between Tra2β and tau exon 10 RNA was detected by autoradiography following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. (Fig.7C,7C, lane 2). The same UV cross-linking-immunoprecipitation experiment was carried out in the presence of an excess amount of a control protein (SRp40) or SRp54 (Fig. (Fig.7C,7C, lanes 3 and 4, respectively). The addition of SRp54, but not SRp40, significantly antagonized the binding of Tra2β to the tau exon 10 RNA. These observations indicate that SRp54 competes with Tra2β in binding to the purine-rich enhancer region in tau exon 10 and suppresses exon 10 inclusion by antagonizing Tra2β.
Deletion of the purine-rich element eliminates SRp54-mediated suppression of tau exon 10 inclusion.
To test whether the purine-rich element in exon 10 is a functionally important SRp54 binding site, we deleted the AAGAAG sequence in tau exon 10. As shown in Fig. Fig.8,8, either wild-type tau exon 10 (A2, lane 1) or a mutant tau exon 10 with the AAGAAG sequence deleted (A2delBS, lane 3) or nonspecific RNA control (lane 2) was incubated with SRp54-Flag in the UV cross-linking-immunoprecipitation experiment. Deletion of this purine-rich element significantly reduced the binding of SRp54 to the tau exon 10 transcript (Fig. (Fig.8A,8A, compare lane 3 to lane 1), although a low level of residual binding was detectable (Fig. (Fig.8,8, lane 3). This suggests that the AAGAAG sequence is a major binding site for SRp54 inside exon 10.
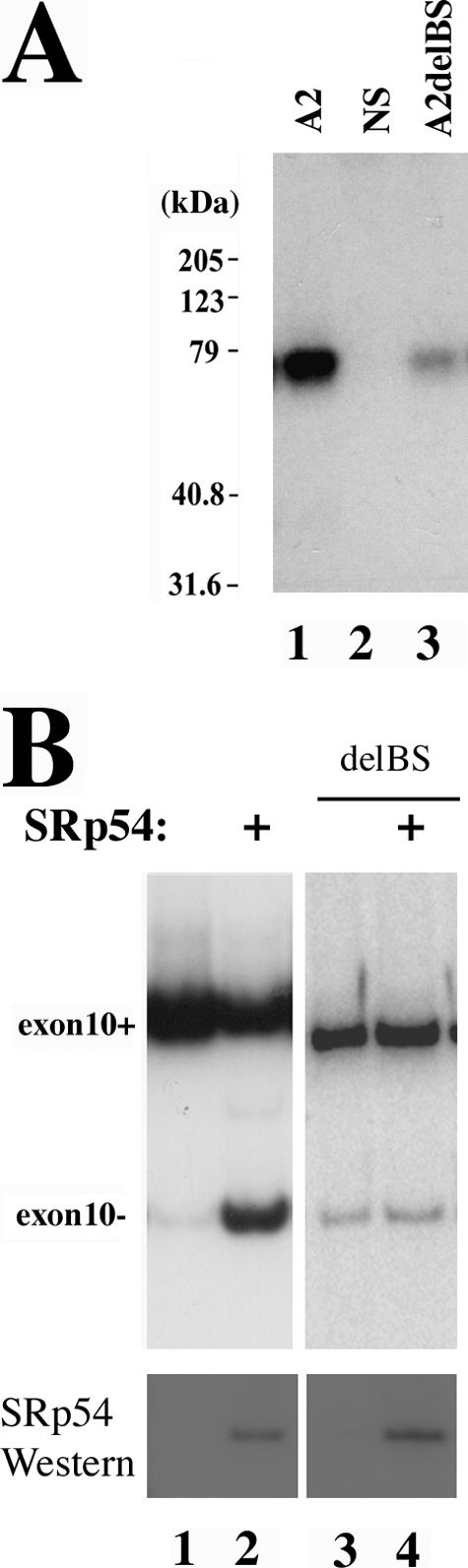
Deletion of the AAGAAG sequence in exon 10 reduces SRp54 binding and eliminates the activity of SRp54 in suppressing exon 10 inclusion. A. UV cross-linking-coupled immunoprecipitation was carried out as for Fig. Fig.7B7B using SRp54-Flag with 32P-labeled tau exon 10 (A2) transcript (lane 1) or nonspecific control RNA (lane 2) or A2 RNA with AAGAAG deleted (A2delBS, lane 3). B. Deletion of the AAGAAG sequence eliminated the activity of SRp54 in suppressing exon 10 inclusion. Cells were transfected with the original tau exon 9-10-11 (DDPAC) construct (lanes 1 and 2) or the minigene containing the deletion of AAGAAG (delBS, lanes 3 and 4) together with the control vector or SRp54-Flag (lanes 2 and 4). tau exon 10 splicing was examined using RT-PCR with tau exon 9 and exon 11 primers. The bottom panel shows that the expression of SRp54 was equivalent, as detected by Western blotting using anti-Flag antibody.
The tau minigene containing the deletion of the purine-rich element was tested for exon 10 splicing. In the cells transfected with the original tauEx9-10-11 DDPAC minigene, SRp54 transfection led to a significant increase in exon 10-skipping splicing product (Fig. (Fig.8B,8B, compare lane 2 with lane 1). When the AAGAAG element was deleted, the basal level of exon 10 skipping was increased (Fig. (Fig.8B,8B, lane 3), suggesting that a splicing activator(s) interacting with this element, such as Tra2β, may play a dominant role in exon 10 splicing. When cotransfected with SRp54, this mutant minigene with the deletion of the AAGAAG element showed no response to SRp54 (Fig. (Fig.8B,8B, compare lane 3 to lane 4). Western blotting showed that a similar level of SRp54 was expressed in cells expressing the original DDPAC tau minigene (lane 2) and the mutant minigene with the deletion of the AAGAAG element (lane 4). These results indicate that the deletion of the AAGAAG element in exon 10 eliminates the response of the tau minigene to SRp54 and that the purine-rich sequence inside exon 10 is a major element responsible for SRp54 activity in suppressing exon 10 inclusion.
DISCUSSION
A large number of genes in the human genome undergo alternative splicing in their functionally important regions. Understanding how these alternative splicing events are regulated by cis-acting sequence elements and trans-acting splicing factors is a major challenge in functional genomic studies. In the past two decades, a number of alternative splicing regulators have been identified. Most known mammalian splicing factors have been identified by biochemical approaches, with their functional activity in splicing established by in vitro assays.
We have established a tau minigene system to study mechanisms regulating tau exon 10 alternative splicing. This model system for alternative splicing has several features. tau exon 10 alternative splicing regulation is clearly important for neuronal function. Both intronic and exonic mutations in the tau gene can disrupt the balance of tau4R/tau3R isoforms and lead to the development of tauopathy. tau exon 10 alternative skipping/inclusion represents a general paradigm for alternative splicing. A large number of genes important for neuronal function utilize alternative exon skipping/inclusion as a mechanism to generate functionally distinct protein products. Such exon skipping/inclusion represent a common mode for alternative splicing regulation in neural tissues. The establishment of a tau3RDDPAC-GFP reporter gene and the expression cloning strategy enabled us to identify splicing regulators based on their functional activity in regulating tau exon alternative splicing. Our expression cloning strategy should be generally applicable for identifying trans-acting factors involved in alternative splicing regulation of other genes.
Molecular mechanisms controlling tau exon 10 alternative splicing are not well understood. Several cis-acting regulatory elements have been identified that influence tau exon 10 alternative splicing. Many of the tau intronic mutations are near the splice donor site (5′ splice site) of the intron following exon 10. Biochemical and structural studies support the model that intronic mutations near the 5′ splice site following exon 10 destabilize a stem-loop structure and lead to enhanced U1 snRNP interaction with the 5′ splice site of exon 10, and thus an increase in exon 10 inclusion and an increase in the ratio of tau4R to tau3R (16, 19, 21, 41). Intronic splicing silencer and other regulatory elements have also been proposed to function as linear regulatory elements to modulate the splice site selection (9, 11). Mutations may also disrupt the function of these splicing regulatory elements. In addition to intronic mutations, several exonic mutations, including N279K, Del280K, L284L, N296N, and S305N, also affect exon 10 splicing (reviewed in references 1, 10, and 25).
A number of known splicing regulators have been tested using a chimeric tau minigene (13). An hnRNP protein, hnRNPE2, was isolated by the yeast three-hybrid assay and found to have a moderate activity in stimulating exon 10 splicing in a chimeric tau splicing cassette (4). The overexpression of known splicing factors influences exon 10 splicing when cotransfected with a chimeric tau minigene (13, 44). The effects of these splicing factors on tau exon 10 splicing have not been demonstrated in the context of the native tau gene splice sites. Two protein kinases have been reported to affect tau exon 10 splicing. Overexpression of cdc2-like kinase 2 represses exon 10 inclusion (17), whereas inhibition of GSK-3β kinase activity increases exon 10 splicing (18). Our previous study identified Tra2β as a protein stimulating exon 10 inclusion by interacting with the AG-rich splicing enhancer inside exon 10 (22). The role of Tra2β in regulating tau exon 10 splicing has also been confirmed by other groups (8, 28, 44). In this study, we report the development of a new strategy using an expression cloning to systematically screen for factors important for tau exon 10 skipping. Using this expression cloning method to screen the human fetal brain cDNA library, we identified SRp54 as a repressor for tau exon 10 inclusion. Interestingly, SRp54 acts by competing with Tra2β in binding to the purine-rich element in exon 10.
SRp54 was originally isolated as a nuclear protein with speckled intranuclear distribution (6). Our previous work demonstrated that SRp54 is an essential splicing factor of the SR protein superfamily (48). Consistent with this, the SRp54 gene is required for viability in Caenorhabditis elegans (30). In the current study, we report the identification of SRp54 using our expression cloning strategy based on its effect in stimulating tau exon 10 skipping. Down-regulation of SRp54 using RNAi increases tau exon 10 inclusion. The high level of expression of SRp54 in the fetal brain and its reduced expression in the adult brain correlate with the tau exon 10 alternative splicing pattern changes, supporting a role of the endogenous SRp54 protein in regulating tau exon 10 splicing during development. It is worth noting that tau exon 10 splicing regulation is species specific. Rodent tau genes express predominantly exon 10-containing tau4R in the adult, rather than producing an equal ratio of tau3R and tau4R, as in the human adult brain. At present, it is not clear how SRp54 expression in the human brain changes during neural development or whether SRp54 expression varies in different brain regions. These important questions await further investigations and require the development of better reagents, such as SRp54-specific antibodies suitable for immunohistochemical staining.
SR proteins play important roles in both constitutive splicing and alternative splicing. They are characterized by two structural motifs: an RNA recognition motif of the RNP type in the N terminus and a serine-arginine-rich domain at the C terminus. SR proteins play versatile roles in regulating alternative splicing, acting as splicing activators or splicing repressors. A large number of studies have reported the role of SR proteins in activating pre-mRNA splicing and promoting exon inclusion (reviewed in references 3, 5, 12, 15, 40, and 45). However, several SR domain-containing proteins have also been reported to have splice-repressing activity, including SR proteins and SR-related proteins such as SRrp86 and SRp38 (for examples, see references 2, 7, 24, 27, 36-38, and 46).
It remains unclear exactly what features determine whether an SR domain-containing protein acts as a splicing enhancer to stimulate exon inclusion or as a repressor to promote exon skipping. It is likely that combinatorial effects of protein-RNA and protein-protein interactions play important roles in determining the activity of a particular splicing regulator. Clearly, SRp54 and Tra2β have different repertoires of protein interaction partners. SRp54 interacts with several SR domain-containing proteins (48), PUF60 (34), and RNPS1 (35). Tra2β, on the other hand, binds SF2/ASF, SRp55, SRp30c, SAF-B, and hnRNP G (32, 39, 42, 44, 47). In tau exon 10 splicing, our previous study and results from other groups show that Tra2β activates exon 10 inclusion (8, 22, 28, 44). In this study, we show that in transfected cells, SRp54 antagonizes the activity of Tra2β. Our UV cross-linking-coupled immunoprecipitation experiments indicate that SRp54 competes with Tra2β for binding to the purine-rich exonic element (Fig. (Fig.7C).7C). This provides a mechanism by which SRp54 represses tau exon 10 inclusion.
It is interesting that exon 10 skipping increased when the AAGAAG sequence was deleted from the tau minigene (Fig. (Fig.8B,8B, compare the exon 10+/exon 10- transcript ratio in lane 3 with that in lane 1). This suggests that a splicing activator(s) interacting with this purine-rich element, such as Tra2β, plays a dominant role over a splicing repressor(s), such as SRp54, that interacts with the same element. Nonetheless, when this purine-rich element was deleted, the repression of exon 10 splicing by SRp54 was lost, demonstrating that interaction of SRp54 with this AAGAAG element is necessary for exon 10 splicing repression by SRp54 and that the residual binding of SRp54 to sequences outside this element is not important for such repression (Fig. (Fig.88).
Sequence analyses and database searches show that mammalian SRp54 proteins are highly similar in peptide sequences, whereas Drosophila and nematode SRp54 protein sequences are more divergent from that of mammalian ones (B. Yu and J. Y. Wu, unpublished data). The Drosophila homologue of SRp54 binds C-rich pyrimidine tracts in short introns and promotes the binding of U1 and U2 RNPs to the flanking splice sites (26). It has been proposed that SRp54 mediates interactions across the intron and across the exon by interacting with U2AF65 and SR proteins (26). Our current data show that SRp54 interacts with the purine-rich exonic sequence and suppresses exon 10 inclusion. The peptide sequence differences in the fly and human SRp54 proteins may explain the differences in their RNA binding properties.
Identification of critical regulators for tau exon 10 alternative splicing not only helps us understand the regulatory mechanisms underlying tau alternative splicing but also provides useful information for designing future therapeutic approaches to correct the tau splicing defects. For example, SRp54, identified in this study, is capable of stimulating tau exon 10 skipping and reversing the tau4R/tau3R ratio to the normal 1:1 ratio in cells expressing the DDPAC tau splicing mutant gene. This suggests that activation or enhancement of SRp54 expression in the adult brain may have therapeutic potential in FTDP-17 patients who carry tau splicing mutations that cause excessive exon 10 inclusion.
Acknowledgments
We thank N. Chaudary for generously providing the antibody against SRp54, Z. H. Jiang for help in the initial phase of the work, and Bill Nash for excellent technical assistance. We are grateful to members of the Wu laboratory for helpful discussions and/or critical reading of the manuscript.
This work was supported by grants from NIH (AG17518, GM07967, and EY014576), The Society for Progressive Supranuclear Palsy, the Muscular Dystrophy Association (J.Y.W.), and by a Scholar award from the Leukemia Society of America (J.Y.W.).
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.00739-06
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1592875?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Alternative splicing in neurodegenerative disease and the promise of RNA therapies.
Nat Rev Neurosci, 24(8):457-473, 19 Jun 2023
Cited by: 27 articles | PMID: 37336982
Review
A Proteome-Wide Effect of PHF8 Knockdown on Cortical Neurons Shows Downregulation of Parkinson's Disease-Associated Protein Alpha-Synuclein and Its Interactors.
Biomedicines, 11(2):486, 08 Feb 2023
Cited by: 0 articles | PMID: 36831023 | PMCID: PMC9953648
Alternative splicing of HSPA12A pre-RNA by SRSF11 contributes to metastasis potential of colorectal cancer.
Clin Transl Med, 12(11):e1113, 01 Nov 2022
Cited by: 8 articles | PMID: 36394206 | PMCID: PMC9670187
Identification of HnRNPC as a novel Tau exon 10 splicing factor using RNA antisense purification mass spectrometry.
RNA Biol, 19(1):104-116, 31 Dec 2021
Cited by: 8 articles | PMID: 34965173 | PMCID: PMC8786334
Alternative Splicing of Human Telomerase Reverse Transcriptase (hTERT) and Its Implications in Physiological and Pathological Processes.
Biomedicines, 9(5):526, 09 May 2021
Cited by: 8 articles | PMID: 34065134 | PMCID: PMC8150890
Review Free full text in Europe PMC
Go to all (37) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NM_004768
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA.
Genes Cells, 9(2):121-130, 01 Feb 2004
Cited by: 49 articles | PMID: 15009090
Arginine/serine-rich protein interaction domain-dependent modulation of a tau exon 10 splicing enhancer: altered interactions and mechanisms for functionally antagonistic FTDP-17 mutations Delta280K AND N279K.
J Biol Chem, 281(5):2460-2469, 23 Nov 2005
Cited by: 34 articles | PMID: 16308321
Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2 beta.
J Biol Chem, 278(21):18997-19007, 20 Mar 2003
Cited by: 79 articles | PMID: 12649279 | PMCID: PMC2140226
Regulation of alternative splicing of tau exon 10.
Neurosci Bull, 30(2):367-377, 14 Mar 2014
Cited by: 53 articles | PMID: 24627328 | PMCID: PMC5562657
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NEI NIH HHS (1)
Grant ID: R01 EY014576
NIA NIH HHS (2)
Grant ID: AG 17518
Grant ID: R01 AG017518
NIGMS NIH HHS (1)
Grant ID: GM 07967





