Abstract
Free full text

Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development
Abstract
The Notch ligand Jagged1 (Jag1) is essential for vascular remodeling and has been linked to congenital heart disease in humans, but its precise role in various cell types of the cardiovascular system has not been extensively investigated. We show that endothelial-specific deletion of Jag1 results in embryonic lethality and cardiovascular defects, recapitulating the Jag1 null phenotype. These embryos show striking deficits in vascular smooth muscle, whereas endothelial Notch activation and arterial-venous differentiation appear normal. Endothelial Jag1 mutant embryos are phenotypically distinct from embryos in which Notch signaling is inhibited in endothelium. Together, these results imply that the primary role of endothelial Jag1 is to potentiate the development of neighboring vascular smooth muscle.
The development and remodeling of the vasculature involves a number of complex processes, and the Notch signaling pathway has been shown to be one critical determinant (1–6). Notch is a short-range signaling pathway that occurs between membrane-bound receptors and ligands expressed on adjacent cells. Binding of ligands induces a proteolytic cleavage of the Notch receptor, releasing its intracellular domain (ICD). This truncated form of Notch then translocates to the nucleus where it forms an active transcriptional complex with the DNA-binding protein CSL [also known as CBF1, Su(H), Lag-1, RBP-J] and the coactivator Mastermind-like (MAML) (7).
Mammals express four Notch receptors (Notch 1–4) and five ligands [Jagged (Jag) 1 and Jag2 and Dll (Delta-like) 1, Dll3, and Dll4]. Targeted disruption of several of these signaling components results in embryonic lethality associated with cardiovascular defects (1, 2, 4–6). However, the precise roles of each signaling component are only beginning to be teased apart. The emerging data suggest that Notch plays multiple, distinct roles in cardiovascular development. Studies performed in zebrafish and later in mice demonstrated a requirement for Notch in the specification of arterial and venous fate in the developing endothelium, and inhibition of Notch in endothelium results in arterial-venous malformations (5, 8, 9). In addition, endothelial Notch signaling plays a critical role in angiogenesis during both development and disease (10–16).
Another critical function for Notch is in vascular smooth muscle development. In vitro studies have shown that Notch can either promote or inhibit smooth muscle gene expression, depending on context (17–20). However, in vivo studies show that in the context of development, Notch plays a critical role in promoting vascular smooth muscle differentiation. Smooth muscle precursors derive from multiple sources during embryogenesis. Smooth muscle of the developing aortic arch arteries is derived from the cardiac neural crest, whereas vascular smooth muscle elsewhere in the thorax and abdomen derives from lateral plate mesoderm (21). We previously demonstrated a cell-autonomous requirement for Notch in neural crest precursors during aortic arch smooth muscle differentiation. Inhibition of Notch signaling in the neural crest results in congenital heart defects, including pulmonary artery stenosis and aortic arch patterning defects (22). However, the identity of the Notch ligand in this process was thus far unknown.
Two Notch ligands, Jag1 and Dll4, are prominently expressed in the vasculature. Targeted disruption of each of these genes in mice results in embryonic lethality associated with cardiovascular defects, suggesting that both play essential, nonredundant functions (1, 2, 5, 6). Dll4 has recently emerged as the critical ligand in Notch signaling between adjacent endothelial cells, negatively regulating blood vessel growth during both development and tumor angiogenesis (10–16). Jag1, on the other hand, has been less extensively studied. Jag1 knockout mice die between embryonic day (E) 10.5 and E11.5 with defects in yolk sac and embryonic vasculature (6). However, the mechanism by which loss of Jag1 leads to embryonic lethality is unclear, particularly as Jag1 is expressed in multiple parts of the cardiovascular system, including endothelial cells, vascular smooth muscle, and the cardiac outflow tract (22–24).
The importance of Jag1 in human disease is evident from its role in Alagille syndrome, a congenital disorder linked to mutations in the JAG1 gene (25, 26). One of the principal findings in Alagille syndrome is congenital heart disease, especially pulmonary artery stenosis, and vascular disease including a predisposition to intracranial bleeding (27, 28). Therefore, a better understanding of the role of Jag1 in cardiovascular development promises to provide insight into the pathogenesis of Alagille syndrome and other forms of congenital heart and vascular diseases.
In this study, we show that endothelial-specific deletion of Jag1 results in embryonic lethality and cardiovascular defects, similar to the gross defects reported for the complete Jag1 knockout. Furthermore, we show that expression of vascular smooth muscle markers is severely diminished in the endothelial-specific Jag1 mutant embryos. Conversely, our data suggest that Notch signaling in the endothelium remains intact in the absence of Jag1. Together, these findings suggest that a primary role of endothelial Jag1 is to promote vascular smooth muscle differentiation.
Results and Discussion
To elucidate the role of Jag1 in endothelial cells, we generated endothelial-specific Jag1 knockout mice. We crossed a conditional allele of Jag1 (Jag1flox) (29) with Tie2-Cre (30), which we, and others, have shown to be specific for endothelial cells and some hematopoietic cells during development [supporting information (SI) Fig. 5]. We were unable to detect any conditional knockouts of 44 live births resulting from Tie2-Cre+;Jag1flox/+ by Jag1flox/flox crosses, indicating embryonic lethality. At E10.5 Tie2-Cre+;Jag1flox/flox mutant embryos were present in expected Mendelian ratios but were readily distinguishable from their littermates by the absence of large blood vessels in the yolk sac (Fig. 1 A and B). In addition, the mutant embryos were smaller and showed signs of cardiovascular failure, including pericardial effusions, dilated, blood-filled vessels, and localized hemorrhage (Fig. 1 D and E). By E11.5, all of the mutant embryos were necrotic and being resorbed. This phenotype is indistinguishable from that seen in global Jag1 knockouts that we generated by crossing with CMV-Cre transgenic mice to mediate ubiquitous recombination of the Jag1flox allele (Fig. 1 C and F). Detailed information on the distribution of genotypes in offspring from Tie2-Cre and CMV-Cre Jag1flox crosses is available in SI Tables 1 and 2. To investigate the patterning of the vasculature in Jag1 mutants, we performed whole-mount immunostaining for the endothelial marker platelet-endothelial cell adhesion molecule (PECAM). At E10.5 the blood vessels in the head region of both endothelial-specific and global Jag1 mutant embryos appeared abnormal and were less finely branched when compared with littermate controls (Fig. 1 G–I). These defects were mild and were not evident at E9.5 (data not shown), so it is possible that they may be secondary to developmental delay and not caused by a primary role for Jag1 in vessel patterning. Together, the phenotypes we observed were similar to those previously reported in complete Jag1 null embryos (6).
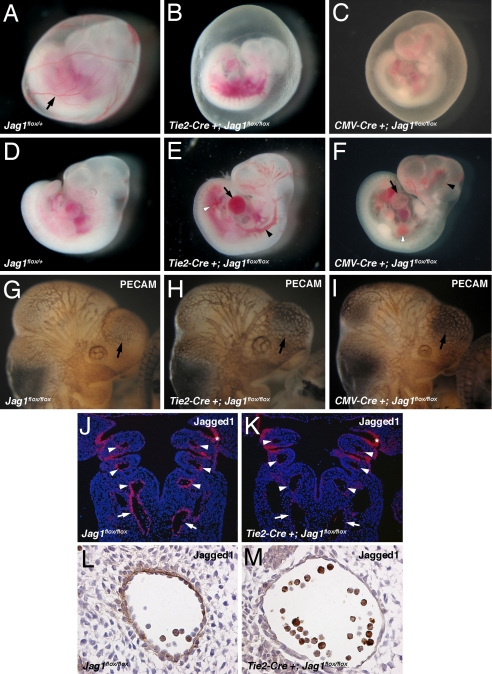
Endothelial-specific Jag1 mutants display cardiovascular defects. (A–C) E10.5 embryos with yolk sacs. Control yolk sacs (A) display prominent blood vessels (arrow) that are absent from both endothelial-specific (B) and global (C) Jag1 mutants. (D–F) E10.5 control (D), endothelial Jag1 mutant (E), and global Jag1 mutant (F) embryos demonstrating cardiovascular defects in mutants including pericardial effusions (arrows in E and F), dilated blood vessels (black arrowheads in E and F), and hemorrhage (white arrowheads in E and F). (G and H) Whole-mount PECAM immunostaining of E10.5 control (G), endothelial Jag1 mutant (H), and global Jag1 mutant (I) embryos. Mutant embryos show a less intricate vascular network over forebrain vesicles (arrows). (J and K) In situ hybridizations for Jag1 on frontal sections through E10.5 control (J) and mutant (K) embryos. Mutant embryos show loss of Jag1 expression in the dorsal aortae (arrows) and aortic arch arteries (arrowheads), but not pharyngeal endoderm (*). (L and M) Immunostaining for Jag1 on sections through the dorsal aorta of E10.5 control (L) and mutant (M) embryos, showing loss of endothelial Jagged1 protein in mutants. (Magnifications: A–F, ×40; G–K, ×100; L and M, ×400.)
In situ hybridizations for Jag1 on sections through Tie2-Cre+;Jag1flox/flox embryos at E10.5 revealed loss of Jag1 mRNA expression in arteries of the mutant embryos, including the dorsal aorta and the aortic arch arteries. In contrast, normal Jag1 expression was maintained in other tissues, including the pharyngeal endoderm (Fig. 1 J and K). Immunohistochemistry confirmed that Jag1 protein was specifically deleted from the endothelium in Tie2-Cre+;Jag1flox/flox embryos (Fig. 1 L and M). We therefore conclude that Jag1 expression in endothelial cells is essential for cardiovascular development, and that the embryonic lethality in Jag1 null embryos can be accounted for by loss of Jag1 expression in the endothelium.
We previously demonstrated a critical role for Notch in differentiation of vascular smooth muscle precursors, and in vitro studies suggest that Jag1 may play a role in smooth muscle development (20). However, an in vivo requirement for this Notch ligand during smooth muscle differentiation has not been investigated to our knowledge. We hypothesized that endothelial Jag1 expression might promote smooth muscle development. In control embryos at E10.5, the smooth muscle marker SM22α is prominently expressed around the dorsal aorta and in neural crest-derived smooth muscle of the aortic arch arteries (Fig. 2A–A″). Smooth muscle α actin (αSMA), another marker that is expressed at a slightly later time point in smooth muscle development, is expressed in the dorsal aorta of control embryos at E10.5 (Fig. 2 C and C′). In contrast, both SM22α and αSMA are dramatically down-regulated in Tie2-Cre+;Jag1flox/flox mutants at this time point (Fig. 2 B–B″ and D and D′), whereas endothelial markers remained unperturbed (Fig. 2 E and F). It is unlikely that the loss of smooth muscle markers in the mutant embryos could be secondary to cardiovascular collapse, as SM22α expression in the dorsal aorta was severely diminished at E9.5 when mutant embryos are indistinguishable from their littermates (data not shown).
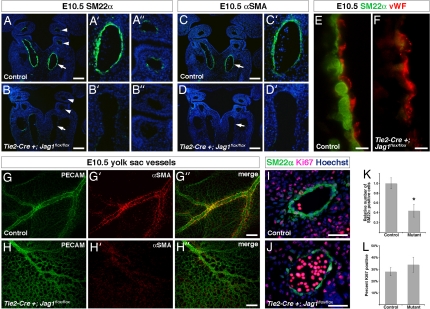
Vascular smooth muscle development in endothelial-specific Jag1 mutants. (A–A″ and B–B″) Immunostaining for SM22α (green) on frontal sections through E10.5 control (A–A″) and mutant (B–B″) embryos. Control embryos show prominent SM22α expression in dorsal aorta (arrow in A, higher magnification in A′) and a few cells in the aortic arch arteries (arrowheads in A, higher magnification in A″). Mutant embryos show significant loss of SM22α in dorsal aorta (arrows in B and B′) and aortic arch arteries (arrowheads in B and B″). (C, C′, D, and D′) Immunostaining for αSMA (green) on frontal sections through E10.5 control (C and C′) and mutant (D and D′) embryos. Control embryos express αSMA in the dorsal aorta (arrows in C and C′), whereas mutant embryos lack αSMA expression (arrows in D and D′). (E and F) High-magnification images of the endothelial-smooth muscle interface in the dorsal aorta of E10.5 control (E) and mutant (F) embryos, immunostained for SM22α (green) and von Willebrand factor (vWF, red). (G–G″ and H–H″) E10.5 yolk sacs whole-mount immunostained for PECAM (green) and αSMA (red). Control yolk sacs (G–G″) show large blood vessels with prominent αSMA expression, whereas mutant yolk sacs (H–H″) show fewer large blood vessels that were abnormal in appearance and lacked αSMA expression. (I and J) Representative images of SM22α, Ki67 coimmunostaining on sections through the dorsal aorta of E10.5 control (I) and mutant (J) embryos. (K) Quantification of total number of SM22α-positive cells in the dorsal aortae of control and mutant embryos. (L) Quantification of percentage of SM22α-positive cells that coexpress Ki67 in the dorsal aortae of control and mutant embryos. Error bars indicate 1 SD. *, P < 0.001. (Scale bars: A–D′, 100 μm; E and F, 5 μm; G–H″, 200 μm; I and J, 50 μm.)
We also examined vascular smooth muscle development in the yolk sac vasculature by whole-mount immunostaining for αSMA in combination with the endothelial marker PECAM. Control yolk sacs contained many prominent blood vessels that stained strongly for αSMA. Tie2-Cre+;Jag1flox/flox yolk sacs showed far fewer large vessels. Notably, these vessels failed to express αSMA (Fig. 2 G–G″ and H–H″). We therefore conclude that endothelial Jag1 expression is required for smooth muscle development in both embryonic and yolk sac blood vessels. Subsequent loss of vascular wall integrity and cardiovascular collapse is the likely cause of lethality in these embryos.
Notch has been implicated in smooth muscle proliferation and survival in addition to differentiation (31). Therefore, loss of vascular smooth muscle cells in these mutants could be caused by defective differentiation of smooth muscle precursors or decreased proliferation or increased apoptosis of smooth muscle cells. To distinguish between these possibilities, we performed coimmunostaining for SM22α and Ki67, a marker of proliferating cells. Whereas endothelial-specific Jag1 mutant embryos showed a significant reduction in the total number of SM22α-positive cells, there was no significant difference in the number of cells that were Ki67-positive (Fig. 2 I–L), suggesting that loss of smooth muscle was not caused by diminished proliferation. We also investigated smooth muscle cell survival by using TUNEL assays, which failed to reveal significant levels of apoptosis in either control or mutant embryos (data not shown).
These results suggest that endothelial Jag1 affects differentiation of adjacent smooth muscle. We also examined endothelial cells in the endothelial Jag1 mutants. In loss-of-function models for endothelial Notch signaling, blood vessels lose arterial specification and assume a venous phenotype. This change is associated with loss of EphrinB2 expression, which has been shown to be a direct target of Notch (5, 8, 32). As veins develop a thinner vascular smooth muscle layer than arteries, we were interested in whether or not loss of arterial specification may explain the smooth muscle phenotype we see in conditional Jag1 mutants. To analyze endothelial Notch activation, we performed immunostaining for the ICD of Notch1, which is the predominant receptor in endothelial cells but shows minimal activation in vascular smooth muscle cells (33). This staining revealed nuclear expression in the endothelial layer of both control and Tie2-Cre+;Jag1flox/flox embryos (Fig. 3 A and B). Quantification of this staining revealed no significant difference in the total number of Notch1 ICD-positive endothelial cells between control and mutant embryos (Fig. 3C). As this antibody is specific for Notch1, we cannot exclude the possibility that activation of other Notch receptors may be disrupted in the mutant embryos. In addition to Notch1, Notch4 is expressed the endothelium (4). However, Notch4 expression is not required for embryonic development (4), so it is unlikely that loss of activation of this receptor would result in the observed phenotype. We also examined the expression of the Notch effector EphrinB2, which was expressed in the dorsal aorta but not the cardinal vein in both control and Tie2-Cre+;Jag1flox/flox embryos (Fig. 3 D and E). These results do not reveal any major disruptions in endothelial Notch activation or arterial specification in the endothelial Jag1 mutants.
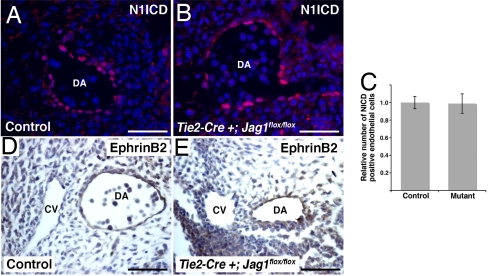
Endothelial Notch1 activation and EphrinB2 expression in endothelial-specific Jag1 mutants. (A and B) Immunostaining for Notch1 ICD (N1ICD, red) with Hoechst nuclear counterstaining (blue) on sections through the dorsal aorta (DA) of E10.5 control (A) and mutant (B) embryos. (C) Quantification of number of endothelial cells with positive nuclear staining for N1ICD in control and mutant embryos. Error bars indicate 1 SD. (D and E) Immunostaining for EphrinB2 (brown) showing expression in the dorsal aorta (DA) but not the cardinal vein (CV) of both control (D) and mutant (E) embryos. (Scale bars: 50 μm.)
To further investigate the possibility that Jag1 may play a role in endothelial Notch activation, we asked whether the phenotypes resulting from loss of Jag1 were similar to those associated with loss of endothelial Notch signaling. Several mutant mouse models, including endothelial Notch1 and CSL knockouts, display a characteristic phenotype. In addition to arterial specification defects, these embryos demonstrate severe defects in remodeling of the vascular plexus of the yolk sac and constriction of the major blood vessels of the embryo proper. The result of these defects is typically embryonic lethality at E9.5 (4, 5, 34). To generate our own endothelial loss-of-function model, we used a mouse in which a dominant negative form of MAML (DNMAML) can be activated in a tissue-specific manner by using Cre recombinase (35). This model is a potent and specific means of inhibiting signaling by all four mammalian Notch receptors in vivo (22, 35, 36). We activated DNMAML specifically in endothelial cells by crossing with Tie2-Cre transgenics. The resulting Tie2-Cre+;DNMAML embryos displayed multiple abnormalities that strongly resemble those reported in other endothelial-specific Notch mutants (Fig. 4).
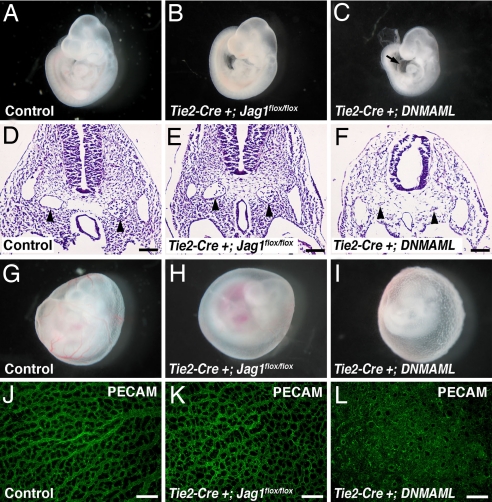
Comparison of phenotypes of endothelial-specific Jag1 and endothelial-specific DNMAML mutants. (A–C) E9.5 embryos. Tie2-Cre+;Jag1flox/flox mutants (B) are grossly indistinguishable from controls (A), whereas Tie2-Cre+;DNMAML mutants (C) are developmentally delayed and show pericardial effusions (arrow in C). (D–F) Hematoxylin and eosin-stained sections through E9.5 embryos with dorsal aortae indicated by arrowheads. Dorsal aortae in Tie2-Cre+;Jag1flox/flox mutants are morphologically normal, whereas Tie2-Cre+;DNMAML mutant dorsal aortae are atretic. (G–I) E9.5 yolk sacs showing loss of large blood vessels in Tie2-Cre+;Jag1flox/flox mutants and abnormal rough texture in Tie2-Cre+;DNMAML mutants. (J–L) E9.5 yolk sacs whole-mount immunostained for PECAM (green). Tie2-Cre+;Jag1flox/flox yolk sac capillaries appear similar to controls, whereas Tie2-Cre+;DNMAML yolk sac vessels form a markedly abnormal, highly fused plexus. Images in A–C and G–I were photographed at ×40 magnification. (Scale bars: D–F, 100 μm; J–L, 200 μm.)
Comparing the phenotypes of Tie2-Cre+;Jag1flox/flox and Tie2-Cre+;DNMAML embryos revealed a number of differences. Whereas Tie2-Cre+;DNMAML embryos showed severe developmental delay and pericardial effusions at E9.5, Tie2-Cre+;Jag1flox/flox embryos were indistinguishable from their littermates at this time point (Fig. 4 A–C). Cross-sections through the embryos showed severely narrowed dorsal aortae in the Tie2-Cre+;DNMAML mutants, whereas the dorsal aortae were grossly normal in the Tie2-Cre+;Jag1flox/flox mutants (Fig. 4 D–F). Examination of the yolk sac vasculature at E9.5 also revealed differences between Tie2-Cre+;DNMAML and Tie2-Cre+;Jag1flox/flox embryos. As previously mentioned, the yolk sacs of Tie2-Cre+;Jag1flox/flox embryos contained very few large vessels when compared with controls. The Tie2-Cre+;DNMAML yolk sacs also lacked large vessels, but had a rough texture that was not evident in the Tie2-Cre+;Jag1flox/flox yolk sacs (Fig. 4 G–I). This phenotype indicates a severely underdeveloped vasculature that is characteristic of a lack of angiogenic remodeling. We examined the yolk sac vessels in more detail with whole-mount immunostaining for the endothelial marker PECAM, followed by confocal microscopy. The fine capillary network of the Tie2-Cre+;Jag1flox/flox yolk sacs was not significantly different from control yolk sacs. In contrast, the vasculature of the Tie2-Cre+;DNMAML yolk sacs showed no evidence of remodeling from the primary vascular plexus into a fine capillary network (Fig. 4 J–L). By E10.5, all Tie2-Cre+;DNMAML embryos were necrotic, indicating that they die between E9.5 and E10.5, 1 full day earlier than Tie2-Cre+;Jag1flox/flox embryos.
Our results clearly demonstrate a vital function for Jag1 in endothelium, where it is required for vascular smooth muscle development. We favor a model in which endothelial Jag1 signals directly to Notch receptors on vascular smooth muscle cells, although indirect signaling cannot be excluded. A direct signaling model is supported by our previous results demonstrating a requirement for Notch activity in neural crest-derived vascular smooth muscle differentiation (22), but the specific Notch receptor responsible for Jag1-mediated signaling in smooth muscle remains to be identified. Observations from our group and others have shown that Notch1 and Notch4 are the predominant Notch receptors expressed by vascular endothelium, whereas Notch2 is expressed by smooth muscle precursors in the neural crest and Notch3 is expressed by vascular smooth muscle cells (3, 22, 23, 37, 38).
Our data also suggest that defects associated with loss of endothelial Notch signaling cannot be accounted for by loss of endothelial Jag1 alone. Although we cannot completely rule out the ability of endothelial Jag1 to signal to adjacent endothelial cells, we were unable to demonstrate any such activity. The expression of activated (nuclear) Notch1 ICD in endothelium was unchanged by loss of Jag1, arterial identity was preserved, and a direct Notch target in endothelium, EphrinB2, was expressed at normal levels. Also, inhibition of Notch signaling in endothelium produces a more severe vascular phenotype than does deletion of Jag1. Therefore, it is likely that a Notch ligand other than Jag1 plays a more dominant role to mediate Notch signaling between endothelial cells. The most obvious candidate is Dll4, which is prominently expressed in the endothelium and has been shown to act by promoting arterial specification and inhibiting angiogenesis (5, 11–15). In fact, several reports demonstrate that loss of Dll4 can result in defects remarkably similar to those that we observe in Tie2-Cre+;DNMAML embryos (1, 2, 5).
Our suggestion that Jag1 acts by signaling to adjacent smooth muscle precursors to promote their differentiation, whereas Dll4 signals to adjacent endothelial cells to influence angiogenesis and arterial specification, is supported by in vitro studies showing that Notch-dependent activation of the promoter of the smooth muscle myosin heavy chain gene occurs only when cells are stimulated with Jag1, but not with Dll4 (20). The idea that different ligands may have differing downstream effects is an emerging theme in the Notch signaling field (39, 40).
Human mutations in JAG1 result in Alagille syndrome, a congenital disease associated with significant cardiovascular pathology (25, 26). We suggest that diminished Jag1 expression on endothelial cells results in abnormal smooth muscle development, which may be responsible for the pulmonary artery stenosis that is a frequent finding in Alagille syndrome patients. Consistent with this finding, we have previously shown that inhibition of Notch in neural crest cells, which act as smooth muscle precursors in the pulmonary artery, results in pulmonary artery stenosis and other congenital heart defects reminiscent of those seen in Alagille syndrome (22). Defects in smooth muscle development may also be responsible for other vascular pathologies seen in patients with Alagille syndrome, such as a predisposition to intracranial bleeding (28).
Materials and Methods
Mice.
Tie2-Cre (30) and CMV-Cre (JAX) mice were genotyped by using previously described Cre-specific primers (22). Jag1flox and DNMAML mice were genotyped as described (22, 29). All mice were maintained on mixed genetic background. Animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Immunostaining.
Antibodies used for immunohistochemistry and immunofluorescence were anti-Jagged1 rabbit polyclonal H-114 (Santa Cruz Biotechnology), anti-Notch1 ICD Val-1744 rabbit polyclonal (Cell Signaling Technology), anti-EphrinB2 goat polyclonal (R&D Systems), anti-PECAM rat monoclonal MEC 13.3 (BD Pharmingen), anti-αSMA mouse monoclonal 1A4 (Sigma), anti-SM22α goat polyclonal (Abcam), anti-Ki67 rabbit monoclonal (Vector Laboratories), and anti-von Willebrand factor rabbit polyclonal (Sigma). Immunostaining was performed on paraformaldehyde-fixed, paraffin-embedded sections. Detailed protocols are available (41).
For whole-mount immunostaining, tissues were fixed for 2 h (yolk sacs) or overnight (embryos) in 4% paraformaldehyde and stained with anti-PECAM and anti-αSMA antibodies (as above) followed by secondary detection with Alexa Fluor-488- or -568-conjugated secondary antibodies (Molecular Probes) or HRP-conjugated secondary antibodies (Abcam). Yolk sacs were flat-mounted in 90% glycerol and analyzed by confocal microscopy.
Cell Counting and Statistics.
To quantify immunostaining data, fluorescent images were overlaid by using Adobe Photoshop, and cell counting was performed with ImageJ software. Cell counts were obtained from multiple transverse sections through the dorsal aortae from the level of the aortic arch arteries to the cardiac inflow tract. Counts were averaged for each embryo, and graphed values represent the means of these values obtained from multiple embryos. For SM22α and Ki67 staining, ≈20–25 sections were analyzed from each of five control and mutant embryos. For Notch1 ICD staining, ≈10 sections were analyzed from each of four control and mutant embryos.
ACKNOWLEDGMENTS.
We thank Andrea Stout for assistance with confocal microscopy. This work was supported by a Developmental Biology Training Grant from the University of Pennsylvania (to F.A.H.), the W.W. Smith Charitable Trust, and National Institutes of Health Grants P01 HL075215 and RO1 HL61475 (to J.A.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709663105/DC1.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0709663105
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2538864?pdf=render
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/105/6/1955
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/105/6/1955
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/105/6/1955.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The N-acetylglucosaminyltransferase Radical fringe contributes to defects in JAG1-dependent turnover and signaling of NOTCH3 CADASIL mutants.
J Biol Chem, 300(10):107787, 19 Sep 2024
Cited by: 0 articles | PMID: 39303912 | PMCID: PMC11525139
Overexpression of miR-199b-5p in Colony Forming Unit-Hill's Colonies Positively Mediates the Inflammatory Response in Subclinical Cardiovascular Disease Model: Metformin Therapy Attenuates Its Expression.
Int J Mol Sci, 25(15):8087, 25 Jul 2024
Cited by: 0 articles | PMID: 39125657 | PMCID: PMC11311364
Akt3 activation by R-Ras in an endothelial cell enforces quiescence and barrier stability of neighboring endothelial cells via Jagged1.
Cell Rep, 43(3):113837, 24 Feb 2024
Cited by: 0 articles | PMID: 38402584
eNOS plays essential roles in the developing heart and aorta linked to disruption of Notch signalling.
Dis Model Mech, 17(1):dmm050265, 22 Jan 2024
Cited by: 0 articles | PMID: 38111957 | PMCID: PMC10846539
Progress to Clarify How <i>NOTCH3</i> Mutations Lead to CADASIL, a Hereditary Cerebral Small Vessel Disease.
Biomolecules, 14(1):127, 18 Jan 2024
Cited by: 1 article | PMID: 38254727 | PMCID: PMC10813265
Review Free full text in Europe PMC
Go to all (210) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Loss of Jagged1 in mature endothelial cells causes vascular dysfunction with alterations in smooth muscle phenotypes.
Vascul Pharmacol, 145:107087, 02 Jul 2022
Cited by: 11 articles | PMID: 35792302
Notch activation of Jagged1 contributes to the assembly of the arterial wall.
Circulation, 125(2):314-323, 06 Dec 2011
Cited by: 101 articles | PMID: 22147907 | PMCID: PMC3260393
Vascular smooth muscle Notch signals regulate endothelial cell sensitivity to angiogenic stimulation.
J Biol Chem, 286(15):13741-13753, 23 Feb 2011
Cited by: 26 articles | PMID: 21349836 | PMCID: PMC3075718
Anti-Jagged-1 immunotherapy in cancer.
Adv Med Sci, 67(2):196-202, 11 Apr 2022
Cited by: 3 articles | PMID: 35421813
Review
Funding
Funders who supported this work.
NHLBI NIH HHS (3)
Grant ID: R01 HL061475
Grant ID: P01 HL075215
Grant ID: R01 HL61475





