Abstract
Free full text

Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers
Abstract
Here we provide a mechanism for specific, efficient transcription of the TNF gene and, potentially, other genes residing within multigene loci. We identify and characterize highly conserved noncoding elements flanking the TNF gene, which undergo activation-dependent intrachromosomal interactions. These elements, hypersensitive site (HSS)−9 and HSS+3 (9 kb upstream and 3 kb downstream of the TNF gene, respectively), contain DNase I hypersensitive sites in naive, T helper 1, and T helper 2 primary T cells. Both HSS-9 and HSS+3 inducibly associate with acetylated histones, indicative of chromatin remodeling, bind the transcription factor nuclear factor of activated T cells (NFAT)p in vitro and in vivo, and function as enhancers of NFAT-dependent transactivation mediated by the TNF promoter. Using the chromosome conformation capture assay, we demonstrate that upon T cell activation intrachromosomal looping occurs in the TNF locus. HSS-9 and HSS+3 each associate with the TNF promoter and with each other, circularizing the TNF gene and bringing NFAT-containing nucleoprotein complexes into close proximity. TNF gene regulation thus reveals a mode of intrachromosomal interaction that combines a looped gene topology with interactions between enhancers and a gene promoter.
TNF, a critical cytokine involved in the immune response and inflammation, is secreted by multiple classes of cells, including T cells (1, 2). Transcription of the TNF gene is regulated in a cell type- and stimulus-specific fashion through the recruitment of specific sets of transcription factors and coactivators to an ≈200-bp promoter region, forming distinct nucleoprotein complexes known as enhanceosomes (3–8). For example, in T cells, TNF regulation by antigen receptor engagement or calcium influx depends on nuclear factor of activated T cells (NFAT)p binding to conserved DNA motifs in the promoter (3, 5, 9–11).
The TNF gene resides in a locus with the lymphotoxin (LT) α and β genes, their coding regions occupying ≈12 kb of genomic DNA on human chromosome 6 and mouse chromosome 17. The TNF and LT-α genes are in the same transcriptional orientation; LT-β is in the opposite orientation (12–15). The intergenic regions of the TNF/LT locus are generally poorly conserved, but they do contain short, highly conserved noncoding sequences (16). Among these is the TNF promoter itself (12–15, 17), which shows almost complete conservation among primate species (18) and complete conservation in humans in the proximal region critical for transcriptional regulation (19).
Long-distance interactions between regulatory elements in their native chromatin context comprise a critical aspect of eukaryotic gene regulation. The presence of widely separated (up to hundreds of kilobases) cis-acting regulatory sequences at native gene loci provided the initial evidence for functional interaction between distal regulatory elements (20). Studies that have identified and analyzed DNase I hypersensitive (DH) sites (indicative of altered DNA accessibility and thus chromatin remodeling), histone modifications such as acetylation [which favors “open” or transcriptionally active chromatin configuration (21)], and conserved noncoding sequence elements (22) at native loci further supported a model whereby looping out of the DNA between remote functional elements facilitates protein–protein interactions involved in transcriptional regulation (23–26). Recently, direct evidence for long-range interactions between distal regulatory elements in vivo has come from the chromosome conformation capture (3C) assay, which assesses the spatial proximity of DNA-bound proteins in their native chromatin context (27, 28).
Here we have characterized the chromatin configuration of the TNF/LT locus in T cells, which reveals previously undescribed features of intrachromosomal interactions involved in the activation of TNF gene transcription. We identify and characterize hypersensitive site (HSS)−9 and HSS+3, two regions of high sequence conservation between mice and humans that contain DH sites and associate with hyperacetylated histones in primary murine T cells. HSS-9 and HSS+3 bind to NFATp in vitro and in vivo and function as enhancers of NFAT-dependent transcriptional activation mediated by the TNF promoter. Strikingly, in activated T cells, HSS-9 and HSS+3 undergo intrachromosomal interactions that place them in close proximity to the TNF promoter and to each other, in a configuration that potentially underlies selective activation of the TNF gene within the TNF/LT locus. These results may provide a model for how single genes are selectively transcribed in gene-dense regions.
Results
A Specific DH Pattern at the TNF/LT Locus in Primary T Lymphocytes.
To examine chromatin remodeling at the TNF/LT locus, we performed DH assays using primary murine T lymphocytes that were either naive or cultured under T helper 1 (Th1) or Th2 polarizing conditions in vitro (Fig. 1). We detected four HSS within the 10.5-kb region upstream of the first TNF intron (Fig. 1A). We designated each HSS band based on its approximate position relative to the first exon of TNF. The smallest band, HSS-0.8, maps to a region 0.8 kb upstream of the first exon, corresponding to the TNF promoter (Fig. 1 A and C). This is consistent with previous findings that this region is physically accessible to regulatory proteins (5–8) and associated with acetylated histones (29). The genomic DNA fragment corresponding to HSS-0.8 is small in the BglII digest (Fig. 1A) and, in the XbaI digest (Fig. 1B), lies near the edge of the probe; this technical limitation, or a relatively weak hypersensitivity of this region, may contribute to the weak intensity of the HSS-0.8 bands. The next two bands, HSS-3.5 and HSS-4, coincide with the LT-α promoter (Fig. 1 A and C). The most prominent DH site, HSS-9, is located ≈9 kb upstream of the TNF gene and 5 kb upstream of the LT-α gene (Fig. 1 A and C). We also found one strong DH site, HSS+3, ≈3 kb downstream of the start site of TNF transcription (Fig. 1 B and C).
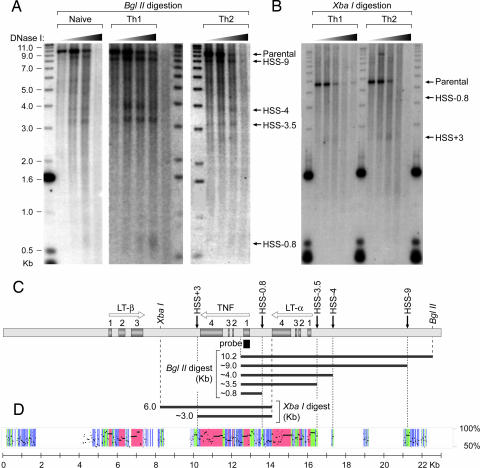
TNF/LT locus DH profiles in naive, Th1, and Th2 subsets of primary murine T lymphocytes. (A and B) Nuclei from naive T cells and Th1 and Th2 cells were digested with increasing amounts of DNase I. Purified DNA was subsequently digested with BglII (A) or XbaI (B) followed by resolution by agarose gel electrophoresis (molecular weight markers in kb), Southern blot analysis, hybridization with a probe corresponding to the first exon of TNF, and visualization by autoradiography. HSS found in T cells are indicated with arrows and designated by their approximate positions relative to the first TNF exon. Migration of digested genomic DNA fragments is consistently greater than expected relative to the marker, most likely because of a greater amount of total DNA (≈15 μg) than marker (10 pg). (C) Positions and directions of transcription of the TNF, LT-β, and LT-α genes in the ≈23-kb locus are shown by white arrows; exons are shown by numbered gray boxes; and the Southern blotting probe (186 bp) is shown by a black box. Positions of the parental BglII and XbaI fragments and the DNase I digestion products are indicated. (D) Sequence conservation between murine and human TNF/LT loci using PipMaker (www.bx.psu.edu) (32) as described in SI Fig. 6, showing strongly and moderately conserved noncoding regions (green and blue, respectively) and exons (red).
Strikingly, we found that all five DH sites occurred in highly conserved noncoding regions [Fig. 1D and supporting information (SI) Fig. 6]. Furthermore, inspection of the 1.2-kb HSS-9 region, which did not overlap any previously known regulatory region, revealed two consensus binding motifs for the NFAT protein family (30) at positions −8,394 (NFAT-8,394) and −8,914 (NFAT-8,914) relative to the TNF transcription start site (SI Fig. 7). NFAT-8,394 also matched a consensus binding site for transcription factors of the NF-κB family (31). Furthermore, we found eight consensus NFAT binding sites in HSS+3 (SI Fig. 8). Two of these sites, NFAT+2,840 and NFAT+2,856, overlap sites that were previously shown to bind NF-κB in EMSAs using the isolated DNA motifs as probes (32, 33). The conservation of these putative NFAT binding motifs between human and mouse suggested that they played an important role in TNF gene regulation.
NFATp and NF-κB p50/p65 Bind to HSS-9 and HSS+3 in Vitro.
To determine whether NFAT binds to the putative consensus sequences in the HSS-9 and HSS+3 regions, we performed quantitative DNase I in vitro footprinting using recombinant NFATp. Analysis of region I of HSS-9 (see Fig. 2 and SI Fig. 7) revealed that NFATp bound strongly to NFAT-8,394 and with lower affinity to three additional sites: NFAT-8,232, NFAT-8,304, and NFAT-8,466. NF-κB p50/p65 bound only to NFAT-8,394 (Fig. 2 A and B). Region II of HSS-9 contains only one potential NFAT site, NFAT-8,914 (SI Fig. 7), which we note does not match an NF-κB consensus binding site. NFATp in nuclear extracts from primary T lymphocytes bound to NFAT-8,914 (SI Fig. 9). Thus, NFATp binds to five sites within HSS-9.
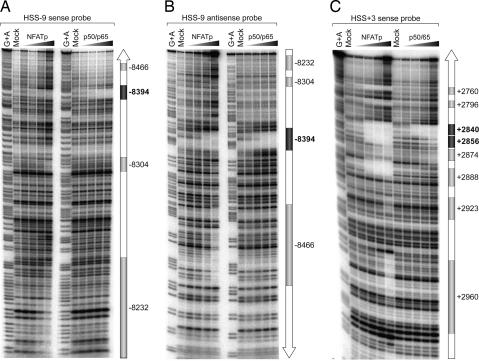
NFATp binds to multiple sites in HSS-9 and HSS+3. Shown is quantitative DNase I footprinting analysis using sense (A) and antisense (B) probes spanning region I (306 bp) of HSS-9 and a sense probe (C) spanning HSS+3 (307 bp) and increasing amounts (4 ng, 20 ng, or 100 ng) of recombinant NFATp or NF-κB p50/p65. Positions of the NFAT (gray boxes) and NFAT/NF-κB (dark gray boxes) binding sites (relative to the TNF transcription start site) are indicated.
Furthermore, DNase I footprinting of HSS+3 revealed that NFATp binds to all eight consensus NFAT binding sites in HSS+3 (at +2,960, +2,923, +2,888, +2,874, +2,856, +2,840, +2,796, and +2,760) with varying affinities. By contrast, NF-κB p50/p65 only bound with high affinity to NFAT+2,840 and with lower affinity to NFAT+2,856 (Fig. 2C).
NFATp Binding Is Inducible and Histone Acetylation Is Increased at HSS-9 and HSS+3 in Activated Primary CD4+ T Cells in Vivo.
To determine whether NFATp and/or NF-κB bind HSS-9 and HSS+3 in vivo, and to determine whether there is an increase in histone acetylation at these regions upon T cell activation, we performed ChIP assays using antibodies specific for NFATp, NF-κB p50, NF-κB p65, and acetylated forms of histones H3 (Ac-H3) and H4 (Ac-H4). The NFATp-specific antibody selectively immunoprecipitated DNA from region I and region II in HSS-9 (Fig. 3A) and from HSS+3 (Fig. 3C) in stimulated cells, indicating that NFATp is inducibly recruited to these regions in vivo. By contrast, neither the p50 nor the p65 subunit of NF-κB was inducibly recruited to regions I and II of HSS-9 in the ChIP assay (Fig. 3A). In the case of HSS+3, p65 binding was strongly inducible, whereas p50 binding was slightly inducible (Fig. 3C).
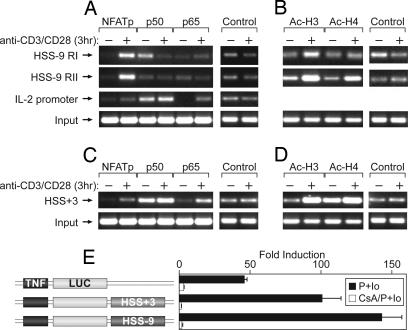
HSS-9 and HSS+3 bind NFATp in murine primary CD4+ T cells in vivo and function as enhancers of TNF transcription. (A–D) ChIP analysis in unstimulated (−) or anti-CD3/CD28-stimulated (+) mouse CD4+ T cells. Chromatin was immunoprecipitated with the indicated antibodies to transcription factors NFATp, NF-κB p50, and NF-κB p65 (A and C) or the acetylated forms of histones H3 and H4 (B and D) and amplified with primers specific for regions I and II of HSS-9, HSS+3, and the IL-2 promoter (SI Figs. 7 and 8). Nonimmune IgG antibody and 10 times the amount of the input DNA used for the specific antibodies (to detect background) were used for PCR controls for nonspecific binding. (E) 68-41 cells were transiently transfected with the indicated reporter constructs. After mock stimulation or P+I stimulation (P+Io) in the presence or absence of cyclosporin A (CsA) pretreatment, luciferase activity was normalized to Renilla luciferase activity. Error bars represent standard deviation from at least four independent experiments.
Basal levels of DNA precipitated by both Ac-H3 and Ac-H4 were increased by cellular stimulation (Fig. 3 B and D), indicating that acetylation occurs, or is increased, after T cell activation, consistent with a conformation that allows transcription factors access to DNA.
HSS-9 and HSS+3 Function as Enhancers of TNF Transcription.
To test whether HSS-9 or HSS+3 could function as regulatory elements, we prepared luciferase reporter constructs under the control of the TNF promoter with the 1.2-kb HSS-9 fragment or the 0.5-kb HSS+3 fragment downstream of the luciferase gene and transiently transfected them into the murine T cell line 68-41, where we obtained the same results in DH and ChIP assay as we did using primary T cells (data not shown). The addition of HSS-9 or HSS+3 to the minimal TNF promoter enhanced its activity by 2- and 3-fold, respectively (Fig. 3E). Furthermore, pretreatment of the cells with cyclosporin A, which binds to the phosphatase calcineurin and inhibits NFAT translocation to the nucleus (30) but has no effect on NF-κB binding (34), inhibits the augmentation of transcription of TNF promoter reporters bearing HSS-9 or HSS+3 (Fig. 3E), which would not be expected if these enhancers were NFAT-independent. Consistent with these findings, we found that TNF production is decreased in anti-CD3/CD28-stimulated Th1 and Th2 cells from NFATp-deficient mice relative to their wild-type littermates (SI Fig. 10), corroborating a role for NFATp acting at these distal elements in a physiological setting.
Activation-Dependent Intrachromosomal Interactions Among HSS-9, HSS+3, and the TNF Promoter in T Cells.
To test the hypothesis that HSS-9 and/or HSS+3 played a role in the activation of TNF gene transcription via a long-range interaction with the TNF enhanceosome, we next performed a 3C assay in mock- or phorbol 12-myristate 13-acetate plus ionomycin (P+I)-stimulated 68-41 T cells. We used the restriction enzyme BstY I to digest the TNF/LT locus and the Gapd locus, which was used as a negative control because the Gapd gene is constitutively expressed in all cell types and has no known secondary chromatin conformation (35–37) (Fig. 4A, SI Fig. 11, and SI Materials and Methods). For each of the major DNA restriction fragments spanning regulatory regions of interest in the TNF/LT locus (A through F in Fig. 4A), primers close to the BstY I site were designed to allow optimal detection of the potential ligation product between distal elements that physically interact (Fig. 4A and SI Fig. 12; primers A1 and A2 for fragment A, B1 and B2 for fragment B, etc.). Primer pairs yielding optimal amplification were determined empirically (SI Fig. 13).
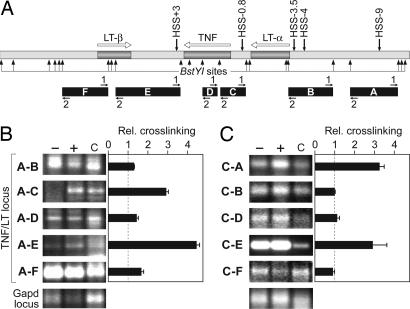
3C assay. (A) Genomic DNA products of BstY I digestion (arrows), with areas of interest (HSS and gene promoters) labeled A–F and PCR primers numbered 1 and 2. (B and C) Relative cross-linking frequencies (±SEM) between the indicated fragments and fragment A (HSS-9) using primers A2-B2, A1-C2, A2-D2, A1-E1, and A2-F2 (B) or fragment C (TNF promoter) using primers C2-A1, C1-B1, C1-D1, C1-E2, and C1-F1 (C) with chromatin from unstimulated (−) or P+I-stimulated (+) cells or genomic DNA control (c). PCR products in B and C are from independent PCR plates, and PCR products within each of these two sets were amplified simultaneously; for each pair of genomic fragments, similar results were obtained by using the other possible primer pair combinations (SI Fig. 13B).
We modified previous 3C protocols so that purified genomic DNA, digested with BstY I and randomly ligated at high concentrations, was the control template in our 3C assay. Thus, we were able to optimize conditions to assay both the TNF and the Gapd loci in their native genomic context, with the same genomic control DNA template, and did not have to enrich the amount of TNF or Gapd DNA fragments in the genomic DNA control template for PCR amplification or introduce exogenous DNA for our controls (see SI Materials and Methods).
We first measured the cross-linking frequency of fragment A (containing HSS-9), to fragments B (LT-α promoter), C (TNF promoter), D (TNF gene middle exon–intron region), E (containing HSS+3), and F (LT-β promoter and first exon). We detected an inducible increase in cross-linking frequency between fragments A and C and between fragments A and E in stimulated 68-41 cells, indicating intrachromosomal interactions between HSS-9 and the TNF promoter and between HSS-9 and HSS+3, respectively (Fig. 4B). We next performed the 3C assay using fragment C (containing the TNF promoter) as the primary target. We detected an inducible association between fragments C and A and C and E, but not between C and B, D, or F, confirming a specific intrachromosomal interaction between the TNF promoter and HSS-9 and between the TNF promoter and HSS+3 (Fig. 4C). We did not detect any specific secondary chromatin structure in the TNF/LT locus in unstimulated T cells (Fig. 4 B and C, SI Fig. 13B, and data not shown). Taken together, these results demonstrate intrachromosomal interactions between the TNF promoter and HSS+3, between the TNF promoter and HSS-9, and between HSS+3 and HSS-9 in vivo as depicted in the model in Fig. 5.
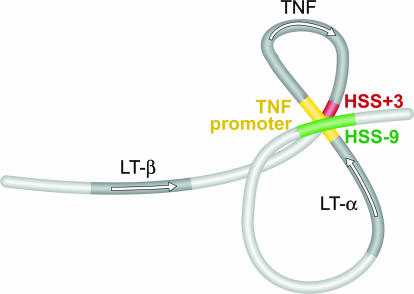
A model of intrachromosomal interactions at the TNF/LT locus in activated T cells. Simultaneous intrachromosomal interactions at the TNF/LT locus (TNF promoter–HSS+3, TNF promoter–HSS-9, and HSS+3–HSS-9) would both juxtapose NFAT-binding regulatory regions and selectively circularize the TNF gene (see SI Fig. 14).
Discussion
Two basic modes of intrachromosomal interactions promote transcriptional activation in eukaryotes (38). One mode, observed in yeast (39, 40) and mammalian mitochondrial rDNA (41), is gene looping, that is, formation of a simple loop that brings the 5′ promoter and 3′ termination sequence of a single active gene into close proximity. This has been proposed to promote “recycling” of RNA polymerase after one round of transcription and coordination of mRNA processing and transcription. A second mode of intrachromosomal interaction brings distal conserved noncoding elements (generally within locus control regions) together with core promoters of a set of genes clustered in a specific locus by looping out the intervening DNA (23–26). This has been observed at several mammalian gene loci, including β-globin (28, 42–46), Th2 (35, 37), and IFN-γ (36). Other studies have linked intrachromosomal looping to transcriptional repression (47–49). At the maternal allele of the Igf2/H19 locus (48–51), for example, intrachromosomal interactions place the Igf2 gene into an “inactive chromatin loop,” presumably sequestering it from active enhancers (49).
The analysis of the TNF/LT locus presented here reveals a mode of intrachromosomal interaction that combines gene circularization with long-range promoter–enhancer interactions as depicted in the model in Fig. 5. In activated T cells, we have demonstrated physical interactions between the TNF promoter and HSS-9, between the TNF promoter and HSS+3, and between HSS+3 and HSS-9. This is consistent with the formation of a double-looped structure bringing NFAT-containing nucleoprotein complexes formed at the TNF promoter (the enhanceosome) into close proximity with the NFAT-containing nucleoprotein complexes formed at HSS+3 and at HSS-9. Furthermore, this structure juxtaposes the initiation (the promoter) and termination (HSS+3) regions of the TNF gene, a topology that would be expected to promote reinitiation of transcription. To our knowledge, this is the first example of activation-induced circularization of a RNA polymerase II-dependent gene in a eukaryote other than yeast. Finally, the LT-α promoter does not interact with HSS+3 or HSS-9 in the 3C assay (Fig. 4), consistent with the fact that the induction of the LT-α promoter fused to a luciferase reporter gene in response to P+I (≈8-fold) is increased only slightly (by ≈1.5-fold) by the addition of HSS+3 or HSS-9 (data not shown). By contrast, the TNF promoter exhibits ≈45-fold induction after P+I treatment that is further increased 2- to 3-fold by the addition of HSS+3 or HSS-9 (Fig. 3E). Taken together with the topology of the TNF/LT locus derived from the 3C assay, we conclude that HSS+3 and HSS-9 specifically enhance TNF gene transcription during T cell activation.
HSS-9 and HSS+3 are highly conserved noncoding sequences that, based on our DH and ChIP assays, are associated with an open chromatin configuration, both of which are common features of elements involved in long-range interactions in the native chromatin context. We have also shown that, like the TNF promoter itself (3, 5, 9), HSS-9 and HSS+3 bind to the transcription factor NFATp and regulate calcineurin- and NFAT-dependent TNF gene transcription in T cells. Given that certain transcription factors and other regulatory proteins are critical for establishing higher-order chromatin configurations (35, 43, 45–47, 49, 50), it is likely that the chromosome configuration of the TNF/LT locus in T cells depends on NFATp, potentially involving interactions between NFATp proteins and associated factors bound at widely separated motifs in HSS-9, HSS+3, and the TNF promoter. Furthermore, given our previous characterization of the formation of distinct TNF enhanceosomes, in both NFATp-dependent and NFATp-independent fashions (3–8), specific higher-order chromatin structures at the TNF/LT locus may be linked to cell-type and inducer-specific regulation of the TNF gene.
It remains to be determined whether single- and/or double-looped intrachromosomal configurations at the TNF/LT locus represent static or dynamic populations, or whether single-looped structures are present as intermediates in the formation of the double-looped structure. However, the sum total of intrachromosomal interactions that we observe as shown in Fig. 5, specifically the interactions between the TNF promoter and HSS-9, between the TNF promoter and HSS+3, and between HSS+3 and HSS-9, could not result from the exclusive formation of any one of the single-looped structures shown in SI Fig. 14.
In conclusion, our data indicate that TNF activation involves a combination of both circularization of the TNF gene itself and looping that involves distal enhancers and the TNF promoter. Thus, this study suggests a model for gene regulation in mammalian loci containing several genes arranged head-to-tail (or head-to-head) in close proximity. At these loci, we image that circularization of a specific gene within the cluster (directed by protein–protein interactions and areas of open chromatin conformation) could selectively favor expression of that gene in response to a specific stimulus. Our results thus present a potential molecular mechanism by which a eukaryotic gene located in a gene-dense chromosomal locus can achieve transient and high levels of transcription without interfering with the expression of neighboring genes.
Materials and Methods
DH Analysis.
Assays were performed, based on methods described previously (52, 53), in primary T lymphocytes. Th1 and Th2 cells were derived in vitro as described elsewhere (54, 55). DNA samples from DNase I-treated (0, 1, 3, 5, and 10 μg/ml) nuclei were digested with BglII or XbaI and analyzed by Southern blotting using a probe corresponding to the coding region of the first exon of the TNF gene.
DNase I Footprinting.
Radiolabeled fragments of the HSS-9 (307 bp) and HSS+3 (306 bp) regions were incubated with recombinant NFATp and NF-κB p50/p65 proteins and processed as described previously (4, 7).
ChIP Assays.
Purified cross-linked chromatin was prepared as described previously (5) and immunoprecipitated with 5 μg of antibodies against Ac-H3 (Lys-9/14), Ac-H4 (Lys-8), NFATp, NFATc, p50 (NF-κB), p65 (NF-κB), and (as a negative control) rabbit IgG (anti-NFATc; ABR Affinity Bioreagents; other antibodies were from Santa Cruz Biotechnology).
Luciferase Reporter Assays.
In the pGL3-Basic vector (Promega), the mouse TNF promoter (−200 to +178 bp relative to the transcription start site of the TNF gene) was inserted upstream of the luciferase gene with the HSS-9 enhancer (1,342 bp) or HSS+3 enhancer (448 bp) downstream of the luciferase gene as indicated in Fig. 3E. The 68-41 T cell line was maintained, transfected by using DEAE-dextran, and treated with P+I and cyclosporin A and luciferase assays performed as described previously (6).
3C Assays.
The 3C protocol for mammalian cells (28) was modified as described in SI Materials and Methods. Briefly, 50 × 107 murine 68-41 T cells were split into two aliquots and mock-stimulated or stimulated with 20 ng/ml phorbol 12-myristate 13-acetate and 1 μM ionomycin (P+I) for 3 h. The cells were fixed with 2% formaldehyde for 30 min at room temperature in a buffer containing 10 mM Tris·HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA, quenched with glycine (125 mM final concentration), washed in cold PBS, and lysed in a buffer containing 0.25% Triton X-100, 0.5% Nonidet P-40, 10 mM Tris·HCl (pH 8.0), and 1 mM EDTA. Nuclei were pelleted and digested with BstY I (New England BioLabs) for 3 h at 60°C (inactivated at 80°C for 20 min). Chromatin samples were diluted to 5 ng/μl and ligated for 1 h at 16°C by using T4 ligase (New England BioLabs). The cross-links were then reversed by incubation at 65°C overnight in the presence of 0.2 mg/ml proteinase K and 0.5% SDS. The DNA was purified by phenol-chloroform extraction and ethanol precipitation. PCR primers are shown in SI Fig. 12. PCR, control template preparation, and PCR primer optimization were performed as shown in SI Fig. 13. PCR products were run on 2.5% agarose gels and quantified by using a Bio-Rad Molecular Imager. Template preparation was repeated at least three times independently. For each prepared template, PCR was repeated at least four times independently.
Acknowledgments
We thank Anne O'Garra and Margarida Saraiva (National Institute for Medical Research, London, U.K.) for the gift of DNase-treated Th1, Th2, and naive T cells and for assistance with the preparation of T cell subsets and the analysis of the NFATp-deficient mice. We also thank Dimitris Thanos (Academy of Athens, Athens, Greece) for recombinant proteins, Laurie Glimcher (Harvard School of Public Health, Boston, MA) for the NFATp-deficient mouse strain, Karolina Maciag for technical assistance, and Renate Hellmiss-Peralta for graphic artwork. This work was supported by National Institutes of Health Grant RO1GM076685 (to A.E.G.).
Abbreviations
| 3C | chromosome conformation capture |
| DH | DNase I hypersensitive or DNase I hypersensitivity |
| HSS | hypersensitive site |
| LT | lymphotoxin |
| NFAT | nuclear factor of activated T cells |
| P+I | phorbol 12-myristate 13-acetate plus ionomycin |
| Thn | T helper n. |
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708210104/DC1.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0708210104
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2040403?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
SIKs Regulate HDAC7 Stabilization and Cytokine Recall in Late-Stage T Cell Effector Differentiation.
J Immunol, 211(12):1767-1782, 01 Dec 2023
Cited by: 0 articles | PMID: 37947442
Polymorphisms in Lymphotoxin-Alpha as the "Missing Link" in Prognosticating Favourable Response to Omega-3 Supplementation for Dry Eye Disease: A Narrative Review.
Int J Mol Sci, 24(4):4236, 20 Feb 2023
Cited by: 2 articles | PMID: 36835647 | PMCID: PMC9965360
Review Free full text in Europe PMC
Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9.
Nat Commun, 12(1):5711, 29 Sep 2021
Cited by: 31 articles | PMID: 34588447 | PMCID: PMC8481236
Epigenetic strategies to boost CAR T cell therapy.
Mol Ther, 29(9):2640-2659, 06 Aug 2021
Cited by: 21 articles | PMID: 34365035 | PMCID: PMC8417511
Review Free full text in Europe PMC
Identification of a Distal Locus Enhancer Element That Controls Cell Type-Specific TNF and LTA Gene Expression in Human T Cells.
J Immunol, 205(9):2479-2488, 25 Sep 2020
Cited by: 3 articles | PMID: 32978279 | PMCID: PMC7576113
Go to all (54) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
NFAT is well placed to direct both enhancer looping and domain-wide models of enhancer function.
Sci Signal, 1(13):pe15, 01 Apr 2008
Cited by: 7 articles | PMID: 18385038
Identification of a Distal Locus Enhancer Element That Controls Cell Type-Specific TNF and LTA Gene Expression in Human T Cells.
J Immunol, 205(9):2479-2488, 25 Sep 2020
Cited by: 3 articles | PMID: 32978279 | PMCID: PMC7576113
Chromatin profiling across the human tumour necrosis factor gene locus reveals a complex, cell type-specific landscape with novel regulatory elements.
Nucleic Acids Res, 36(15):4845-4862, 24 Jul 2008
Cited by: 19 articles | PMID: 18653526 | PMCID: PMC2528168
Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation.
Adv Immunol, 118:37-128, 01 Jan 2013
Cited by: 39 articles | PMID: 23683942 | PMCID: PMC4118600
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: R01 GM076685
Grant ID: R01 GM 076685




