Abstract
Free full text

A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria.
Associated Data
Abstract
Bacterial 16S ribosomal RNA (rRNA) genes contain nine “hypervariable regions” (V1 – V9) that demonstrate considerable sequence diversity among different bacteria. Species-specific sequences within a given hypervariable region constitute useful targets for diagnostic assays and other scientific investigations. No single region can differentiate among all bacteria; therefore, systematic studies that compare the relative advantage of each region for specific diagnostic goals are needed. We characterized V1 - V8 in 110 different bacterial species including common blood borne pathogens, CDC-defined select agents and environmental microflora. Sequence similarity dendrograms were created for hypervariable regions V1 – V8, and for selected combinations of regions or short segments within individual hypervariable regions that might be appropriate for DNA probing and real-time PCR. We determined that V1 best differentiated among Staphylococcus aureus and coagulase negative Staphylococcus sp. V2 and V3 were most suitable for distinguishing all bacterial species to the genus level except for closely related enterobacteriaceae. V2 best distinguished among Mycobacterial species and V3 among Haemophilus species. The 58 nucleotides-long V6 could distinguish among most bacterial species except enterobacteriaceae. V6 was also noteworthy for being able to differentiate among all CDC-defined select agents including Bacillus anthracis, which differed from B. cereus by a single polymorphism. V4, V5, V7 and V8 were less useful targets for genus or species-specific probes. The hypervariable sequence-specific dendrograms and the “MEGALIGN” files provided online will be highly useful tools for designing specific probes and primers for molecular assays to detect pathogenic bacteria, including select agents.
1. Introduction
Sequence analysis of the 16S ribosomal RNA (rRNA) gene has been widely used to identify bacterial species and perform taxonomic studies (Choi et al., 1996; Clarridge, 2004; Munson et al., 2004; Petti et al., 2005; Schmalenberger et al., 2001). Bacterial 16S rRNA genes generally contain nine “hypervariable regions” that demonstrate considerable sequence diversity among different bacterial species and can be used for species identification (Van de Peer et al., 1996). Hypervariable regions are flanked by conserved stretches in most bacteria, enabling PCR amplification of target sequences using universal primers (Baker et al., 2003; Lu et al., 2000; McCabe et al., 1999; Munson et al., 2004). Numerous studies have identified 16S rRNA hypervariable region sequences that identify a single bacterial species or differentiate among a limited number of different species or genera (Becker et al., 2004; Bertilsson et al., 2002; Choi et al., 1996; Clarridge, 2004; Kataoka et al., 1997; Lu et al., 2000; Marchesi et al., 1998; Maynard et al., 2005; Rothman et al., 2002; Yang et al., 2002). Rapid approaches that detect certain species-specific sequences within a single hypervariable region are also in common use (Bertilsson et al., 2002; Stohr et al., 2005; Varma-Basil et al., 2004; Yang et al., 2002).
Unfortunately, 16S rRNA hypervariable regions exhibit different degrees of sequence diversity, and no single hypervariable region is able to distinguish among all bacteria. Molecular diagnostic methods, such as real-time PCR (Selim et al., 2005; Varma-Basil et al., 2004; Yang et al., 2002) or melting temperature analysis (Skow et al., 2005) generally use fluorescent probes that hybridize to relatively short amplicons. This places additional limits on the size of the DNA sequence that can be used for bacterial species identification in these assay formats. Given the increasing importance of real-time PCR to medical diagnostics, it is surprising that few studies have focused on matching short segments of hypervariable 16S rRNA gene regions with the common pathogenic or environmental bacteria that can be differentiated by each segment. Furthermore, no investigations to date have applied this analysis to determine the 16S rRNA gene sequences best suited for identifying both common human pathogens and “select agents” of bioterrorism.
The purpose of the current study was to simplify the development of specific probe or primer-based PCR assays for detecting common bacterial pathogens and select agents. Our aim was to identify the most appropriate 16S rRNA hypervariable region targets for genus and species-specific probes or primers and to experimentally confirm a portion of these results. We used the neighbor joining method to create dendrograms of each 16S rRNA hypervariable segment in 113 available sequences of 110 different bacterial species, including common blood borne pathogens and select agents. Our results demonstrate that it is possible to distinguish among almost all of these pathogens using a small number of hypervariable gene segments. We also provide criteria for selection of specific target sequences based on the desired purposes of the assays and provide recommendations for probes and primer pairs that would be useful in these assays.
2. Materials and Methods
2.1. Sequence retrieval and phylogenetic analysish
The 113 16S rRNA gene sequences analyzed in this study included the sequences from 110 different bacterial species commonly detected in human infections including pneumonia, abscesses, blood stream infections and sepsis (Kumar et al., 2006) as well as most other known pathogenic bacteria including select agents, and common contaminants of clinical samples (Sanford, 2003) (Table 1). Additional 16S rRNA sequences were included from three different strains of Escherichia coli and two different strains of Mycobacterium tuberculosis. The 16S rRNA gene sequences corresponding to each of these 110 bacterial species were retrieved from Genbank database or TIGR CMR, RNA gene resource (http://www.tigr.org/tigr-scripts/CMR2/rna_form.spl?db=CMR). The Genbank accession numbers and the TIGR CMR sequence names are provided in Table 1. For bacteria with multi-copy 16S rRNA genes, sequences retrieved from the TIGR CMR were first aligned among themselves to check for sequence heterogeneity. The sequence with the maximum copy number in each species was then chosen to represent that bacterium. For sequences retrieved from the Genbank database, all available16S rRNA sequences larger than 1,000 base pairs were analyzed for each bacterium. Sequences with ambiguous residues were eliminated; the remaining sequences were used to generate a consensus sequence, which was used in subsequent analyses. The complete 16S rRNA sequences were aligned for all 113 16S rRNA gene sequences using MEGALIGN 6.1 sequence analysis software (Lasergene V6, DNASTAR) using the ClustalW analysis, which performs alignments using the Neighbour-Joining method. Hypervariable regions flanked by conserved stretches were identified based on this initial alignment and then each individual hypervariable region from all the 113 sequences was separately re-aligned using the same software. Sequence similarity dendrograms were produced for each hypervariable gene alignment using MEGALIGN using the neighbor-joining method to generate the dendrograms. Hypervariable stretches, in which numerous 16S rRNA gene sequences contained “gaps” were aligned using Clustal V.
Table 1
Bacterial 16S rRNA gene sequences used in this study
| Bacteria | Accn no./ TIGR CMR seq. designation |
|---|---|
| Acinetobacter calcoaceticus | AJ888984 |
| Actinomyces israelii | X82450 |
| Arcanobacterium pyogenes | X79225 |
| Aerococcus viridans | AY707778 |
| Actinomyces meyeri | X82451 |
| Burkholderia cepacia | AB211325 |
| B. mallei | BMAA_Bm16SC |
| B. pseudomallei | NTBP01_Bp16SA |
| Brucella melitensis | NTBM01_Bm16SA |
| B. suis | GBR_Bs16SA |
| Bordetella parapertussis | NTBP02_Bp16SA |
| Borrelia burgdorferi B31 | GBB_Bb16SA |
| Bacillus anthracis | GBA_Ba16SA |
| B. cereus | NTBC01_Bc16SA |
| B. subtillis | NTBS01_Bs16SA |
| Bacteroides fragilis | M11656 |
| Bordetella pertussis | NTBP03_Bp16SA |
| Clostridium botulinum | AF105402 |
| C. difficile | AF072474 |
| C. septicum | U59278 |
| C. acetobutylicum | NTCA01_Ca16SA |
| C. perfringens | NTCP03_Cp16SA |
| C. diptheriae | NTCD01-Cd16SA |
| C. tetani | NTCT02-Ct16SA |
| Corynebacterium jeikeium | X84250 |
| Campylobacter jejuni | NTCJ01_Cj16SA |
| Chlamydia trachomatis | NTCT01_16SrRNA-1 |
| C. pneumoniae | NTCP05_Cp16SA |
| Coxiella burnetti | GCB_Cb16SA |
| Enterococcus gallinarum | AJ301833 |
| E. faecalis | AJ291732 |
| E. faecium | AJ291732 |
| Enterobacter aerogenes | AB004750 |
| Escherichia coli BL21 | AJ605115 |
| E. coli K12 | NTEC01_16SrRNA-1 |
| E. coli O157H7 | NTEC02_Ec16SA |
| Fusobacterium sulci | AJ006963 |
| F. alocis | AJ006962 |
| F. equorum | AJ295750 |
| F. naviforme | AJ006965 |
| F. nucleatum | AJ810275/NTFN01_Fn16SA |
| F. periodonticum | AJ810271 |
| F. prausnitzii | AJ413954 |
| F. necrophorum | X74407 |
| Francisella tularensis | AJ698865 |
| Haemophilus paraphrophilus | AY365451 |
| H. parahaemolyticus | AJ290758 |
| H. ducreyi | AF525028/NTHD01_Hd16SA |
| H. parainfluenzae | AJ290755 |
| H. aphrophilus | AY365453 |
| H. influenzae | AY613778/GHI_HIrrnA16S |
| Helicobacter pylori | NTHP01_Hp16SA |
| Klebsiella pneumoniae | Y17657 |
| Listeria grayi | X98526 |
| L. monocytogenes | AJ535697 |
| Leptospira interrogans | AM050580 |
| Legionella pneumophila | NTLP02_NT02LP0369 |
| Moraxella catarrhalis | U10876 |
| Mycobacterium avium | AJ536037 |
| M. intracellulare | AJ536036 |
| M. fortuitum | AJ416915 |
| M. gordonae | AJ581472 |
| M. kansasii | AJ536035 |
| M. scrofulaceum | AF480604 |
| M. bovis | NTMB01_Mb16SA |
| M. leprae | NTML01_Ml16SA |
| M. tuberculosis CDC1551 | GMT_Mt16SA |
| M. tuberculosis H37Rv | NTMT02_NT02MT1437 |
| Neisseria gonorrhoeae | AJ239304 |
| N. mucosa | AJ239282 |
| N. lactamica | AJ239313 |
| N. meningitidis | GNM_NMrrnaA16S |
| Nocardia brasiliensis | AF430038 |
| N. pseudobrasiliensis | X84855 |
| Oligella urethralis | AY513496 |
| Proteus vulgaris | X07652 |
| P. mirabilis | AJ301682 |
| Peptostreptococcus micros | AF542231 |
| Propionibacterium acnes | NTPA02_NT02PA0587 |
| Pasteurella multocida | NTPM01_Pm16SA |
| Pseudomonas aeruginosa | AY268175 |
| Rickettsia prowazekii | NTRP01_Rp16SA |
| R. rickettsii | U11021 |
| Rhodococcus equi | X80594 |
| Shigella dysenteriae | X96966 |
| S. flexneri | NTSF02_Sf16SD |
| Salmonella paratyphi | X80682 |
| S. enterica typhi | NTST04_St16SA |
| S. typhimurium | NTST01_St16SA |
| Serratia marcescens | AJ233431 |
| Streptococcus uberis | U41048 |
| S. pyogenes | NTSP03_Sp16SA |
| S. pneumoniae | BSP_Sp16SA |
| S. agalactiae | GBS_Sa16SA |
| S. mutans | NTSM02_Sm16SA |
| S. gordonii | D38483 |
| S. mitis | GMI_Sm16SA |
| S. salivarius | AY188352 |
| Staphylococcus aureus | SACOL_Sa16SA |
| S. epidermidis | NTSE02_Se16SA |
| S. saprophyticus | NTSS03_NT03SS0724 |
| S. caprae | AY346310 |
| S. haemolyticus | D83367 |
| S. hominis | AJ717375 |
| S. intermedius | D83369 |
| S. lugdunensis | AB009941 |
| S. schleiferi | D83372 |
| S. simulans | D83373 |
| S. warneri | L37603 |
| Treponema pallidum | GTP_Tp16SA |
| Vibrio cholerae | X76337 |
| Yersinia enterocolitica | Z75316 |
| Yersinia pestis | NTYP02_Yp16SA |
2.2. Bacterial Isolates, DNA isolation and quantification
The thirty strains including five select agents representing twenty four bacterial genera, that were used to experimentally confirm the performance of PCR primers and probes are shown in Table 2. The bacterial cultures used in these experiments included both standard laboratory strains and de-identified patient isolates. Bacterial DNA was extracted using Phenol–Chloroform as described previously (Maloy, 1989). DNA was isolated from Bacillus anthracis, Yersinia pestis, Burkholderia mallei and Francisella tularensis in a biosafety level III laboratory certified to work with select agents (registration number 20011016-798; entity number C20031133-0125).
Table 2
Bacterial strains used in this study
| Bacteria | Strain | Bacteria | Strain |
|---|---|---|---|
| Aerococcus viridans | ATCC 700406 | Moraxella cattarhalis | ATCC 8176 |
| Arcanobacterium pyogenes | ATCC 49698 | Mycobacterium tuberculosis | H37Rv |
| Bacillus anthracis | Vollum and Sterne | Oligella urethralis | ATCC 17960 |
| Bacteroides uniformis | ATCC 8492 | Proteus vulgaris | ATCC 49132 |
| Burkholderia mallei | ATCC 23344 | Pseudomonas aeruginosa | ATCC 27853 |
| Clostridium difficile | ATCC 9689 | Neisseria gonorrhoeae | ATCC 49226 |
| Campylobacter jejuni | ATCC 33291 | Neisseria lactamica | ATCC 23971 |
| Corynebacterium pseudodiphtheriticum | ATCC 10700 | Neisseria mucosa | ATCC 69695 |
| Enterococcus galinarium | ATCC 700425 | Serratia marcescens | Clinical isolate |
| Escherichia coli O157:H7 | ATCC 35150 | Streptococcus agalactiae | ATCC 12386 |
| Francisella tularensis | Live vaccine strain | Streptococcus pyogenes | ATCC 19615 |
| Klebsiella pneumoniae | Clinical isolate | Streptococcus pneumoniae | Clinical Isolate |
| Haemophilus influenzae | ATCC 49247 | Staphylococcus aureus | ATCC 25923 |
| Haemophilus parahemolyticus | ATCC 10014 | Staphylococcus epidermidis | Clinical Isolate |
| Listeria grayi | ATCC 25401 | Yersinia pestis | CO92 |
2.3. Primer and probe design
Universal PCR primers complementary to four conserved regions flanking two hypervariable sequences (V3 and V6) were designed using Primer Select software (Lasergene V6, DNASTAR). The primers were designed so that at least the 10-most 3’ nucleotides were complimentary to highly conserved 16 rRNA sequences. A molecular beacon probe complementary to the hypervariable region V6 was designed as described previously (Tyagi and Kramer, 1996). The sequences of all primers and the molecular beacon are shown in Table 3.
Table 3
Primers and probes used in the study
| Primer | ||||
|---|---|---|---|---|
| Name | Sequencea | Amplified hypervariable region | ||
| V3F | 5’ CCAgACTCCTACGGGAGGCAG 3’ (334-354) | V3 (334-537) b | ||
| V3R | 5’ CGTATTACCGCGGCTGCTG 3’ (519-537) | |||
| V6F | 5’ TCGAtGCAACGCGAAGAA 3’ (961-78) | V6 (986-1043) | ||
| V6R | 5’ ACATtTCACaACACGAGCTGACGA 3’ (1062-85) | |||
| Molecular Beacon probec | ||||
| Name | Sequence | Target region | ||
| SEP-V6 probe | TxR-5’tgcgcCTAGAGGGGTCAGAGGATgcgca 3’- BHQ2 | 1005-1022 * | ||
2.4. Real-time and end-point detection PCR
For the Staphylococcus epidermidis specific molecular beacon assay, real time PCR was carried out in a Smart Cycler II (Cepheid, Sunnyvale, California, USA) real time PCR thermal cycler. Each tube was loaded with 25μl final volume of a solution containing 1X Amplitaq Gold polymerase buffer, 0.03 U/μl of Amplitaq Gold polymerase enzyme (Applied Biosystems, California, USA), 4 mM MgCl2, 250 μM dNTP mix, 0.5 pmol of each primer and 5ng/μl of the molecular beacon (Biosearch Technologies, California, USA) and the appropriate amount of chromosomal DNA (106 to 1 genome equivalents) or an equal volume of water (for no DNA control reactions). PCR amplification were carried out as follows: Initial denaturation at 95°C for 10 min, followed by 45 cycles each of 95°C for 20 s, 52°C for 30s and 72°C for 30 s. Data were collected during the annealing step for analysis. End-point detection PCR was carried out in the thermal cycler Gene Amp PCR system 9700 (Applied Biosystems, California, USA). Reaction compositions were identical as described in case of the real time PCR assay, except that the reaction did not contain any molecular beacons, the annealing temperature was 55°C and 10 ng of DNA was used as templates in the assay. The PCR products for the end point assays were visualized under UV light by ethidium bromide staining after agarose gel electrophoresis. Prior to both real time and end-point detection PCR assays, the PCR cocktail (without any template DNA added) was digested with 0.1U/μl of AluI restriction enzyme (Invitrogen Life Technologies, Carlsbad, California, USA) to degrade any endogenous DNA present in the recombinant Taq Polymerase and then heat inactivated as described (20).
2.5. DNA sequencing and analysis
The V3 and V6 hypervariable regions amplified from a representative set of 19 bacteria including Arcanobacterium pyogenes, Bacillus anthracis, Burkholderia mallei, Clostridium difficile, Campylobacter jejuni, Escherichia coliO157H7,Klebsiella pneumoniae, Haemophilus influenzae, Listeria grayi, Moraxella catarrhalis, Oligella urethralis, Proteus vulgaris, Pseudomonas aeruginosa, Neisseria gonorrhoeae, Serratia marcescens, Streptococcus pyogenes, S. pneumoniae, Staphylococcus aureus and Yersinia pestis were subjected to DNA sequencing to confirm and correlate the sequence information with the in-silico data analysis.
3. Results
3.1. Hypervariable segment specific dendrograms
Previous alignments of bacterial 16S rRNA gene sequences have revealed nine separate hypervariable regions, which we term V1 – V9 in concordance with previous nomenclature (Van de Peer et al., 1996). We aligned the 113 16S rRNA gene sequences from all the 110 bacterial species used in this study to confirm this observation and to define the borders of each hypervariable region for the bacterial species of interest (Supplementary file 1; available online). Our alignment confirmed the presence of all the nine hypervariable regions in the bacterial species for which the complete 16S rRNA gene sequence was available. The nine hypervariable regions spanned nucleotides 69-99, 137-242, 433-497, 576-682, 822-879, 986-1043, 1117-1173, 1243-1294 and 1435-1465 for V1 through V9 respectively [numbering based on the E. coli system of nomenclature (Brosius et al., 1978), if not mentioned otherwise]. Complete sequence data were available for the regions V1 – V8 for all 113 sequences included in this study. Sequence data for V9 was either incomplete or not available for 15 of the 113 sequences analyzed; and we excluded the V9 region from further analysis. The bacteria Pseudomonas aeruginosa (Accn no. AY268175), Rhodococcus equi (Accn no. X80594) and Salmonella paratyphi (Accn no. X80682) contained ambiguous nucleotide residues in V1; and Arcanobacterium pyogenes (Accn no. X79225) and Clostridium septicum (Accn no. U59278) contained ambiguous residues in V8. The ambiguous regions of these bacterial species were also eliminated from the analysis. Alignments for the entire 16S rRNA gene for all the 113 sequences and for the hypervariable regions V3 and V6 are provided as supplementary files 1, 2 and 3 respectively at http://njms.umdnj.edu/departments/medicine/infectious_diseases/our_research.cfm.
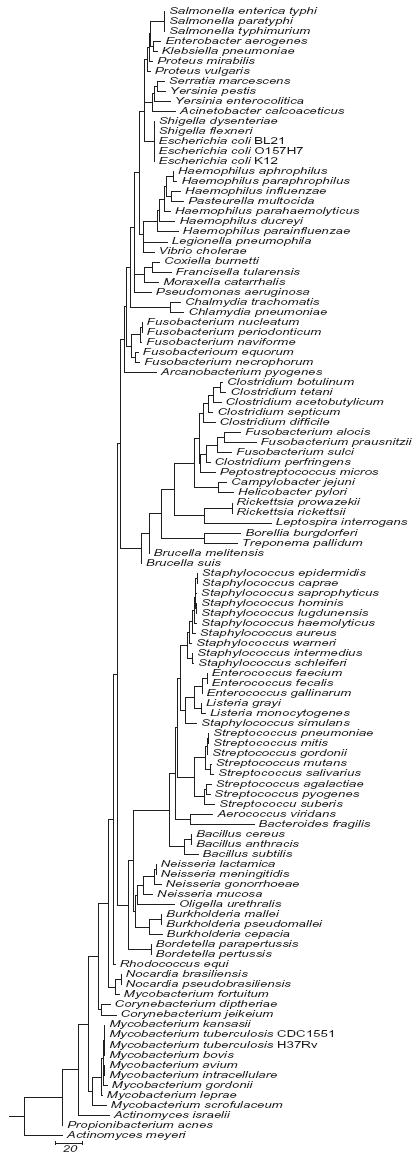
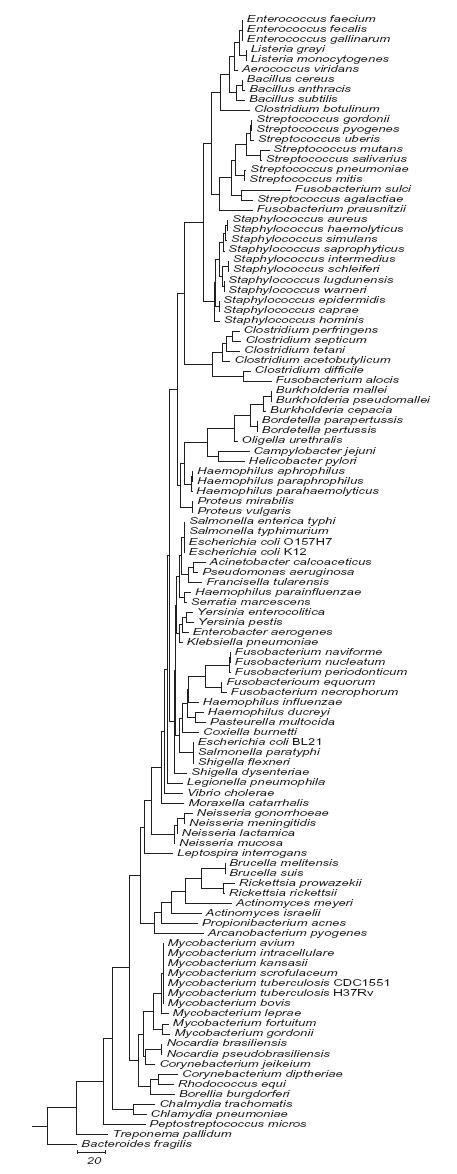
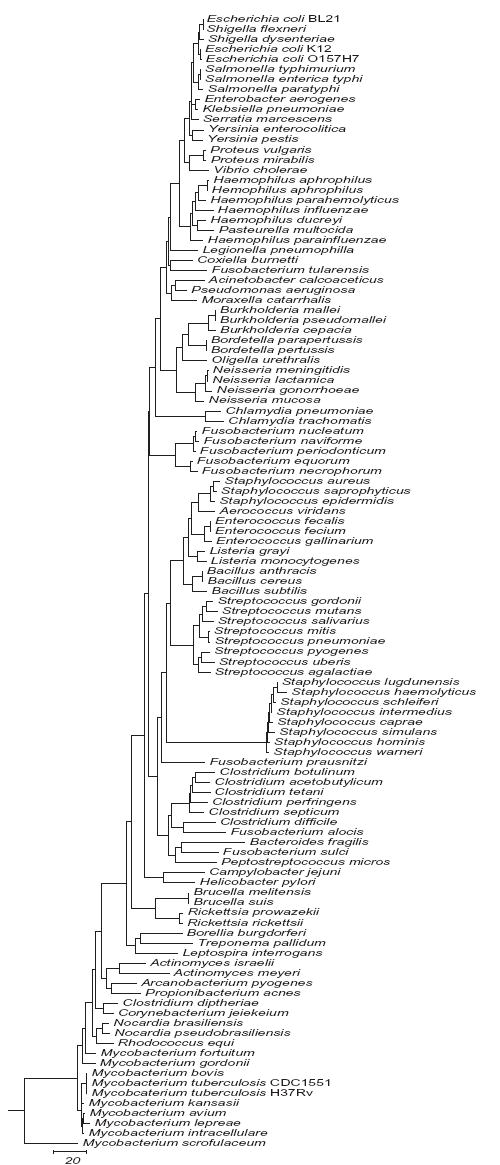
We analyzed each hypervariable region for its potential to distinguish among the 110 bacterial species in this study. A separate sequence similarity dendrogram was generated for each region. Representative dendrograms for regions V3 and V6 and a dendrogram derived from concatenating V2, V3 and V6 are shown in Figs. 1a, 1b and 1c respectively. The complete series of dendrograms used in this analysis are available on line at http://njms.umdnj.edu/departments/medicine/infectious_diseases/our_research.cfm as supplimentary figs. 1 to 6. A bacterial species with a unique DNA sequence (compared to the entire 110 bacterial species data set) was identified by its sole occupancy on a dendrogram branch. Bacterial species with identical DNA sequences in a given region were identified by the presence of multiple species on a single dendrogram branch. The degree of sequence similarity (or number of sequence differences) between any pair of bacteria could also be determined by calculating the sum of horizontal distances between each pair of bacterial species as measured on the dendrogram.
3.2. Region V1 (nucleotides 69-99)
The dendrogram of V1 (supplementary figs. 1) revealed that this region could be used to distinguish common pathogenic Streptococcus sp. and to differentiate between Staphylococcus aureus and coagulase negative Staphylococcus (CONS) species. An alignment of a shorter 28 nucleotide-long region within V1 spanning nucleotides 70 to 97 (numbering based on S. aureus 16S rRNA gene) for 122 sequences from 31 Staphylococcus species showed that the S. aureus 16S rRNA gene sequence contained at least 4 unique single nucleotide polymorphisms (SNPs) in this short region compared to other CONS species at positions (numbering based on S. aureus 16S rRNA gene) 73, 80, 89 and 90 (except for S. equorum and S. schleiferi which were identical at position 73 and S. lentus, S. pulvereri and S. sciuri, which were identical at positions 80, 89 and 90). S. aureus differed from the latter three CONS species at the positions 73 and 76 in V1; therefore, this short sequence is ideal for designing S. aureus specific probes.
3.3. Region V2 (nucleotides 137-242)
The dendrogram of V2 (supplementary fig. 2) revealed that this region could distinguish all of the 110 bacterial species in this study to the genus level except for (i) most Escherichia sp. and Shigella sp. and (ii) K. pneumoniae and E. aerogenes. This region was also able to distinguish among the common Staphylococcal and Streptococcal pathogens and among Clostridium, Haemophilus and Neisseria species. V2 also appeared to be the best target for distinguishing among Mycobacterial species. V2 has an average length of 100 bp, which is a relatively large target region for DNA probes. A separate analysis of three 15 – 35 bp regions (nucleotides 137-168, 182-194 and 205-242) within V2 revealed that the 13 nucleotides spanning 182-194 contained the maximum SNP variations between mycobacterial and haemophilus species and the region spanning 205-242 contained most of the SNP variation between the Staphylococcal, Streptococcal, Haemophilus, Fusobacterium, Clostridium and Neisseria species as well as other bacterial genus analyzed.
3.4. Region V3 (nucleotides 433-497)
The dendrogram of V3 (Fig. 1a), demonstrated that V3 was similar to V2 in its ability to distinguish among the 110 bacteria to the genus level. Examination of shorter regions within V3 revealed that nucleotides 456 through 479 contained the maximum number of SNPs between most bacterial species, producing almost the same dendrogram (not shown) as the full V3 sequence except for identities among Mycobacterium fortuitum, Nocardia sp., Propionibacterium acnes and Rhodococcus equi. V3 appeared to be better than V2 in distinguishing between the closely related enterobacteriaceae K. pneumoniae and E. aerogenes, and the SNP variation among different Haemophilus species in this region was also more than that in V2. V3 is shorter than V2 (65 nucleotides versus 106 nucleotides) and PCR amplification of V3 with universal primers specific to flanking sequences would result in a smaller amplicon compared to V2. The smaller amplicon size may be preferred in some real time PCR assays (Chakravorty et al., 2006).
3.5. Region V6 (nucleotides 986-1043)
Although the V6 region was only 58 bp in length, it showed considerable sequence variability and was able to distinguish among most bacterial species except for the closely related enterobacteriaceae Escherichia sp., Shigella sp. and Salmonella sp. (Fig. 1b). V6 appeared to be the best target region for assays designed to distinguish between B. anthracis and B. cereus due to a SNP at the position 988 in the B. anthracis 16S rRNA gene. This SNP was the only 16S rRNA polymorphism detected between these two closely related bacteria that was present in all the copies of their 16S rRNA genes (B. anthracis 11 copies and B. cereus 12-13 copies) (Fogel et al., 1999; Klappenbach et al., 2001). An examination of 11 CDC classified “select” agents B. anthracis, B. mallei, B. pseudomallei, Coxiella burnetti, Clostridium botulinum, C. perfringens, E. coli O157H7, F. tularensis, Rickettsia prowazekii, R. rickettsii and Y. pestis demonstrated that V6 contained unique DNA sequences that were specific for each select agent species except for E. coli O157H7. E. coli O157H7 could not be distinguished from Salmonella sp. in this region. An inspection of shorter overlapping 20-30 bp sequences in the V6 region (998-1032 and 1018-1043) revealed that nucleotides 998-1032 provided a better target for species and genus specific probes than 1018-1043 because B. anthracis and B. cereus were identical and the different Neisseria species were indistinguishable in the second region.
3.6. Regions V4, V5, V7 and V8 (nucleotides 576-682, 822-879, 1117-1173 and 1243-1294)
These four hypervariable regions (supplementary figs. 3, 4, 5 and 6) particularly V5, were less suitable for species identification due to a higher degree of sequence conservation compared to the other hypervariable regions. However, V7 may be useful for designing probes to detect most select agents. We found that B. anthracis and B. cereus differed by two SNPs in this region. However, the SNP differences were present only in three and five respectively out of the 11 copies of the B. anthracis 16S rRNA genes (Ueda et al., 1999). This might result in a less sensitive assay, as the specific probes designed to this region might not bind to all of the amplified targets.
3.7. Combining hypervariable regions
Our dendrogram analysis implied that combining V2, V3 and V6 together would provide sufficient sequence diversity to identify all the 110 bacterial species in this study to the level of the bacterial genus. Species level identification would also be possible for most but not all of the bacterial species. We therefore individually concatenated the V2, V3 and V6 sequences of each of the 110 species in this study and then generated a new dendrogram of the concatenated sequence (Fig 1c). All of the 110 bacterial species clustered within their genus on independent branches except for E. coli BL21 and S. flexneri using this approach, indicating that species-level discrimination would be possible for 97 of the 110 bacterial species. The bacteria that could be detected only to genus level using these three regions were: (i) Salmonella (spp. typhimurium / enterica typhi) (ii) B. mallei and B. pseudomallei (iii) B. parapertussis and B. pertussis (iv) Enterococcus faecalis and E. faecium (v) Brucella (spp. melitensis / suis) (vi) M. bovis and M. tuberculosis.
3.8. Universal primers and PCR sensitivity and specificity
Each hypervariable region of the 16S rRNA gene is flanked by a conserved sequence (Baker et al., 2003); this has made it possible to design “universal” PCR primers that can amplify 16S rRNA hypervariable regions from a large number of different bacterial species (Baker et al., 2003). We designed universal primers to conserved regions flanking hypervariable regions V3 and V6 based on our alignment data (Table 3). The universal primers flanking the region V3 are similar but not identical to those described previously (Baker et al., 2003). V6 is the shortest hypervariable region with the most nucleotide diversity in the sequences among bacteria analyzed. Paired with V3, most of the bacterial species analyzed here can be differentiated, as evident from their individual dendrograms (Figs. 1a and 1b). We amplified the hypervariable regions V3 and V6 from 30 bacteria, which included 20 of the most common pathogens described here, and 5 select agents (Table 2) using V3 and V6 primers. Amplicons were produced from all the bacteria tested, with the expected amplicon sizes (204 bp and 125 bp respectively) (Fig 2a and b). The small differences in the band sizes observed in some bacteria after amplification of V3 (Fig 2a) is consistent with the sequence data showing partial V3 deletions in many bacteria. In order to demonstrate that the V6 primers could be used for assays that were both sensitive and specific, we repeated the V6 PCR assay, this time using a S. epidermidis species-specific molecular beacon as a reporter molecule (SEP V6 probe; Table 3). The results show that S. epidermidis DNA could be detected with a sensitivity of 10 molecules added to the initial reaction, while control DNA from the closely related S. aureus species (which differs by three SNPs in the molecular beacon target region) was not detected even when 1,000,000 molecules of S. aureus DNA were added to the initial reaction (Fig. 3). It should be noted that universal primer-based assays would be most relevant in studies of sterile body fluids. Tests of the environment or of contained body sites would require that more specific primer sets be developed.
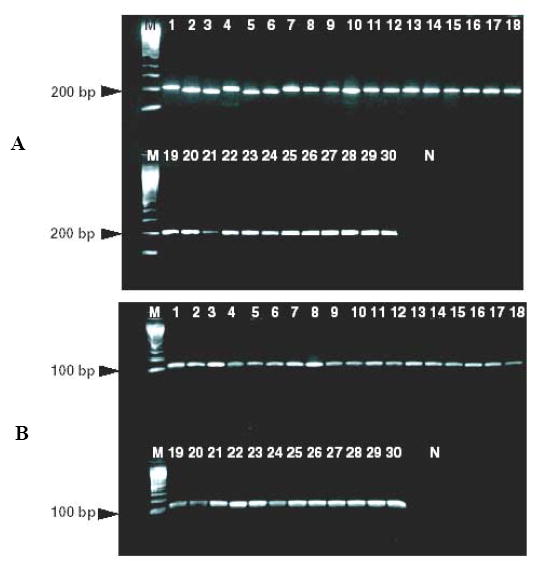
PCR amplification of 16S rRNA hypervariable regions of from 30 bacterial species using universal primers. Lane M: 100 kb molecular weight DNA marker; Lanes 1-30: Aerococcus viridans, Arcanobacterium pyogenes; Clostricium difficile; Bacillus anthracis, Bacteroides uniformis, Campylobacter jejuni, Burkholderia mallei, Corynebacterium pseudodiptheriticum, Enterococcus galinarium, Escherichia coli O157:H7, Francisella tularensis, Klebsiella pneumoniae, Haemophilus influenzae, Haemophilus parahemolyticus, Listeria grayi, Moraxella cattarhalis, Mycobacterium tuberculosis, Oligella urethralis, Proteus vulgaris, Pseudomonas aeruginosa, Neisseria gonorrhoeae, Neisseria lactamica, Neisseria mucosa, Serratia marcescens, Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis and Yersinia pestis. A: universal primers amplifying V3; B: universal primers amplifying V6.
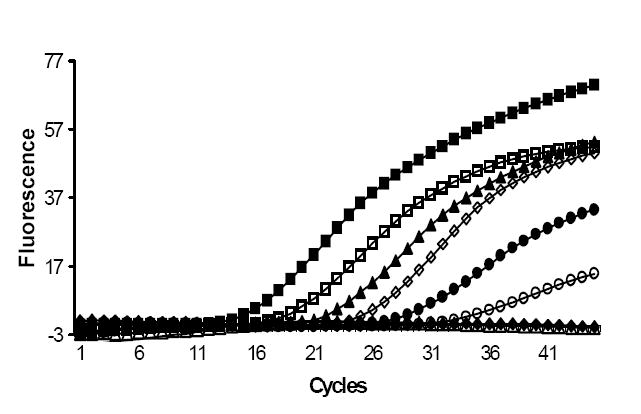
Real time PCR plot showing amplification of S. aureus and S. epidermidis genomic DNA using universal primers to V6 and a Staphylococcus epidermidis specific molecular beacon. The number of bacterial genomes added to each PCR reaction are indicated.
 106 genomes
106 genomes
 105 genomes
105 genomes
 104 genomes
104 genomes
 103 genomes
103 genomes
 100 genomes
100 genomes
 10 genomes
10 genomes
 1 genome ― DNA negative control
1 genome ― DNA negative control
 S. aureus 106 genomes
S. aureus 106 genomes
3.9. DNA sequence analysis
We sequenced the V3 and V6 regions of a diverse set of 19 bacteria (out of the 30 bacteria shown in Table 2), including most bacterial select agents and common human pathogens. The V3 sequences exactly matched the downloaded sequences used in our analysis. The V6 sequences matched the downloaded sequences by 98% – 99%. Dendrograms generated for the V3 and V6 regions using the sequence data were identical to those obtained using the downloaded sequences (data not shown). These results establish the real-life utility of our in silico analysis.
4. Discussion
Sequence analysis of conserved “housekeeping” genes such as the bacterial 16S rRNA gene are increasingly being used to identify bacterial species in clinical practice and scientific investigations (Clarridge, 2004, Petti et al., 2005). In the case of 16S rRNA analysis, species identification is easiest when most or the entire gene can be sequenced. However, DNA sequencing is impractical in medical diagnostics where speed is often of the essence. Species-specific sequences can be identified very rapidly in assays that combine nucleic acid amplification and a sequence-specific probe of the amplified product. These approaches are usually only able to query short DNA sequences; therefore, it is important to identify the regions within the target gene that supply the most taxonomic information in the smallest stretch of nucleotides. Additional benefits of small amplicon size may include increased assay sensitivity and applicability to archival specimens (Chakravorty et al., 2006: Marchetti et al., 1998).
The aim of the current study was to characterize each of the hypervariable segments of the 16S rRNA gene in 110 bacterial species and to identify short hypervariable DNA sequences that would be most beneficial for identifying specific pathogens among the entire 110 bacterial species study set. The hypervariable regions most appropriate for designing probes to identify Streptomyces sp. have been described previously (Kataoka et al., 1997; Stackebrandt et al., 1991). Another study showed that the initial 500-1500 bp of the 16S rRNA gene sequence was sufficient to discriminate among 100 bacteria (Clarridge, 2004). Schmalenberger et al. studied the utility of the 16S rRNA hypervariable regions to detect 13 bacterial species using SSCP PCR techniques and inferred that region V4-V5 would be useful (Schmalenberger et al., 2001). However, unlike our investigation, none of these previous studies focused on the relative benefits of each hypervariable region and none of these studies examined the range of pathogenic, commensal and environmental bacteria that would likely be encountered in a medical diagnostic assay.
The dendrogram analysis that we used in this study can be individually tailored to analyze portions of a hypervariable region, and thereby assess the potential discriminatory power of any assay target. Two or more regions or portions of regions can be also concatenated and then subjected to dendrogram analysis to investigate the discriminatory power of multiplexed assays. This process is further simplified by the availability of the MEGALIGN files online for the full length 16S rRNA gene and the two most relevant hypervariable regions V3 and V6 for PCR-based diagnostics of these 110 bacteria, included as supplementary materials to this study. The alignment files for the other hypervariable segments can be provided by the authors to interested investigators upon request.
Our investigation demonstrates that the hypervariable regions V2 (nucleotides 137-242), V3 (nucleotides 433-497) and V6 (nucleotides 986-1043) contain the maximum nucleotide heterogeneity and the maximum discriminatory power for the 110 bacterial species analyzed. The hypervariable region V6 (986-1043) is the shortest hypervariable region with the maximum degree of sequence heterogeneity. We also determined that V1 is the best target for discriminating between S. aureus with potentially pathogenic CONS. V2 and V3 appear to be excellent targets for speciation among the common Staphylococcal and Streptococcal pathogens as well as Clostridium and Neisseria species, with V2 especially useful for speciation of Mycobacterium sp. and for designing specific probes to detect E. coli O157H7. V3 appears to be especially useful for speciation of Haemophilus sp. V6 is the best target for the development of specific probe-based PCR assays to identify and distinguish the CDC select agents that are potential bio-terrorism agents. We created universal PCR primers for V3 and V6 to separetely amplify each hypervariable region in all 110 bacterial species in this study. Both primer sets appear to work effectively in PCR assays using a wide range of bacterial DNA. We also demonstrate the utility of the V6 primers and a molecular beacon probe designed using the dendrogram data and MEGALIGN file in a sensitive and specific real-time PCR assay to detect S. epidermidis and differentiate it from S. aureus. We confirmed our in silico analysis experimentally by sequencing the V3 and V6 regions from 19 bacteria representing a variety of clinically important genera, thereby demonstrating the utility of this type of study for designing a 16S rRNA gene based bacterial diagnostic assay. The dendrogram analysis and the sequence alignment files included in this study should be useful tools to design probes and assays that target 16S rRNA genes. Additional sequences from other bacteria may also be added to the existing MEGALIGN files to perform comparative alignments among an even broader range of bacterial species. Primers such as the ones described in this study could be combined with a number of probes such as molecular beacons or TaqMan probes (Yang et al., 2002) depending on the desired goal of the assay. This procedure will enable the creation of relatively rapid, simple and sensitive multiplexed species-identification assays.
Supplementary Material
01
02
03
04
05
06
07
08
09
Acknowledgments
This work was supported by Public Health Service grant AI-056689 from the National Institutes of Health and Department of Defense grant DAMD 17-01-1-0787 from the United States Army Medical Research Material Command.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–55. [Abstract] [Google Scholar]
- Becker K, Harmsen D, Mellmann A, Meier C, Schumann P, Peters G, von Eiff C. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J Clin Microbiol. 2004;42:4988–95. [Europe PMC free article] [Abstract] [Google Scholar]
- Bertilsson S, Cavanaugh CM, Polz MF. Sequencing-independent method to generate oligonucleotide probes targeting a variable region in bacterial 16S rRNA by PCR with detachable primers. Appl Environ Microbiol. 2002;68:6077–86. [Europe PMC free article] [Abstract] [Google Scholar]
- Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978;75:4801–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Chakravorty S, Pathak D, Dudeja M, Haldar S, Hanif M, Tyagi JS. PCR amplification of shorter fragments from the devR (Rv3133c) gene significantly increases the sensitivity of tuberculosis diagnosis. FEMS Microbiol Lett. 2006;257:306–11. [Abstract] [Google Scholar]
- Choi BK, Wyss C, Gobel UB. Phylogenetic analysis of pathogen-related oral spirochetes. J Clin Microbiol. 1996;34:1922–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Clarridge JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–62. [Europe PMC free article] [Abstract] [Google Scholar]
- Fogel GB, Collins CR, Li J, Brunk CF. Prokaryotic Genome Size and SSU rDNA Copy Number: Estimation of Microbial Relative Abundance from a Mixed Population 1999 [Abstract] [Google Scholar]
- Kataoka M, Ueda K, Kudo T, Seki T, Yoshida T. Application of the variable region in 16S rDNA to create an index for rapid species identification in the genus Streptomyces. FEMS Microbiol Lett. 1997;151:249–55. [Abstract] [Google Scholar]
- Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001;29:181–4. [Europe PMC free article] [Abstract] [Google Scholar]
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Cheang M. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–1596. [Abstract] [Google Scholar]
- Lu JJ, Perng CL, Lee SY, Wan CC. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol. 2000;38:2076–2080. [Europe PMC free article] [Abstract] [Google Scholar]
- Maloy S. Experimental techniques in bacterial genetics. Jones and Bartlett; Boston: 1989. [Google Scholar]
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymock D, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. [Europe PMC free article] [Abstract] [Google Scholar]
- Marchetti G, Gori A, Catozzi L, Vago L, Nebuloni M, Rossi MC, Esposti AD, Bandera A, Franzetti F. Evaluation of PCR in detection of Mycobacterium tuberculosis from formalin-fixed, paraffin-embedded tissues: comparison of four amplification assays. J Clin Microbiol. 1998;36:1512–1517. [Europe PMC free article] [Abstract] [Google Scholar]
- Maynard C, Berthiaume F, Lemarchand K, Harel J, Payment P, Bayardelle P, Masson L, Brousseau R. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl Environ Microbiol. 2005;71:8548–8557. [Europe PMC free article] [Abstract] [Google Scholar]
- McCabe KM, Zhang YH, Huang BL, Wagar EA, McCabe ER. Bacterial species identification after DNA amplification with a universal primer pair. Mol Genet Metab. 1999;66:205–211. [Abstract] [Google Scholar]
- Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–3029. [Europe PMC free article] [Abstract] [Google Scholar]
- Petti CA, Polage CR, Schreckenberger P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J Clin Microbiol. 2005;43:6123–6125. [Europe PMC free article] [Abstract] [Google Scholar]
- Rothman RE, Majmudar MD, Kelen GD, Madico G, Gaydos CA, Walker T, Quinn TC. Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay. J Infect Dis. 2002;186:1677–1681. [Abstract] [Google Scholar]
- Sanford JP. Hyde Park; Vermont, USA: 2003. The Sanford Guide to Antimicrobial Therapy, Antimicrobial Therapy Incorporated. [Google Scholar]
- Schmalenberger A, Schwieger F, Tebbe CC. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl Environ Microbiol. 2001;67:3557–3563. [Europe PMC free article] [Abstract] [Google Scholar]
- Selim AS, Boonkumklao P, Sone T, Assavanig A, Wada M, Yokota A. Development and assessment of a real-time PCR assay for rapid and sensitive detection of a novel thermotolerant bacterium, Lactobacillus thermotolerans, in chicken feces. Appl Environ Microbiol. 2005;71:4214–4219. [Europe PMC free article] [Abstract] [Google Scholar]
- Skow A, Mangold KA, Tajuddin M, Huntington A, Fritz B, Thomson RB, Jr, Kaul KL. Species-level identification of staphylococcal isolates by real-time PCR and melt curve analysis. J Clin Microbiol. 2005;43:2876–2880. [Europe PMC free article] [Abstract] [Google Scholar]
- Stackebrandt E, Witt D, Kemmerling C, Kroppenstedt R, Liesack W. Designation of Streptomycete 16S and 23S rRNA-based target regions for oligonucleotide probes. Appl Environ Microbiol. 1991;57:1468–1477. [Europe PMC free article] [Abstract] [Google Scholar]
- Stohr K, Hafner B, Nolte O, Wolfrum J, Sauer M, Herten DP. Species-specific identification of mycobacterial 16S rRNA PCR amplicons using smart probes. Anal Chem. 2005;77:7195–7203. [Abstract] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. [Abstract] [Google Scholar]
- Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181:78–82. [Europe PMC free article] [Abstract] [Google Scholar]
- Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. [Europe PMC free article] [Abstract] [Google Scholar]
- Varma-Basil M, El-Hajj H, Marras SA, Hazbon MH, Mann JM, Connell ND, Kramer FR, Alland D. Molecular beacons for multiplex detection of four bacterial bioterrorism agents. Clin Chem. 2004;50:1060–1062. [Abstract] [Google Scholar]
- Yang S, Lin S, Kelen GD, Quinn TC, Dick JD, Gaydos CA, Rothman RE. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. Journal of Clinical Microbiology. 2002;40:3449–3454. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.mimet.2007.02.005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2562909?pdf=render
Citations & impact
Impact metrics
Article citations
Comparative analyses on nitrogen removal microbes and functional genes within anaerobic-anoxic-oxic and deoxidation ditch sewage-treating processes in Wuhan and Xi'an cities, China.
Front Microbiol, 15:1498681, 30 Oct 2024
Cited by: 0 articles | PMID: 39539698 | PMCID: PMC11557530
Biochemical characterization of <i>Mycobacterial</i> RNA polymerases.
J Bacteriol, 206(10):e0025624, 24 Sep 2024
Cited by: 0 articles | PMID: 39315796
Molecular characteristics and antimicrobial susceptibility profiles of bovine mastitis agents in western Türkiye.
J Vet Sci, 25(5):e72, 01 Sep 2024
Cited by: 0 articles | PMID: 39363660 | PMCID: PMC11450396
Effects of Differently Processed Tea on the Gut Microbiota.
Molecules, 29(17):4020, 25 Aug 2024
Cited by: 0 articles | PMID: 39274868 | PMCID: PMC11397556
Review Free full text in Europe PMC
The impact of lactic acid bacteria inoculation on the fermentation and metabolomic dynamics of indigenous Beijing douzhi microbial communities.
Front Microbiol, 15:1435834, 30 Jul 2024
Cited by: 0 articles | PMID: 39139380 | PMCID: PMC11319256
Go to all (520) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 69 of 69)
- (2 citations) ENA - U59278
- (2 citations) ENA - X79225
- (2 citations) ENA - AJ291732
- (1 citation) ENA - AY346310
- (1 citation) ENA - X98526
- (1 citation) ENA - D83373
- (1 citation) ENA - AJ239282
- (1 citation) ENA - AF430038
- (1 citation) ENA - X76337
- (1 citation) ENA - AY268175
- (1 citation) ENA - L37603
- (1 citation) ENA - AB004750
- (1 citation) ENA - Y17657
- (1 citation) ENA - AJ239313
- (1 citation) ENA - D38483
- (1 citation) ENA - X80594
- (1 citation) ENA - AJ535697
- (1 citation) ENA - D83372
- (1 citation) ENA - AJ581472
- (1 citation) ENA - AY707778
- (1 citation) ENA - U41048
- (1 citation) ENA - AJ717375
- (1 citation) ENA - AF105402
- (1 citation) ENA - AY613778
- (1 citation) ENA - X84250
- (1 citation) ENA - D83369
- (1 citation) ENA - D83367
- (1 citation) ENA - M11656
- (1 citation) ENA - AB211325
- (1 citation) ENA - AF542231
- (1 citation) ENA - AJ295750
- (1 citation) ENA - AJ413954
- (1 citation) ENA - AF072474
- (1 citation) ENA - AJ810271
- (1 citation) ENA - X74407
- (1 citation) ENA - AJ536035
- (1 citation) ENA - AY188352
- (1 citation) ENA - U10876
- (1 citation) ENA - AJ536036
- (1 citation) ENA - X80682
- (1 citation) ENA - AJ536037
- (1 citation) ENA - AM050580
- (1 citation) ENA - AJ239304
- (1 citation) ENA - X96966
- (1 citation) ENA - AF525028
- (1 citation) ENA - AJ416915
- (1 citation) ENA - U11021
- (1 citation) ENA - AB009941
- (1 citation) ENA - AJ290755
- (1 citation) ENA - AJ810275
- (1 citation) ENA - Z75316
- (1 citation) ENA - AY365453
- (1 citation) ENA - AY365451
- (1 citation) ENA - AJ301682
- (1 citation) ENA - AJ233431
- (1 citation) ENA - X84855
- (1 citation) ENA - AJ888984
- (1 citation) ENA - AJ290758
- (1 citation) ENA - AY513496
- (1 citation) ENA - X82451
- (1 citation) ENA - X82450
- (1 citation) ENA - AJ301833
- (1 citation) ENA - AJ006965
- (1 citation) ENA - AJ006963
- (1 citation) ENA - AJ006962
- (1 citation) ENA - AJ698865
- (1 citation) ENA - AF480604
- (1 citation) ENA - X07652
- (1 citation) ENA - AJ605115
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform.
BMC Microbiol, 17(1):23, 21 Jan 2017
Cited by: 68 articles | PMID: 28109256 | PMCID: PMC5251215
Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples.
PLoS One, 11(2):e0148047, 01 Feb 2016
Cited by: 70 articles | PMID: 26829716 | PMCID: PMC4734828
Determining the most accurate 16S rRNA hypervariable region for taxonomic identification from respiratory samples.
Sci Rep, 13(1):3974, 09 Mar 2023
Cited by: 21 articles | PMID: 36894603 | PMCID: PMC9998635
[16S rRNA gene sequence analysis for bacterial identification in the clinical laboratory].
Rinsho Byori, 61(12):1107-1115, 01 Dec 2013
Cited by: 4 articles | PMID: 24605544
Review
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: AI-056689
Grant ID: R01 AI080653
Grant ID: U01 AI056689