Abstract
Background & aims
Expansion and patterning of the endoderm generate a highly ordered, multiorgan digestive system in vertebrate animals. Among distal foregut derivatives, the gastric corpus, antrum, pylorus, and duodenum are distinct structures with sharp boundaries. Some homeodomain transcription factors expressed in gut mesenchyme convey positional information required for anterior-posterior patterning of the digestive tract. Barx1, in particular, controls stomach differentiation and morphogenesis. The Nirenberg and Kim homeobox gene Bapx1 (Nkx3-2) has an established role in skeletal development, but its function in the mammalian gut is less clear.Methods
We generated a Bapx1(Cre) knock-in allele to fate map Bapx1-expressing cells and evaluate its function in gastrointestinal development.Results
Bapx1-expressing cells populate the gut mesenchyme with a rostral boundary in the hindstomach near the junction of the gastric corpus and antrum. Smooth muscle differentiation and distribution of early regional markers are ostensibly normal in Bapx1(Cre/Cre) gut, but there are distinctive morphologic abnormalities near this rostral Bapx1 domain: the antral segment of the stomach is markedly shortened, and the pyloric constriction is lost. Comparison of expression domains and examination of stomach phenotypes in single and compound Barx1 and Bapx1 mutant mice suggests a hierarchy between these 2 factors; Bapx1 expression is lost in the absence of Barx1.Conclusions
This study reveals the nonredundant requirement for Bapx1 in distal stomach development, places it within a Barx1-dependent pathway, and illustrates the pervasive influence of gut mesenchyme homeobox genes on endoderm differentiation and digestive organogenesis.Free full text

Role of the homeodomain transcription factor Bapx1 in mouse distal stomach development
Abstract
Background & Aims
Expansion and patterning of the endoderm generate a highly ordered, multi-organ digestive system in vertebrate animals. Among distal foregut derivatives, the gastric corpus, antrum, pylorus and duodenum are distinct structures with sharp boundaries. Some homeodomain transcription factors expressed in gut mesenchyme convey positional information required for anterior-posterior patterning of the digestive tract. Barx1, in particular, controls stomach differentiation and morphogenesis. The NK homeobox gene Bapx1 (Nkx3-2) has an established role in skeletal development but its function in the mammalian gut is less clear.
Methods
We generated a Bapx1Cre knock-in allele to fate map Bapx1-expressing cells and evaluate its function in gastrointestinal development.
Results
Bapx1-expressing cells populate the gut mesenchyme with a rostral boundary in the hindstomach, near the junction of the gastric corpus and antrum. Smooth muscle differentiation and distribution of early regional markers are ostensibly normal in Bapx1Cre/Cre gut, but there are distinctive morphologic abnormalities near this rostral Bapx1 domain: the antral segment of the stomach is markedly shortened and the pyloric constriction is lost. Comparison of expression domains and examination of stomach phenotypes in single and compound Barx1 and Bapx1 mutant mice suggest a hierarchy between these two factors; Bapx1 expression is lost in the absence of Barx1.
Conclusions
This study reveals the non-redundant requirement for Bapx1 in distal stomach development, places it within a Barx1-dependent pathway, and illustrates the pervasive influence of gut mesenchyme homeobox genes on endoderm differentiation and digestive organogenesis.
INTRODUCTION
Mechanisms responsible for organizing the mammalian stomach into fundus, corpus, and antral-pyloric segments are poorly understood. Corpus epithelium typically carries numerous oxyntic and zymogenic cells that produce acid and digestive enzymes, respectively.1 The distal stomach, which encompasses the antrum and pylorus, lacks these cell lineages but is marked in mouse and man by presence of endocrine cells that secrete gastrin and mucous cells that produce mucin 6.1 Muscle cells in the outer pylorus create a sphincter that controls passage of food into the duodenum.
The digestive tract differentiates in response to signals from adjacent mesenchyme.2 Expression of homeobox genes is often segmental along the anterior-posterior axis of the developing gut and may be especially important in relaying rostro-caudal position.3 Clustered Hox genes, for example, are expressed in the gut in overlapping domains, reminiscent of patterns observed along the skeletal axis;4, 5 they are implicated in regional identity and in formation of intestinal sphincters and the cecum.6–10 Homeodomain proteins participate in mesoderm-endoderm signaling.
The homeobox gene Barx1 is confined to embryonic stomach mesenchyme and required for proper stomach development.10–13 In its absence the stomach is markedly small, abnormally shaped, lacks a pyloric constriction, shows mixing of cells from different segments, and carries intestinal villi distally.11, 12 Some homeobox genes regulate fibroblast growth factor (FGF) expression in the hindgut,10 and overexpression of NKX2.5 in chick embryos inhibits Wnt5a and Bmp4 expression during formation of the hindstomach (gizzard) and pylorus;14, 15 Barx1 acts in part by limiting the duration of Wnt signaling in early stomach development.11, 12 Many other factors that regulate genetic and tissue interactions in stomach development remain unknown.
The homeodomain of mammalian Nkx3-2 (Bapx1) shares ~87% identity with Drosophila BAGPIPE, a NK2 sub-family member that specifies gut smooth muscle in flies.16 Viral misexpression studies in the chicken suggest that Bapx1 functions in development of the gizzard, a muscular, keratinized structure in the posterior stomach.15 In mouse embryos, Bapx1 mRNA appears first in lateral plate mesoderm, adjacent to gut endoderm, around embryonic day (E) 8.5.17 Bapx1 knockout mice were therefore predicted to have gut musculature defects, but the intestine in three separate mutant lines is largely intact18–20 and investigation has centered on Bapx1’s role in spleen and skeletal development. One group commented on abnormal gastro-duodenal morphology20 without investigating molecular details. Although the nature and possible reasons for the defect are unknown, Bapx1 is cited as being required to generate pyloric sphincter muscle.21
We created a targeted mouse line that marks Bapx1-expressing cells and eliminates gene activity. Here we report that Bapx1 is necessary for proper antral-pyloric morphogenesis and development of antral-type epithelium. We also show that Bapx1 expression in the distal stomach requires Barx1. These studies reveal a focal requirement for Bapx1 in hindstomach organogenesis and outline a transcriptional hierarchy in mammalian stomach development.
MATERIALS and METHODS
Mouse gene targeting
A λ-phage clone from a 129/Sv mouse genomic library was provided by Drs. K-I. Yoshiura and J. Murray, University of Iowa. A 3.6-kb BglII-SacII fragment containing 5′ flanking sequences and the first 46 codons of Bapx1 exon 1 served as the 5′-homology arm; a 1.6-kb SmaI fragment containing codon 112 through the end of exon 2 served as the 3′-homology arm. A PGK-NeoR cassette and Cre recombinase cDNA were inserted in frame with Bapx1 coding sequence at the SacII restriction site (Fig. 1A). The construct was electroporated into AB2.2 embryonic stem cells. Two targeted cell lines were used to produce chimeras and Bapx1+/Cre mice. For Southern genotyping, the probe was a [α-32P] dCTP-labeled BglII-SacI fragment from the 3′ segment of the gene, which identifies 5-kb and 7.5-kb bands for wild-type and Cre knock-in alleles, respectively (Fig. 1B). To demonstrate correct targeting at the 5′ end, we used primers complementary to the Cre insert (CRE3′: GCCGCATAACCAGTGAAACAGCATTGC) and to genomic DNA ~4.5 kb 5′ to the Bapx1 gene (GTTATGAGTGACAGCCTGGGACG) to amplify a 5.7-kb DNA fragment in the targeted allele (Suppl. Fig. 1). Identity of this fragment was confirmed by BglII/ClaI digestion and sequence analysis using internal primers GGTTTCAAAATGAGGCTC and CATGTATGAATGTGTGGAACCTGG. For subsequent genotyping we used CRE3′ along with CTCGTTCTCTTCGCTCAGGGCTGAG and CCAGGCGATCCTCAACAAGAAGAGGG in a coupled PCR reaction with a 56°C annealing step (Fig. 1C). Bapx1 targeted mice were maintained on the C57BL/6 background.
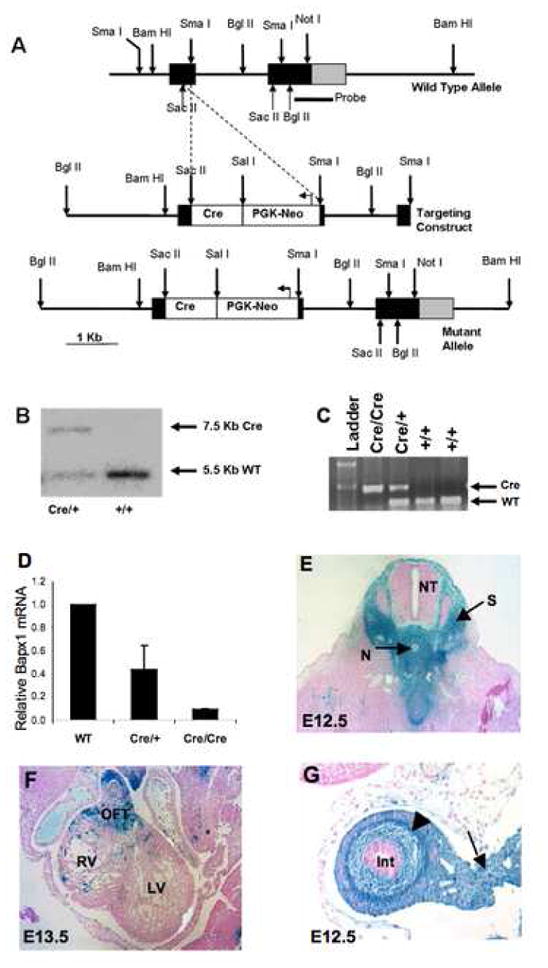
(A) Molecular strategy to generate ES cells with Cre recombinase targeted to the first Bapx1 exon between the SacII and SmaI sites. (B) Mice were genotyped by Southern hybridization using probes corresponding to the 3′ half of exon 2 and part of the 3′ untranslated region. Subsequent generations were genotyped by PCR (C). (D) Confirmation that Bapx1 expression is extinguished in Bapx1Cre/Cre mice. qRT-PCR on fetal and newborn tibial RNA, expressing Bapx1 mRNA content relative to Gapdh mRNA in wild-type (WT, n=10), Bapx1Cre/+ (HET, n=14) and Bapx1Cre/Cre (KO, n=10) samples. (E–G) Bapx1Cre/+ mice were crossed with the ROSA26R reporter strain and progeny were examined for Cre-expressing cells and their descendants by β-galactosidase activity. In E12.5 embryos, staining was detected in sites of documented Bapx1 expression or function, including condensing vertebrae (E) and gut mesenchyme (arrowhead) and mesentery (arrow) (G). Staining also appeared in E13.5 cardiac outflow tract (F) and, later, throughout the developing skeleton (see Suppl. Fig. 1). OFT, cardiac outflow tract; RV, right ventricle; LV, left ventricle; CC, central canal; NT, neural tube; N, notochord; S, sclerotome.
Expression analyses
β-galactosidase activity was determined on whole-mount preparations using published methods.22 For histology, embryos were embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin. Bapx1Cre/+ and Barx1+/− mice were intercrossed to obtain compound homozygotes. Organs from crosses with Nkx2.5-GFP transgenic mice, described previously23, were visualized under a Leica MZ FLIII fluorescent dissecting microscope.
RNA was reverse transcribed using SuperScript™ (Invitrogen). cDNA was detected by PCR using Bapx1 primers agatgtcagccagcgtttc and gcagaggcgagcaggtcggc. Fetal stomach lysates were resolved by SDS-PAGE. Binding of Bapx1 mouse antiserum H00000579-A01 (Abnova, 1:500) was detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse Ab.
Embryos were fixed overnight in 4% paraformaldehyde at 4°C. 8-μm thick paraffin sections were dried, deparaffinized in xylenes, and rehydrated. For antigen retrieval, slides were immersed in 10 mM sodium citrate, pH 6.0, and treated in a pressure cooker for 2 minutes at 15 psi. Endogenous peroxidase activity was blocked with 3% H2O2 in methanol for 15 min and non-specific antibody (Ab) binding with 5% Fetal Bovine Serum for 1 h at 25°C. Primary Ab: SMαA (Sigma A2547 (1A4), 1:1000), PGP9.5 (Chemicon AB1761, 1:2500), H/K-ATPase (D032-3; MBL International; Woburn, MA; 1:1000), intrinsic factor (gift from Dr. D. Alpers, Washington University; St. Louis, MO; 1:24,000), Pdx1, (1:6000, gift of Dr. C. Wright, Vanderbilt University, TN), BMP4, (1:300, Chemicon MAB1049), Nkx 2.5 (1:200, SC14033, Santa Cruz), were applied for 3 h at 25°C or overnight at 4°C. After treating with anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch), then avidin-biotin complex solution (Vector Laboratories) for 1 h at 25°C, color reactions were developed in 0.05% 3,3′-diaminobenzidine and 0.1% H2O2. Slides were counterstained with hematoxylin. Alcian blue staining was by standard methods.
For in situ hybridization, tissues were fixed in 4% paraformaldehyde (Sigma-Aldrich), dehydrated, embedded in paraffin, and sectioned at 6-μm thickness. Deparaffinized, rehydrated sections were treated with Proteinase K (Roche) and 0.1M triethanolamine before hybridization with probes generously provided by T. Lufkin (Bapx1), A. McMahon (Ihh), and R. Harvey (Nkx2.5). After overnight hybridization at 63°C, slides were washed for 2 h in decreasing concentrations of SSC from 2X to 0.2X at 63 °C, then incubated in 5% serum in PBS followed by digoxigenin Ab (Roche; 1:2000) at 4° C overnight. Slides were equilibrated in 100 mM NaCL; 100 mM Tris, pH 9.5; 50 mM MgCl2 and stained (NBT/BCIP tablets, Roche) for 2–4 h.
For expression arrays, RNA was extracted from the distal stomach of E18.5 Bapx1Cre/Cre embryos and wild-type littermates using the RNeasy kit (Qiagen). After confirmation of RNA quality, samples were processed and hybridized to Codelink mouse bioarrays (Amersham Biosciences). Raw data were normalized on a log2 scale and filtered to reduce noise. Differential gene expression and functional gene groupings were analyzed using MatchMiner (http://discover.nci.nih.gov/matchminer/), GoMiner (http://discover.nci.nih.gov/gominer/) and GeneSpring (Agilent Technologies) software and are deposited in the GEO database (acc. # _______).
RESULTS
Tracing Bapx1 expression
We used homologous recombination to replace Bapx1 at codon 46 (exon 1) with in-frame Cre cDNA (Bapx1Cre, Fig. 1A). Two independent mutant lines showed evidence for correct gene targeting (Fig. 1B,C) loss of Bapx1 mRNA (Fig. 1D), and no material effect on expression of the two flanking genes (Suppl. Fig 1C). We crossed these mice with ROSA26 reporter mice, where a floxed translation-stop sequence restricts LacZ gene expression to Cre-expressing cells and their progeny.24 β-galactosidase (β-gal/LacZ) activity first appeared in Bapx1Cre;ROSA26R embryos at E9.5 in gut mesoderm and weakly in somites (Fig. 2A,B; data not shown). Between E10.5 and E12.5, β-gal activity was prominent in the splanchnic mesoderm, somites, calvarium, Meckel’s cartilage, and spleen anlage (Figs. 1E, 2C–D, Suppl. Fig. 2A–G). By E13.5 Bapx1 expression was evident in the cardiac outflow tract (Fig. 1F) and condensing cartilage of the ribs, skull (Suppl. Fig. 2I–N), and long bones. Staining in the digestive tract was confined to mesodermal derivatives and excluded from endoderm at all stages (Figs. 1G and 2E,F). These findings agree with previous reports of Bapx1’s role in developing skeleton and spleen17–20 and establish the fidelity of Bapx1Cre mice to mark Bapx1-expressing cells and elucidate Bapx1 function in other organs.
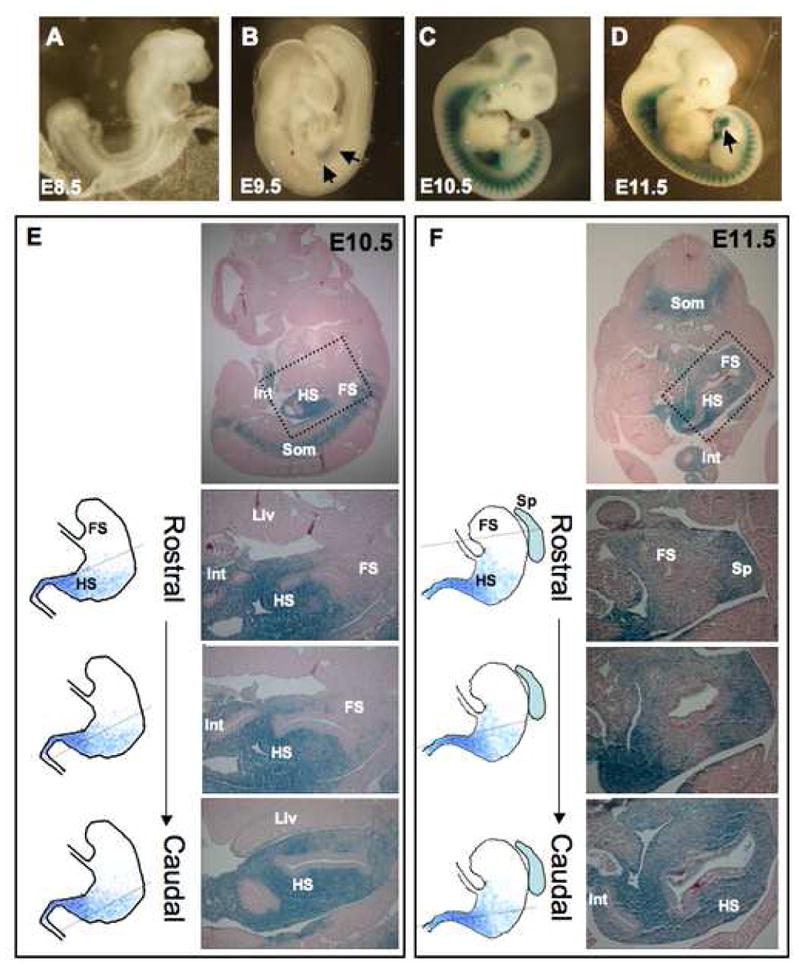
(A–D) Whole-mount LacZ staining of Bapx1Cre/+; ROSA26R embryos identifies Cre-expressing cells and their descendants. Staining is not apparent through E8.5 (A) and first appears near the stomach-intestine boundary by E9.5 (B, arrows). By E10.5, LacZ activity is evident in all anterior somites and in gut and craniofacial mesenchyme (C). Expression is readily apparent in mesenchyme of herniated E11.5 intestine (D, arrow). (E,F) Microscopic examination exposes a rostral limit of LacZ activity in the hindstomach at E10.5 (E) and E11.5 (F): intestine (Int) and hindstomach (HS) are stained strongly, whereas forestomach (FS) activity is much reduced or absent. Som, somites; Liv, liver; Sp, spleen. Sections from embryos at each stage are arranged in rostral to caudal sequence from areas corresponding to those outlined by dotted boxes. Rostral sections show minimal LacZ activity in the forestomach compared to the caudal (hindstomach) tissue or spleen (Sp). Schemas for the domain of Bapx1 gene expression in relation to stomach and spleen anatomy are depicted next to each section, with a dotted line indicating the approximate plane of section.
Definition of gene expression along the long axis of the embryonic stomach is confounded by rotation of the organ, from an initial lie parallel to the body’s anterior-posterior axis to a final position that is nearly perpendicular. We examined serial embryo sections with the attention required to distinguish the stomach’s antero-posterior and radial axes. LacZ expression in Bapx1Cre/+ embryonic gut initiated in the distal stomach. Staining at E10.5 was intense in the caudal foregut and stomach-intestine junction but absent from the rostral foregut and stomach (Fig. 2E, sections from the same embryo in a rostral to caudal series). At E11.5, expression remained evident in the hindstomach but faint or absent in forestomach (Fig. 2F), extended into the full length of intestine, and included the spleen anlage (Fig. 2F). LacZ staining involved all cells in the full thickness of the mesenchyme (Suppl. Fig. 3). In older embryos, β-gal activity was present in much of the stomach, with a persistent caudal-to-rostral gradient (data not shown).
Early chick embryos express Bapx1 in the prospective gizzard (posterior stomach) but not the proventriculus (anterior stomach).15 Our Cre-based lineage analysis in mice confirmed Bapx1 expression in tissues with known functions and disclosed an anterior boundary previously unappreciated in mammalian stomach. The rostral limit of earliest Bapx1 expression corresponds roughly to the junction between corpus and antrum.
Abnormal stomach development in Bapx1Cre homozygotes
Crosses between Bapx1+/Cre mice yielded null mutants in Mendelian proportions until E18.5 (25.6% Bapx1+/+, 48% Bapx1Cre/+ and 26.4% Bapx1Cre/Cre). Mutant homozygotes typically died 1 to 3 days after birth, and ~90% of weanlings were wild-type or heterozygote. Perinatal lethality, similar to that reported with other Bapx1-null alleles,17–20 likely reflects skeletal malformation (Suppl. Fig. 2O,P). The spleen was absent or markedly hypoplastic in Bapx1Cre/Cre mice, as judged grossly (Fig. 3B) and by expression of a Nkx2.5-GFP transgene (Suppl. Fig. 4), a marker of the developing spleen.12, 25
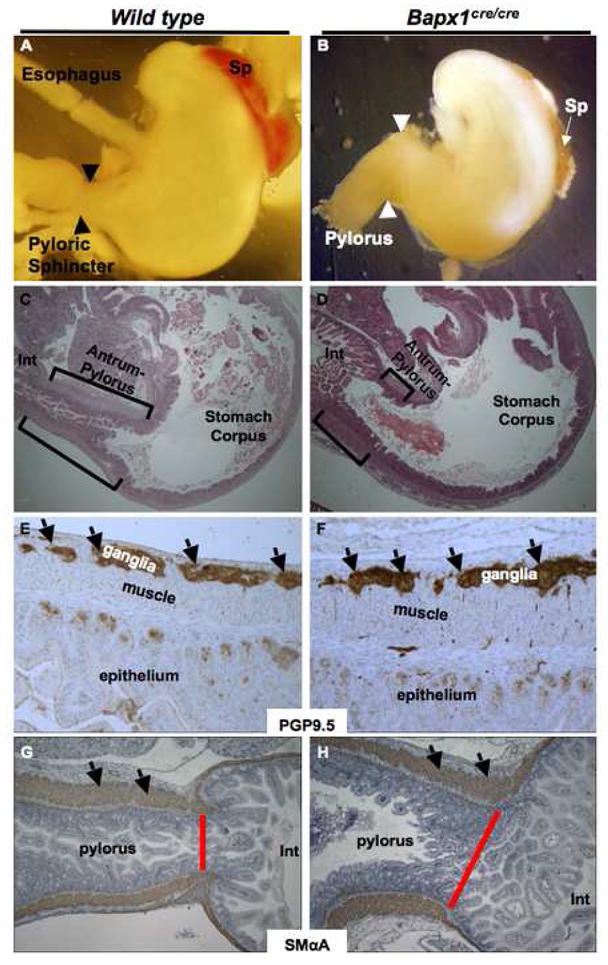
(A–D) Gross and microscopic evidence for hindstomach defects in Bapx1Cre/Cre mice. Whole-mount views (A,B) and hematoxylin/eosin-stained histologic sections (C,D) of E16.5 and neonatal specimens, respectively. Marked truncation of the antral-pyloric segment of the stomach (brackets) and lack of the pyloric constriction (arrowheads) are readily evident. The spleen (Sp) is also absent or markedly hypoplastic in Bapx1cre/cre mice. (E–H) Immunohistochemical analysis of PGP9.5 (E,F) and smooth muscle α-actin (G,H) in Bapx1cre/cre neonatal hindstomach indicates ostensibly intact enteric nerve and smooth muscle differentiation, respectively. Arrows point to immunostaining of ganglia (E,F) or smooth muscle (G,H). Red bars demarcate the pylorus and highlight the marked difference in width between control and mutant samples.
Bapx1Cre/Cre stomachs were modestly reduced in size. Nearly all of this reduction occurred in the distal segment, which was also dilated and lacked constriction at the gastro-duodenal junction, the site of the pyloric sphincter (Fig. 3A,B). Histologic examination confirmed distal dilatation and revealed severe shortening of the antral-pyloric segment (Fig. 3C,D). Expression and distribution of PGP9.5, an enteric nervous system marker,26, 27 and smooth-muscle α-actin were intact (Fig. 3E–H).
Epithelia in the gastric body and antrum have distinctive features. Specialized, Alcian blue-avid mucous cells found at the base of antral gland units are normally absent from the corpus; conversely, the antrum lacks chief and oxyntic cells, the dominant lineages in the body.1 Normal hindstomach hence corresponds to the Alcian blue-staining region between the zone of chief and parietal cells in the corpus and the villous duodenal epithelium (Suppl. Fig. 5A). Bapx1Cre/Cre stomachs carried few, and in many cases, no glandular units with basal Alcian blue avidity (Fig. 4A,E vs 4B,F), whereas intestinal goblet cells stained readily with Alcian blue (Fig. 4F). Additionally, the distance between H/K-ATPase- (Fig. 4C,G vs 4D,H) or gastric intrinsic factor- (GIF, Suppl. Fig. 5B–E) expressing cells in the stomach body and the villous intestinal epithelium was markedly reduced. These corpus lineage markers frequently abutted the intestine (Fig. 4H, Suppl. Fig. 5E), indicating diminution or loss of mature antral character. Scrutiny of Alcian blue, H/K-ATPase, and GIF stains revealed normal cell composition in distal corpus glands and absence of mixed corpus-antral units (Suppl. Fig. 5F).
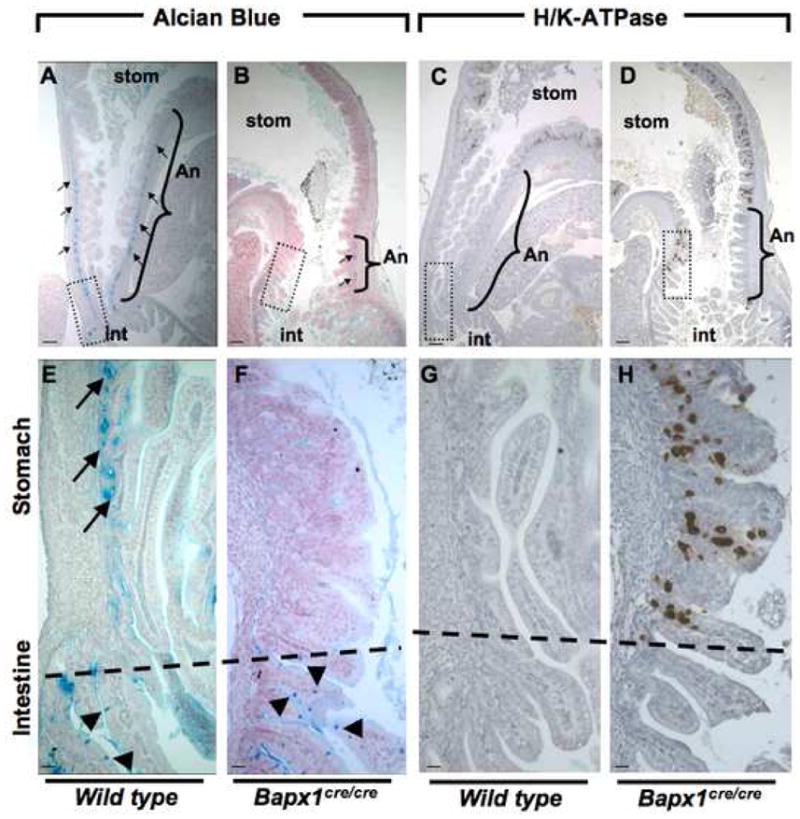
Critical examination of antral Alcian blue staining and of the gastric corpus marker H/K-ATPase. (A–D) Low-power microscopic views of the gastro-duodenal junction in wild-type (A,C) and Bapx1Cre/Cre (B,D) neonates. Brackets mark the region expressing antral-pyloric markers along the greater curvature (An, antrum-pylorus), separating the stomach (stom) corpus from intestine (int). Boxes outline regions of the gastro-duodenal junction shown at higher magnification in panels E–H. Alcian blue staining in mutant antral epithelium, which marks characteristic mucous cells at the gland base (arrows) in wild-type mice (A,E), is significantly reduced along the greater curvature and missing from the lesser curvature. Intestinal goblet cells (arrowheads), which also stain with Alcian Blue, are unaffected. Conversely, H/K-ATPase immunostaining marks parietal cells in wild-type gastric corpus and is absent from normal antral-pyloric mucosa (C,G). In Bapx1Cre/Cre mice, H/K-ATPase-expressing cells reside immediately adjacent to the intestine (D,H), again disclosing loss of antral mucosa. Similarly, expression of Intrinsic Factor, a marker of corpus-resident zymogenic cells, abuts the intestine along the lesser curvature and comes close to the intestine along the greater curvature in Bapx1Cre/Cre stomach (Suppl. Fig. 3). Bars A–D, 300um; E–H, 60um.
Thus, absence of Bapx1 leads to significant hindstomach truncation and loss of the pyloric constriction. The normal appearance of gastric smooth muscle (Fig. 3H) and ostensibly normal intestine and gastric corpus point to a localized defect in antral-pyloric development. Although histologic examination sometimes gave the impression that antral hypoplasia was more severe along the lesser than the greater curvature of the stomach (e.g., Fig. 4C,D), most samples lacked such disparity (e.g., Fig. 3C,D, Suppl. Fig. 6B), which we attribute to subtle variation in tissue orientation. Indeed, objective quantitation of multiple samples confirmed that both aspects of the antrum were affected (Suppl. Fig. 6A).
Molecular correlates of Bapx1 in hindstomach development
Indian Hedgehog (Ihh) mRNA is enriched in fetal mouse corpus and antrum, whereas Sonic hedgehog (Shh) is enriched in the forestomach.28 In a sign that early patterning is preserved in Bapx1Cre/Cre stomach, the boundaries of Ihh (Fig. 5A,B) and Shh (data not shown) expression were intact at E11.5. Expression of the homeobox gene Pdx1 is normally limited to the antral-pyloric segment, providing a reliable marker of this stomach region.29 Pdx1 expression also was similar in Bapx1Cre/Cre and wild-type stomach early in development (E11.5 and E14.5 shown in Fig. 5C–F). Consistent with the observation of antral hypoplasia, the Pdx1 expression domain was substantially smaller at E18.5 (data not shown). However, the typical transition in staining pattern between antrum and corpus, and the symmetry across greater and lesser curvatures, were preserved. Antral hypoplasia in the absence of Bapx1 hence occurs on the background of correct anterior-posterior stomach patterning.
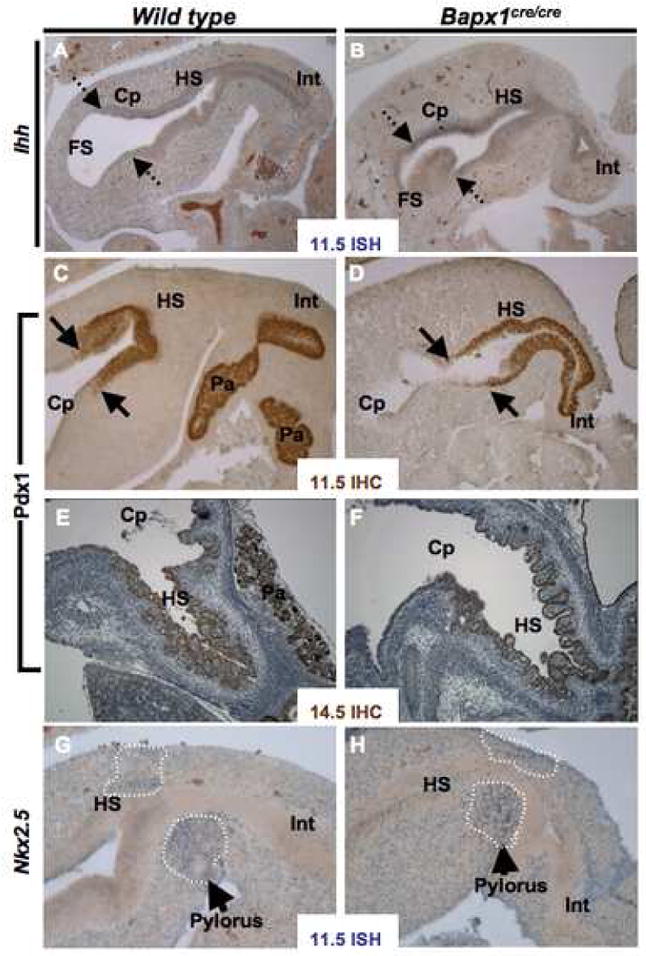
(A–F) Markers of hindstomach endoderm, Ihh mRNA (A,B) and Pdx1 protein (C–F), showed no difference in expression between wild-type and Bapx1Cre/Cre stomachs at E11.5 (A–D) or E14.5 (E,F). Dashed arrows mark the rostral limit of Ihh expression, which is much stronger in hindstomach (HS) and corpus (Cp) compared to forestomach (FS); solid arrows mark the anterior boundary of PDX1 expression, at the corpus-antral junction. PDX1 is also expressed in proximal intestine (Int) and pancreas (Pa), as seen especially clearly in the control samples (C,E). (G–H) Expression of the chick hindstomach and pyloric determinant Nkx2.5 mRNA in Bapx1Cre/Cre and wild-type stomachs at E11.5. Nkx2.5 expression, which marks developing pyloric sphincter muscle (arrows, dashed lines), was unaffected by Bapx1 loss. Similar results were observed by immunostaining (E14.5, data not shown). ISH, in situ hybridization; IHC, immunohistochemistry.
In chick embryos, Nkx2.5 and Bapx1 are expressed in the distal stomach (gizzard), while Bmp4 and Wnt5a appear in the proximal proventriculus and are excluded from the gizzard.15, 30 Nkx2.5 may regulate pyloric sphincter development, and forced Bapx1 expression in the proventriculus inhibits endogenous Bmp4 expression.15 In mouse embryos, by contrast, we observed Bmp4 expression throughout stomach and intestinal mesenchyme (data not shown); Nkx2.5 mRNA and protein were also expressed widely in mesoderm at the gastroduodenal junction but clearly enriched in pyloric sphincter muscle, as predicted (Fig. 5G and Suppl. Fig. 2). Levels and distribution of both Nkx2.5 (Fig. 5H, Suppl. Fig. 4) and Bmp4 (data not shown) were unaltered in Bapx1Cre/Cre stomach, indicating that Bapx1 loss does not interfere with their expression.
Next we surveyed changes in Bapx1Cre/Cre antral gene expression, using microarray analysis followed by qRT-PCR confirmation of representative results (data not shown). As early stomach pattern seems intact, we reasoned that antra from older embryos would better reveal aberrant gene expression. The distal stomach in Bapx1Cre/Cre embryos at E18.5 showed an increase in corpus-specific H/K-ATPase and Gif transcripts and a corresponding decrease in antrum-specific Muc6 mRNA (Suppl. Table 1A). These changes are consistent with the loss of antral, and distal extension of corpus, character. Considering functional gene classes (Gene Ontology), we noted increased expression of transcripts in groups related to epithelial-mesenchymal transition and regulation of endocytosis, whereas groups associated with Smad proteins, nuclear protein import, and vesicle membranes were expressed at lower levels (Suppl. Table 1B). These molecular changes in distal Bapx1Cre/Cre stomach represent an unknown combination of additional regional markers and possible underpinnings of antral hypoplasia.
Bapx1 may function downstream of Barx1 to mediate antral-pyloric development
Bapx1 is co-expressed in embryonic hindstomach mesenchyme with Barx1, although the domain of Barx1 expression encompasses nearly the whole stomach (Fig. 6A). Both genes influence differentiation of overlying stomach endoderm and formation of the pyloric sphincter; the antral segment is abbreviated in Bapx1Cre/Cre mice (Figs. 3,,4)4) and likely lost in Barx1−/− mice.12 We crossed mice to produce compound homozygote mutants, which we studied immediately after birth because Barx1−/− mice die of respiratory failure in the perinatal period.12 Stomach anomalies in Barx1−/−; Bapx1Cre/Cre and Barx1−/−;Bapx1+/+ neonates were identical (Fig. 6B,C); there was no worsening of the isolated Barx1 mutant phenotype, which is more severe than the Bapx1Cre/Cre antral defect.
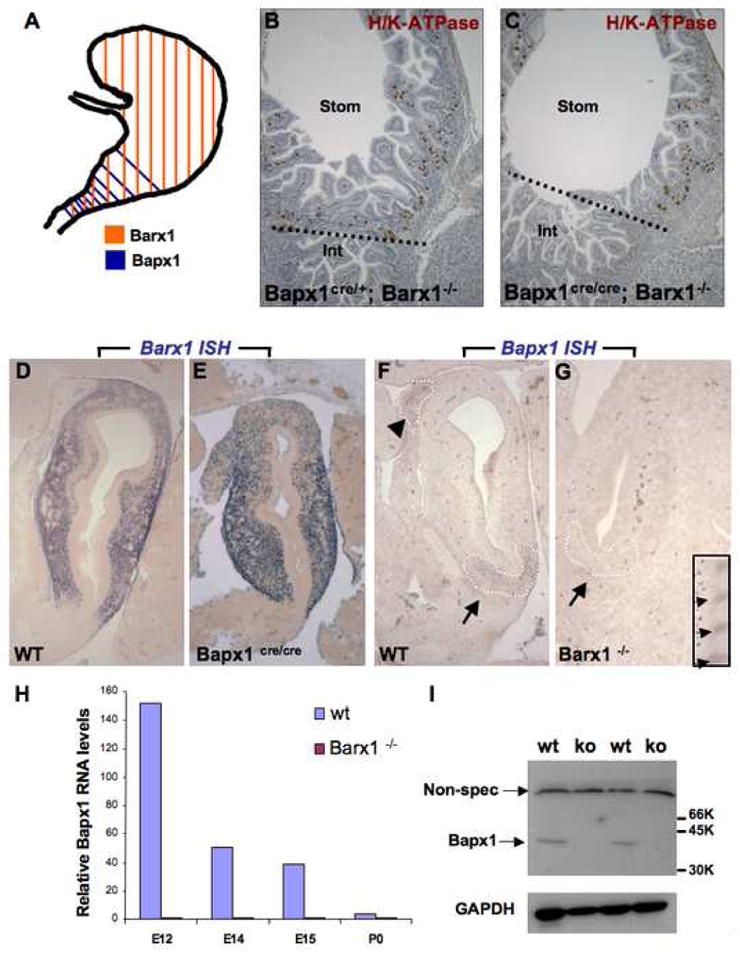
(A) Schematic depiction of Barx1 and Bapx1 expression in mid-gestation mouse stomach mesenchyme. Early Bapx1 expression is restricted to the antrum and pylorus (blue) and overlaps with Barx1 expression, which extends throughout the stomach (orange). (B,C) Further loss of Bapx1 does not worsen the stomach phenotype in Barx1−/− mice. Stomachs from Barx1−/−; Bapx1Cre/+ (B) and Barx1−/−;Bapx1Cre/Cre (C) neonates are shown. H/K-ATPase immunostaining reveals that a defined parietal cell-depleted region representing the antrum is absent with loss of Barx1 regardless of whether Bapx1 is functional or not. Other markers such as Cdx2 and Alcian blue staining gave similar results (data not shown). (D–G) In situ mRNA analysis confirms the overlapping schema depicted in panel A and reveals an expression hierarchy between Barx1 and Bapx1. Barx1 mRNA, which is expressed throughout fetal stomach mesenchyme, is unaltered in level or distribution with loss of Bapx1 (D–E), whereas Bapx1 expression in the embryonic hindstomach is severely compromised with Barx1 loss (F,G; arrows and dashed outlines mark hindstomach mesenchyme). Bapx1 expression was also observed in wild-type spleen (F, arrowhead and dashed outline), but loss of this signal occurred selectively in hindstomach; somites, another prominent site of Bapx1 expression (G inset, small arrows), were unaffected. (H,I) Confirmation of loss of Bapx1 expression by qRT-PCR and immunoblotting. RNA was isolated from stomachs dissected at multiple developmental stages and normalized to Gapdh levels (H) or Bapx1 protein levels were examined in E13.5 stomachs by immunoblot analysis (I). Bapx1 expression was virtually undetectable by either method in Barx1−/− stomachs.
To further evaluate the relationship between these co-expressed factors, we investigated gene expression in each individual knockout strain. Levels and distribution of Barx1 mRNA were not reduced or altered in fetal Bapx1Cre/Cre stomachs and may even increase slightly (Fig. 6D–E, Suppl. Table 1A). Thus, Barx1 does not require Bapx1 for its expression and acts either upstream or independent of Bapx1. Conversely, we detected Bapx1 transcripts in wild-type hindstomach and spleen (Fig. 6F, arrow and arrowhead) but not in the caudal Barx1−/− stomach (Fig. 6G, arrow), indicating that hindstomach Bapx1 expression requires Barx1 function. Bapx1 expression was equally robust in wild-type and Barx1−/− somites (Fig. 6G inset), ruling out trivial reasons for lack of a stomach signal. Quantitative RT-PCR and immunoblot analyses confirmed that Bapx1 mRNA levels were markedly reduced or absent in Barx1−/− stomachs (Fig. 6H,I). These observations collectively suggest that Bapx1 expression depends on Barx1 and that antral dysmorphogenesis in Barx1−/− stomachs might potentially reflect the attendant Bapx1 deficiency.
DISCUSSION
Organogenesis requires positional cues to specify cell and tissue types correctly. Homeobox genes play a vital role in regulating developmental processes and imparting positional identity.31, 32 We used homologous recombination to drive Cre expression from the mouse Bapx1 locus, thus creating a new null allele to define expression and study gene function in the developing gut. Bapx1+/Cre;ROSA26R mice confirmed Bapx1 expression domains reported previously in cartilage and spleen and revealed that early in digestive tract development, Bapx1-expressing cells and their progeny are confined to the intestine and prospective hindstomach. In line with this observation, Bapx1Cre/Cre mice show significant shortening of the antral segment and virtual apposition of the gastric body to the duodenum. Pdx1 and Ihh, two posterior markers, show correct regional expression, implying that certain elements of early stomach patterning are preserved. Thus, Bapx1Cre/Cre hindstomach defects seem to reflect a failure of proper expansion and morphogenesis of the antral-pyloric segment. As the affected region corresponds to that where Bapx1 expression initiates in the digestive tract, we infer that Bapx1 activity is uniquely responsible for these aspects, even if the precise molecular mechanism is presently unknown.
Despite ostensibly normal smooth muscle differentiation and preserved expression of Nkx2.5, a gene implicated in chick gizzard development, 33 Bapx1Cre/Cre mice also lack normal pylorus morphology. Mice deleted for nearly the full Hoxd gene cluster lose multiple gastrointestinal valves, including the pyloric sphincter, with associated changes in regional smooth muscle and mucosa;9 the pyloric constriction is also missing in Barx1−/− mice.12 These findings may be relevant to hypertrophic pyloric stenosis, a common congenital disorder.34 Future efforts should aim to understand how these homeobox genes interact to generate the pyloric sphincter.
The stomach corpus and intestine developed normally, indicating that the antrum-pylorus is the only gut segment that requires Bapx1 for proper development. Alternatively, Bapx1 may function redundantly with other homeobox genes elsewhere. Less likely, abnormal hindstomach development could reflect dysmorphogenesis of the spleen and pancreas. Around E8.5 in mouse development, Bapx1 mediates lateral growth of the splanchnic mesodermal plate and coupled leftward growth of the dorsal pancreas, associated with control of Fgf10 expression.35 However, anomalies akin to those we identify in Bapx1Cre/Cre stomach are not seen with a wide range of defects in spleen and pancreas development.12, 25 In chondrocytes, Bapx1 serves both proliferative and anti-apoptotic roles,19, 36 and one reason the antrum and pylorus may develop aberrantly in its absence is if hindstomach progenitors are disadvantaged relative to anterior cells programmed for corpus differentiation. Immunostaining for cleaved caspase 3 did not reveal excess apoptosis in E11.5 hindstomachs (data not shown).
Forced Bapx1 expression in the chick proventriculus (forestomach) suppresses Bmp4 and Wnt5a expression and region-specific differentiation; conversely, forced expression of Bapx1-VP16, which artificially converts a presumed repressor into a transcriptional activator, promotes occasional expression of BMP4 and Wnt5a in the gizzard.15, 37 By contrast, Bmp4 expression does not appear to be compartmentalized in the mammalian stomach, nor did we detect aberrant expression of Bmp4 or Nkx2.1 in the mutant organ. Thus, despite similarities in Bapx1 expression and function in developing posterior stomach, phenotypes and affected pathways in chick and mouse seem different. These could reflect different mechanisms to create the keratinized avian gizzard versus the glandular mammalian antrum.
Although Barx1 and Bapx1 appear in different compartments in the developing spleen and their mutant phenotypes in that organ are distinct,12, 18, 19, 21, 38 their expression in distal stomach mesenchyme is overlapping. Absence of Barx1 markedly disrupts stomach development, producing aberrant morphogenesis, intestinal homeosis, and pyloric sphincter agenesis. Additional loss of Bapx1 does not worsen this phenotype, and the greater severity of antral- pyloric defects in the Barx1 mutant hints at actions upstream of Bapx1. Indeed, Bapx1 expression is virtually lost in Barx1-null stomach and its absence could potentially account for some part of the Barx1−/− phenotype in the distal organ. Mice with tissue-specific loss of a third stomach transcription factor, the nuclear hormone receptor COUP-TFII, also show a mild patterning defect.39 Besides expansion and disorganization of circular smooth muscle and enteric neurons, the margin between forestomach and corpus is shifted anteriorly and the glandular stomach accordingly occupies a larger relative space. Although expansion of the corpus is a common feature of the two phenotypes, they occur at opposite ends, anteriorly in the case of COUP-TFII deficiency and posteriorly in Bapx1Cre/Cre animals.
Barx1 is expressed throughout stomach mesenchyme, whereas Bapx1 is initially confined to the caudal region. Thus, although Barx1 seems to be required for stomach Bapx1 expression, it cannot be sufficient to restrict expression to the hindstomach; other factors may promote Bapx1 expression caudally or repress it rostrally. We are presently investigating Barx1’s role in COUP-TFII expression. Our results meanwhile implicate Barx1 and Bapx1 within an essential pathway for mammalian hindstomach development.
Acknowledgments
Grant support: National Institutes of Health grants P01HL49953 and P01HL067155 (to RJS); R01CA095608 (to WEZ); R01DK061139 (to RAS); and the Center for Environmental and Rural Health, Texas A&M, P30 ES09106 (WEZ and RJS). Molecular Endocrinology Training Program grant T32DK07696 (MNS); training grant T32DK07477 and a fellowship from the Crohn’s and Colitis Foundation of America (MPV); UNCF/Merck Graduate Fellowship Program and Robert C. McNair Foundation (KAM).
ACKNOWLEDGMENTS and AUTHOR ATTRIBUTION
We thank Renee Braun and Wei Yu for expert technical assistance; Xuan Chi for Nkx2.5-GFP transgenic mice; and D. Alpers, C. Wright, T. Lufkin, A. McMahon, and R. Harvey for antibodies and probes.
Footnotes
Financial disclosures: None
MPV, MNS, KAM, B-MK and YZ performed the research; all authors participated in design and interpretation of the studies and analysis of the data; MPV and RAS wrote the paper with input from WEZ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1053/j.gastro.2009.01.009
Read article for free, from open access legal sources, via Unpaywall:
http://www.gastrojournal.org/article/S0016508509000158/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Pseudogenization of NK3 homeobox 2 (Nkx3.2) in monotremes provides insight into unique gastric anatomy and physiology.
Open Biol, 14(7):240071, 03 Jul 2024
Cited by: 1 article | PMID: 38955222 | PMCID: PMC11285752
Integration of Single-Cell RNA Sequencing and Bulk RNA Sequencing to Identify an Immunogenic Cell Death-Related 5-Gene Prognostic Signature in Hepatocellular Carcinoma.
J Hepatocell Carcinoma, 11:879-900, 16 May 2024
Cited by: 0 articles | PMID: 38770169 | PMCID: PMC11104445
Beneficial islet inflammation in health depends on pericytic TLR/MyD88 signaling.
J Clin Invest, 134(14):e179335, 17 Jun 2024
Cited by: 0 articles | PMID: 38885342 | PMCID: PMC11245159
Generation of human gastric assembloids from primary fetal organoids.
Pediatr Surg Int, 40(1):6, 24 Nov 2023
Cited by: 0 articles | PMID: 37999863 | PMCID: PMC10673726
Pericytes modulate islet immune cells and insulin secretion through Interleukin-33 production in mice.
Front Endocrinol (Lausanne), 14:1142988, 09 Mar 2023
Cited by: 2 articles | PMID: 36967785 | PMCID: PMC10034381
Go to all (50) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Control of stomach smooth muscle development and intestinal rotation by transcription factor BARX1.
Dev Biol, 405(1):21-32, 06 Jun 2015
Cited by: 22 articles | PMID: 26057579 | PMCID: PMC4529797
Independent functions and mechanisms for homeobox gene Barx1 in patterning mouse stomach and spleen.
Development, 134(20):3603-3613, 12 Sep 2007
Cited by: 46 articles | PMID: 17855428
Targeted disruption of the homeobox transcription factor Bapx1 results in lethal skeletal dysplasia with asplenia and gastroduodenal malformation.
Genes Cells, 5(6):499-513, 01 Jun 2000
Cited by: 66 articles | PMID: 10886375
Barx1 and evolutionary changes in feeding.
J Anat, 207(5):619-622, 01 Nov 2005
Cited by: 19 articles | PMID: 16313395
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA095608
Grant ID: R01CA095608
NHLBI NIH HHS (4)
Grant ID: P01 HL049953
Grant ID: P01HL067155
Grant ID: P01HL49953
Grant ID: P01 HL067155
NIDDK NIH HHS (7)
Grant ID: T32 DK007696
Grant ID: R01 DK061139
Grant ID: T32 DK007477
Grant ID: R01 DK061139-04S1
Grant ID: R01DK061139
Grant ID: T32DK07477
Grant ID: T32DK07696
NIEHS NIH HHS (2)
Grant ID: P30 ES009106
Grant ID: P30 ES09106





