Abstract
Free full text

Amerindian Helicobacter pylori Strains Go Extinct, as European Strains Expand Their Host Range
Abstract
We studied the diversity of bacteria and host in the H. pylori-human model. The human indigenous bacterium H. pylori diverged along with humans, into African, European, Asian and Amerindian groups. Of these, Amerindians have the least genetic diversity. Since niche diversity widens the sets of resources for colonizing species, we predicted that the Amerindian H. pylori strains would be the least diverse. We analyzed the multilocus sequence (7 housekeeping genes) of 131 strains: 19 cultured from Africans, 36 from Spanish, 11 from Koreans, 43 from Amerindians and 22 from South American Mestizos. We found that all strains that had been cultured from Africans were African strains (hpAfrica1), all from Spanish were European (hpEurope) and all from Koreans were hspEAsia but that Amerindians and Mestizos carried mixed strains: hspAmerind and hpEurope strains had been cultured from Amerindians and hpEurope and hpAfrica1 were cultured from Mestizos. The least genetically diverse H. pylori strains were hspAmerind. Strains hpEurope were the most diverse and showed remarkable multilocus sequence mosaicism (indicating recombination). The lower genetic structure in hpEurope strains is consistent with colonization of a diversity of hosts. If diversity is important for the success of H. pylori, then the low diversity of Amerindian strains might be linked to their apparent tendency to disappear. This suggests that Amerindian strains may lack the needed diversity to survive the diversity brought by non-Amerindian hosts.
Introduction
Humans coevolved with their microbiomes [1]–[3] and persistent human microbes can be markers of human migrations. Viruses such as HPV [4], Hepatitis G [5], RNA retrovirus HTLV-1 [6] and the bacterium Helicobacter pylori [7]–[9] show signs of coevolution with their human host, reflecting the serial founding African-origin model of human evolution due to successive human migrations that consisted of only a subset of the genetic variation available at the source location [10], [11].
The original human migration out of Africa occurred approx. 60,000 years ago, towards the Middle East and thereafter independently to Europe and Asia [12]. The Americas were populated by humans of East Asian ancestry that crossed the Bering Strait, about 15 thousand years ago. These first Americans suffered a genetic bottleneck [13]–[15], and the reduced genetic diversity in Amerindians is evidenced in the absolute dominance of the O blood group among Amerindians ( Fig. 1 ), their low heterozygosity and the reduced number of mitochondrial DNA haplogroups [16], [17]. However, the diversity of Amerindians has increased through genetic inflow from Europeans and Africans over the last 500 years, leading to an increasing Mestizo population.
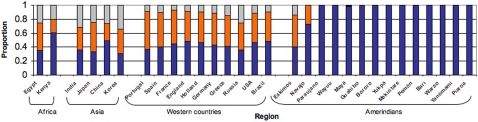
The blue, orange and grey bars represent respectively O, A and B allele frequencies [34]–[43]. The remarkable dominance of O blood groups among Amerindians from South America affects the ABO blood group recognition by H. pylori strains.
H. pylori is a human gastric indigenous bacteria, in spite of being a risk factor for gastric cancer and peptic ulcer disease [18], [19]). A congruent divergence of H. pylori and their human hosts suggests that the quality of host resources influences the diversity of indigenous bacteria and their adaptations. In host-parasite interactions, a restricted gene flow in the host leads to local microbial adaptation [20], [21]. An example of local adaptation is the differing affinity of H. pylori strains to bind blood group antigens expressed in the human gastric mucosa: European H. pylori strains bind all three (A, B and O) blood group antigens, but an important proportion of strains from the are dominantly O blood group Amerindians have higher affinity for O blood antigen [22].
Environmental diversity allows different species to occupy different niches, sustaining coexistence and species diversity. There is a strong body of evidence for the loss of diversity by reduction of niches. In addition to landscape mosaicism, temporal variability also affects diversity. Simulations predict that while spatial variability increases diversity, the strongest increase occur at intermediate levels of temporal variability [23]. When the environment of a species is provided by another species, there is a set of age-changing mosaic niches provided by each individual host for the life of the host. Since microbes circulate in a dynamic population of hosts, the microbe diversity is held at the host population –and not individual- level. The studies that have provided the basis for many ecology theories have come from the field of plant ecology. Plant ecological studies have shown that diversity of plant associated herbivores increases with diversity of plant species [24] but also with population genotypic diversity, within a single species [25]. As such, a direct association between diversities in host and microbiome microbes could be expected. To test this hypothesis, we determined intra-population genetic diversity of strains of the highly diverse gastric bacteria H. pylori and of humans from the Americas and other geographical populations relevant to its peopling.
Results
We determined the intra-population genetic diversity of multilocus sequences of 7 housekeeping genes from 131 strains of the gastric bacteria H. pylori and of 2,232 sequences of the human mtDNA hypervariable segment I. H. pylori strains were selected from human populations relevant to the ancestry of the people of the Americas, with East Asians providing a reference group for Amerindians: 131 H. pylori isolates were from Amerindians, Spanish, West-Africans, South American Mestizos and Koreans. Based on their DNA multilocus sequences, the strains were assigned to one of 6 geographical bacterial populations: hpEastAsia (subpopulations hspAmerind, hspMaori and hspEAsia), hpAsia2, hpEurope, hpNEAfrica, hpAfrica1 (hspWAfrica, hspSAfrica) and hpAfrica2 [7], [26]. Selected human sequences were from similar locations to those where H. pylori strains had been isolated. A total of 1,148 sequences of human mtDNA hypervariable segment I from non-Bantu Africans, Spanish, Koreans, Amerindians (Guahibo, Huitoto, Inuit) and Latin American Mestizos were retrieved from Genbank and from published work. Bacterial and human genetic distance matrixes were generated and compared using non-parametric statistics.
Strain genetic diversity
The bacterial strains were assigned to populations according to their multilocus DNA sequences: those from African hosts yielded only hpAfrica1, those from Spanish yielded hpEurope and those from Koreans yielded hspEAsia ( Table 1 ). However, Huitoto and Guahibo Amerindians yielded both hspAmerind and hpEurope strains, and Mestizos yielded hpEurope and hpAfrica1, but not hspAmerind. The least and most diverse strains of H. pylori populations were hspAmerind and hpEurope, respectively ( Figure 2A ). Nonetheless, when grouping the strains by host, strain diversity in Amerindians increased to the levels found in Spanish and Mestizo hosts ( Figure 2B ), consistently with the circulation of mixed strains ( Table 1 ) and with the remarkable mosaicism reflected in the multilocus sequences ( Figure 3 ). As stated previously [7], [26], the ancestry patterns of modern hpEurope strains revealed extensive recombination between the two ancestral populations ancestral Europe1 and ancestral Europe2. Spanish H. pylori ( Figure 3 ) further include components from ancestral hpAfrica1, which possibly reflects the role Africa has played in shaping the Spanish modern human gene pool. Traces of African bacterial ancestry were also detected in strains from Mestizos and Amerindians. The observed mosaicism reflects extensive recombination and results in lower genetic structure.
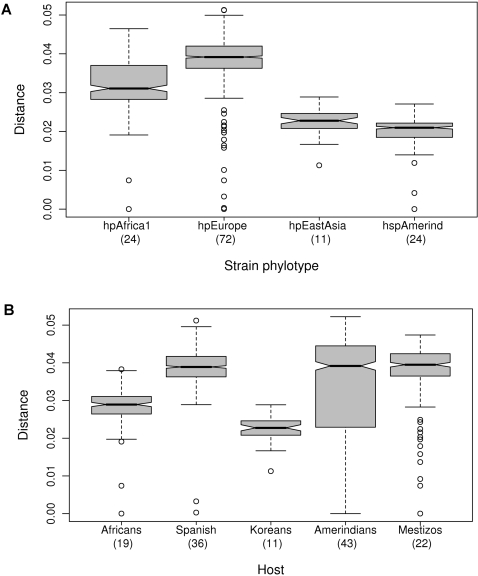
Differences in pairwise distances among strains in Fig. 2A were significant (Kruskal-Wallis test, p<2.2×10−16, Wilcoxon and Bonferroni; p<10−14) with a decreasing order hpEurope>hpAfrica1>hpEastAsia>hspAmerind. When grouped by the human host from which each strain was isolated (Fig 2B), strain diversity in Amerindians was as high as in Spanish and Mestizos (with no significant differences among them; Wilcoxon Pairwise comparison p>0.7), with a decreasing order Spaniards =
= Amerindians
Amerindians =
= Mestizos>Africans>Koreans. The waist of the dress-like box is the median with the waist side openings indicating the 95% interval for the median; the top represents the 3rd quartile and the bottom the 1st quartile. The interval in dashed lines represents a maximum of 1.5× interquartile range and the open circles are outliers. Permutation tests confirmed group differences in strain diversity.
Mestizos>Africans>Koreans. The waist of the dress-like box is the median with the waist side openings indicating the 95% interval for the median; the top represents the 3rd quartile and the bottom the 1st quartile. The interval in dashed lines represents a maximum of 1.5× interquartile range and the open circles are outliers. Permutation tests confirmed group differences in strain diversity.
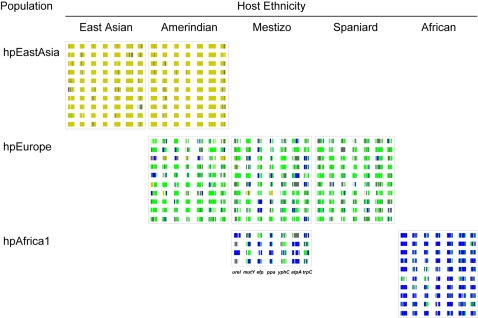
The ancestral source of each polymorphic nucleotide is shown by a vertical line for each of the seven gene fragments in the multilocus analysis of 10 representative strains from each group (see the legend below Mestizo strains). Individual nucleotides were derived from ancestral Europe1 (grey), ancestral Europe2 (green), ancestral Africa1 (blue) and ancestral EastAsia (yellow). Nucleotides not assigned with >50% probability to any one population are indicated by white lines. African and European components can be observed in hpEurope strains from Spaniards and Mestizos, as well as in hpAfrica1 from Mestizos, while African hpAfrica1 strains and Amerindian hspAmerind strains tested were largely homogeneous.
Table 1
| Location/source of human population | Bacterial population | ||||
| No. of strains | hpAfrica1 | hpEurope | hpEAsia | hspAmerind | |
| African | 19 | 19 | |||
| Senegal | 5 | 5 | |||
| Burkina Faso | 14 | 14 | |||
| Europe | 36 | 36 | |||
| Spain | 36 | 36 | |||
| East Asian | 11 | 11 | |||
| Korea | 11 | 11 | |||
| Amerindian | 43 | 19 | 24 | ||
| Inuit (Eskimo) | 13 | 4 | 9 | ||
| Athabaskan (Na-Dene) | 6 | 6 | |||
| Huitoto | 16 | 12 | 4 | ||
| Piaroa | 3 | 3 | |||
| Guahibo | 5 | 3 | 2 | ||
| Mestizo | 22 | 5 | 17 | ||
| Colombia | 12 | 1 | 11 | ||
| Venezuela | 10 | 4 | 6 | ||
| All | 131 | 24 | 72 | 11 | 24 |
Multilocus sequences of the strains were from 7 housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI and yphC). While only the expected populations were identified in African, Spanish and Korean hosts, strains assigned to more than one population were observed among Amerindians and Mestizos.
Human genetic diversity
Host genetic diversity increased in this study in the order: Amerindians<Spaniards<Koreans<Africans<Mestizos (Kruskal-Wallis p<10−14, Figure S4). However, measurement of human diversity through mtDNA underestimates paternal-linked diversity. Mestizos diversity has probably been introduced by both maternal (Amerindian and African) and paternal (European and African) gene flow, but the remarkably low diversity in the Spanish mtDNA may not reflect the true variability of human genetic diversity introduced in Spain by paternal lines.
Discussion
Human evolution has been shaped by the early forms of life in the planet, the microbes. Indeed, evolution of all eukaryotes occurred in a bacterial planet, and microbes found niches on surfaces, invaginations and guts of humans, as well. Helicobacters found a niche in the gastric pouch of mammals [27], including humans. Helicobacter pylori has colonized the human stomach since the beginning of the human history [26], and maintained a tuned evolution with its host, reflecting in its genome some genetic, linguistic and cultural traits of the human population. It can therefore be a useful model to determine how humans modulate the diversity of their microbiomes.
Genetic divergence and convergence
Although most of the data analyzed in this study was already available, they had previously not been analyzed in terms of the associations between genetic diversity of humans and their indigenous microbial population. There are many genetic studies that confirm that human intrapopulation genetic diversity decreases with geographic distance from Africa, and that the Amerindians suffered a genetic bottleneck [12]–[17]. The finding that a low-diversity human population was associated with a low-diversity bacterium could be due to increased genetic drift (on a populations of low effective size), and/or selection by low diversity blood groups on the H. pylori population.
Genetic isolation and drift as well as selection may explain the radiation of humans into different groups, and the concomitant divergence of H. pylori into geographical types, whereby both host and bacteria show patterns of isolation by distance [26]. However, the modern world is increasingly bringing genetic influx into each of the human groups, both at the human and microbiome level. Indigenous microbes that once diverged during most of human history are currently in the process of converging, which erase phylogeographic signals. How this process is occurring is important because some human-evolved bacteria are relevant to human health (for example H. pylori is implicated in gastric diseases), and changes in the dynamics of its coevolution with humans might lead to changes in disease patterns.
Genetic diversity and fitness
There might be greater local genetic variation in parasite populations relative to their hosts. In this case, parasite populations might increase their genetic structure by locally adapting to their hosts [28]. Consistently, mosaic strains would be expected to be genetically more diverse and more generalist. This might explain why hpAfrica1 strains from Mestizos are more diverse than those from African hosts (Kruskal-Wallis p<10−14; Figure S1 ). Recombination obviously requires coexistence of different strains in a single stomach, which we indeed have found to be frequent in patients from Venezuela [29].
Diversity optimizes niche partitioning. In the gastric context, strain mosaicism (intra-genomic diversity) and strain diversity (inter-genomic variants) is likely to be a key factor in the hpEurope strains host range expansion (in both Mestizos and Amerindians). Meanwhile, hspAmerind strains seem to lack the diversity that would be required to survive in the high host variability brought by Mestizos. Displacement of hspAmerind by hpEurope strains could occur by two different means: 1) Strain competition during colonization, in which “generalist” strains will have better chance to colonize diverse niches than “specialist” strains [as in the case of O binder Amerindian strains described above, which would have less success in colonizing non-O blood group Mestizos [22]]; 2) Strain subversion by transformation, in which DNA from hpEurope strains is taken up and recombinant strains become increasingly European. The high component of European ancestry in the mosaic strains from Mestizos and Amerindians, and the relatively low Amerindian component in Mestizos ( Figure 3 ) support this second hypothesis.
The consumption of antibiotics and other drugs (such as proton pump inhibitors that reduce gastric acidity) might also be currently affecting the bacterial populations by increasing selection pressure in favor of particular variants, thus shaping the genome of indigenous microbes in modern humans. Modern human admixture results not only in increased human diversity but also in increased microbial diversity of the human microbiome. In the long term, however, one can predict that maximum genetic distances will be succeeded by homogenization of the genomic structure in strain variants (all mosaic or recombinant strains) and humans (all Mestizos) and decreased within population genetic distances brought by further recombination and lack of isolation, as illustrated in Figure 4 . Since no influx of genetic variation is expected from outside the global village, low genetic flow in both microbes and hosts will eventually result in decreased human and microbiome diversity.
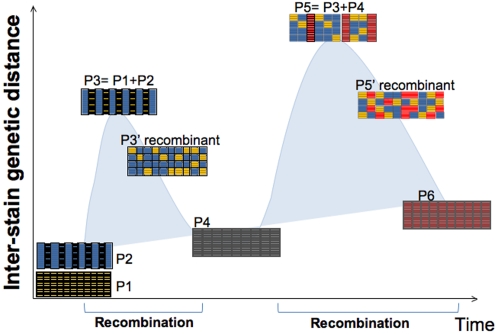
In the context of our hypothesis, a low diversity H. pylori population (P1) arose from co-evolution with the isolated Amerindian host population. With the introduction of new H. pylori strains (P2) the new population formed (P3) is more diverse than any of the source populations alone. Selection acts and strains recombine with the consequent homogenization of the population (P3′). The cycle is repeated when new populations (P4) arrive. Given time and isolation (no gene flow), population diversity will be reduced (P6). Based on their mosaic structure and high genetic distances, it seems that current H. pylori from Amerindians and Mestizos are in one of the intermediate states (P3′ or P5′). Arrows indicate introduction of new populations.
To our knowledge, Wirth et al. [30] were the first to analyze both human DNA and H. pylori sequences from 50 patients in Ladakh in northern India. Our results support the need of a study with a true large scale population-based approach involving human and microbe samples from the same hosts in order to better measure the extent in which human populations shape the genomes of H. pylori. As new technology optimizes cost and time of sequencing, there can be new studies performed in individual Homo sapiens and his individual microbiome, using more genes. These studies will be crucial to understand the coevolution of humans and their microbes.
Conclusions
In the analysis of differences in genetic diversity in human groups and the concomitant diversity of a human indigenous microbe, our study provides support to the hypothesis that the distribution of genetic diversity in humans determines the genetic diversity of indigenous microbes. Specifically, we found genetic evidence of i) a decreased diversity of H. pylori strains in the human group with the least genomic diversity; ii) an increased diversity in strains from Mestizos, hybrid strains with mosaic structure assigned to the hpEurope group; iii) a host-range expansion of these hpEurope strains into Mestizos and Amerindian hosts, confirming that niche diversity widens the sets of resources for colonizing species.
Materials and Methods
H. pylori strains
We selected strains from Amerindians and other human populations relevant to the peopling of the Americas. These included, Spanish, West-Africans, Mestizos, and East Asians who provide a reference group for Amerindians.
We analyzed multilocus sequences from 7 housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI and yphC) [7] in 131 H. pylori isolates cultured from 19 Africans (14 from Burkina Fasso, 5 from Senegal), 36 Spanish, 11 Koreans, 43 Amerindians (13 Inuit, 6 Athabaskan, 16 Huitoto, 5 Guahibo and 3 Piaroa), and 22 Mestizos (12 from Colombia and 10 from Venezuela). Sequences were available in http://www.mlst.net/ and the EMBL database except for 14 sequences experimentally obtained in this work, from 8 Amerindians (5 Guahibos, 3 Piaroas) and 6 Venezuelan Mestizos, now available in GenBank (accession numbers EU878038–EU878135) and in the MLST database in Oxford (http://www.mlst.net/).
Strain population assignment was performed as described by Falush et al [7], using the “no admixture” model in STRUCTURE2.0 [31], the proportion of nucleotides being derived from ancestral population was estimated using the “linkage” model in Structure2.0 as described [7], [26].
Human mtDNA sequences
A total of 1,148 sequences of human mtDNA hypervariable segment I were retrieved from Genbank from published work (Table 2), from humans close to the hosts from which H. pylori strains had been cultured: non-Bantu Africans, Spanish, Koreans, Amerindians (Guahibo, Huitoto, Inuit) and South American Mestizos.
Table 2
| Host group | Number of mtDNA sequences | References |
| Africans | 53 | [44] |
| Hausa (non-Bantu) | 13 | |
| Sierra Leone (non-Bantu) | 40 | |
| Europeans-Spanish | 718 | |
| Andalusia | 130 | Genbank; [45]–[47] |
| North Eastern | 118 | [48] |
| Catalans | 61 | [45]–[47] |
| Baleares | 222 | [49] |
| Galicia | 92 | [50] |
| Canary Island, | 54 | [46] |
| Spain various | 41 | [46], [47] |
| East Asians-Koreans | 64 | [51] |
| East Asians- Amerindians | 143 | |
| Guahibo | 66 | GenBank |
| Huitoto | 7 | [52] |
| Inuit (Eskimo) | 70 | [53] |
| Mestizos a | 170 | [54] |
| African haplotypes | 58 | |
| European haplotypes | 59 | |
| Amerindian haplotypes | 53 | |
| TOTAL | 1,148 |
Statistical analyses
Genetic distances (pairwise distances) of H. pylori isolates and human mtDNA were analyzed by strain ancestral phylogroups and by human host groups. Distances were calculated using the Kimura 2-parameter model in MEGA3 [32]. Nonparametric tests (Kruskal-Wallis rank sum, Wilcoxon test with Bonferroni adjustment for multiple comparisons) were used to compare strains by bacterial population and by host. In addition, these comparisons were performed using permutation tests based on 5000 permutations. All statistical analyses were performed using R statistical software [33].
Supporting Information
Figure S1
Intrapopulation genetic distance of H. pylori strains hpAfrica1 and hpEurope, by host source. hpAfrica1 strains from Mestizos were much more diverse than those from Africans (Kruskal-Wallis p<10−14). In contrast, neither Mestizos nor Amerindians increased the already high variability hpEurope strains from Spain. Medians are represented as the waist of the dress-like box, and the waist side openings indicate the 95% interval for the median. Above and below the median are the 3rd and 1st quartile respectively. The interval in dashed lines represents a maximum of 1.5× interquartile range and the open circles are outliers. Outliers are mostly low pairwise distances, indicating that similar pair of strains are less common than distant strains.
(0.24 MB TIF)
Figure S2
Genetic distances between mtDNA sequences of diverse human groups. Differences between median distances were significant for each of the human groups (Kruskal-Wallis test p<2.2×10−15; Wilcoxon with Bonferroni adjustment p<10−14). Permutation tests with 5,000 permutations confirmed human group differences (p =
= 0). Distances decreased in the order: mestizos>Africans>Koreans>Spanish> Amerindians. For explanation of the box plot see Figure S1.
0). Distances decreased in the order: mestizos>Africans>Koreans>Spanish> Amerindians. For explanation of the box plot see Figure S1.
(18.39 MB DOC)
Figure S3
Genetic distances of human mtDNA sequences within Africans, Spanish, Koreans and groups of Amerindians and Mestizos. South American Amerindians studied have a degree of admixture as indicated by their higher genetic diversity than the Inuit. The Mestizos with African or Amerindian haplotypes have increased diversity in relation to Mestizos with European haplotypes. For explanation of the box plot see Figure S1.
(0.26 MB TIF)
Acknowledgments
We are grateful to J. Bertrand Petit for providing Spanish sequences of human mtDNA. We thank Gladys Ramos for her help with Figure 3.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by UPR intramural FIPI grant 880314. B.L. was supported by ERA-Net grant BMBF 0313930B to Mark Achtman.
References
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0003307
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0003307&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pone.0003307
Article citations
Pathogenomics of Helicobacter pylori.
Curr Top Microbiol Immunol, 444:117-155, 01 Jan 2023
Cited by: 2 articles | PMID: 38231217
Review
Peptic Ulcer and Gastric Cancer: Is It All in the Complex Host-Microbiome Interplay That Is Encoded in the Genomes of "Us" and "Them"?
Front Microbiol, 13:835313, 25 Apr 2022
Cited by: 1 article | PMID: 35547123 | PMCID: PMC9083406
Review Free full text in Europe PMC
Identification of New Helicobacter pylori Subpopulations in Native Americans and Mestizos From Peru.
Front Microbiol, 11:601839, 14 Dec 2020
Cited by: 6 articles | PMID: 33381095 | PMCID: PMC7767971
A 500-year tale of co-evolution, adaptation, and virulence: Helicobacter pylori in the Americas.
ISME J, 15(1):78-92, 02 Sep 2020
Cited by: 17 articles | PMID: 32879462 | PMCID: PMC7853065
Phylogenetic analysis of clinical strains of Helicobacter pylori isolated from patients with gastric diseases in Tibet.
Ann Transl Med, 7(14):320, 01 Jul 2019
Cited by: 2 articles | PMID: 31475190 | PMCID: PMC6694268
Go to all (36) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - EU878135
- (1 citation) ENA - EU878038
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Comparative study between Helicobacter pylori and host human genetics in the Dominican Republic.
BMC Evol Biol, 19(1):197, 01 Nov 2019
Cited by: 4 articles | PMID: 31675915 | PMCID: PMC6823972
Helicobacter pylori genotyping from American indigenous groups shows novel Amerindian vacA and cagA alleles and Asian, African and European admixture.
PLoS One, 6(11):e27212, 03 Nov 2011
Cited by: 19 articles | PMID: 22073291 | PMCID: PMC3207844
Phylogeographic evidence of cognate recognition site patterns and transformation efficiency differences in H. pylori: theory of strain dominance.
BMC Microbiol, 13:211, 19 Sep 2013
Cited by: 5 articles | PMID: 24050390 | PMCID: PMC3849833
The Story of Helicobacter pylori: Depicting Human Migrations from the Phylogeography.
Adv Exp Med Biol, 1149:1-16, 01 Jan 2019
Cited by: 7 articles | PMID: 31016625
Review





