Abstract
Free full text

Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae
Abstract
A synthetic genetic array was used to identify lethal and slow-growth phenotypes produced when a mutation in TRM6, which encodes a tRNA modification enzyme subunit, was combined with the deletion of any non-essential gene in Saccharomyces cerevisiae. We found that deletion of the REX1 gene resulted in a slow-growth phenotype in the trm6-504 strain. Previously, REX1 was shown to be involved in processing the 3′ ends of 5S rRNA and the dimeric tRNAArg-tRNAAsp. In this study, we have discovered a requirement for Rex1p in processing the 3′ end of tRNAiMet precursors and show that precursor tRNAiMet accumulates in a trm6-504 rex1Δ strain. Loss of Rex1p results in polyadenylation of its substrates, including tRNAiMet, suggesting that defects in 3′ end processing can activate the nuclear surveillance pathway. Finally, purified Rex1p displays Mg2+-dependent ribonuclease activity in vitro, and the enzyme is inactivated by mutation of two highly conserved amino acids.
INTRODUCTION
The production of mature non-coding RNA, such as tRNA and rRNA, does not result from transcription alone, but also requires a number of post-transcriptional processing and modification reactions. For example, tRNAs are transcribed as precursors that have extra nucleotides at both the 5′ and 3′ ends—and sometimes introns as well—that need to be removed (1). Furthermore, tRNAs acquire abundant nucleoside modifications, including base and ribose methylations, base isomerizations and base deaminations, that are introduced post-transcriptionally by numerous enzymes (2).
While 5′ leader removal, carried out by RNase P, is a conserved process among diverse organisms, 3′ end processing has been found to be a more complex and varied process, with differences not only between organisms, but also between the different tRNAs in a single organism (3,4). In Saccharomyces cerevisiae, both endonucleases and exonucleases that process the 3′ ends of tRNA in vitro have been found (5). A model for tRNA 3′ end processing has been proposed in which the Lhp1 protein binds the 3′ end of a precursor tRNA and promotes processing by endonucleolytic cleavage, while in the absence of Lhp1p, precursors are instead processed by exonucleases (6). However, for some tRNA precursors, no change in 3′ end processing is seen in strains lacking Lhp1p (6,7). This observation, and the fact that LHP1 is not an essential gene, suggests tRNA 3′ end processing in yeast can occur through multiple pathways (8).
The nucleases involved in tRNA 3′ processing in S. cerevisiae are generally unknown, including the endonuclease that cleaves tRNAs bound by Lhp1, or have been identified but not fully characterized. Several exo- and endonucleases that process the 3′ ends of tRNAs in vitro have been purified from yeast, but the corresponding genes were not identified, and the role of these enzymes in tRNA processing in vivo is unknown (5). tRNase Z, an endonuclease found in all three domains of life that removes the 3′ trailer from tRNAs, has been identified in yeast (TRZ1) and found to be essential for viability (9,10). While in vitro endonuclease activity for this enzyme has been observed using human precursor tRNAArg as a substrate (11), the function of yeast Trz1p in vivo is not known. Lastly, a ribonuclease encoded by the REX1 gene has been shown to trim the 3′ ends of some intron-containing tRNAs and a tRNAArg that is produced from a dimeric transcript also encoding tRNAAsp (12,13).
Previously, REX1 was named RNA82 and described as a nuclease involved in processing 5S rRNA (14). In an rna82.1 mutant strain, 5S rRNA was found to have an extended 3′ end, with up to 13 extra nucleotides detected in pulse-labeling experiments, and as many as three additional nucleotides found in steady-state RNA samples (14). In addition, the dimeric tRNAArg-tRNAAsp transcript was not fully processed in an extract from rna82.1 cells, as the 3′ end of tRNAArg was found to retain nucleotides normally removed in an extract from a wild-type strain (15). Rex1p also appears to process the 3′ ends of some tRNAs that contain introns, such as tRNATyr(GUA) and tRNALys(UUU) (13). With the availability of protein sequence data, Rex1p has been classified as a member of the DEDD superfamily of exonucleases (16), although this activity has not been demonstrated using purified Rex1 protein.
Previously, we identified and characterized Trm6p and Trm61p from S. cerevisiae, the two subunits of a tRNA modification enzyme that methylates A58 in the TΨC loop of tRNAs (17). In this study, we have found that a synthetic slow-growth phenotype results when REX1 is deleted in a trm6-504 mutant strain. We demonstrate that Rex1p has a role in initiator tRNAMet (tRNAiMet) 3′ end processing and show that tRNAiMet precursors, particularly those with extended 3′ trailers, accumulate in a trm6-504 rex1Δ strain. We show that loss of Rex1p results in polyadenylation of tRNAiMet, as well as the previously identified substrates of Rex1p—5S rRNA and tRNAArg-tRNAAsp. Rex1p purified from yeast displays ribonuclease activity when mature or precursor tRNAiMet is provided as a substrate and mutation of amino acids conserved among DEDD exonucleases eliminates this activity.
MATERIALS AND METHODS
Yeast strains
REX1 was deleted in strains Y200 and Y190 (18) by insertion of the kanamycin resistance gene. PCR amplification of the KanMX4 cassette with REX1 flanking sequence was performed using primers JA396 and JA397. Yeast strains were transformed as described (19) with the PCR product and allowed to grow in Yeast extract–Peptone–Dextrose (YPD) liquid media at 30°C for 3 h before being plated to YPD with 200 μg/ml geneticin. Transformants were replica printed to YPD with geneticin and the resulting colonies screened for the correct insertion of KanMX4 by yeast colony PCR (20) with primer sets JA370 and JA398, JA385 and JA398 and JA399 and JA398. Deletion of REX1 in Y190 was named Y386 and in Y200 was named Y387. Y200, Y190, Y386 and Y387 transformed with IMT4 (p108) and imt4-3 (p467) are strains Y450-453 and Y454-457, respectively. Similarly, strains carrying the empty plasmid pRS316 (21) are strains Y468-471. Y200, Y190, Y386 and Y387 strains carrying the empty plasmid YCplac33 (22) are Y266, Y265, Y439 and Y402, respectively. For purification of wild-type (Wt) and mutant Rex1p, Y386 was transformed with pAV101, to create Y438, and p532, to create Y495. Strain H2457 with high-copy plasmids carrying the IMT1-4 genes has been described (23).
Plasmid construction
IMT4 was previously cloned into the HindIII site of pRS316 as a HindIII fragment isolated from plasmid C-50 (24), giving pJA108. Mutagenesis of the 3′ end of IMT4 was carried out using Quik Change site-directed mutagenesis (Stratagene) with pJA108 as a template and mutagenic primers JA546 and JA547. The correct sequence of the mutant, p467, was confirmed by DNA sequencing. A yeast shuttle vector containing REX1 under the control of a galactose-inducible promoter, pAV101, was kindly provided by Dr. Ambro van Hoof. Alanine substitutions at residues D229 and E231 were introduced using Quik Change site-directed mutagenesis (Stratagene), with primers JA616 and JA617 and confirmed by DNA sequencing, giving p532.
In vitro T7 RNA polymerase transcription constructs were created in pUC18 (25), except the mature tRNAiMet construct (HG300), which was obtained as a gift from Dr. Henri Grosjean. IMT3 with approximately 500-bp of upstream sequence and 600 bp of downstream sequence was amplified from yeast genomic DNA using primers JA571 and JA572 and cloned into the BamHI sites of pRS316 (21) as a BamHI fragment. The correct sequence of this plasmid (p480) was confirmed by DNA sequencing. For T7 transcription, IMT3 was amplified from p480 using PCR with primers JA602, which includes a T7 promoter, and JA603. The PCR product was digested with SalI and BamHI and ligated to pUC18 that had been digested with SalI and BamHI, giving p522. The IMT4 T7 transcription construct was created similarly by amplifying tRNAiMet sequence from HG300 using PCR with primers JA602 and JA618. The PCR product was digested with SalI and BamHI and ligated to pUC18 that had been digested with SalI and BamHI, giving p528.
SGA analysis
trm6-504 genetic interactions were identified using a high-throughput, array-based method known as Synthetic Genetic Array (26–28). First, the trm6-504 mutation was introduced into strain Y5563 [C. Boone lab, (29)] using two PCR products; one containing trm6-504 with sequence at the 3′ end complementary to the URA3 promoter (made using primers JA269 and JA270), and a second product encoding URA3MX6 with sequence at the 3′ end complementary to sequence downstream of trm6-504. [amplified using primers JA271 and JA272 with the template p4348 (28)]. Y5563 was co-transformed with these two DNA fragments and transformants were selected on Sc−URA. The integration of URA3 at the correct location was confirmed by PCR and the presence of trm6-504 was verified by a temperature sensitive phenotype; this strain was then crossed with a set of ~5000 viable haploid gene deletion strains (29). A series of robotic arraying procedures enabled selection of haploid double mutant meiotic progeny, which were subsequently examined for defects in colony growth at 26°C and 30°C (29). Approximately 200 candidate synthetic lethal or slow growth interactions were identified in the high-throughput screen.
RNA isolation
Total RNA from yeast was purified using the MasterPure Yeast RNA Purification Kit (Epicentre). When using the MasterPure Kit, cultures were grown to OD600 = 0.6 − 0.8 and cells were pelleted from 1.5–3 ml of culture.
Northern blot analysis
Total RNA was separated on 8% or 10% polyacrylamide gels containing 8 M urea and electrophoresis was carried out in 1 × TBE at 450 V. Gels containing radioactive samples, such as those used in activity assays, were dried prior to visualization using phosphorimaging (GE Healthcare). Gels were first fixed for 15 min in a 15% methanol, 5% acetic acid solution, and then dried at 72°C for 55 min on a slab gel dryer (Thermo Scientific). For northern blotting, gels were soaked for 10 min in 0.5 × TBE and samples transferred to Biodyne B nylon membrane (Pall Corporation) using a semi dry electroblotter (Owl Separation Systems) at 400 mA for 60 min. Following transfer, the RNA was cross-linked to the membrane using a UV stratalinker (Stratagene). After being probed, blots were stripped with 800 ml of boiling hot 1% SDS, and then tested for residual signal by exposure to a phosphorimager screen overnight prior to being probed again.
Purification of poly (A)+ RNA
Yeast strains were grown to an O.D.600 of 0.5–0.6 in 250 ml YPD at 30°C. Cells were harvested by centrifugation at 6000 × g for 5 min at 4°C, washed with 1 × phosphate buffered saline and frozen at − 80°C. Purification of total and poly (A) + RNA was then carried out as described (18) using 50 mg of oligo (dT) cellulose (New England Biolabs). Poly (A) + RNA was eluted with 2 ml elution buffer and collected as 0.5 ml fractions The OD260 of each fraction was determined and fractions having the greatest amounts of RNA were pooled. Samples were then extracted with AE (50 mM sodium acetate, pH 5.0 and 10 mM EDTA) buffer-saturated phenol and the aqueous layer extracted again with chloroform: isoamylalcohol (24:1). RNA was precipitated with 1/10 volume 3 M sodium acetate (pH 5.3) and 3 volumes of ethanol at −20°C overnight. After precipitation, poly (A) + RNA was collected by centrifugation at 14 000 rpm for 15 min at 4°C. Pellets were washed with 70% ethanol and allowed to air dry. The poly (A) + RNA was resuspended in 15 μl of nuclease free water. For northern blotting, equal amounts of poly (A) + RNA were loaded based on the different inputs of total RNA used for oligo (dT) selection. The total RNA concentration in each sample was determined by UV spectroscopy and the sample that had the highest amount of total RNA set to 100%. All other samples were calculated as a percentage of this sample and the amount of poly (A) + RNA loaded onto the gel was increased based on the difference in the total RNA input. For example, if one of the samples had only 65% as much total RNA as the sample set at 100%, then 35% more poly (A) + RNA was run for this sample, thereby accounting for the lower amount of total RNA used for oligo (dT) cellulose purification for this sample.
Purification of Rex1p
A 25 ml culture of Y438 or Y495 grown at 30°C in synthetic complete medium lacking uracil (Sc−URA) was used to inoculate a 1 l culture in Sc−URA lacking glucose and supplemented with 1% galactose and 2% sucrose (ScGal/Suc−URA). Cells were grown at 30°C, harvested at OD600 = 0.8 − 1, and the pellets frozen on dry ice and stored at −80°C. To purify Rex1p, the cell pellet was thawed on ice and resuspended in breaking buffer [20 mM HEPES, pH 7, 150 mM NaCl, 1× complete protease inhibitor cocktail (Roche)]. Cells were disrupted using a French press (Thermo IEC) at a pressure of 20 000 psi and the lysate subjected to centrifugation at 3000g, 4°C, for 10 min. This clarified extract was then subjected to centrifugation at 70 000g, 4°C, for 30 min, and the clarified supernatant added to anti-FLAG M2 agarose (Sigma) pre-equilibrated in wash buffer (20 mM HEPES, pH 7.0, 150 mM NaCl). After incubation with gentle mixing at 4°C for 2 h, the resin was collected by centrifugation at 1000 × g, 4°C, for 5 min and the supernatant discarded. The resin was washed with 10 ml wash buffer, collected again, and the supernatant discarded. The resin was then transferred to a 10 ml column and washed with 10 ml wash buffer. Rex1p was eluted with 1 ml breaking buffer containing 150 ng/μl FLAG peptide (Sigma), and collected in five fractions. The fractions were analyzed by SDS-PAGE followed by Coomassie staining or Western blotting with anti-FLAG M2 antibody (Stratagene).
Rex1p in vitro assays
tRNA substrates were transcribed in vitro using T7 RNA polymerase. DNA templates were linearized by restriction digest using BstNI for HG300 and DraI for p522 and p528. The in vitro transcription reaction (100 μl) consisted of 1× T7 RNA polymerase buffer (New England Biolabs), 1.25 mM rNTPs, 10 mM DTT, 20–40U RNasin ribonuclease inhibitor (Promega), ~400 ng DNA template, and 250U T7 RNA polymerase (New England Biolabs). After incubation at 37°C for 3 h, 1U RQ1 DNase was added and samples incubated at 37°C for 20 min. RNA was extracted with AE buffer-saturated phenol, pH 5, ethanol precipitated, and resuspended in nuclease free water. After separation on a 10% denaturing polyacrylamide gel, tRNA was visualized by methylene blue staining and gel purified. tRNAs were then dephosphorylated and 5′ end labeled using 32P-ATP. Scintillation counting was used to determine the concentration of labeled tRNA. Activity assays were carried out in 20 mM HEPES, pH 7, 5 mM MgCl2 and 100 mM NaCl at 30°C. Reaction products were separated on a 10% denaturing polyacrylamide gel and visualized using phosphorimaging or autoradiography.
RESULTS
trm6-504 and rex1Δ exhibit a synthetic genetic interaction
Trm6p and Trm61p comprise the m1A58 tRNA methlytransferase in S. cerevisiae, and we have previously shown that a trm6-504 mutant, which lacks m1A58 from all tRNA, has reduced levels of tRNAiMet when grown at 30°C (23). Synthetic genetic array (26) analysis was conducted to identify synthetic lethal (SGA) and synthetic slow-growth interactions between trm6-504 and nonessential yeast genes. Genes encoding proteins already known to be involved in tRNA modification and processing were detected in the array, including tRNA modification enzymes that act on tRNAiMet. In addition, genes encoding proteins of unknown function, as well as proteins with known functions but no established roles in tRNAiMet production, were identified (Anderson,J.T., unpublished data). One of the genes of known function was RNH70/REX1, which encodes a nuclease that has been shown to be involved in rRNA and tRNA maturation.
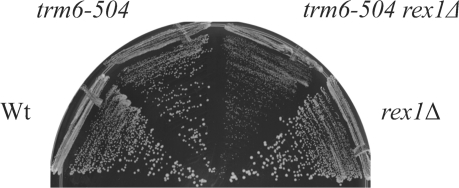
Because REX1 is not known to process tRNAiMet, we were intrigued by the SGA findings. According to the SGA analysis, a trm6-504 rex1Δ double-mutant strain exhibited a severe slow-growth phenotype; however, we did not observe a consistent slow-growth phenotype in a trm6-504 rex1Δ strain after sporulation and tetrad dissection of the strain generated in the array. Therefore, we deleted REX1 in the trm6-504 strain (Y190) we have studied extensively (18). A trm6-504 strain grows slowly at 30°C, and we were unable to see an additional growth defect due to loss of REX1 when a trm6-504 rex1Δ double-mutant strain was grown in a rich medium (YPD) at 30°C. However, we found that trm6-504 rex1Δ double-mutant cells grown on synthetic dextrose minimal medium supplemented with histidine and uracil (SD+His+Ura), conditions that resemble those in the SGA screen, at 30°C or room temperature, consistently displayed a modest growth defect as compared to the trm6-504 strain (Figure 1).
Initiator tRNAMet processing is altered in a trm6-504 rex1Δ double mutant strain
Although a trm6-504 strain grows slowly at 30°C (23) and exhibits reduced levels of mature tRNAiMet, raising the level of initiator tRNAMet by introducing a high-copy (hc) plasmid carrying IMT4, which encodes tRNAiMet, restores robust growth (23). Because the SGA and our experiments revealed that the trm6-504 rex1Δ double mutant grew slower than the trm6-504 mutant, we wanted to determine if there was less mature tRNAiMet in the trm6-504 rex1Δ strain than in the trm6-504 strain. Total RNA was extracted from Wt (Y200), trm6-504 (Y190), trm6-504 rex1Δ (Y386) and rex1Δ (Y387) strains grown at 30°C and separated by electrophoresis using a denaturing 8% polyacrylamide gel. Northern analysis was performed using a probe (JA11, Table 1) that detects both precursor tRNAiMet, which contains 5′ leader and 3′ trailer sequence, and mature tRNAiMet (Figure 2A). While we did not observe an obvious difference in the amount of mature tRNAiMet between trm6-504 and trm6-504 rex1Δ strains, we noticed that a subset of the tRNAiMet precursors accumulated to greater levels in the trm6-504 rex1Δ lane than in the Wt, trm6-504, or rex1Δ lanes (Figure 2A). The same blot was probed for 5S rRNA as a loading control and small differences were observed between samples. Strains in which REX1 has been deleted have slightly longer 5S rRNA than a Wt strain (Figure 2A), in agreement with what has been reported previously (12,14).
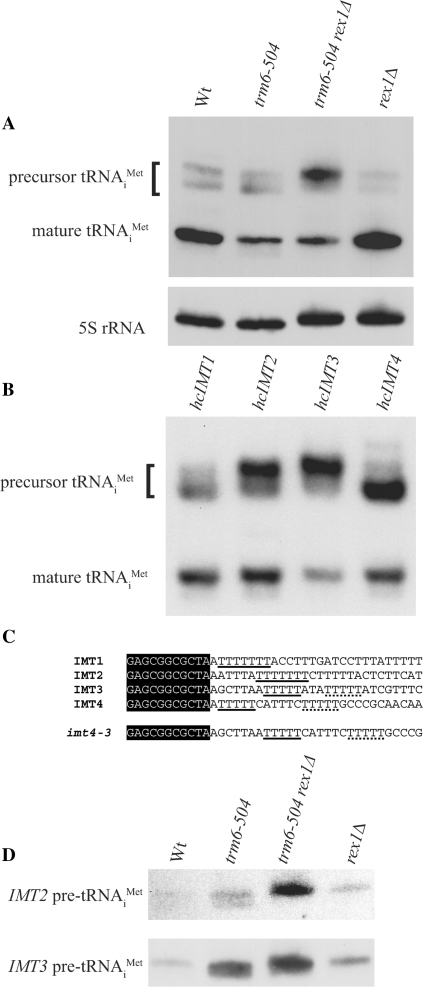
A trm6-504 rex1Δ strain accumulates tRNAiMet encoded by IMT2 and IMT3. (A) Total RNA (5 μg) isolated from Wt (Y200), trm6-504 (Y190), trm6-504 rex1Δ (Y386) and rex1Δ (Y387) strains was separated by electrophoresis using an 8% denaturing polyacrylamide gel. Northern analysis was performed with radiolabeled oligonucleotides that hybridize to tRNAiMet (JA11) or 5S rRNA (JA99). Results were visualized by autoradiography. (B) Northern analysis of total RNA (10 μg) isolated from a trm6-504 strain (H2457) carrying one of the four IMT genes on a high-copy plasmid. The blot was probed for tRNAiMet (JA11) and the results were visualized by autoradiography. (C) An alignment of the tRNAsiMet encoded by the four IMT genes from S. cerevisiae. A portion of the sequence found in the mature tRNA is shown in a black background. The 3′ trailer sequences are shown in black text, with the putative RNA Polymerase III termination sites underlined. A downstream terminator IMT4 is underlined with a dashed line. The imt4-3 mutant has an alteration in the length of the 3′ trailer preceding the first transcriptional terminator. (D) Northern analysis of total RNA (20 μg) performed as described in (A), probing with oligonucleotides complementary to the 5′ leader of IMT2- or IMT3-encoded tRNAiMet (JA72 or JA48, respectively).
Table 1.
Oligonucleotides used for northern analysis
| Name | Target | Sequence (5′→3′) |
|---|---|---|
| JA 11 | tRNAiMet | TCGGTTTCGATCCGAGGACATC AGGGTTATGA |
| JA 48 | 5′ IMT3 | ACGGCGCTTAACTTTTATG |
| JA 66 | 5′ IMT4 | GGCGCTTAGCCAACTTG |
| JA 72 | 5′ IMT2 | GGCGCTGCTAAATCATGAG |
| JA 99 | 5S rRNA | TCGCGTATGGTCACCCACTACA |
| JA 555 | tRNAVal(CAC) | GAAGGCAACGTGATAGCCGC |
| JA 557 | dimeric pre-tRNAArg | AGAAACAAAGCACTCACGAT |
From previous work, we know that the four forms of precursor tRNAiMet migrate at two different positions on a denaturing 8% polyacrylamide gel due to differences in the length of the 3′ trailer (Figure 2B) (23). The slower-migrating species consists of tRNAiMet precursors encoded by IMT2 and IMT3, which have 6–7 nucleotides between the end of the mature tRNAiMet sequence and a RNA Polymerase III (Pol III) termination signal, a stretch of five or more thymines (30) (Figure 2B and C), creating a long trailer (LT). tRNAiMet precursors encoded by IMT1 and IMT4 migrate more quickly, having only one nucleotide, a short trailer (ST), before the predicted Pol III termination signal (Figure 2B and C). As seen by Northern analysis, it is the slower-migrating species that is more intense in a trm6-504 rex1Δ double mutant strain (Figure 2A), suggesting that tRNAiMet precursors encoded by IMT2 and IMT3 accumulate, while those encoded by IMT1 and IMT4 do not. To test this hypothesis, we probed a Northern blot of total RNA from the Wt (Y200), trm6-504 (Y190), trm6-504 rex1Δ (Y386) and rex1Δ (Y387) strains specifically for IMT2 and IMT3 using probes that hybridize to their unique 5′ leader sequences (Figure 2D). Both IMT2- and IMT3-encoded precursors (LT) were more abundant in a trm6-504 rex1Δ strain than in a Wt, trm6-504, or rex1Δ strain (Figure 2D). In addition, it appeared there was a small increase in the amount of IMT2- and IMT3-encoded precursor (LT) tRNAiMet in the rex1Δ strain compared to the Wt strain, suggesting Rex1p is involved in processing tRNAiMet whether or not it lacks m1A58. Levels of IMT3-encoded precursor tRNAiMet were also elevated in a trm6-504 strain, although to a lesser degree than seen in the trm6-504 rex1Δ strain (Figure 2D). Finally, IMT4-encoded precursor did not accumulate in a trm6-504 rex1Δ strain (data not shown). These data suggest that Rex1p is required to efficiently process tRNAiMet precursors with longer 3′ trailing sequences (LT) and is not as crucial for processing of precursors with short 3′ trailing sequences (ST).
Because IMT2 and IMT3 tRNAiMet precursors accumulate in a trm6-504 rex1Δ double mutant strain, we reasoned that precursor tRNAiMet encoded by IMT1 or IMT4 would also accumulate if the 3′ trailer sequence was lengthened. Mutagenesis was carried out on a plasmid carrying IMT4 (pJA108), and the short 3′ trailer (ST) of IMT4 was replaced with the longer trailer (LT) of IMT3. Therefore, the precursor tRNAiMet transcribed from this imt4-3 construct (p467) has the 5′ leader of IMT4 and the 3′ trailer of IMT3 preceding the first Pol III termination sequence (Figure 2C). To ensure that this mutant tRNA was functional, the imt4-3 plasmid was tested for its ability to promote growth of a trm6-504 strain at 30°C, a temperature at which this strain has a slow-growth phenotype (23). While the trm6-504 strain carrying hcIMT4 showed vigorous growth, a strain carrying an empty vector grew slowly (Figure 3A). The trm6-504 strain carrying the imt4-3 plasmid also showed robust growth (Figure 3A), suggesting that functional tRNAiMet is produced from the imt4-3 plasmid.
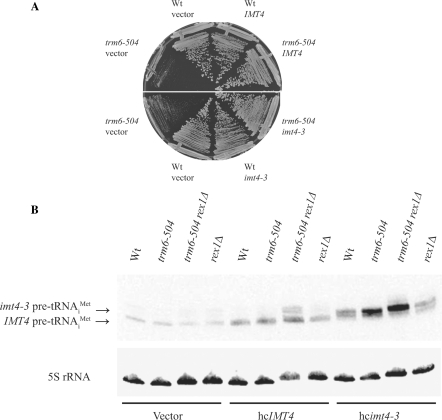
Changes in the 3′ trailer length influence tRNAiMet accumulation in a trm6-504 rex1Δ strain. (A) Wt (Y200) or trm6-504 (Y190) strains carrying vector (YCplac33), hcIMT4, or hcimt4-3 were grown at 30°C for 2 days on SC−URA. (B) Total RNA (20 μg) isolated from Wt (Y200), trm6-504 (Y190), trm6-504 rex1Δ (Y386) and rex1Δ (Y387) strains carrying empty vector (YCplac33), hcIMT4 (pJA108) or hcimt4-3 (p467) was subjected to Northern analysis. The blot was probed with a radiolabeled oligonucleotide complementary to the 5′ leader of IMT4 (JA66), and the results visualized using phosphorimaging. The blot was stripped, probed again with an oligonucleotide that hybridizes to 5S rRNA (JA99), and analyzed by phosphorimaging.
Total RNA was prepared from Wt (Y200), trm6-504 (Y190), trm6-504 rex1Δ (Y386) and rex1Δ (Y387) strains containing empty plasmid or plasmids with IMT4 or imt4-3. Northern analysis was performed using probes that hybridize to the 5’ leader of IMT4 or to 5S rRNA, which serves as a loading control. All of the strains carrying the empty plasmid had approximately the same amount of IMT4-encoded precursor tRNAiMet, as did all of the strains carrying the IMT4 plasmid (Figure 3B). However, among the strains expressing imt4-3-encoded precursor tRNAiMet, there was an increase in precursor in the trm6-504 strain and an even greater increase in the trm6-504 rex1Δ strain (Figure 3B). This result indicates that the length of the 3′ trailer sequence influences whether or not precursor tRNAiMet is influenced by Rex1p for processing.
We note that a trm6-504 rex1Δ strain carrying the high-copy IMT4 plasmid shows accumulation of a precursor species that is longer than IMT4-encoded precursor tRNAiMet. It is likely that this is precursor tRNAiMet created when Pol III fails to stop transcribing at the first termination sequence and instead reads through to a second downstream terminator (Figure 2C). In fact, the sequences CAAAAC and CATATC, which are similar to the sequence that follows the first IMT4 terminator (Figure 2C) have been shown to allow transcriptional read-through by yeast RNA Pol III in vitro (31). As this species would have a longer trailer, it is consistent with our other results that this precursor tRNAiMet accumulates in the trm6-504 rex1Δ strain.
Increased polyadenylation of tRNAiMet occurs in a trm6-504 rex1Δ double mutant strain
Previously, we found that the hypomethylated tRNAiMet from a trm6-504 strain activates a nuclear tRNA surveillance pathway that results in its polyadenylation and degradation (18). We wondered if activation of the surveillance pathway had changed in a trm6-504 rex1Δ strain. As polyadenylated tRNAiMet cannot be detected in total RNA samples, we isolated both total RNA and poly(A) + RNA from Wt (Y200), trm6-504 (Y190), trm6-504 rex1Δ (Y386) and rex1Δ (Y387) strains. Northern analysis was performed using a probe that detects both mature and precursor tRNAiMet. In samples of total RNA, precursor tRNAiMet accumulated in a trm6-504 rex1Δ strain (Figure 4A), and to a lesser extent in a trm6-504 or rex1Δ strain, as observed previously (Figure 2A), while no substantial differences (1.5-fold at most) were seen when detecting a loading control (data not shown). In poly(A) + RNA samples, poly(A) + tRNAiMet was present in the trm6-504 strain, but not the Wt strain, consistent with published data (Figure 4A) (18). Poly(A) + tRNAiMet was also detected in the rex1Δ strain, a finding that suggests inefficient 3′ end processing of tRNAiMet activates the nuclear tRNA surveillance pathway. In the trm6-504 rex1Δ strain, a greater amount of poly(A) + tRNAiMet was evident, approximately four times that found in the trm6-504 or rex1Δ strain (Figure 4A and B). This finding implies tRNAiMet that lacks m1A58 and has an unprocessed 3′ end is especially susceptible to polyadenylation, and, presumably, degradation. Furthermore, polyadenylation of another m1A58-containing tRNA—tRNAVal(CAC)—occurs in the absence of Rex1p (Figure 4C). tRNAVal(CAC) precursors detected in total RNA samples also accumulated in strains lacking Rex1p (Figure 4C), and we note that tRNAVal(CAC) is encoded by two genes in which a Pol III terminator is located 4 or 12 nucleotides downstream of the end of the mature tRNAVal(CAC) sequence.
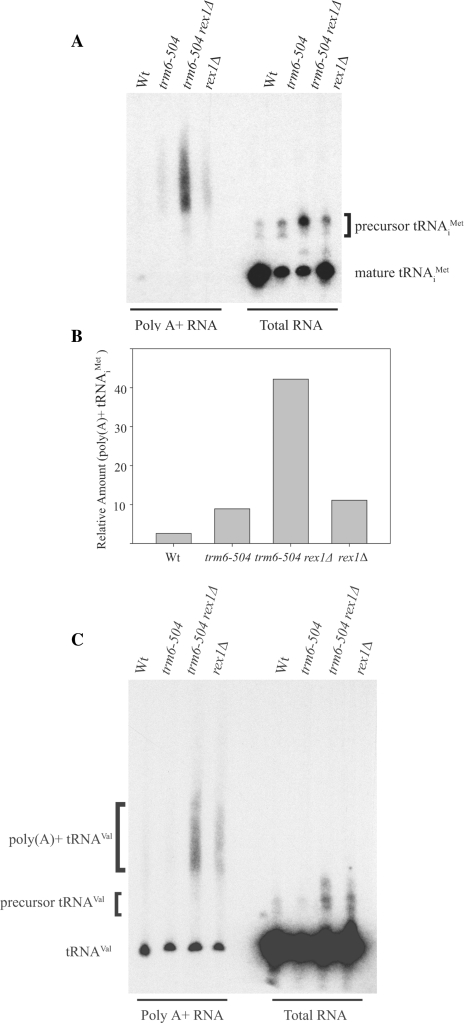
An increase in the level of polyadenylated tRNAiMet is seen in a trm6-504 rex1Δ strain. (A) Total RNA (5 μg) and poly(A) + RNA (3–4 μg, normalized to the total RNA yield, see ‘Methods’ section) was separated on a 6% denaturing polyacrylamide gel. Northern analysis was performed with a radiolabeled oligonucleotide complementary to tRNAiMet (JA11). (B) A bar graph of the relative amounts of poly(A) + tRNAiMet in the Wt, trm6-504, trm6-504 rex1Δ and rex1Δ strains, as seen in (A). Quantitation was performed using ImageQuant TL software (GE Healthcare). (C) The blot in (A) was stripped and probed for tRNAVal(CAC) (JA555). Results were visualized by autoradiography for both blots. The mature tRNA in the poly(A) + lanes represents tRNA that was not eliminated by oligo-(dT) selection.
Loss of Rex1p results in polyadenylation of 5S rRNA and tRNAArg
Prior to the discovery of the nuclear RNA surveillance pathway, 5S rRNA from an rna82.1 (rex1) mutant strain had been found to have a poly(A) tail up to 20 A's in length (14). This led us to test whether 5S rRNA, and the other known processing substrate of Rex1p, tRNAArg-tRNAAsp, would be polyadenylated in a rex1Δ strain. Northern analysis of poly(A) + RNA performed with a probe for 5S rRNA showed poly(A) + 5S rRNA was present in rex1Δ and trm6-504 rex1Δ strains, but not in a Wt or trm6-504 strain (Figure 5A). There was no obvious difference between the amount of poly(A) + 5S RNA in the trm6-504 rex1Δ strain and the rex1Δ strain. Therefore, unlike what is seen for tRNAiMet, polyadenylation of 5S RNA is a result of a loss of Rex1p and does not seem to be influenced by the presence or absence of trm6-504. This result is expected, since 5S RNA is not a Trm6p substrate, and would not be expected to exhibit an altered structure due to the absence of m1A.
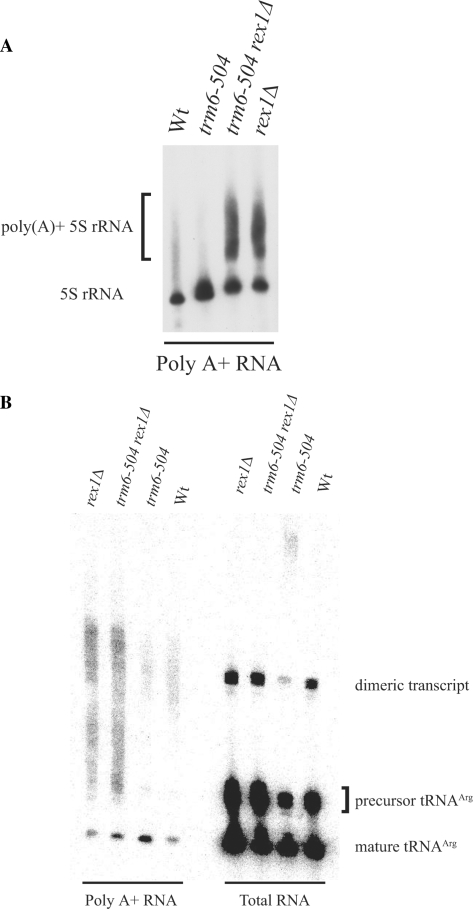
Loss of Rex1p results in polyadenylation of 5S rRNA and tRNAArg from the dimeric tRNAArg-tRNAAsp transcript. (A) Total and poly (A) + RNA were analyzed as described for Figure 4. The blot was probed for 5S rRNA (JA99) and the results visualized by autoradiography. (B) Northern analysis was performed using a labeled oligonucleotide (JA557) that hybridizes to the 3′ end of tRNAArg and the linker sequence between the two tRNAs. Phosphorimaging was used to analyze results. The presence of mature forms of RNAs in the poly(A) + lanes represent RNAs not eliminated during oligo-(dT) selection.
The tRNAArg-tRNAAsp dimeric transcript is processed by a series of endonucleolytic and exonucleolytic steps to produce two mature tRNAs (32). While endonucleolytic cleavages separate the tRNAs from each other and generate the mature 5′ ends, exonucleolytic processing by Rex1p is believed to produce the mature 3′ end of tRNAArg, and possibly tRNAAsp (12,15,32). To look at possible polyadenylation of tRNAArg-tRNAAsp, we conducted Northern analysis with a probe that hybridizes to the 3′ end of tRNAArg and the 10 nt linker region between the two tRNAs. In total RNA samples, we observed greater amounts of tRNAArg that were not fully processed in the trm6-504 rex1Δ and rex1Δ strains (Figure 5B). This result is consistent with previous studies that showed a rex1Δ strain contains 3′ extended tRNAArg precursors encoded by the dimeric tRNAArg-tRNAAsp transcript (12). 5S rRNA was detected as a loading control and no significant variations (1.3-fold at most) were seen between the total RNA samples (data not shown). The dimeric transcript was also detected in total RNA samples, but was less abundant in a trm6-504 strain; instead, much larger RNAs, appearing as a smear near the top of the membrane, were detected. While we can not explain this observation at present, we hope to understand this result through future studies of RNA processing in a trm6-504 strain. From the poly(A) + samples, we could see that polyadenylation of tRNAArg was occurring in the trm6-504 rex1Δ and rex1Δ strains, and to approximately the same extent (Figure 5B). In addition, we note that some of the poly(A) + RNA detected is large enough to be the dimeric tRNAArg-tRNAAsp transcript. We conclude that tRNAArg and possibly the dimeric tRNAArg-tRNAAsp transcript are not efficiently processed at their 3′ ends when Rex1p is absent and become polyadenylated. This is in contrast to the polyadenylation of tRNAiMet seen in a trm6-504 mutant where we believe an altered tRNAiMet structure, due to the absence of m1A58, activates the RNA surveillance pathway. While tRNAArg has m1A58 in a Wt strain, tRNAArg was not found to be polyadenylated when lacking this modification, as in the trm6-504 strain (Figure 5B). These data indicate that inefficiently processed precursor tRNAs can activate the RNA surveillance pathway.
Rex1p exhibits tRNA 3′ end processing activity
A 60 kDa exonuclease capable of removing the 3′ trailer from a tRNA substrate was previously purified from S. cerevisiae extracts (5); given that Rex1p is predicted to be 63 kDa, it is likely that this exonuclease was Rex1p. Rex1p, along with Rex2p, Rex3p and Rex4p, possesses four acidic amino acids, spread over three Exo motifs, which are highly conserved among the DEDD family of exoribonucleases (Figure 6A) (16). We used a galactose-inducible promoter to drive over-expression of FLAG-tagged Rex1p in a trm6-504 rex1Δ strain, and purified Rex1p using affinity chromatography (Figure 6B). Purified protein was confirmed as Rex1p based on size and Western blotting with anti-FLAG antibodies (data not shown). In order to be confident that any observed exonuclease activity belonged to Rex1p and not a contaminant, we also designed and purified a D229A/E231A mutant Rex1p. This mutant has alanine substituted for the highly conserved DE residues found in the ExoI motif of DEDD exoribonucleases (Figure 6A). For RNase T from Escherichia coli, which is also a DEDD exonuclease, it has been shown that an alanine substitution at either of these positions inactivates the enzyme (Figure 6A) (33).

Rex1p displays tRNA 3′end processing activity in vivo and in vitro. (A) A diagram of Rex1p showing the conserved amino acids found in the three Exo motifs. An alignment of Rex1, 2, 3 and 4, and E. coli RNase T protein sequence is shown underneath to illustrate the conservation of the D × E sequence found in the ExoI domain. (B) Wt and mutant FLAG-tagged Rex1p were purified from yeast (strains Y438 and Y495, respectively) using affinity chromatography. Purified protein was subjected to SDS-PAGE and visualized by Coomassie staining. The positions of molecular weight standards (Broad Range Protein Marker, New England Bioloabs) are indicated. (C) Gel purified 32P- 5′ end labeled tRNAsiMet (~ 10 pM) were incubated in buffer alone or with Wt or mutant Rex1p (~7.5 nM) at 30°C for 10 min. Reaction products were separated on a 10% denaturing polyacrylamide gel and visualized by autoradiography. The positions of RNAs of known length (Decade Marker, Ambion) are indicated (in nucleotides). (D) Wt Rex1p (~7.5 nM) was incubated with labeled ‘IMT3’ tRNAiMet (~20 pM) for known periods of time from 0.5–7 min, as indicated. Control reactions with ‘IMT3’ tRNAiMet lacking either enzyme or Mg2+ are shown, as well as a control reaction lacking enzyme that contained the ‘mature’ tRNAiMet. After separation on a denaturing 10% polyacrylamide gel, results were visualized using phosphorimaging. (E) Northern analysis of total RNA (10 μg) isolated from a trm6-504 rex1Δ strain carrying an empty vector (pRS316), or a plasmid with galactose-inducible Wt REX1 (pAV101) or mutant rex1 (p532), grown under non-inducing or inducing conditions. The blot was probed with a radiolabeled oligonucleotide complementary to tRNAiMet (JA11) and the results visualized using autoradiography.
To assay the activity of Rex1p, several tRNAiMet substrates were prepared in vitro using T7 RNA polymerase—a ‘mature’ tRNAiMet which ends in CCA, an ‘IMT3’ tRNAiMet which has the seven nucleotides trailer of IMT3 and a stretch of five uridines, and an ‘IMT4’ tRNAiMet, which ends with the one nucleotide trailer of IMT4 and a stretch of five uridines. tRNAiMet substrates were labeled with 32P at the 5′ end and incubated with purified Wt or mutant Rex1p. Reaction products were separated on a denaturing polyacrylamide gel and visualized by exposure of the dried gel to a phosphorimager screen. All three substrates were shortened at their 3′ ends in the presence of Wt Rex1p, but were unchanged when incubated in buffer alone or in the presence of the mutant Rex1p (Figure 6C). This result illustrates the nuclease activity of Rex1p, and demonstrates that this activity is lost when two highly conserved amino acids are mutated. All three substrates were converted to products similar in length, meaning that more nucleotides were removed from the ‘IMT3’ substrate than from the ‘IMT4’ substrate, and more from the ‘IMT4’ substrate than the ‘mature’ substrate. We note that the ‘IMT3’ substrate was not fully digested during the 10 min incubation period. Despite gel purification, this substrate consisted of two bands, and the slower-migrating species was consistently more resistant to trimming by Rex1p.
To visualize the exonuclease activity of Rex1p, we monitored product formation over time. The ‘IMT3’ substrate was incubated with Wt Rex1p for a period of 0.5, 1, 3 or 7 min. In addition, control reactions consisting of the ‘IMT3’ substrate without added MgCl2 and the ‘IMT3’ and ‘mature’ substrates without Rex1p, were carried out for 7 min. Reaction products were separated on a denaturing 10% polyacrylamide gel and visualized by phosphorimaging. Exonucleolytic processing of the ‘IMT3’ substrate by Rex1p was observed, as illustrated by the intermediates that can be seen (Figure 6D). Processing was not observed in the control reaction lacking Mg2+ (Figure 6D), demonstrating that Mg2+ is required for Rex1p activity, which is not unexpected since other DEDD family members require divalent cations (16).
In addition to our in vitro studies, we investigated the effects of Wt and mutant Rex1p expression in vivo. Total RNA was isolated from trm6-504 rex1Δ strains containing empty vector, REX1, or the rex1 mutant, grown in either synthetic complete medium lacking uracil (Sc−URA), which does not induce REX1 expression, or Sc−URA lacking glucose and supplemented with galactose and sucrose (ScGal/Suc−URA), which induces REX1 expression. Northern analysis performed using a probe that detects both precursor and mature tRNAiMet showed that induction of Wt Rex1p prevented the accumulation of tRNAiMet precursors observed previously, whereas induction of the mutant Rex1p did not prevent accumulation of precursor tRNAiMet (Figure 6E). A loading control did not show substantial differences (1.5-fold at most) between samples (data not shown). We conclude that the exoribonuclease activity of Rex1p is responsible for processing hypomethylated tRNAiMet precursors in vivo.
DISCUSSION
We have shown that tRNAiMet precursors accumulate in a trm6-504 rex1Δ double-mutant strain—in particular those encoded by IMT2 and IMT3. Compared to IMT1 and IMT4, the IMT2 and IMT3 genes have an extended 3′ trailer sequence preceding the Pol III termination signal. We found that increasing the length of the 3′ trailer preceding the terminator in IMT4 from one to seven nucleotides resulted in its accumulation in a trm6-504 rex1Δ strain. One explanation for these results is that Rex1p may specifically process tRNAiMet precursors with long 3′ trailers, and not those with short 3′ trailers. Alternatively, in the absence of Rex1p, other ribonucleases in the cell may only be capable of efficiently processing those tRNAiMet precursors with short trailers. In support of this interpretation, we found that a trm6-504 rex1Δ strain continued to accumulate tRNAiMet precursors despite over-expression of LHP1 (data not shown). As increased levels of Lhp1p have been proposed to promote endonucleolytic processing (6), there does not appear to be an endonuclease that can compensate for the loss of Rex1p function. In E. coli, both redundancy and substrate preference have been observed for exonucleases that process the 3′ ends of tRNAs. While an E. coli strain lacking five exonucleases—RNases II, D, T, BN and PH—is not viable, growth is restored when any one of these enzymes is introduced, indicating that these exonucleases have overlapping abilities (34). In addition, each exonuclease is able to process some precursor tRNA species efficiently, and others very poorly, suggesting that different groups of tRNAs are most effectively processed by different nucleases (4).
Currently, the only precursor tRNAs known to be polyadenlyated and degraded by the nuclear surveillance pathway are hypomodified tRNAiMet in S. cerevisiae and a mutant tRNASer in Schizosaccharomyces pombe (18,35,36). We have shown that tRNAiMet and tRNAVal(CAC) are polyadenylated in a rex1Δ strain, suggesting that tRNAs with unprocessed 3′ ends are also substrates for the nuclear surveillance machinery. The finding that tRNAiMet lacking m1A58 and containing a 3′ extension is apparently more susceptible to polyadenylation suggests that these features influence the efficiency of recognition by the nuclear surveillance machinery. In addition, 5S rRNA and the dimeric tRNAArg-tRNAAsp transcript were polyadenylated in strains lacking Rex1p, suggesting these RNAs are also subject to degradation by the nuclear surveillance pathway when their 3′ ends are not efficiently processed. This phenomenon may not be restricted to 3′ end processing defects, since the polyadenylated species observed also retains the 5′ leader sequence. This could indicate that a loss of m1A in tRNAiMet perturbs 5′ end processing by RNaseP, and that this too activates the surveillance pathway. Since it is widely accepted that RNaseP cleavage initiates processing of precursor tRNAs (32,37), this would explain why increased levels of polyA + tRNAiMet are seen in strains in which both 5′ and 3′ end processing are slowed. We conclude that RNAs with unprocessed 3′ ends can now be added to the current list of substrates for the nuclear surveillance pathway.
The role of Rex1p in RNA processing is similar to that of the non-orthologous enzyme RNase T from E. coli, which is also a 3′–5′ exonuclease and a member of the DEDD exonuclease superfamily (16,38). RNase T is required for maturation of 5S rRNA, and, like a rex1Δ strain, an E. coli strain lacking RNase T contains 5S rRNA with extra nucleotides at the 3′ end (39). RNase T is also involved in tRNA maturation, and has been found to trim the 3′ ends of tRNA precursors (40). The crystal structures of DEDD exonucleases, such as RNase T and RNase D, reveal that four highly conserved acidic amino acids (DEDD) coordinate two divalent metal ions involved in catalysis (41,42). Consistent with these findings, we observed Rex1p activity to be dependent on the presence of Mg2+ and found that alanine substitutions of two conserved acidic amino acids (DE) inactivated the enzyme. The specificity of both Rex1p and RNase T for 5S rRNA and tRNAs suggests there are features common to both substrates that can be recognized by these enzymes. In yeast, the 3′ end of both precursor tRNAiMet and 5S rRNA consists of a double-stranded stem followed by several unpaired nucleotides. For both RNAs the single stranded 3′ end would be expected to have a poly(U) tail due to transcription of the RNA Pol III termination signal, and we have detected poly(U) tails on the IMT3-encoded precursor tRNAiMet that accumulates in a trm6-504 rex1Δ strain (data not shown). Interestingly, in the dimeric tRNAArg-tRNAAsp transcript, the linker region between the two tRNAs is also U rich. Therefore, a double-stranded stem and a single-stranded U-rich sequence may be important for Rex1p to recognize its substrate, and future studies are needed to explore this possibility. While Rex1p was recently shown to be involved in the 3′ end processing of some intron-containing tRNAs (13), both tRNAiMet and tRNAVal do not contain introns, indicating that the presence an intron is not required for recognition by Rex1p.
While the results of our in vivo experiments suggested Rex1p may only process tRNAiMet precursors with long 3′ trailers, Rex1p processed the 3′ ends of tRNAsiMet with both long and short trailers in our in vitro assay. Exonuclease activity was also observed when a tRNAiMet ending in CCA was used as a substrate. The absence of other nucleases and proteins involved in tRNA processing in the in vitro assay may allow Rex1p to remove nucleotides from tRNAiMet substrates that would normally be processed more efficiently by other enzymes in vivo. Furthermore, the tRNAiMet substrate ending in CCA would become aminoacylated in vivo, which would presumably block exonucleolytic degradation, as has been shown for RNase T (38). It should be noted that the tRNA substrates used in this assay were transcribed in vitro and lack modifications, including m1A58. The ability of Rex1p to process these unmodified tRNAs in vitro is consistent with our observation that more precursor tRNAiMet accumulates in a rex1Δ strain where tRNAiMet also lacks m1A58.
We have shown Rex1p to have Mg2+-dependent 3′→5′exonuclease activity in vitro, and these results are consistent with previous findings for a 60 kDa protein purified from yeast (5). In yeast, the mature 3′ ends of tRNAiMet (prior to CCA addition), tRNAsArg (from the dimeric transcript, prior to CCA addition) and 5S rRNA all consist of a double-stranded stem with a 3′ overhang of a single nucleotide. This indicates that Rex1p may function by binding single-stranded 3′ trailer sequences and trimming the RNA within one nucleotide of a double-stranded stem. Further studies to identify the products generated by Rex1p in vitro are needed to determine whether or not this is true. In addition, we currently do not fully understand the influence of the m1A58 modification on 3′ end processing. Rex1p appears to be able to process hypomethylated precursor tRNAiMet to some degree, as we see less accumulation in a trm6-504 strain than a trm6-504 rex1Δ strain. We propose that in the absence of both m1A58 and Rex1p activity, processing of precursor tRNAsiMet with long 3′ trailers is carried out very inefficiently by other nucleases, resulting in the accumulation and polyadenylation of tRNAsiMet we have observed. However, when m1A58 is present, processing by other nucleases appears to be more effective, even in the absence of Rex1p, as we observe only a slight increase in IMT2- and IMT3-encoded precursor tRNAsiMet in a rex1Δ strain. A more thorough characterization of nucleases known to be involved and identification of additional nucleases involved in tRNA 3′ end processing may allow us to better understand the relationship between tRNA modification and 3′ end maturation.
FUNDING
National Institute of General Medical Sciences (GM066791 to J.T.A., GM069949 to J.T.A.); National Institutes of Health Bethesda, Maryland; Genome Canada through the Ontario Genomics Institute, Toronto, Ontario (2004-OGI-3-01 to C.B.); Canadian Institutes of Health Research Ottawa, Ontario, (GSP-41567 to C.B.). Funding for open access charge: National Institute of General Medical Sciences, National Institutes of Health Bethesda, Maryland (GM069949).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Ambro van Hoof, Michelle Steiger and Richard Maraia for critical reading of this manuscript.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkn925
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/37/1/298/3411878/gkn925.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/37/1/298
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/37/1/298
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/37/1/298.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/153833467
Article citations
tRNA Modifications and Dysregulation: Implications for Brain Diseases.
Brain Sci, 14(7):633, 25 Jun 2024
Cited by: 0 articles | PMID: 39061374 | PMCID: PMC11274612
Review Free full text in Europe PMC
Advances in the Structural and Functional Understanding of m<sup>1</sup>A RNA Modification.
Acc Chem Res, 08 Feb 2024
Cited by: 4 articles | PMID: 38331425 | PMCID: PMC10882958
Different modification pathways for m1A58 incorporation in yeast elongator and initiator tRNAs.
Nucleic Acids Res, 51(19):10653-10667, 01 Oct 2023
Cited by: 5 articles | PMID: 37650648 | PMCID: PMC10602860
The life and times of a tRNA.
RNA, 29(7):898-957, 13 Apr 2023
Cited by: 33 articles | PMID: 37055150 | PMCID: PMC10275265
Review Free full text in Europe PMC
N1-methyladenosine modification in cancer biology: Current status and future perspectives.
Comput Struct Biotechnol J, 20:6578-6585, 25 Nov 2022
Cited by: 44 articles | PMID: 36467585 | PMCID: PMC9712505
Review Free full text in Europe PMC
Go to all (49) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae.
Genes Dev, 18(11):1227-1240, 14 May 2004
Cited by: 358 articles | PMID: 15145828 | PMCID: PMC420349
Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA.
RNA, 12(3):508-521, 23 Jan 2006
Cited by: 142 articles | PMID: 16431988 | PMCID: PMC1383588
Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation.
RNA, 14(6):1214-1227, 02 May 2008
Cited by: 68 articles | PMID: 18456844 | PMCID: PMC2390804
3' processing of eukaryotic precursor tRNAs.
Wiley Interdiscip Rev RNA, 2(3):362-375, 01 May 2011
Cited by: 82 articles | PMID: 21572561 | PMCID: PMC3092161
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: GM066791
Grant ID: GM069949