Abstract
Free full text

Nucleoporins Prevent DNA Damage Accumulation by Modulating Ulp1-dependent Sumoylation Processes
Associated Data
Abstract
Increasing evidences suggest that nuclear pore complexes (NPCs) control different aspects of nuclear metabolism, including transcription, nuclear organization, and DNA repair. We previously established that the Nup84 complex, a major NPC building block, is part of a genetic network involved in DNA repair. Here, we show that double-strand break (DSB) appearance is linked to a shared function of the Nup84 and the Nup60/Mlp1–2 complexes. Mutants within these complexes exhibit similar genetic interactions and alteration in DNA repair processes as mutants of the SUMO-protease Ulp1. Consistently, these nucleoporins are required for maintenance of proper Ulp1 levels at NPCs and for the establishment of the appropriate sumoylation of several cellular proteins, including the DNA repair factor Yku70. Moreover, restoration of nuclear envelope-associated Ulp1 in nucleoporin mutants reestablishes proper sumoylation patterns and suppresses DSB accumulation and genetic interactions with DNA repair genes. Our results thus provide a molecular mechanism that underlies the connection between NPC and genome stability.
INTRODUCTION
The nuclear envelope is the physical barrier between the nucleus and the cytoplasm in all eukaryotic cells, and, for that reason, it plays a fundamental role in the exchange of molecules between the two compartments. Indeed, the traffic of all soluble materials occurs through nuclear pore complexes (NPCs), which are evolutionarily conserved, large, multiprotein assemblies located at the fusion points between the outer and the inner nuclear membranes (Suntharalingam and Wente, 2003). Beside this canonical role, a growing body of evidence suggests that the function of nuclear pore proteins—or nucleoporins—is not limited to nucleocytoplasmic transport. Multiple connections between the nuclear periphery and different aspects of intranuclear metabolism have been highlighted over the past years. Indeed, NPCs seem to play a crucial role in defining the transcriptionally active domains within the genome. In budding yeast, activated genes were shown to be physically recruited to nuclear pores (Casolari et al., 2005; Taddei et al., 2006). Association of active loci with the nuclear periphery requires several proteins of the nuclear envelope, including nucleoporins, but it also requires chromatin-modifying complexes and mRNA export factors (Brickner and Walter, 2004; Menon et al., 2005; Cabal et al., 2006; Dieppois et al., 2006; Luthra et al., 2006). Furthermore, this process may help to define heterochromatin domains (Schmid et al., 2006, and references therein). Such a phenomenon may be conserved in metazoans, as suggested by the association of the dosage compensation complex with nucleoporins in flies (Mendjan et al., 2006). In parallel, NPCs and other nuclear envelope-associated proteins may play a more global role in chromosomal organization within the eukaryotic nucleus. Indeed, budding yeast telomeres are tethered in four to eight foci at the nuclear periphery, and this anchoring requires the two redundant Sir4-Esc1 and Ku pathways (Andrulis et al., 2002; Taddei et al., 2004). Ku is a conserved dimer of the Yku70 and Yku80 proteins in yeast, which plays a crucial role in DNA repair through non-homologous-end joining (NHEJ), a double-strand break (DSB) repair pathway that is alternative to homologous recombination (HR). Telomere tethering at the nuclear periphery promotes, in turn, the establishment of a transcriptionally repressed state at subtelomeric loci. However, the contribution of nucleoporins to the anchoring of telomeres at the nuclear periphery and to subtelomeric transcriptional repression has led to a controversial debate (Galy et al., 2000; Feuerbach et al., 2002; Hediger et al., 2002; Therizols et al., 2006). Two subsets of nucleoporins seem to be specifically involved in connecting NPCs with nuclear metabolism and/or organization. On one hand, a leading role has emerged for nuclear basket proteins, possibly reflecting their strategic location at the nuclear side of NPCs. Among them, the yeast Nup60 nucleoporin and the associated myosin-like proteins, Mlp1 and Mlp2, as well as their orthologues in metazoans, Nup153 and Tpr/MTor, respectively, have been implicated in various nuclear processes (Feuerbach et al., 2002; Hediger et al., 2002; Casolari et al., 2005; Mendjan et al., 2006). In particular, it was demonstrated that Nup60/Mlp1–2 maintain the SUMO-protease Ulp1 at the nuclear envelope, thereby preventing clonal lethality (Zhao et al., 2004). On the other hand, the Saccharomyces cerevisiae Nup84 complex, a symmetrically localized and essential scaffold of NPCs, plays a crucial role in telomere tethering at the nuclear periphery, and in some aspects of transcriptional regulation, including subtelomeric repression (Galy et al., 2000; Menon et al., 2005; Therizols et al., 2006). This evolutionarily conserved complex is composed, in yeast, of Nup133, Nup84, the C-terminal domain of Nup145, Nup85, Nup120, Seh1, and a fraction of Sec13 (Lutzmann et al., 2002). Recently, we uncovered a strong functional link between the Nup84 complex and DNA repair in budding yeast (Loeillet et al., 2005). Inactivation of representative members of the Nup84 complex led to synthetic lethality when combined with deletions of genes of the RAD52 epistasis group, which is required for DSB repair through HR. Moreover, mutants of the Nup84 complex were highly sensitive to DNA-damaging treatments, such as ionizing irradiation or clastogen chemicals (Loeillet et al., 2005, and references therein), a phenotype conserved in Aspergillus nidulans (De Souza et al., 2006). Double-strand breaks, which are first recognized by the MRX complex, coalesce into nuclear foci, where, depending on the cell cycle phase, they are further engaged for repair either via the NHEJ or the HR pathway (Lisby and Rothstein, 2005). In the latter, Rad52 is subsequently recruited within the repair foci. We previously observed an increased occurrence of Rad52-containing DNA repair foci in the nup133Δ mutant (Loeillet et al., 2005), suggesting that inactivation of the Nup84 complex triggers an accumulation of unrepaired DNA breaks. It was subsequently demonstrated that Nup84 complex constituents are specifically required for DNA repair in subtelomeric regions (Therizols et al., 2006).
However, the molecular mechanism that underlies the connections between the Nup84 nuclear pore complex and DNA repair remained to be investigated. In this report, we establish that in addition to the Nup84 complex, the Nup60 nucleoporin is required to prevent DSB accumulation. We show that this function is ensured by the maintenance of proper levels of the Ulp1 SUMO-protease at the nuclear envelope. We further demonstrate that nucleoporins mutants affect the sumoylation status of some cellular proteins, including the DNA repair factor Yku70, which was recently shown to be modified by SUMO (Zhao and Blobel, 2005). Our results thus demonstrate the involvement of Ulp1 as a downstream effector connecting two specific nucleoporin complexes with DNA repair processes.
MATERIALS AND METHODS
Yeast Strains and Plasmids
The genotypes and origins of the strains used are listed in Supplemental Table 1. All strains are isogenic to S288c, except RAD52-yellow fluorescent protein (YFP), leu2-K::URA3-ADE2::leu2-k, and YKU70-myc–tagged strains that are W303 derivatives. Most strains were obtained by successive crosses between single-gene deletants obtained from EUROSCARF (Frankfurt, Germany), and green fluorescent protein (GFP)- or monomeric red fluorescent protein (mRFP)-tagged BY derivatives. Auxotrophy marker conversion was performed using the KanMX::URA3 modifier (Loeillet et al., 2005). For scoring genetic interactions, nup133Δ and nup60Δ strains harboring the haploid-specific marker P2LEU2 (Loeillet et al., 2005) were crossed to MAT α haploids from the EUROSCARF deletion collection. Genotypes were checked by polymerase chain reaction (PCR) amplification (the sequences of the primers used are available upon request). Yeast growth in standard YPD or SC media, gene induction by galactose, transformation, mating, and sporulation were performed as described previously (Loeillet et al., 2005; Palancade et al., 2005). Except when indicated, cells were grown at 30°C.
Plasmids used in this study are listed in Supplemental Table 2.
Cell Imaging
Live cell imaging was performed as described previously (Loeillet et al., 2005; Palancade et al., 2005) by using a three-dimensional (3D)-epifluorescence microscope driven by MetaMorph 6.2.6 software (Molecular Devices, Sunnyvale, CA). Images were further processed using Adobe Photoshop CS (Adobe Systems, Mountain View, CA). For statistical analyses of Rad52 foci, 3D-projections of Z-stack images (n = 11; plane spacing, 0.4 μm) of live cells were used, and foci were scored by visual inspection. Quantification of the number of foci per nucleus was thereby determined for the whole cell population (circular diagrams and histogram in Figure 5C). To quantify Rad52 foci occurrence for each stage of the cell cycle, cells were staged as G1 (unbudded), S (small-budded), or G2/M (large-budded) based on differential interference contrast (DIC) images. Within each of these subpopulations, the percentages of cells exhibiting at least one Rad52 focus were quantified (histograms). In vivo nuclear import assays were performed essentially as described previously (Timney et al., 2006). Briefly, exponentially growing cells expressing the appropriate nuclear localization signal (NLS)–GFP fusion protein were metabolically poisoned in the presence of 2-deoxyglucose and sodium azide to equilibrate the reporter between the nuclear and cytoplasmic compartments. Nuclear import was induced in the presence of glucose-containing medium, and images were acquired every 20 s. Nuclear intensities were determined using MetaMorph, and they were plotted as a function of time; the slope of the linear portion of the resulting import curve (t <5 min) was normalized to the initial cytoplasmic concentration of the reporter to calculate the absolute import rate.
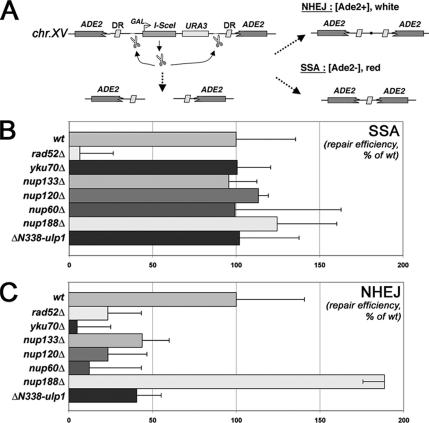
DNA repair efficiencies in nucleoporin and ulp1 mutants. (A) Schematic figure that summarizes the principle of the assay is shown. DR, direct repeats, mediating homologous recombination. (B and C) Absolute SSA and NHEJ values were calculated as described in Materials and Methods, and they represent the number of colonies grown in galactose-relative to those grown in glucose-containing medium. The results of three independent experiments are shown along with the corresponding SD, and they are normalized to 100 for the wt value.
Protein Extraction and Analysis
Whole-cell lysates were obtained from exponentially growing cells by bead-beating in 50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% NP-40, 2.5 μg/ml aprotinin, 2.5 μg/ml pepstatin, 5 μg/ml leupeptin, and 2.5 mM phenylmethylsulfonyl fluoride. Lysates were further supplemented with an equal volume of protein sample buffer and clarified by centrifugation at 10,000 × g for 10 min. Immunoprecipitation of the Yku70–13myc protein under denaturing conditions and detection of the sumoylated Yku70 were performed as described previously (Zhao and Blobel, 2005). Western blotting was performed according to standard procedures using the following primary antibodies: anti-GFP (Roche Diagnostics, Mannheim, Germany), anti-NOP1 A66 (Tollervey et al., 1991), anti-Myc (9E10; Sigma-Aldrich, St. Louis, MO), and anti-SUMO (Zhao and Blobel, 2005). Detection was performed using enhanced chemiluminescence, and quantification was achieved through MetaMorph as described in the corresponding figure legends.
DNA Repair Assays
NHEJ/single-strand annealing (SSA) assay was performed essentially as described previously (Karathanasis and Wilson, 2002). Cells were grown to mid-log phase in synthetic medium lacking uracil to maintain the I-SceI cassette, and they were plated in synthetic medium supplemented with 40 μg/ml adenine and 2% glucose or galactose. NHEJ and SSA efficiencies were calculated as the number of [Ade2+] and [Ade2−] colonies, respectively, formed on galactose medium relative to the number of colonies grown on glucose. Recombination rate was determined as the frequency of deletions of the chromosomal leu2-k::ADE2-URA3::leu2-k system based on two 2.16-kb leu2 repeats. For each genotype, the average and standard deviations were based on the median values obtained from four to six fluctuations tests made with two to three different transformants by using six independent colonies per fluctuation test (Piruat and Aguilera, 1996).
RESULTS
DSB Accumulation Is a Common Phenotype of Mutants of the Nup84 and Nup60 Complexes
We previously reported that, unlike NUP133 deletion, the nup133ΔN separation-of-function mutation, which mainly impairs NPC distribution, does not lead to an increased level of DNA repair foci (Loeillet et al., 2005). To broaden this study, we investigated whether the DSB accumulation observed in the nup133Δ mutant was linked to another canonical NPC function, or to a more specific function of the Nup84 complex in DNA damage prevention and/or repair. We therefore analyzed Rad52 foci appearance in other mutants of the Nup84 complex (i.e., nup120Δ and nup84Δ), nucleoporin mutants showing defects in mRNA export and nuclear pore distribution (i.e., nup159-1, nup82Δ108; Belgareh et al., 1998), mutants with impaired protein transport (i.e., ntf2-1, kap121-34, Corbett and Silver, 1996; Leslie et al., 2002) and a mutant primarily affected in mRNA export (i.e., sac3Δ; Fischer et al., 2002). As observed previously for the nup133Δ mutant, the other analyzed mutants of the Nup84 complex (nup120Δ and nup84Δ) exhibited DNA repair foci accumulation (Figure 1A), in a cell-cycle independent manner (Figure 1Ab) and with an unusual rate of multiple foci per cell (Figure 1Ac). In contrast, mutations within most nuclear pore or nuclear transport components, which affect NPC distribution, protein import, or mRNA export, did not increase the amount of Rad52 foci (Figure 1A), even when analyzed at restrictive temperatures (data not shown). One notable exception was the nup60Δ nucleoporin mutant. Indeed, this mutant exhibited elevated accumulation of DNA repair foci with an enhanced occurrence of multiple foci, even more pronounced than in the Nup84 complex mutants (Figure 1A). Because Nup60 is known to anchor Mlp1 and Mlp2 at the NPC, we also analyzed Rad52 foci in the corresponding mutants. Although no defects were observed upon deletion of either MLP1 or MLP2, an increased occurrence of Rad52 foci was observed in the mlp1Δ mlp2Δ double mutant (Zhao, unpublished data). These results thus indicated that DSB accumulation is unlikely to result from primary defects in nuclear transport or NPC distribution, but rather that it is linked to a shared function of the Nup84 and Nup60/Mlp1–2 complexes. As a complementary approach, we used a chromosomal reporter designed to monitor spontaneous homologous recombination events between repeats (i.e., the leu2k-ADE2-URA3-leu2k construct, in which the loss of the ADE2 and URA3 prototrophy markers is used as readout; Piruat and Aguilera, 1996). This assay revealed that homologous recombination per se was not impaired in nup133Δ, nup120Δ, and nup60Δ mutants (as also shown in Figure 5). Rather, these mutants presented exacerbated levels of recombination (Figure 1B), a result consistent with an increased occurrence of spontaneous breaks. Indeed, DSBs accumulating within the chromosomal reporter region can serve as starting points for homologous recombination and hence trigger hyperrecombination. Together, these data suggest that mutants of both the Nup84 and the Nup60–Mlp1/2 complexes exhibit an increased occurrence of DNA double-strand breaks.
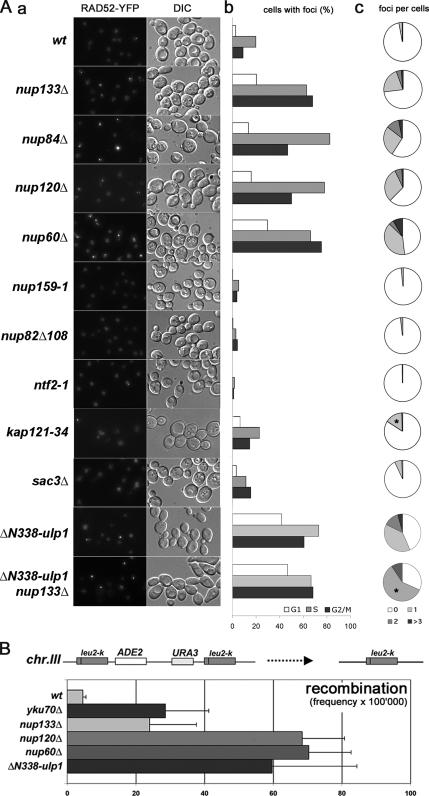
DNA repair foci analysis in nuclear pore and nucleocytoplasmic transport mutants. (A) a, fluorescence microscopy analysis of Rad52-YFP–expressing strains grown at 30°C. The DIC images are also shown. b, quantification of Rad52–YFP foci occurrence for each phase of the cell cycle. c, quantification of the number of Rad52–YFP foci per nucleus. The results of one representative experiment among three is shown, where at least 300 cells were counted for each strain. *, apparent increase in the number of foci in kap121-34 and ΔN338-ulp1 nup133Δ mutants is caused by accumulation of cells in G2/M (~60% in both mutants compared with 25–30% in wt cells). (B) Hyperrecombination assay in nucleoporins, ulp1, and yku70Δ mutants. Recombination frequencies were calculated for the indicated strains as described in Materials and Methods.
Nucleoporin and ulp1 Mutants Exhibit Similar Genetic Interaction Profiles and Repair Phenotypes
Another DNA repair-related characteristic of the Nup84 complex mutants is their synthetic lethality or synergistic interaction with mutants that have defects in DNA replication and/or repair, mainly in the Rad52 pathway (Loeillet et al., 2005). We therefore analyzed genetic interactions between the nup60Δ mutant and mutants with defects in different pathways of DNA repair. The genetic interaction profile of the nup60Δ mutant strikingly resembled that of mutants of the Nup84 complex, such as nup133Δ (Table 1). Indeed, the nup60Δ mutant showed a strongly impaired cell growth when combined with mutants of the RAD52 pathway or with rad27Δ, but not when they were associated with mutants with defects in NHEJ (yku70Δ) or DNA damage bypass (rad6Δ). The growth phenotypes of the double mutants were more pronounced with nup133Δ, which could be attributed to a poorer fitness of the nup133Δ single mutant. Finally, the mlp1Δ mlp2Δ double mutant exhibited colethality when combined with mutants carrying deletions of RAD52 or RAD27 (Supplemental Figure 1). The genetic interaction profiles of nup133Δ and nup60Δ were reminiscent of the profile described previously for a thermosensitive allele of ULP1, ulp1-I615N, which was also shown to accumulate DNA damage (Table 1; Soustelle et al., 2004). ULP1 encodes a SUMO-protease that is located at NPCs (Li and Hochstrasser, 2003; Panse et al., 2003). Analysis of the ΔN338-ulp1 mutant (Zhao et al., 2004), which lacks its nuclear envelope localization domain (Li and Hochstrasser, 2003; Panse et al., 2003), revealed a strong hyperrecombination phenotype (Figure 1B) and an increased occurrence of Rad52 foci (Figure 1A). The extent of DNA repair foci accumulation was comparable in the ulp1 and nucleoporin mutants, and it was not significantly enhanced when the ΔN338-ulp1 mutation was combined with NUP133 deletion. Finally, our systematic synthetic lethal screening of the collection of nonessential gene deletions using the nup133Δ mutation as a bait, although confirming previously described interactions of the Nup84 complex with several genes involved in DNA repair (Loeillet et al., 2005), also identified SLX5 and SLX8, two genes involved in regulating DNA repair and sumoylation-dependent processes (Mullen et al., 2001; Wang et al., 2006). Further analyses indicated that the combined deletion of NUP60 and SLX8 also leads to a strong synergistic phenotype (Table 1). Together, these data suggest that the Nup84 complex, Nup60/Mlp1–2, and Ulp1 could be involved in a common pathway that prevents the accumulation of unrepaired DNA damage through sumoylation-dependent processes.
Table 1.
Summary of the genetic interactions between nup133Δ, nup60Δ, or ulp1-I615N and DNA repair pathways mutants
| Mutants assayed | nup133Δa,b | nup60Δb | ulp1-I615Nc |
|---|---|---|---|
| rad50Δ | SL | S | SL |
| mre11Δ | SL | S | SL |
| rad52Δ | SL | S | SS |
| rad51Δ | SS | S | SS |
| rad54Δ | SL | SS | SS |
| rad55Δ | SS | S | SS |
| srs2Δ | SS | S | SS |
| yku70Δ | V | V | V |
| rad6Δ | V | V | V |
| rad27Δ | SL | SS | SL |
| slx5Δ | SS | n.d. | n.d. |
| slx8Δ | SS | SS | n.d. |
nupΔ mutants were tested for genetic interactions with mutants in which different aspects of DNA metabolism are affected, including DSB resection (rad50Δ and mre11Δ), homologous recombination (rad52Δ, rad51Δ, rad54Δ, rad55Δ, and srs2Δ), non-homologous end-joining (yku70Δ), and ubiquitination-mediated DNA damage bypass (rad6Δ), replication (rad27Δ), and DNA stability/sumoylation processes (slx5Δ and slx8Δ). Genetic interactions were scored upon tetrad analysis of double mutant growth as described previously (Loeillet et al., 2005). SL, synthetic lethal; SS, strong synergic; S, synergic; V, viable; n.d., not determined.
Nuclear Envelope Localization and Stability of Ulp1 Requires Both Nup60 and the Nup84 Complex
Previously published data indicated that Ulp1 is targeted to the nuclear pore basket through interaction with Mlp1–2 and that destabilization or mislocalization of Ulp1 could account for the clonal lethality occurring in the mlp1Δ mlp2Δ mutant (Zhao et al., 2004). To determine whether the Nup84 complex could also be involved in this process, we analyzed the subcellular localization of an Ulp1–GFP fusion protein, expressed under the control of its endogenous promoter, in mutants of the Nup84 complex. As reported previously for Mlp1–2 and associated proteins (Galy et al., 2004; Zhao et al., 2004; Palancade et al., 2005), Ulp1 was asymmetrically distributed along the nuclear envelope in wild-type (wt) cells (Figure 2A). Strikingly, deletions in nucleoporins of the Nup84 complex (i.e., nup133Δ, nup120Δ) led to a major decrease in the level of NPC-associated Ulp1, with only a faint residual signal in clustered NPCs. In contrast, the signal of Nup49-GFP, a classical nuclear pore marker, was increased in NPCs clusters, an expected effect of NPC aggregation in these mutants (Figure 2A). Quantitative Western blot analysis revealed a fivefold decrease of the Ulp1 protein levels in the Nup84 complex mutants (Figure 2, B and C). Similarly, deletion of NUP60 led, as described previously (Zhao et al., 2004), to the mislocalization and destabilization of Ulp1 (Figure 2, A–C). In contrast, Ulp1–GFP localization and stability were not affected in the nup159-1 mutant or in the absence of Nup188, a structural nucleoporin that does not belong to the Nup84 complex. Notably, NPC levels of Ulp1 were partially restored upon MG132 treatment of drug-responsive (erg6Δ) nup133Δ or nup60Δ derivatives (Supplemental Figure 2), thus indicating that proteasome-mediated degradation of Ulp1 contributes to its decreased levels at NPCs. Therefore, Ulp1 targeting and/or stabilization at NPCs specifically involve both Nup60 and the Nup84 complex. Consistent with this finding, analysis of the global pattern of sumoylation in the nup120Δ and nup60Δ mutants revealed that the sumoylation status of several proteins was affected. A similar effect was also observed in the ulp1 mutant lacking its N-terminal NPC localization domain (Figure 2D). In both nucleoporins and ΔN338-ulp1 mutants, either increased or reduced sumoylation level of some proteins was observed (see bands indicated by arrowheads and stars, respectively). These effects may reflect the consequences both of Ulp1 degradation (which causes an overall decrease in Ulp1 activity) and mislocalization (which leads to an increased Ulp1 activity in the nucleoplasm) in these mutants (see Discussion).
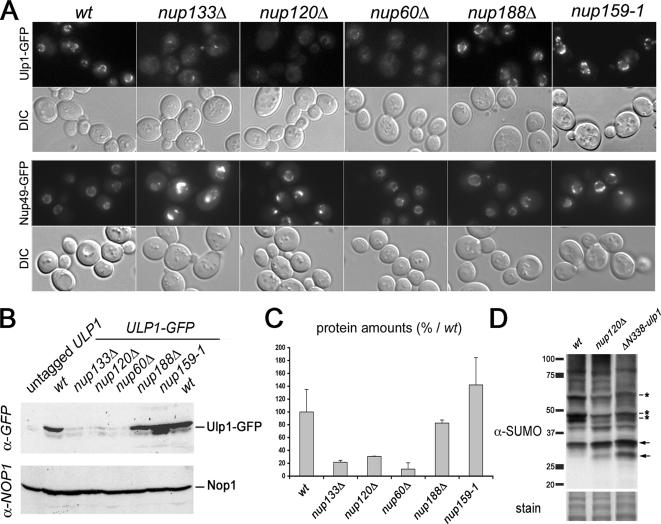
The Nup84 complex and Nup60 are required for Ulp1 localization and stabilization and for the establishment of proper sumoylation patterns. (A) Fluorescence microscopy analysis of ULP1-GFP and NUP49-GFP localization in wild-type and nucleoporin mutant strains. The DIC images are also shown. Cells were grown at 30°C, except for the nup159-1 mutant, which was grown at 22°C to visualize NPC aggregation (Belgareh et al., 1998). (B) Whole cell lysates from ULP1-GFP–tagged strains were analyzed by Western blot by using an anti-GFP antibody and an anti-NOP1 (nucleolar protein) antibody as a loading control. The positions of the Ulp1–GFP fusion protein and of Nop1 are indicated. (C) Quantification of the amounts of Ulp1 in the different nucleoporin mutants. Serial dilutions of the samples used in B were analyzed by Western blotting and the integrated intensities of the Ulp1–GFP bands were measured using MetaMorph software. The amounts are expressed as a percentage of the wt value. (D) Total sumoylated proteins from wt, nup120Δ and ΔN338-ulp1 strains were detected using an anti-SUMO antibody. Similar amounts of proteins were loaded for each lane, as indicated by the amido-black staining of the same blot (“stain”). Molecular weights (kilodaltons) are indicated. SUMO-conjugates that are decreased in the mutants are indicated by stars, and the increased conjugates are indicated by arrows.
Karyopherin-mediated Pathways Do Not Account for the Decreased Levels of Ulp1 at NPCs in Nup84 or Nup60/Mlp1–2 Complexes Mutants
We next aimed at understanding the molecular mechanisms underlying the requirement of both Nup84 and Nup60/Mlp1–2 complexes for the localization and/or stabilization of Ulp1 at the nuclear envelope. Because it has been previously suggested that Nup145-C, a subunit of the Nup84 complex, may anchor Nup60 at the NPCs (Feuerbach et al., 2002), we first analyzed the localization of Nup60 in mutants of the Nup84 complex. Deletion of NUP133, NUP120, or NUP145-C led to the accumulation of Nup60-GFP within the NPC clusters, but it did not otherwise affect its NPC localization (Figure 3A), even at restrictive temperatures (data not shown). Similarly, Mlp1 and Mlp2 became concentrated within the NPC clusters in nup133Δ and nup120Δ mutants (Palancade et al., 2005; data not shown). Conversely, deletion of NUP60 did not affect Nup133 localization (data not shown). Thus, the NPC localization of the Nup84 and Nup60/Mlp1–2 complexes seem to be two unrelated processes. Previous studies demonstrated an involvement of the Kap121 and Kap95/Kap60 karyopherins in the localization of Ulp1 at the nuclear pores (Panse et al., 2003; Makhnevych et al., 2007). We thus asked whether the phenotype of the Nup84 complex mutants could be the indirect consequences of an alteration in these karyopherin-mediated pathways. Analysis of Kap95–GFP and Kap121–GFP fusion proteins revealed that both karyopherins were properly localized at NPCs in nup133Δ and nup60Δ mutant cells (Figure 3B). In addition, analysis of the static distribution and import kinetics of Kap60/Kap95-dependent (cNLS–GFP) or Kap121-dependent (pNLS–GFP) reporters did not reveal any significant import defect in nup120Δ or nup60Δ strains (Supplemental Figure 3, A and B), even though a mild import defect was observed for the pNLS–GFP reporter in the nup133Δ strain. Finally, analysis of Rad52–YFP localization did not reveal an increased occurrence of DNA repair foci in the kap121-34 mutant strain (Figure 1A; of note, the mild increase seen in Figure 1Ac is due to the accumulation of these cells in G2). These data prompted us to compare the respective effects of karyopherin and nucleoporin mutants on the localization and cellular levels of Ulp1 tagged with GFP. In agreement with a previous study (Panse et al., 2003), karyopherin inhibition, achieved by expressing a dominant-negative form of the Yrb4 importin, or by inactivation of the kap121-34-thermosensitive mutant at semirestrictive temperature (30°C), impaired the nuclear envelope localization of mildly overexpressed GFP–Ulp1 (i.e., under the control of the NOP1 promoter; Supplemental Figure 3C). However, these treatments did not significantly affect the localization of Ulp1 tagged with GFP at its cognate locus (Supplemental Figure 3C, compare with Figure 2A). Although the increased fading at higher temperature of the GFP variant used in this study did not enable us to observe any significant change in kap121-34 cells shifted to 37°C, a similar analysis, recently performed using Ulp1 tagged with the GFP+ variant, revealed that shifting the kap121-34 strain to 37°C causes a decreased level of NPC-associated Ulp1 and a corresponding increase in its cytoplasmic level (Makhnevych et al., 2007). However, Western blot analysis revealed that, unlike in the nucleoporin mutants, Ulp1-GFP was not destabilized in the kap121-34 mutant even after a 3-h shift to 37°C. Rather, a slower migrating form of Ulp1-GFP was observed in kap121-34 cells (Figure 3C), suggesting the occurrence of posttranslational modification(s) of Ulp1 in this karyopherin mutant. Together, these data indicate that karyopherin and nucleoporin mutants differentially affect Ulp1 localization and stability at NPCs. The phenotypes of the nucleoporin mutants are therefore not merely caused by a defect in these karyopherin-mediated pathways. This mildly overexpressed GFP–Ulp1 fusion protein has been shown to be targeted to the nuclear envelope in an mlp1Δ mlp2Δ ulp1Δ mutant (Zhao et al., 2004). We therefore expressed this construct in the nup133Δ, nup120Δ, and nup60Δ mutants, and we analyzed the localization of GFP-Ulp1 relatively to a core nucleoporin, Nic96, which was tagged with mRFP. In all strains, Ulp1 localization at the nuclear envelope was restored (Figure 3D; data not shown). Interestingly, the overexpressed Ulp1 fusion protein no longer exhibited its asymmetrical distribution within the nuclear envelope (compare Supplemental Figure 3C, left and right). Moreover, unlike Nic96–mRFP, its localization was no longer restricted to the nuclear pores as the GFP labeling spread out of the NPC aggregates in the nup133Δ strain (Figure 3D), indicating that mildly overexpressed GFP–Ulp1 may be ectopically targeted to areas of the nuclear envelope laying between the pores. This result may reflect the ability of this overexpressed fusion protein to directly interact with membranes, an hypothesis consistent with its partial mislocalization to the plasma membrane after karyopherin inhibition (Panse et al., 2003; Supplemental Figure 3C), or, alternatively, the presence of yet uncharacterized additional inner nuclear envelope tether(s) (discussed in Makhnevych et al., 2007). Together, our data demonstrate that although the Nup84 and Nup60/Mlp1–2 complexes are required for the maintenance of endogenous Ulp1 levels at nuclear pores, their requirement for Ulp1 localization and/or stabilization can be bypassed by overexpressing a GFP–Ulp1 fusion protein.
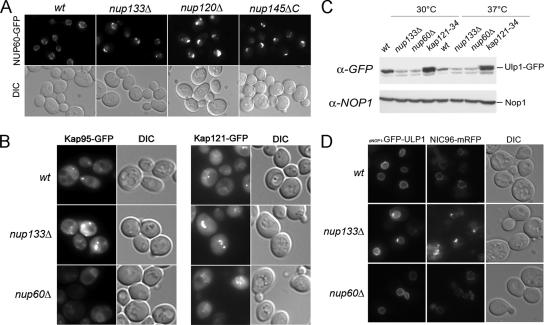
Decreased levels of Ulp1 at NPCs in Nup84 or Nup60/Mlp1–2 complexes mutants are not merely due to defects in Kap121 or Kap95 pathways. (A) Fluorescence microscopy analysis of NUP60-GFP localization in wild-type or nucleoporin mutant strains grown at 30°C. The DIC images are also shown. (B) Fluorescence microscopy analysis of KAP95-GFP (left) or KAP121-GFP (right) localization in wild-type or nucleoporin mutant strains grown at 30°C. (C) Ulp1-GFP expression levels were analyzed by Western blot by using an anti-GFP antibody in wt, nup133Δ, nup60Δ, or kap121-34 mutant strains grown at 30°C or shifted for 3 h at 37°C. The anti-NOP1 antibody was used as a loading control. (D) Fluorescence microscopy analysis of wild-type or nucleoporin mutant strains expressing NIC96-mRFP and transformed with the pNOP1-GFP-ULP1 construct. DIC images are also shown.
Restoration of Proper Ulp1 Levels at the Nuclear Envelope Partially Complements the Altered Sumoylation Patterns and the DNA Repair-related Phenotypes of the Nucleoporin Mutants
We next analyzed the consequences of the mild overexpression of this GFP–Ulp1 protein on the global pattern of sumoylation in wild-type and nucleoporin mutant strains either untreated or treated with the DNA-damaging drug methyl methane sulfonate (MMS). MMS treatment modified the sumoylation status of some proteins (Figure 4A), suggesting the involvement of sumoylation in DNA damage-induced signaling pathways. In untreated cells, GFP–Ulp1 overexpression led to a general decrease of SUMO conjugates in both wild type and nucleoporin mutants (data not shown). Overexpression of GFP-Ulp1 in the nupΔ mutants partially restored a normal sumoylation pattern in MMS-treated cells, with a notable increase of two MMS-induced sumoylated bands (Figure 4A, stars). Conversely, the enhanced sumoylation of several proteins in nupΔ, compared with wild-type cells, was corrected upon GFP-ULP1 overexpression (Figure 4A, arrows). These results show that nucleoporin mutants have altered cellular sumoylation patterns and that restoring proper Ulp1 level at the nuclear envelope counteracts this phenomenon. This result prompted us to assess the functional consequences of Ulp1 restoration on the viability of nupΔ mutants, when combined with mutations affecting DNA repair or replication. nup133Δ rad27Δ, nup133Δ rad52Δ, nup60Δ rad27Δ, and nup60Δ rad52Δ double mutants are not viable or strongly impaired in their growth at 30°C (Table 1; Loeillet et al., 2005), but they can be rescued after dissection of the corresponding diploids at 25°C. Overexpression of GFP-Ulp1 complemented the synthetic lethality or the growth defects of each of the four mutants, whereas a control plasmid could not (Figure 4B and Supplemental Figure 4A). Noteworthy, the complementation was specific for the defects that arose from the combination of nup and rad mutations, because ULP1 overexpression did not affect the growth rates of any of the single mutants (data not shown). To determine which feature of the Ulp1 protein was needed for complementation of the nupΔ radΔ colethality, different constructs were tested in this assay (Figure 4B). A plasmid lacking the GFP tag rescued cell growth as efficiently as the GFP–Ulp1 construct, indicating that the rescue is not merely due to stabilization of Ulp1 via the GFP. The catalytically inactive ulp1-C580S mutant (Li and Hochstrasser, 2003) was no longer able to rescue nupΔ radΔ colethality. Rather, this construct became toxic in the nup60Δ rad52Δ mutant and impaired viability of the nup133Δ rad52Δ mutant (Figure 4B), although it did not affect the viability of the wild type or any of the corresponding single mutants (Figure 4B; data not shown). Because this mildly overexpressed protein was properly targeted to the nuclear envelope (data not shown), it could compete at NPCs with the residual levels of endogenous Ulp1 protein that was still present in the nucleoporin mutants. Finally, expression of the ΔN338-ulp1 mutant (Zhao et al., 2004), which lacks its nuclear envelope localization domain (Li and Hochstrasser, 2003; Panse et al., 2003), did not rescue the synergistic growth defects of the double mutants (Figure 4B). However, this construct was poorly expressed (Supplemental Figure 4B); therefore, it did not allow us to discriminate whether restoration of either the expression levels or the localization of Ulp1 contributed to the complementation phenotypes. We next determined whether the DSB accumulation observed in the Nup84 complex or in nup60 mutants was due to a loss of function of Ulp1. To this end, GFP-Ulp1 was overexpressed in wild-type, nup133Δ, and nup60Δ derivatives carrying the RAD52-YFP reporter. In wt cells, mild overexpression of GFP–Ulp1 did not affect the detection and the occurrence of Rad52 foci (Figure 4C) or the level of Rad52 foci assembly after MMS treatment (data not shown). In contrast, overexpression of GFP-Ulp1 reduced the occurrence of Rad52 foci in nup133Δ and nup60Δ cells with a prominent decrease in the number of cells with multiple foci (Figure 4C and Supplemental Figure 4C). Together, our data indicate that restoring an enzymatically active Ulp1 at the nuclear envelope partially complements both the defective sumoylation of some target proteins and some of the DNA repair-related phenotypes of the nucleoporin mutants.
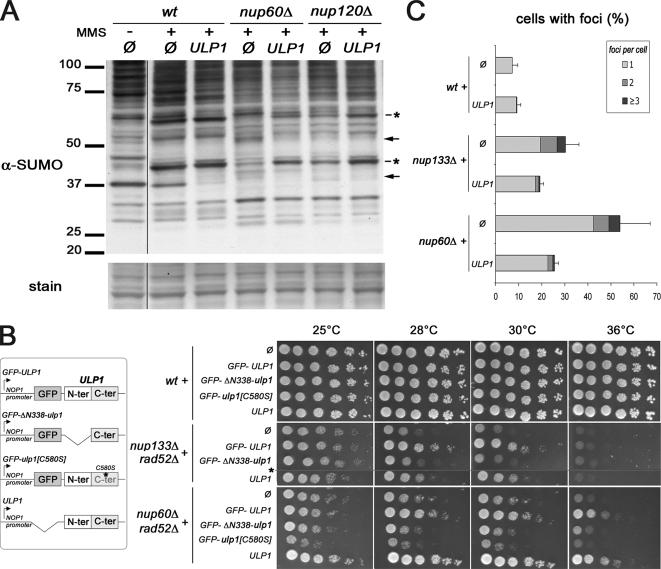
Complementation of nucleoporin mutant phenotypes by restoration of nuclear envelope-associated ULP1. (A) Total sumoylated proteins were detected using an anti-SUMO antibody in whole cell extracts from wt, nup120Δ, or nup60Δ strains that were transformed with the pRS315 (Ø) or the pRS315-NOP1prom-GFP-ULP1 (ULP1) plasmids and treated (+), or not (−), for 2 h with 0.3% MMS. Similar amounts of proteins were loaded for each lane, as indicated by the amido-black staining (“stain”) of the same blot. Molecular weights (kilodaltons) are indicated. Stars indicate SUMO-conjugates, which were decreased in the mutants and have been restored upon ULP1 expression, whereas arrows indicate the conjugates, which were increased in the mutants and reduced upon ULP1 expression. (B) wt, nup133Δrad52Δ, and nup60Δrad52Δ mutant strains were transformed with pRS315 (Ø) or pRS315 derivatives expressing the indicated constructs under the control of the NOP1 promoter (schematized on the left). Transformants were spotted as fivefold dilutions on synthetic medium lacking leucine and plates were incubated at 25, 28, 30, or 36°C. *, nup133Δ rad52Δ strain is not viable in the presence of the GFP-ulp1[C580S] construct. (C) Rad52–YFP foci analysis in wt, nup133Δ, and nup60Δ cells transformed with either pRS315 (Ø) or pRS315-NOP1prom-GFP-ULP1 (ULP1). The percentage of cells, which show one, two, or three or more Rad52–YFP foci per nucleus, is indicated. The SD corresponds to the sum of the standard deviations for each of the three subcategories.
Yku70-dependent Processes and Sumoylation Status Are Both Affected in Nucleoporin and ulp1 Mutants
To further characterize the DNA repair-related phenotypes of the nucleoporin and ulp1 mutants, we measured the efficiency of different DNA repair pathways in these mutants. For this purpose, we used a previously described reporter system (Karathanasis and Wilson, 2002) in which an unique I-SceI DSB, induced at the ADE2 locus within chromosome XV, can be repaired either by SSA, e.g., homologous recombination between direct repeats, or by nonhomologous end joining, generating, respectively, ade2- or ADE2 alleles (Figure 5A). Deletants of RAD52 and YKU70, the main players in SSA and NHEJ, respectively, were used as controls. As reported previously, NHEJ was also affected by RAD52 deletion, a finding that possibly reflects a yet unknown function of recombination proteins in facilitating NHEJ (Hegde and Klein, 2000; Karathanasis and Wilson, 2002). Nucleoporin deletions and the ΔN338-ulp1 allele were introduced into the reporter strain and both classes of DSB repair events were quantified. As shown in Figure 5B, RAD52-dependent SSA was not affected in the tested nucleoporin mutants (i.e., nup133Δ, nup120Δ, and nup60Δ), or in the ΔN338-ulp1 mutant. In contrast, NHEJ efficiency was decreased in mutants of the Nup84 complex (i.e., nup133Δ and nup120Δ), in the nup60Δ strain, and in the ulp1 mutant, whereas it was not changed by the absence of Nup188 (Figure 5C). Of note, flow cytometry analysis revealed that the NHEJ defect was not due to a general cell cycle alteration in the nup mutants (data not shown). These results indicate that the Nup84 complex, Nup60, and Ulp1 are specifically required for accurate DSB repair through the Yku70-dependent NHEJ pathway. Because Yku70 has been recently shown to be sumoylated in vitro and in vivo (Zhao and Blobel, 2005), we investigated its sumoylation status in nucleoporins and ulp1 mutants. To this end, a myc-tagged version of Yku70 was immunoprecipitated under denaturing conditions and its sumoylated forms were detected by Western blot analysis. Yku70 sumoylation levels were strongly reduced upon NUP120 and NUP60 deletion (Figure 6, A and B). A similar effect was also observed in the ΔN338-ulp1 mutant (Figure 6A), confirming that mislocalization and/or destabilization of Ulp1 ultimately impairs Yku70 sumoylation (see Discussion). These data indicate that Yku70 is one of the downstream effector of the nucleoporins and Ulp1-dependent pathway in DNA repair. Consistently, YKU70 loss of function caused a hyperrecombinant phenotype (Figure 1D) and led to colethality when combined with RAD52 deletion (albeit at higher temperatures only; Figure 6, C and D). However, yku70Δ cells did not accumulate Rad52 foci (Figure 6E). Thus, only some of the DNA repair-related phenotypes caused by nucleoporin deletions or Ulp1 mislocalization are observed upon YKU70 loss-of-function. Moreover, this indicates that the Rad52 foci accumulating in the nucleoporins mutants represent a read-out of accumulated DSBs that do not solely arise from defects in the NHEJ pathway.
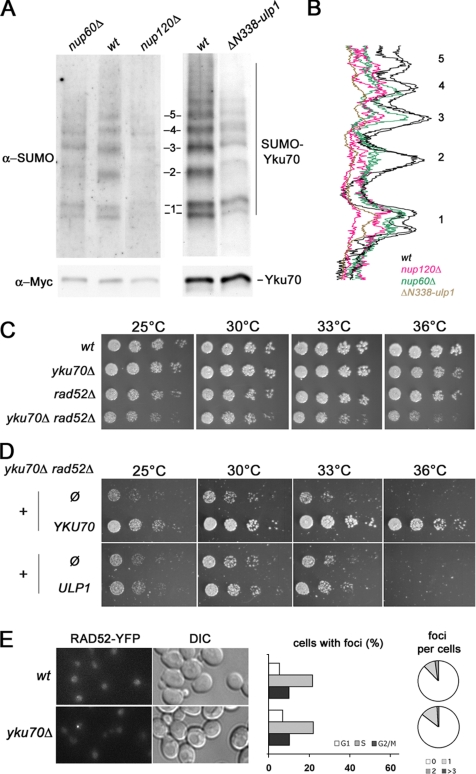
Yku70 sumoylation is altered in nup60Δ, nup120Δ and ΔN338-ulp1 strains. (A) Exponentially growing wt, nup60Δ, nup120Δ, and ΔN338-ulp1 cells expressing Myc-tagged Yku70 at its chromosomal locus were treated for 2 h with 0.3% MMS to facilitate the detection of sumoylated forms of Yku70, as reported previously (Zhao and Blobel, 2005). Unmodified and sumoylated Yku70-Myc were immunoprecipitated under denaturing conditions by using an anti-Myc antibody, and they were detected by Western blot analysis with the anti-Myc (bottom) or anti-SUMO (top) antibodies, respectively. The sumoylated forms of Yku70-Myc represent a very minor fraction of the whole Yku70–Myc population, and they were therefore not detected by the anti-Myc antibody. Control experiments using strains bearing either untagged Yku70 or another myc-tagged open reading frame are provided in Supplemental Figure 5. (B) Line-scan analysis of the sumoylated Yku70 patterns in wt, nup120Δ, nup60Δ (2 independent experiments for these 3 strains), and ΔN338-ulp1 cells were obtained using the MetaMorph software. For each sample, values were normalized to the total amount of immuno-precipitated Yku70 quantified from the anti-Myc Western blot. Numbers (1–5) on the right correspond to the position of the sumoylated bands indicated in A. (C) Segregants of a yku70Δ/+ rad52Δ/+ diploid were spotted as fivefold dilutions on YPD plates, and then they were analyzed for growth at the indicated temperatures. (D) The yku70Δ rad52Δ double mutant strain was transformed with pRS313 (Ø), pRS313-YKU70 (YKU70), pRS315 (Ø), or pRS315-NOP1prom-GFP-ULP1 (ULP1). Transformants were spotted as fivefold dilutions on synthetic medium lacking histidine (top) or leucine (bottom), and growth was analyzed at the indicated temperatures. Growth defects of the double mutant are not complemented by ULP1 overexpression, a result consistent with our model in which Yku70 acts downstream of Ulp1 in the DNA repair processes. (E) Rad52 foci analysis in yku70Δ cells. Left, Fluorescence microscopy analysis of Rad52-YFP–expressing strains grown at 30°C. The DIC images are also shown. Middle, quantification of Rad52–YFP foci occurrence for each phase of the cell cycle. Right, quantification of the number of Rad52–YFP foci per nucleus. The results of one representative yku70Δ strain among four is shown; >150 cells were counted.
DISCUSSION
Maintenance of Proper Ulp1 Levels at the Nuclear Envelope Requires the Nup84 and the Nup60/Mlp1–2 Complexes
In this study, we demonstrate that the Nup84 complex is required, together with Nup60/Mlp1–2, for maintaining proper levels of Ulp1 at NPCs. The asymmetric localization of Ulp1-GFP, when expressed under the control of its endogenous promoter, is strongly reminiscent of the localization described for Mlp1–2 and its interacting partner Pml39 (Galy et al., 2004; Zhao et al., 2004; Palancade et al., 2005). Hence, Mlp1–2 or their binding partners are likely to represent the final tethering site for endogenous Ulp1 in wild-type cells. Because Nup84 complex mutants do not impair Nup60/Mlp1–2 NPC localization, the Nup84 complex could provide an alternative binding site for Ulp1 at NPCs. This issue could not be addressed because all the tested combination of mutations affecting, respectively, the Nup84 and the Nup60-Mlp1/2 complexes led to extremely severe growth defects precluding any phenotypic analysis (Loeillet et al., 2005). Alternatively, Ulp1 may first interact with the Nup84 complex before being targeted to Mlp1–2, thus using a two-step mechanism of delivery as proposed previously for the checkpoint protein Mad1, which was suggested to interact with Nup53 before being anchored to the nuclear basket through Mlp1–2 (Scott et al., 2005). However, our attempts at identifying biochemical interactions between these nucleoporins and Ulp1 have been so far unsuccessful, suggesting that these interactions must be either biochemically unstable, or indirect. Our data also indicate that different mechanisms underlie the decreased levels of NPC-associated Ulp1 observed in the nucleoporin and karyopherin mutants. Indeed, defects in the Kap121 (and to some extent, in the Kap95-Kap60) pathway(s) impair the delivery of Ulp1 to the NPC and/or the nuclear envelope, and they cause the accumulation of Ulp1 in the cytoplasm. In contrast, Ulp1 is destabilized in nup60 or Nup84 complex mutants. In these mutants, impaired tethering of Ulp1 at the nuclear side of NPCs may in turn lead to its release and subsequent proteasome-mediated degradation in the nucleoplasm. Finally, alteration of the Nup84 complex may have a more indirect consequence on Ulp1 metabolism, for example, by regulating yet unknown effectors involved in its degradation. These hypotheses would be consistent with the partial restoration of Ulp1 at NPCs upon treatment of nup133Δ and nup60Δ cells with a proteasome inhibitor.
The Nup84 Complex and Nup60 Regulate Sumoylation Patterns and DNA Repair through Ulp1
Our data indicate that the Nup84 complex and Nup60 mutants, which exhibit decreased levels of Ulp1 at the nuclear envelope, display complex modifications of cellular sumoylation patterns that can be partially suppressed upon restoration of catalytically active Ulp1 at the nuclear envelope. Of note, both increase and decrease in the levels of SUMO-conjugates were observed in these mutants as well as in a strain expressing an N-terminal truncated allele of Ulp1 that lacks its nuclear envelope targeting domain (Figure 2D; Zhao et al., 2004). Consistent with this observation, both the level and the subcellular localization of Ulp1 have previously been shown to be major determinants in its activity toward natural and nonnatural sumoylated substrates. Indeed, disturbing one of these two parameters by overexpressing and/or truncating Ulp1 leads to significant changes in the global pattern of SUMO-protein conjugates (Li and Hochstrasser, 2003; Zhao et al., 2004). Although decreased levels of Ulp1 could lead to an oversumoylation of some of its substrates, mislocalized Ulp1, even in low amounts, could gain access to intranuclear substrates and reduce the amount of their sumoylated forms. For example, Ulp1 was shown to target the genuine substrates of its paralogue Ulp2 when mislocalized from the nuclear periphery (Li and Hochstrasser, 2003). In addition, more indirect effects of the decreased level of Ulp1 at the nuclear envelope, such as altered sumoylation of E3 ligase(s), could potentially also interfere with the sumoylation status of specific substrates. In addition to the sumoylation status of several targets, restoration of catalytically active Ulp1 at the nuclear envelope partially complemented some of the nupΔ repair phenotypes, such as colethality with rad52Δ and rad27Δ or the accumulation of Rad52 foci. However, Ulp1 overexpression poorly suppressed the NHEJ defects of the nucleoporin mutants (Supplemental Figure 4D), and it did not rescue their increased recombination frequency (data not shown). This may reflect the higher sensitivity of this reporter toward properly controlled Ulp1 activity, together with the fact that restoration of Ulp1 functions, as revealed by the aforementioned assays, is only partial. Finally, the growth defects of the nupΔ radΔ strains were enhanced upon mild overexpression of the catalytically inactive form of Ulp1. This indicates that accurate control of the sumoylation levels of some proteins is critical for the viability of nucleoporin mutants in the absence of a functional DNA recombination pathway. Similarly, it was recently reported that a mutant of the Mms21 SUMO-ligase exhibits DNA damage sensitivity (Zhao and Blobel, 2005), whereas the Slx5–Slx8 complex was suggested to be involved both in DNA integrity and sumoylation processes (Wang et al., 2006; Yang et al., 2006). Thus, our data further strengthen the connections between sumoylation and the control of genome integrity (Soustelle et al., 2004; Jacquiau et al., 2005; Motegi et al., 2006).
Multiple Ulp1-dependent Effectors of the Nup84 and Nup60 Complexes Are Required for Genome Integrity
Our study identified Yku70 as one of the targets whose sumoylation is decreased in the nucleoporins mutants as well as in the ΔN338-ulp1 mutant. This decreased sumoylation seems to be associated with a loss of function of Yku70. Indeed, both nucleoporin deletions (nup133Δ, nup120Δ, nup60Δ) and the ΔN338-ulp1 mutant exhibit reduced NHEJ levels and hyperrecombination between direct repeats, which are also observed upon YKU70 deletion (Figure 2). Although a partial loss of function of Yku70 may explain some of the DNA repair-related phenotypes, which are shared by the nucleoporins and ulp1 mutants, YKU70 deletion does not recapitulate all the nupΔ repair phenotypes (Figure 6E). In particular, accumulation of Rad52 foci, a prominent phenotype of nucleoporin and ulp1 mutants, is not observed in yku70Δ cells. This suggests that Ulp1 delocalization or destabilization in the nucleoporin mutants affects the sumoylation status of additional proteins required for DNA maintenance. Interestingly, proteins involved in processes related to DNA maintenance, e.g., DNA replication, repair, and chromatin metabolism, are overrepresented in the SUMO-modified yeast proteomes (Panse et al., 2004; Wohlschlegel et al., 2004; Denison et al., 2005; Hannich et al., 2005; Wykoff and O'Shea, 2005). Among them, PCNA (proliferating cell nuclear antigen), Rad52 (Pfander et al., 2005; Sacher et al., 2006), and members of the RSC chromatin remodeling complex, which is required for NHEJ (Shim et al., 2005), represent attractive, yet far from exclusive, candidates. Noteworthy, other nuclear processes, which are impaired in the Nup84 complex and/or nup60/mlp1–2 mutants, such as telomere tethering to the nuclear periphery, subtelomeric transcriptional repression, or control of telomere length (Feuerbach et al., 2002; Hediger et al., 2002; Therizols et al., 2006), may also depend on the sumoylation status of Yku70 or of additional sumoylated factors involved in telomere capping and repression (for example, Rap1, Sir2-3-4, and Esc1; Wohlschlegel et al., 2004; Hannich et al., 2005). A comprehensive analysis of the SUMO-modified proteome of these nucleoporins mutants may help to further unravel the links between nuclear pore complexes and the different cellular processes in which they are involved.
ACKNOWLEDGMENTS
We thank Anita Corbett, Emmanuelle Fabre, Gérard Faye, Heidi Feldmann, Pierre-Emmanuel Gleizes, David Goldfarb, Martine Heude, Ed Hurt, Won-Ki Huh, Rodney Rothstein, Mike Rout, Roger Tsien, Laurence Vernis-Beringue, Thomas Wilson, and Rick Wozniak for reagents and/or discussion; Sophie Loeillet and Siau-Wei Baï for help with yeast handling; and Alain Nicolas for interest in this work. Many thanks to the Curie Imaging staff and to all members of the Doye, Bornens, and Nehrbass laboratories for valuable comments during the course of this work. This research was supported by a collaborative program between the Institut Curie and the Commissariat à l'Energie Atomique (PIC Paramètres Epigénétiques grant to V.D.), the Ligue Nationale contre le Cancer (Equipe Labelisée 2006 to V.D.), the Association pour la Recherche contre le Cancer (ARC to V.D.), the Ministère de l'Education Nationale, de la Recherche et de l'Enseignement Supérieur (A.C.I. “Jeunes chercheurs” to V.D.), a Cancer Center Support Grant (NCI P30 CA-08478-41 to X.Z.), and the Ministry of Science and Education of Spain (grant SAF2003-00204 to A.A.). B.P. was supported by postdoctoral fellowships from ARC and Fondation Pour la Recherche Médicale.
Abbreviations used:
| DIC | differential interference contrast |
| DSB | double-strand break |
| NHEJ | nonhomologous end joining |
| HR | homologous recombination |
| MMS | methyl methane sulfonate |
| NPC | nuclear pore complex |
| SD | standard deviation |
| SSA | single strand annealing |
| wt | wild type. |
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0123) on May 30, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Andrulis E. D., Zappulla D. C., Ansari A., Perrod S., Laiosa C. V., Gartenberg M. R., Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell Biol. 2002;22:8292–8301. [Europe PMC free article] [Abstract] [Google Scholar]
- Belgareh N., Snay-Hodge C., Pasteau F., Dagher S., Cole C. N., Doye V. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell. 1998;9:3475–3492. [Europe PMC free article] [Abstract] [Google Scholar]
- Brickner J. H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. [Europe PMC free article] [Abstract] [Google Scholar]
- Cabal G. G. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. [Abstract] [Google Scholar]
- Casolari J. M., Brown C. R., Drubin D. A., Rando O. J., Silver P. A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. [Europe PMC free article] [Abstract] [Google Scholar]
- Corbett A. H., Silver P. A. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J. Biol. Chem. 1996;271:18477–18484. [Abstract] [Google Scholar]
- Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell Proteomics. 2005;4:246–254. [Abstract] [Google Scholar]
- De Souza C. P., Hashmi S. B., Horn K. P., Osmani S. A. A point mutation in the Aspergillus nidulans sonBNup98 nuclear pore complex gene causes conditional DNA damage sensitivity. Genetics. 2006;174:1881–1893. [Europe PMC free article] [Abstract] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. [Europe PMC free article] [Abstract] [Google Scholar]
- Feuerbach F., Galy V., Trelles-Sticken E., Fromont-Racine M., Jacquier A., Gilson E., Olivo-Marin J. C., Scherthan H., Nehrbass U. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 2002;4:214–221. [Abstract] [Google Scholar]
- Fischer T., Strasser K., Racz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. [Europe PMC free article] [Abstract] [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. [Abstract] [Google Scholar]
- Galy V., Olivo-Marin J. C., Scherthan H., Doye V., Rascalou N., Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. [Abstract] [Google Scholar]
- Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. [Abstract] [Google Scholar]
- Hediger F., Dubrana K., Gasser S. M. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 2002;140:79–91. [Abstract] [Google Scholar]
- Hegde V., Klein H. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 2000;28:2779–2783. [Europe PMC free article] [Abstract] [Google Scholar]
- Jacquiau H. R., van Waardenburg R. C., Reid R. J., Woo M. H., Guo H., Johnson E. S., Bjornsti M. A. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J. Biol. Chem. 2005;280:23566–23575. [Abstract] [Google Scholar]
- Karathanasis E., Wilson T. E. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics. 2002;161:1015–1027. [Europe PMC free article] [Abstract] [Google Scholar]
- Leslie D. M., Grill B., Rout M. P., Wozniak R. W., Aitchison J. D. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell Biol. 2002;22:2544–2555. [Europe PMC free article] [Abstract] [Google Scholar]
- Li S. J., Hochstrasser M. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 2003;160:1069–1081. [Europe PMC free article] [Abstract] [Google Scholar]
- Lisby M., Rothstein R. Localization of checkpoint and repair proteins in eukaryotes. Biochimie. 2005;87:579–589. [Abstract] [Google Scholar]
- Loeillet S., Palancade B., Cartron M., Thiery A., Richard G.-F., Dujon B., Doye V., Nicolas A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Rep. 2005;4:459–468. [Google Scholar]
- Luthra R., Kerr S. C., Harreman M. T., Apponi L. H., Fasken M. B., Ramineni S., Chaurasia S., Valentini S. R., Corbett A. H. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 2006;282:3042–3049. [Abstract] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. [Europe PMC free article] [Abstract] [Google Scholar]
- Makhnevych T., Ptak C., Lusk C. P., Aitchison J. D., Wozniak R. W. The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol. 2007;177:39–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Mendjan S. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. [Abstract] [Google Scholar]
- Menon B. B., Sarma N. J., Pasula S., Deminoff S. J., Willis K. A., Barbara K. E., Andrews B., Santangelo G. M. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA. 2005;102:5749–5754. [Europe PMC free article] [Abstract] [Google Scholar]
- Motegi A., Kuntz K., Majeed A., Smith S., Myung K. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:1424–1433. [Europe PMC free article] [Abstract] [Google Scholar]
- Mullen J. R., Kaliraman V., Ibrahim S. S., Brill S. J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. [Europe PMC free article] [Abstract] [Google Scholar]
- Palancade B., Zuccolo M., Loeillet S., Nicolas A., Doye V. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell. 2005;16:5258–5268. [Europe PMC free article] [Abstract] [Google Scholar]
- Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 2004;279:41346–41351. [Abstract] [Google Scholar]
- Panse V. G., Kuster B., Gerstberger T., Hurt E. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 2003;5:21–27. [Abstract] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. [Abstract] [Google Scholar]
- Piruat J. I., Aguilera A. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics. 1996;143:1533–1542. [Europe PMC free article] [Abstract] [Google Scholar]
- Sacher M., Pfander B., Hoege C., Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006;8:1284–1290. [Abstract] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U. K. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. [Abstract] [Google Scholar]
- Scott R. J., Lusk C. P., Dilworth D. J., Aitchison J. D., Wozniak R. W. Interactions between Mad1p and the Nuclear Transport Machinery in the Yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:4362–4374. [Europe PMC free article] [Abstract] [Google Scholar]
- Shim E. Y., Ma J. L., Oum J. H., Yanez Y., Lee S. E. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell Biol. 2005;25:3934–3944. [Europe PMC free article] [Abstract] [Google Scholar]
- Soustelle C., Vernis L., Freon K., Reynaud-Angelin A., Chanet R., Fabre F., Heude M. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol. Cell Biol. 2004;24:5130–5143. [Europe PMC free article] [Abstract] [Google Scholar]
- Suntharalingam M., Wente S. R. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. [Abstract] [Google Scholar]
- Taddei A., Hediger F., Neumann F. R., Bauer C., Gasser S. M. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. [Europe PMC free article] [Abstract] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S. M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. [Abstract] [Google Scholar]
- Therizols P., Fairhead C., Cabal G. G., Genovesio A., Olivo-Marin J. C., Dujon B., Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 2006;172:189–199. [Europe PMC free article] [Abstract] [Google Scholar]
- Timney B. L., Tetenbaum-Novatt J., Agate D. S., Williams R., Zhang W., Chait B. T., Rout M. P. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 2006;175:579–593. [Europe PMC free article] [Abstract] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Z., Jones G. M., Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–1509. [Europe PMC free article] [Abstract] [Google Scholar]
- Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. [Abstract] [Google Scholar]
- Wykoff D. D., O'Shea E. K. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell Proteomics. 2005;4:73–83. [Abstract] [Google Scholar]
- Yang L., Mullen J. R., Brill S. J. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 2006;34:5541–5551. [Abstract] [Google Scholar]
- Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA. 2005;102:4777–4782. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao X., Wu C. Y., Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 2004;167:605–611. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Full text links
Read article at publisher's site: https://doi.org/10.1091/mbc.e07-02-0123
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1949349?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102660124
Article citations
Multifaceted roles of SUMO in DNA metabolism.
Nucleus, 15(1):2398450, 17 Sep 2024
Cited by: 0 articles | PMID: 39287196 | PMCID: PMC11409511
Review Free full text in Europe PMC
Interdependence between Nuclear Pore Gatekeepers and Genome Caretakers: Cues from Genome Instability Syndromes.
Int J Mol Sci, 25(17):9387, 29 Aug 2024
Cited by: 0 articles | PMID: 39273335 | PMCID: PMC11394955
Review Free full text in Europe PMC
SUMO protease and proteasome recruitment at the nuclear periphery differently affect replication dynamics at arrested forks.
Nucleic Acids Res, 52(14):8286-8302, 01 Aug 2024
Cited by: 3 articles | PMID: 38917328 | PMCID: PMC11317160
Protocol for machine-learning-based 3D image analysis of nuclear envelope tubules in cultured cells.
STAR Protoc, 5(3):103214, 31 Jul 2024
Cited by: 0 articles | PMID: 39088324 | PMCID: PMC11342193
Loss of FAM111B protease mutated in hereditary fibrosing poikiloderma negatively regulates telomere length.
Front Cell Dev Biol, 11:1175069, 05 Jun 2023
Cited by: 4 articles | PMID: 37342232 | PMCID: PMC10277729
Go to all (104) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality.
J Cell Biol, 167(4):605-611, 01 Nov 2004
Cited by: 99 articles | PMID: 15557117 | PMCID: PMC2172573
A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance.
J Cell Biol, 178(5):813-827, 01 Aug 2007
Cited by: 71 articles | PMID: 17724121 | PMCID: PMC2064546
Sumo-dependent substrate targeting of the SUMO protease Ulp1.
BMC Biol, 9:74, 28 Oct 2011
Cited by: 22 articles | PMID: 22034919 | PMCID: PMC3216068
The dynamics of karyopherin-mediated nuclear transport.
Biochem Cell Biol, 79(5):603-612, 01 Jan 2001
Cited by: 20 articles | PMID: 11716302
Review
 *†
*† 



