Abstract
Background
In eusocial hymenopteran insects, foraging genes, members of the cGMP-dependent protein kinase family, are considered to contribute to division of labor through behavioral caste differentiation. However, the relationship between foraging gene expression and behavioral caste in honeybees is opposite to that observed in ants and wasps. In the previously examined eusocial Hymenoptera, workers behave as foragers or nurses depending on age. We reasoned that examination of a different system of behavioral caste determination might provide new insights into the relationship between foraging genes and division of labor, and accordingly focused on bumblebees, which exhibit size-dependent behavioral caste differentiation. We characterized a foraging gene (Bifor) in bumblebees (Bombus ignitus) and examined the relationship between Bifor expression and size-dependent behavioral caste differentiation.Findings
A putative open reading frame of the Bifor gene was 2004 bp in length. It encoded 668 aa residues and showed high identity to orthologous genes in other hymenopterans (85.3-99.0%). As in ants and wasps, Bifor expression levels were higher in nurses than in foragers. Bifor expression was negatively correlated with individual body size even within the same behavioral castes (regression coefficient = -0.376, P < 0.001, all individuals; -0.379, P = 0.018, within foragers).Conclusion
These findings indicate that Bifor expression is size dependent and support the idea that Bifor expression levels are related to behavioral caste differentiation in B. ignitus. Thus, the relationship between foraging gene expression and behavioral caste differentiation found in ants and wasps was identified in a different system of labor determination.Free full text

Size-dependent foraging gene expression and behavioral caste differentiation in Bombus ignitus
Associated Data
Abstract
Background
In eusocial hymenopteran insects, foraging genes, members of the cGMP-dependent protein kinase family, are considered to contribute to division of labor through behavioral caste differentiation. However, the relationship between foraging gene expression and behavioral caste in honeybees is opposite to that observed in ants and wasps. In the previously examined eusocial Hymenoptera, workers behave as foragers or nurses depending on age. We reasoned that examination of a different system of behavioral caste determination might provide new insights into the relationship between foraging genes and division of labor, and accordingly focused on bumblebees, which exhibit size-dependent behavioral caste differentiation. We characterized a foraging gene (Bifor) in bumblebees (Bombus ignitus) and examined the relationship between Bifor expression and size-dependent behavioral caste differentiation.
Findings
A putative open reading frame of the Bifor gene was 2004 bp in length. It encoded 668 aa residues and showed high identity to orthologous genes in other hymenopterans (85.3-99.0%). As in ants and wasps, Bifor expression levels were higher in nurses than in foragers. Bifor expression was negatively correlated with individual body size even within the same behavioral castes (regression coefficient = -0.376, P < 0.001, all individuals; -0.379, P = 0.018, within foragers).
Conclusion
These findings indicate that Bifor expression is size dependent and support the idea that Bifor expression levels are related to behavioral caste differentiation in B. ignitus. Thus, the relationship between foraging gene expression and behavioral caste differentiation found in ants and wasps was identified in a different system of labor determination.
Background
Animal foraging behavior is a particularly interesting phenomenon from both ecological and evolutionary perspectives [1,2]. Although understanding the genetic basis of individual foraging behavior is important for evolutionary studies, in most case the genes controlling this behavior have not been identified. Candidate genes have, nevertheless, been identified in insects, and genetic analyses of behavioral differences have been conducted. For example, a foraging gene in the fruit fly (Drosophila melanogaster) differentiates behavior via a two-allele system: individuals with an R allele (rovers) show higher activity for food materials than those with a for allele (sitters; [3]). The same locus is also related to sucrose response, resistance to high temperature, and learning [4-6].
Foraging genes, members of the cGMP-dependent protein kinase (PKG) family, are also found in social insects and are related to the division of labor through behavioral caste differentiation in colonies. In honeybees (Apis mellifera), foraging gene (Amfor) activity differs according to worker age, with gene expression levels increasing with increasing age. Young workers with low expression levels (~ 3 weeks from emergence) act as nurses, whereas older workers with high expression levels (≥3 weeks from emergence) act as foragers [7]. Experimental administration of cGMP has been demonstrated to induce a change from nurses to foragers [8]. In contrast, other findings indicate opposite patterns of foraging gene expression among different behavioral castes. Although age-dependent division of labor has also been observed among other social hymenopteran insects, the expression levels of foraging genes were found to be higher in nurses than in foragers in two known instances: the foraging ant (Pogonomyrmex barbatus) and the common wasp (Vespula vulgaris; [9,10]). Furthermore, in the ant (Pheidole pallidula), larger individuals with higher PKG activity are predisposed to defend the nest (defenders), whereas smaller individuals are the foragers with lower PKG activity [11]. Lucas and Sokolowski [11] further demonstrated that the pharmacological activation of PKG increased defense and reduced foraging behavior. This study confirmed that lower expression levels produce foragers even in size-dependent behavioral caste differentiation, whereas higher expression produces defenders rather than nurses. Thus, investigation of other social insects with size-dependent nurse/forager behavioral changes might provide additional supportive evidence for the control of behavioral castes by foraging genes.
Bumblebees (Bombus spp.), which belong to the same family as honeybees, are also social insects but show a different pattern of division of labor. Bumblebees exhibit a unique form of collective nursing, placing several larvae into a single brood chamber and serving food as a lump. Such nursing consequently induces considerable variation in worker size. Worker size is accordingly the major determinant in behavioral castes of bumblebees, although age is also involved to a certain degree. In general, large workers act as foragers and small workers perform as nurses [12,13]. If patterns of gene expression similar to those observed in ants and wasps were to be observed in the size-dependent behavioral caste differentiation system in bumblebees, the relationships between higher (or lower) expression of foraging genes and nurses (or foragers) would be clearer.
We predicted that expression of the foraging gene would be size dependent since there is a size-dependent behavioral caste differentiation system in bumblebees. Thus, in this study, we characterized the foraging gene (Bifor) of the Japanese bumblebee (Bombus ignitus) and examined its expression patterns in workers. The purpose of this study was to examine the relationship between the foraging gene expression level and nurse-forager size difference and to compare the findings with those of previous studies.
Methods
Bombus ignitus is distributed in Japan (Honshu, Shikoku, Kyushu), the Korean Peninsula, and China [14]. Adults (workers) collected in the wild (Sendai, Miyagi Pref., Japan) and those from commercially reared colonies (Agrisect) were used to determine the nucleotide sequences of foraging and ribosomal protein 49 (Rp49) genes. Measurements of body size and mRNA expression analyses were conducted using the commercially reared colonies.
Identification of behavioral castes was achieved by conducting behavioral observations in a colony within an experimental enclosure inside a room (see Additional file 1 for the detailed description for the experimental enclosure and the identification of individual bees). Observations and collection of workers were conducted for two colonies (nests 1 and 2). Nest 1 was observed from September 19 to October 1, 2008, and nest 2 was observed from October 22 to November 3, 2008. In commercially reared colonies, reproductive individuals breed throughout the year; hence, worker individuals can behave as foragers or nurses. The division of labor in such colonies is normally observed in October and November, as is observed in wild colonies during their breeding season.
The observations were made between 7:00 and 10:00, and the bees were individually returned to their colonies until 18:00. After this time, all the workers present outside the colonies were collected and returned to the colonies. Having numbered the bees, they were allowed to independently choose to behave as foragers or nurses for 3 days. Although the bees were placed in relatively small artificial environments, important components required for their foraging behavior (e.g., a sufficient spatial scale for recognition of the direction of food sites and the distance between the nest and food) were provided within the enclosure. We therefore assume that the bees exhibited a normal foraging behavior.
By observing the workers for 3 days or more after numbering, behavioral castes were identified as follows. Workers making three or more trips to collect food within a 3-h observation period were identified as "foragers." Those not making foraging trips during the observation period were defined as "nurses." Foragers were collected at two time periods: while bees visited flowers in the daytime (active foragers) and while they rested in the colony at night (resting foragers). In the daytime, foragers were caught while they were perched on flowers or while they were flying. Nursing behavior could also be observed at night; thus, nurses were collected at night. All the collected bees were immediately fixed in liquid nitrogen and stored at -80°C until use. In this study, head width was used as an indicator of body size.
The expression levels of the foraging gene (Bifor) were determined relative to the expression levels of Rp49. The detailed methods used to determine the nucleotide sequence of Bifor and for real-time PCR analyses are described in Additional file 1.
Results
The full length of the coding region of Bifor was determined as 2004 bp (668 aa). On comparing with the functional region identified in Amfor (two regions in CAP_ED, S_TKc, S_TK_X; Ben-Shahar et al., 2002), Bifor showed high identities with other hymenopteran foraging genes (477 aa: A. mellifera, 99.0%; Nasonia vitripennis, 95.0%; V. vulgaris, 85.3%).
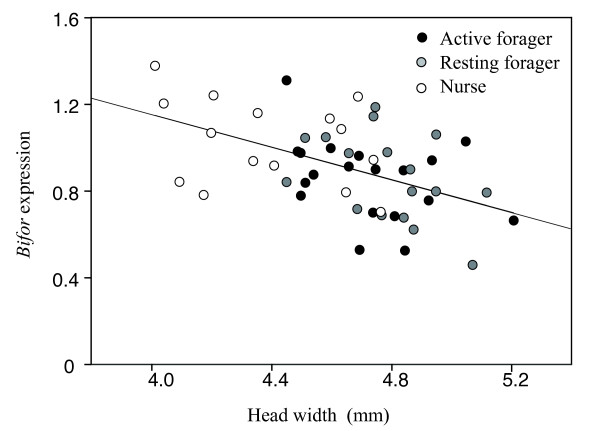
There was a significant difference in head width among foragers (active and resting) and nurses (2-way ANOVA: F2,47 = 15.25, P < 0.0001; Figure Figure1a),1a), but not among the size of the nests (F1,47 = 2.324, P = 0.1341). The head width was significantly wider in the foragers (both active and resting) than in the nurses (Tukey's HSD test: active vs. resting foragers, P = 0.604; active foragers vs. nurses, P < 0.001; resting foragers vs. nurses, P < 0.001). Similarly, there was a significant difference in the level of Bifor expression among active and resting foragers and nurses (2-way ANOVA, F2,47 = 3.931, P < 0.026; Figure Figure1b),1b), but not among the size of the nests (F1,47 = 1.322, P = 0.256). Nurses exhibited a significantly higher level of Bifor expression (relative to the expression level of Rp49) than active foragers (Tukey's HSD test: P = 0.035) and a nearly significantly higher expression than resting foragers (P = 0.0594). A regression analysis of the combined data for all the workers showed that there was a significant negative relationship between Bifor gene expression level and head width [regression coefficient, -0.376 (SE, 0.09), P < 0.001; Figure Figure22].
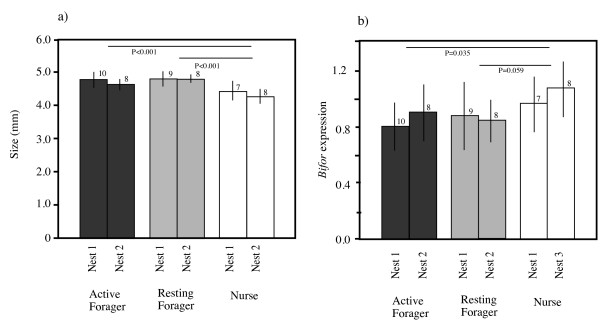
(a) Head width (as an indicator of body size) and (b) Bifor expression levels of active foragers, resting foragers, and nurses. Figures above the bars indicate the number of individuals examined. Vertical lines indicate standard deviation.
Discussion
As found in B. terrestris [13], size-dependent behavioral caste differentiation was confirmed in B. ignitus in this study. Bumblebees also exhibit certain aspects of division of labor according to worker age: young bees remain in colonies, whereas older bees tend to become foragers [15]. However, all young bees act as nurses in the first few days after emergence, and thereafter large-sized bees are likely to switch their functions to foragers, whereas small-sized bees remain nurses [12,16]. Therefore, except for the first few days after emergence, the behavioral caste of bumblebees is determined by worker size. As identification of behavioral caste was conducted at least 3 days after the numbering in this study, we could only demonstrate the size-dependent determination of behavioral castes in bumblebees.
Bifor expression levels were higher in nurses than in foragers, and there was a clear negative relationship between body size and Bifor expression level even within the same behavioral castes (within the foragers, regression coefficient, -0.379 [SE, 0.02], P = 0.018; within the nurse, regression coefficient, -0.251 [SE, 0.19], P = 0.224). Although there was no significant relationship between the expression levels of Rp49 and body size, it is possible that a somewhat weak positive correlation between Rp49 expression levels and body size affected the negative relationship between body size and relative Bifor expression level. However, multiple regression analysis showed that the absolute expression level of Bifor (values not relative to Rp49 expression) was significantly negatively related to size (partial regression coefficient with size, -1.2632, P = 0.0022) and positively related to Rp49 expression (partial regression coefficient with Rp49 = 0.466, P < 0.0001). These findings indicated that Bifor expression levels are negatively correlated to body size irrespective of Rp49 expression levels.
The results of our study indicate that both Bifor expression and behavioral caste differentiation is highly size dependent. They also suggest that in species with size-dependent behavioral caste differentiation, foragers exhibit lower expression of the foraging gene than nurses. This pattern has also been observed in species exhibiting age-dependent division of labor such as P. barbatus and V. vulgaris [9,10], and in size-dependent forager-defender behavioral caste differentiation in Pheidole pallidula [11]. These findings support the hypothesis that a lower expression level of Bifor is associated with foraging behavior and a higher expression level with nursing behavior. A negative relationship between body size and Bifor expression level was observed even within the same labor group. Thus, it can be speculated that Bifor expression might be suppressed with increasing body size, and that there might be a threshold level, below which individuals become foragers. However, further experiments, such as cGMP administration, will be required in order to confirm the relationships among behavioral caste, body size, and foraging gene.
We also found that there was little or no difference in Bifor expression between resting and active foragers, indicating that Bifor expression does not directly affect the foraging activity of bees but that it may be related to the characteristics required for foraging or nursing. Foragers, but not nurses, have strong circadian rhythms [17], which may affect the regulation of many genes. Thus, the foraging behavior observed during the day might have been caused by clock regulation. However, flying behavior is triggered even in the presence of a small amount of light, whereas foraging behavior is adversely affected by light [18]. Furthermore, sensitivity to light depends on body size [18]. Thus, it is possible that Bifor expression might be related to a threshold body size, above which individuals begin to forage in response to light.
Conclusion
B. ignitus exhibits size-dependent behavioral caste differentiation. Bifor expression levels were higher in nurses than in foragers, and there was a clear negative relationship between body size and Bifor expression level even within the same labor group. These findings indicate that Bifor expression is size dependent and support the idea that its expression levels are related to behavioral caste differentiation. Thus, the relationship between foraging gene expression and behavioral caste differentiation previously identified in ants and wasps has been confirmed in another social insect with a different system of labor determination.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YK carried out the behavioral observations, the determination of gene sequences, and real-time PCR analysis. HO assisted with the real-time PCR analysis. JY provided advice on the behavioral and molecular genetic studies. MK supervised the research and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Supplemental material and methods. The detailed descriptions of the methods for Experimental enclosure and identification of individual bees, the nucleotide sequence determination of Bifor and real-time PCR analyses.
Acknowledgements
We thank Dr. N. Matsushima for technical advice during the molecular experiments and Prof. K. Tamura for the use of his laboratory equipment.
References
- Stephens DW, Brown JS, Ydenberg RC. Foraging: Behavior and Ecology. Chigago: The University of Chicago Press; 2007. [Google Scholar]
- Stephens DW, Krebs JR. Foraging theory. Princeton, NJ: Princeton University Press; 1986. [Google Scholar]
- Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, Coulthard A, Pereira HS, Greenspan RJ, Sokolowski MB. Natural Behavior Polymorphism Due to a cGMP-Dependent Protein Kinase of Drosophila . Science. 1997;277:834–836. 10.1126/science.277.5327.834. [Abstract] [CrossRef] [Google Scholar]
- Dawson-Scully K, Armstrong GAB, Kent C, Robertson RM, Sokolowski MB. Natural Variation in the Thermotolerance of Neural Function and Behavior due to a cGMP-Dependent Protein Kinase. PLoS ONE. 2007;8:e773. 10.1371/journal.pone.0000773. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mery F, Belay AT, So AKC, Sokolowski MB, Kawecki TJ. Natural polymorphism affecting learning and memory in Drosophila . Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13051–13055. 10.1073/pnas.0702923104. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Scheiner R, Sokolowski MB, Erber J. Activity of cGMP-dependent protein kinase (PKG) affects sucrose responsiveness and habituation in Drosophila melanogaster . Learning & Memory. 2004;11:303–311. 10.1101/lm.71604. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of Gene Action Across Different Time Scales on Behavior. Science. 2002;296:741–744. 10.1126/science.1069911. [Abstract] [CrossRef] [Google Scholar]
- Ben-Shahar Y, Leung HT, Pak WL, Sokolowski MB, Robinson GE. cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. Journal of Experimental Biology. 2003;206:2507–2515. 10.1242/jeb.00442. [Abstract] [CrossRef] [Google Scholar]
- Ingram KK, Oefner P, Gordon DM. Task-specific expression of the foraging gene in harvester ants. Molecular Ecology. 2005;14:813–818. 10.1111/j.1365-294X.2005.02450.x. [Abstract] [CrossRef] [Google Scholar]
- Tobback J, Heylen K, Gobin B, Wenseleers T, Billen J, Arckens L, Huybrechts R. Cloning and expression of PKG, a candidate foraging regulating gene in Vespula vulgaris . Animal Biology. 2008;58:341–351. 10.1163/157075608X383665. [CrossRef] [Google Scholar]
- Lucas C, Sokolowski MB. Molecular basis for changes in behavioral state in ant social behaviors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6351–6356. 10.1073/pnas.0809463106. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Goulson D. Bumblebees, Their Behaviour and Ecology. Oxford: Oxford University Press; 2003. [Google Scholar]
- Goulson D, Peat J, Stout JC, Tucker J, Darvill B, Derwent LC, Hughes WOH. Can alloethism in workers of the bumblebee, Bombus terrestris, be explained in terms of foraging efficiency? Animal Behaviour. 2002;64:123–130. 10.1006/anbe.2002.3041. [CrossRef] [Google Scholar]
- Takamizawa I. The Japanese Social Wasps and Bees. Nagano: Shinano Shinbun Sha; 2005. [Google Scholar]
- O'Donnell S, Reichardt M, Foster R. Individual and colony factors in bumble bee division of labor (Bombus bifarius nearcticus Handl; Hymenoptera, Apidae) Insectes Sociaux. 2000;47:164–170. 10.1007/PL00001696. [CrossRef] [Google Scholar]
- Pouvreau A. Contribution à l'étude du polyéthisme chez les bourdons, Bombus Latr. (Hymenoptera, Apidae) Apidrogie. 1989;20:229–244. 10.1051/apido:19890305. [CrossRef] [Google Scholar]
- Yerushalmi S, Bodenhaimer S, Bloch G. Developmentally determined attenuation in circadian rhythms links chronobiology to social organization in bees. Journal of Experimental Biology. 2006;209:1044–1051. 10.1242/jeb.02125. [Abstract] [CrossRef] [Google Scholar]
- Kapustjanskij A, Streinzer M, Paulus HF, Spaethe J. Bigger is better: implications of body size for flight ability under different light conditions and the evolution of alloethism in bumblebees. Functional Ecology. 2007;21:1130–1136. 10.1111/j.1365-2435.2007.01329.x. [CrossRef] [Google Scholar]
Articles from BMC Research Notes are provided here courtesy of BMC
Full text links
Read article at publisher's site: https://doi.org/10.1186/1756-0500-2-184
Read article for free, from open access legal sources, via Unpaywall:
https://bmcresnotes.biomedcentral.com/track/pdf/10.1186/1756-0500-2-184
Open access at BioMedCentral
http://www.biomedcentral.com/1756-0500/2/184
Open access at BioMedCentral
http://www.biomedcentral.com/1756-0500/2/184/abstract
Open access at BioMedCentral
http://www.biomedcentral.com/content/pdf/1756-0500-2-184.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Physiological specialization of the brain in bumble bee castes: Roles of dopamine in mating-related behaviors in female bumble bees.
PLoS One, 19(3):e0298682, 13 Mar 2024
Cited by: 0 articles | PMID: 38478476 | PMCID: PMC10936820
Gene expression and epigenetics reveal species-specific mechanisms acting upon common molecular pathways in the evolution of task division in bees.
Sci Rep, 11(1):3654, 11 Feb 2021
Cited by: 10 articles | PMID: 33574391 | PMCID: PMC7878513
The disturbance leg-lift response (DLR): an undescribed behavior in bumble bees.
PeerJ, 9:e10997, 25 Mar 2021
Cited by: 3 articles | PMID: 33828912 | PMCID: PMC8005288
Neural activity mapping of bumble bee (Bombus ignitus) brains during foraging flight using immediate early genes.
Sci Rep, 10(1):7887, 12 May 2020
Cited by: 7 articles | PMID: 32398802 | PMCID: PMC7217898
Managing the risks and rewards of death in eusocial insects.
Philos Trans R Soc Lond B Biol Sci, 373(1754):20170258, 01 Sep 2018
Cited by: 14 articles | PMID: 30012744 | PMCID: PMC6053982
Review Free full text in Europe PMC
Go to all (16) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Gene expression during larval caste determination and differentiation in intermediately eusocial bumblebees, and a comparative analysis with advanced eusocial honeybees.
Mol Ecol, 30(3):718-735, 07 Jan 2021
Cited by: 4 articles | PMID: 33238067 | PMCID: PMC7898649
Role of the foraging gene in worker behavioral transition in the red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae).
Pest Manag Sci, 78(7):2964-2975, 28 Apr 2022
Cited by: 7 articles | PMID: 35419943
Time-course RNASeq of Camponotus floridanus forager and nurse ant brains indicate links between plasticity in the biological clock and behavioral division of labor.
BMC Genomics, 23(1):57, 15 Jan 2022
Cited by: 10 articles | PMID: 35033027 | PMCID: PMC8760764
(Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects.
Insects, 12(6):498, 27 May 2021
Cited by: 7 articles | PMID: 34071806 | PMCID: PMC8229086
Review Free full text in Europe PMC

 1
1