Abstract
Free full text

Role of HIV membrane in neutralization by two broadly neutralizing antibodies
Associated Data
Abstract
Induction of effective antibody responses against HIV-1 infection remains an elusive goal for vaccine development. Progress may require in-depth understanding of the molecular mechanisms of neutralization by monoclonal antibodies. We have analyzed the molecular actions of two rare, broadly neutralizing, human monoclonal antibodies, 4E10 and 2F5, which target the transiently exposed epitopes in the membrane proximal external region (MPER) of HIV-1 gp41 envelope during viral entry. Both have long CDR H3 loops with a hydrophobic surface facing away from the peptide epitope. We find that the hydrophobic residues of 4E10 mediate a reversible attachment to the viral membrane and that they are essential for neutralization, but not for interaction with gp41. We propose that these antibodies associate with the viral membrane in a required first step and are thereby poised to capture the transient gp41 fusion intermediate. These results bear directly on strategies for rational design of HIV-1 envelope immunogens.
HIV-1 infected patients typically generate strong antibody responses to the viral envelope glycoprotein, but these antibodies are usually non-neutralizing or strain-specific, and they fail to prevent initiation or spread of infection (1, 2). Extensive glycosylation, sequence variation, and ligand-induced reorganization of the conserved CD4 and coreceptor binding sites in the envelope protein, as well as potential host immunoregulatory constraints, pose major obstacles to generating broadly reactive neutralizing antibodies (Nabs) (1–4). These considerations may explain why the conventional approaches to vaccination have so far failed to elicit broadly reactive Nabs. They suggest that any successful vaccine will depend on innovative immunogen design based on detailed understanding of HIV-1 molecular immunobiology.
A few broadly reactive neutralizing monoclonal antibodies (mAbs) have been described. These mAbs recognize three important regions of the HIV-1 envelope glycoprotein (5–8). Two of them, designated 2F5 and 4E10, recognize epitopes on the gp41 segment adjacent to the viral membrane: the 20-residue membrane proximal external region (MPER). Crystal structures of Fab fragments from these mAbs in complex with their corresponding core epitope peptides show the epitope conformations (9, 10) (Fig. S1). In the 2F5-bound structure, the gp41 epitope (at the N-terminal end of the MPER) adopts an extended conformation with two overlapping β-turns. On 4E10, the gp41 epitope (toward the C-terminal end of the MPER) is α-helical, contacting the antibody through one face of the helix. In both cases, a relatively long CDR H3 loop in the antigen-combining site presents a hydrophobic surface that does not contact the peptide antigens in the crystal structures. It has been proposed that they interact instead with the viral membrane (9, 10). Both antibodies are also polyspecific and can bind anionic phospholipids (4, 11–13). The significance for HIV-1 neutralization of these documented lipid and membrane interactions has remained uncertain.
In solution, peptides containing the linear epitopes of 2F5 and 4E10 have been shown to adopt a wide range of conformations, including unstructured, α-helical, or β-turn conformations, depending on experimental parameters (14–17). Lack of defined conformations implies that the MPER region is probably flexible when unconstrained. Attempts to use peptides from this region as an immunogen have failed to induce Nabs; it has been suggested that the epitopes have not been presented in a relevant conformation (reviewed in ref. 18). The HIV envelope glycoprotein undergoes large structural rearrangements upon engagement with CD4 and coreceptor. During viral entry, there are at least three distinct conformational states of the HIV-1 envelope protein: a prefusion conformation; a “prehairpin,” extended intermediate; and a postfusion, trimer-of-hairpins conformation (19–22). We have reported biochemical evidence showing that 2F5 and 4E10 neutralize by specifically targeting the prehairpin intermediate conformation of gp41 (23), raising the critical question: how can these antibodies efficiently capture an intermediate state with a finite lifetime?
To dissect the molecular mechanisms of neutralization by 4E10 and 2F5, we have examined the role of their hydrophobic CDR H3 surfaces in HIV-1 membrane interactions, in binding the gp41 target, and in mediating HIV-1 neutralization. We have used a single-chain Fv fragment (scFv) of 4E10 to study the properties of a series of CDR H3 mutants. We find that 4E10 scFv neutralizes almost as potently as the intact IgG, and that it binds weakly, with high off-rates, to synthetic liposomes that mimic the lipid composition of HIV-1 membrane (viral liposomes), as well as to both HIV-1 and SIV virion preparations. These results are consistent with the notion that 4E10 does not target the untriggered, prefusion state of the envelope protein. Alanine substitutions at the multiple hydrophobic positions in the CDR H3 loop eliminate both binding of the 4E10 scFv to membrane and neutralization of HIV-1 but have no effect on the high-affinity interaction with a gp41 protein (gp41-inter) that mimics the prehairpin intermediate (23). We have made similar observations with recombinant 2F5 IgG. Gp41-inter efficiently blocks neutralization of HIV-1 by mAb 4E10 even after preincubation of the antibody with the virus. These results support a model in which 4E10 attaches reversibly to the viral membrane before capturing the triggered, transient form of gp41 induced by receptor binding. Our findings suggest that a lipid component may be required in an immunogen to elicit neutralizing antibodies against the MPER.
Results and Discussion
Mutations in the CDR H3 Loops.
In each of the CDR H3 loops of mAbs 4E10 and 2F5, there are three hydrophobic residues that could contact the virion lipid bilayer. These are W100, W100b, and L100c in 4E10; and L100a, F100b, and V100c in 2F5. We expressed three mutants of a 4E10 scFv with progressively increasing numbers of alanine substitutions: one with a single mutation, 4E10-mut1 scFv (W100A); a second with two, 4E10-mut2 scFv (W100A/W100bA); and a third with three, 4E10-mut3 scFv (W100A/W100bA/L100cA). We expressed a fourth mutant, 4E10-mut4 scFv (I56A/Sl94R), with mutations in the binding site for the gp41 core epitope designed to disrupt gp41 binding (Fig. S1). We likewise generated four mAb mutants using recombinant 2F5 IgG: 2F5-mut1 rIgG (L100aA); 2F5-mut2 rIgG (F100bA); 2F5-mut3 rIgG (L100aA/F100bA); and 2F5-mut4 rIgG (R95A). The first three harbored mutations in residues at the hydrophobic tip of CDR H3 loop and the last, a mutation in the CDR H3 near the gp41 binding site (Fig. S1). All 4E10 scFvs were expressed in E. coli and refolded in vitro; r2F5 IgG and the mutants were produced in 293T cells. As shown in Fig. S2, refolded 4E10 scFv and its mutants were purified by Ni-NTA and eluted as a very sharp peak by gel filtration chromatography from a Superdex 200 column, indicating that the protein preparations were stable and homogenous. As expected, wild-type 4E10 scFv bound the epitope peptide tightly (Fig. S3), consistent with previously published data (11, 23). 4E10-mut1, 4E10-mut2, and 4E10-mut3 scFvs also bound the peptide, with somewhat reduced affinity (Fig. S3), indicating that these proteins were correctly folded and functional and that the hydrophobic residues in the CDR H3 loop do not make major contributions to contacts with gp41, as shown by the crystal structures (9, 10). 4E10-mut4 scFv showed significant binding to the gp41 peptide, although it had the weakest affinity of the four, suggesting that these substitutions are not sufficient to eliminate gp41 epitope binding (Figs. S1 and S3). We obtained similar results with mutations in the CDR H3 loop of r2F5, as summarized in Fig. S4; in that case, the R95A mutation in the peptide epitope site did eliminate detectable interaction with gp41.
Hydrophobic Residues in the CDR H3 Loops Are Required for Membrane Binding.
To assess how the hydrophobic residues in the CDR H3 loops of 4E10 and 2F5 may contribute to binding to the viral membrane, we examined by SPR the interactions of 4E10 scFv and its mutants first with synthetic lipid bilayers, including liposomes that mimic the lipid composition of HIV viral membrane [phosphatidylcholine: phosphatidylethanolamine:phosphatidylserine:sphingomyelin:cholesterol = 9.35:19.25:8.25:18.15:45.00; (24)], as well as liposomes containing cardiolipin and phosphatidylserine (PS), and then with chemically inactivated HIV-1 and SIV virion preparations directly.
When the synthetic viral liposomes were immobilized on the surface of a Biacore L1 sensor chip by a hydrophobic linker, 4E10 scFv and 4E10-mut4 scFv bound with Kd of 0.7 and 1.0 μM, respectively (Fig. 1A and Fig. S5), with high on- and off-rates. 4E10-mut1 scFv bound the viral liposomes more weakly than did the wild-type, and binding by 4E10-mut2 and 4E10-mut3 scFvs was indistinguishable from that by negative control antibodies (Fig. 1A). Similar patterns were observed with these mutants when cardiolipin or PS liposomes were immobilized on the sensor chip (Fig. S6). Although the synthetic lipid bilayers do not mimic physiological membranes in all aspects, the data indicate that the hydrophobic residues in the CDR H3 loop do indeed mediate lipid binding by 4E10.
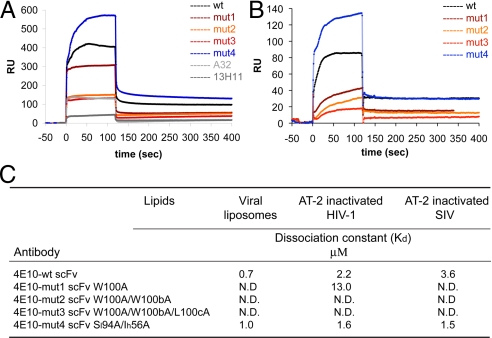
Binding to synthetic lipid bilayer and HIV-1 membrane of the scFv from broadly neutralizing antibody 4E10. Liposomes that mimic the lipid composition of HIV viral membrane containing phosphatidylcholine/phosphatidylethanolamine/phosphatidylserine/sphingomyelin/cholesterol with a ratio of 9.35:19.25:8.25:18.1:45 (24) (A) or AT-2 inactivated virion preparation of HIV-1 ADA (B) were immobilized at similar levels on L1 chips through an alkyl linker. 4E10 scFv and its variants were passed over the surface at a concentration of 8 μM (200 μg/mL). An anti-gp120 mAb, A32, and an anti-gp41 13H11, which show no binding to lipids (4), were also passed over the surface as negative controls. The sensorgrams of 4E10 in B were subtracted from the one obtained with A32. The recorded sensorgrams are shown in black for 4E10 scFv; dark red for 4E10-mut1 scFv; orange for 4E10-mut2 scFv; red for 4E10-mut3 scFv; blue for 4E10-mut4 scFv; light gray for mAb A32; and dark gray for 13H11 Fab. The experiments were repeated at least twice with similar results. Dissociation constants derived from the titration series using either PS-liposomes or HIV-1 virions, shown in Fig. S6, are summarized in (C). The weak affinities of 4E10-mut2 and 4E10-mut3 scFv could not be determined accurately. N.D., not determined.
To evaluate mAb lipid binding more directly related to neutralization, we examined association of 4E10 scFv with viral membranes, using AT-2 (Aldrithiol-2) inactivated HIV-1 ADA or SIV mac239 virions, which are competent in membrane fusion (25). These virion preparations often contain nonviral microvesicles derived from host cells, but SDS/PAGE analysis indicates that a substantial fraction of the total protein of the preparations is indeed viral protein (26). As shown in Fig. 1B and Fig. S4, the binding kinetics (high on- and off-rates) by AT-2 treated HIV-1 and SIV virion preparations were very similar to those observed with synthetic viral liposomes, suggesting that the association detected by SPR was primarily with membranes, not with envelope glycoprotein or any other components on the membrane surface, as SIV does not contain the 4E10 epitope (27) and HIV-1 preparation did not show any enhanced binding. Some binding observed with these preparations may be mediated by microvesicle membranes, which are probably indistinguishable from viral membranes in composition. Among the scFv mutants, 4E10-mut4, with mutations in the gp41 binding site, bound relatively tightly to the HIV-1 virion preparation, with a Kd comparable to that of wild-type 4E10 scFv (Fig. 1 A and C). In contrast, mutations in the CDR H3 loop had much stronger effects. The 4E10-mut1 scFv bound HIV-1 virions weakly, and the affinity of 4E10-mut2 and 4E10-mut3 scFvs for HIV-1 and SIV preparations was too weak to be measured accurately (Fig. 1B and Fig. S6C). These data indicate that the hydrophobic residues in the CDRH3 loop are necessary for the observed interaction of 4E10 scFv with membrane and that multiple residues may contribute to viral lipid binding. We obtained similar results with the r2F5 CDRH3 mutants (Figs. S1 and S4). Lack of lipid binding by 2F5-mut4 rIgG (R95A) suggests that R95, at the base of the 2F5 CDRH3, also influences bilayer association, perhaps by shifting the positions of other arginine residues (e.g., R96 and R100h) or by distorting the conformation of the CDR H3 loop. Similar 2F5 CDRH3 mutants were reported previously, but the conformational homogeneity of the mutants and the recombinant gp41 protein used in the study was not assessed (28). In summary, substitutions that reduce hydrophobicity (and in one case, positive charge) of the CDR H3 loops of 4E10 and 2F5 disrupt binding of these antibodies to lipid bilayers, and probably to HIV-1 viral membranes as well.
Effect of the CDR H3 Loop Mutations on Binding gp41.
Our previous biochemical studies suggest that 2F5 and 4E10 mAbs exert their neutralizing activity by binding the prehairpin intermediate conformation of gp41 (23), consistent with data reported by other groups, showing that both 2F5 and 4E10, like T20, are effective only during a short time interval during the fusion process (29–31). We therefore used the trimeric gp41-inter protein to assess the effect of the substitutions in the CDR H3 loop of mAb 4E10 on binding to its gp41 target. As shown in Fig. 2, 4E10 scFv bound strongly to gp41-inter immobilized on a CM5 chip, with koff = 2.2 × 10−5 s−1 and Kd = 0.19 nM. All three 4E10 scFv mutants with substitutions in the CDR H3 loop showed comparably tight binding (Fig. 2). Thus, in contrast to their effect on binding of 4E10 to HIV-1 viral membrane or phospholipids, substitutions of alanine for the bulky hydrophobic residues in the CDR H3 loops of 4E10 and 2F5 have very little effect on binding of these mAbs to their gp41 envelope target.
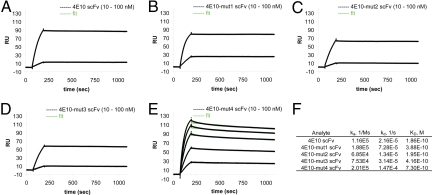
Interaction of 4E10 scFv and its mutants with the HIV-1 gp41 prehairpin intermediate. 92UG-gp41-inter-Fd was immobilized on a CM5 chip. Each of the following analytes was passed over a freshly prepared, gp41-inter surface, as the chip could not be completely regenerated (see Materials and Methods): (A) 4E10 scFv (10 and 100 nM); (B) 4E10-mut1 scFv (10 and 100 nM); (C) 4E10-mut2 scFv (10 and 100 nM); and (D) 4E10-mut3 scFv (10 and 100 nM). (E) 4E10-mut4 scFv at various concentrations (10, 25, 50, 75, and 100 nM) was passed over a single chip regenerated between runs by a solution containing 50 mM NaOH and 2 M NaCl. Binding kinetics were evaluated using BIAevaluation software (Biacore) and a 1:1 Langmuir binding model. The recorded sensorgrams are shown in black and the fits in green. The derived constants are summarized in (F). All experiments were repeated at least three times using different preparations of the proteins and gave essentially the same results.
The interaction of 4E10-mut4 scFv with gp41-inter is still relatively strong, but it has the lowest affinity (0.73 nM) among all of the scFv mutants. In particular, its dissociation rate (1.5 × 10−4 s−1) is seven times greater than that of wild-type 4E10 scFv. Indeed, the dissociation rates of all of the scFvs except 4E10-mut4 are too slow for accurate measurement, and we therefore believe the corresponding estimates of Kd to be upper limits. This conclusion is consistent with the observation that 4E10-mut4 is the only one, among all of the 4E10 scFv proteins tested, that can reproducibly be stripped from the sensor chip during regeneration (see Materials and Methods). The effect of the mut4 changes on epitope affinity may therefore be much greater than the factor of seven estimated here.
The CDR H3 Hydrophobic Residues Are Essential for Neutralizing Activity.
We analyzed the neutralization of two different isolates of HIV-1 [BG1168 and SF162 (32)] by each of the 4E10 scFv mutants, using the TZM/bl pseudovirus infectivity assay (32). The 4E10 scFv neutralizes both viral isolates with IC50s (0.5 and 1.8 μg/mL, respectively), similar to those of the bivalent 4E10 IgG (0.2 and 1.8 μg/mL). The 4E10-mut1 and 4E10-mut4 scFvs also neutralize both viruses, although with lower potency than that of the wild-type scFv, requiring between 10- and 30-fold higher antibody concentrations to achieve 50% neutralization. The 4E10-mut4 scFv neutralized both isolates more potently than did 4E10-mut1 scFv, although its affinity for gp41-inter is slightly lower, indicating a likely role for lipid affinity, which is reduced in mut1. Indeed, both 4E10-mut2 and 4E10-mut3 scFvs, which have little or no detectable lipid binding, failed to neutralize either of the HIV-1 isolates, despite their high affinity for the protein epitope in the MPER of gp41 (Fig. 3A). Mutations in the CDR H3 loop of r2F5 that likewise disrupt lipid binding with minimal impact on binding the epitope peptide also fail to neutralize HIV-1 (Fig. S4). Thus, the neutralizing activity of 4E10 and 2F5 mAbs depends on the hydrophobic residues in their CDR H3 loops and, we infer, on the role of these residues in an interaction with viral membrane.
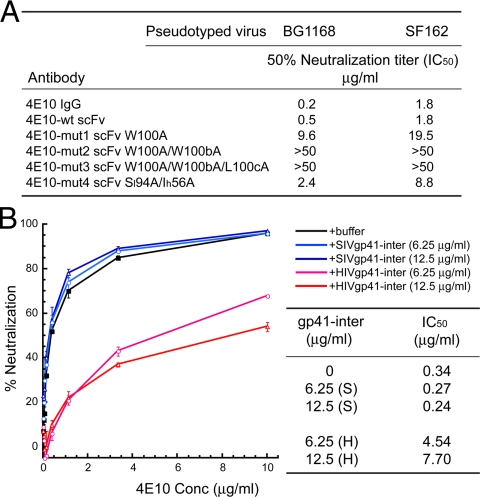
Neutralization by 4E10 variants and blocking of neutralization by gp41-inter. (A) Neutralization determined by reduction of luciferase reporter-gene expression after a single round of infection by pseudotyped HIV-1 viruses in TZM-bl cells. The pseudotyped viruses containing Envs derived from HIV-1 isolates, BG1168 and SF162, were preincubated with serial dilutions of the antibodies in 96-well plates. After mixing and incubation with target TZM-bl cells, the concentration of half-maximal inhibition (IC50) was calculated from the luciferase activities determined by luminescence measurements. (B) Effect of addition of gp41-inter on neutralization by 4E10. Serial dilutions of 4E10 IgG were plated and preincubated with the BG1168 pseudovirus for 1 h at 37 °C, followed by addition of TZM-bl cells, premixed with either HIV-1 or SIV gp41-inter protein. Percent neutralization was determined from luciferase activities and plotted against the concentration of 4E10. Black, neutralization by 4E10 (no gp41-inter added); light blue and blue, neutralization by 4E10 in the presence of SIV gp41-inter (6.25 and 12.5 μg/mL, respectively); light red and red, neutralization in the presence of HIV gp41-inter (6.25 and 12.5 μg/mL, respectively). IC50 values were read directly from the plots. The experiments were repeated twice and gave the same results.
How does lipid binding contribute to neutralization? The affinity of 4E10 for membrane is at least three orders of magnitude lower than its affinity for the gp41 protein epitope (Figs. 1 and and2),2), yet loss of this weak binding to viral membrane leads to complete loss of neutralizing activity. It thus appears that the viral lipid bilayer has a role in neutralization in addition to providing extra contacts for the antibody. One possibility is that attachment to viral membrane by 4E10 is a first, required step in neutralization. This association would concentrate the antibody on the surface of virion and give it a kinetic head start for capturing a transiently-exposed gp41 epitope (23). Without first attaching to the viral surface, 4E10 mAbs may not be able to find their gp41 targets during the restricted interval of the fusion-promoting structural transition.
To test this hypothesis, we examined whether gp41-inter can block neutralization of HIV-1 by 4E10 mAb even after preincubation of the antibody with the virus. Binding to viral membrane is rapidly reversible; binding to the gp41 epitope, effectively irreversible. If the two contacts occur together on the virion surface, then the gp41 interaction should prevent gp41-inter from competing for an antibody already associated with the virion. On the other hand, if 4E10 mAb first attaches to viral membrane and “waits” for triggering of gp41 by CD4 and coreceptor to access gp41, then gp41-inter should inhibit neutralization, either by capturing the antibody that dissociates from the membrane or by binding the antibody attached to the membrane and blocking interaction with gp41. Control experiments showed that gp41-inter alone neither inhibits nor enhances viral infectivity, but gp41-inter effectively blocks neutralization of HIV-1 by 4E10 mAb at a relatively low concentration even after preincubation of the antibody with the virus for 1 h at 37 °C (Fig. 3B). As a negative control, SIV gp41-inter protein, which does not bind 4E10, has no effect on 4E10-mediated neutralization of HIV-1 (Fig. 3B). These data support our conclusion that the attachment of 4E10 to HIV membrane is a first step in neutralization, before docking of the antibody to its protein target on gp41.
Molecular Mechanism of HIV-1 Neutralization by 4E10 and 2F5.
The results described above provide strong evidence for participation of the viral membrane in neutralization of HIV-1 by the two MPER-directed, broadly reactive neutralizing antibodies, 4E10 and 2F5. The antibodies bind weakly and reversibly to AT-2 inactivated HIV-1 or SIV virions, in agreement with our previous conclusion that 4E10 and 2F5 do not target the native, prefusion form of the envelope glycoprotein, but rather target only the receptor-triggered, prehairpin intermediate conformation (20, 23). We propose the two-step mechanism outlined in Fig. 4. In step 1, the antibodies attach to the viral membrane through their long, hydrophobic CDR H3 loops. Before engagement of gp120 with CD4 and coreceptor, gp41 maintains a native, prefusion conformation, in which the MPER is either poorly exposed or inappropriately configured for recognition by 4E10 and 2F5 (23), and the membrane-bound antibody, concentrated on the virion surface, remains in rapid, reversible equilibrium with antibody in solution (Fig. 1). Once triggered, gp41 undergoes a cascade of conformational changes that lead to the prehairpin intermediate, with the fusion peptide inserted into the target-cell membrane and the transmembrane segment anchored in the viral membrane. This conformation is the target of T-20/enfuvirtide; the estimated exposure time for the T-20 binding site is about 15 min (33). Thus, 4E10 and 2F5 have a relatively narrow window of opportunity to capture their MPER target. Preconcentration of the antibody on the virion surface accelerates this capture. Once docked tightly and effectively irreversibly onto the gp41 epitope (Fig. 2), the antibody can inhibit the further structural rearrangements of gp41 required for membrane fusion. The slow dissociation rate of the antibody-gp41 complex may be critical, as dissociation could allow gp41 to proceed toward fusion. The higher dissociation rate (with respect to wild-type) of the gp41-inter complex with 4E10-mut4 scFv may account for its 5-fold lower neutralization potency (Fig. 2).
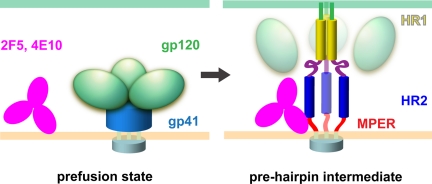
Two-step model for neutralization of HIV-1 by MPER-directed antibodies. HIV envelope glycoprotein undergoes a large structural transition (from the prefusion state to the prehairpin intermediate conformation) after sequential binding of receptor, CD4, and coreceptor, CXCR4 or CCR5, to gp120. Dissociation of gp120 and insertion of the fusion peptide of gp41 into the target cell membrane lead to formation of a prehairpin intermediate. Antibody is shown in pink; gp120, in green; and the untriggered gp41 in blue. After conformational changes, the N-terminal fusion peptide of gp41 is shown in green; heptad repeat 1 (HR1) in yellow; the C-C loop in purple; heptad repeat 2 (HR2) in dark blue; MPER in red and the transmembrane segment in cyan. Step1: MPER-directed antibody (e.g., 4E10 or 2F5) attaches to the viral membrane through its long, hydrophobic CDR H3 loop. The MPER is either concealed or configured inappropriately for antibody recognition until the virus encounters CD4 and coreceptor. Step 2: Once triggered by association of gp120 with CD4 and coreceptor, gp41 undergoes a cascade of conformational changes, leading to the prehairpin intermediate. Antibodies such as 4E10 or 2F5 have only a brief interval within which their MPER target is exposed. Once the antibody has docked onto the gp41 epitope, it can prevent the further structural rearrangements of gp41 that are required for membrane fusion.
Our results suggest that HIV-1 membrane is an important participant in binding and neutralization by 4E10 and 2F5. It is therefore reasonable to assume that a lipid component may be required for an immunogen to mimic all of the structural components required for neutralization and to induce 4E10- and 2F5-like antibody responses. The data presented here bring us closer to a complete molecular picture of neutralization by MPER-directed neutralizing antibodies and will guide experiments designed to elicit them.
Materials and Methods
Expression Constructs and Protein Production.
Details of the expression constructs and protein production of 4E10 scFv, recombinant 2F5 IgG and their mutants are described in SI Text.
SPR for Soluble Proteins.
Unless otherwise specified, all experiments were performed in duplicate with a Biacore 3000 or T-100 instrument (Biacore) at 25 °C in HBS-EP running buffer [10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% (vol/vol) Surfactant P20]. Immobilization of ligands to CM5 and SA chips (Biacore) followed standard procedures recommended by the manufacturer. The final immobilization levels were 350–400 RU for gp41-inter and 100 RU for 4E10 epitope peptide to avoid rebinding events (23). For kinetic measurements, sensorgrams were obtained by passing various concentrations of an analyte over the ligand surface at a flow rate of 50 μL/min, with a 2-min association phase and a 15-min dissociation phase. When possible, the sensor surface was regenerated between each experiment with two 10 s injections of 50 mM NaOH and 2M NaCl, or 10 mM HCl, at a flow rate of 100 μL/min. Repeated efforts failed to identify suitable regeneration conditions for interactions between gp41-inter and all 4E10 scFv proteins except 4E10-mut4 scFv, presumably due to the extremely tight binding. In these cases, two concentrations of the analyte were passed over two separate surfaces with the same immobilization level of the ligand without need for regeneration. Identical injections over blank surfaces were subtracted from the data for kinetic analysis. Binding kinetics was evaluated with BIAevaluation software (Biacore).
Viral Liposomes and AT-2 (Aldrithiol-2) Inactivated Virion Preparations.
Liposomes that mimic the lipid composition of HIV-1 viral membrane (24) were made from phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), brain sphingomyelin (SM), and cholesterol (CH) at a molar ratio of 9.35:19.25:8.25:18.15:45.00 following a procedure described previously (11). AT-2 inactivated virion preparations, HIV-1 ADA-M (Lot P4107) and SIVmac239 (Lot P4149), were kindly provided by Dr. Jeff Lifson at the Biological Products Core of the AIDS and Cancer Virus Program, SAIC-Frederick, Inc., National Cancer Institute. These preparations were purified by sucrose gradient banding, but often contain substantial amounts of contaminating microvesicles derived from nonvirion membranes (34, 35). The matching microvesicles were also purified from the uninfected culture of the same batch of cells as a control (36). Binding properties of 2F5, 4E10 and other control antibodies to the microvesicles were found to be identical to those with either HIV or SIV, in agreement with our inference that the interactions observed by SPR reflected the interactions of the antibodies with membrane lipids, and not with other components in the virion preparations.
SPR for Liposomes and Virions.
All measurements using liposomes or virions by SPR were carried out on a Biacore 3000 instrument, and data analyses were performed using the BIAevaluation 4.1 software (Biacore). As described previously (11), liposomes and virions were captured on a Biacore L1 sensor chip, which uses an alkyl linker for capturing liposomes/virions through interactions with lipids. The frozen stocks of Aldrithiol (AT-2) inactivated virions (1,000-fold concentrated culture supernatants derived from HIV-1 ADA or SIV mac239 isolate) were thawed on ice, and then diluted in PBS buffer (pH 7.4). The samples were vortexed and briefly sonicated (three cycles of 15 s sonication at 70 W power output with a 30 s resting pulse between cycles) in a bath sonicator (Misonix Sonicator 3000) to disperse aggregates. Mild vortexing and sonication have previously been shown to not disrupt HIV-1 mAb, sCD4 or lipid mAb binding to epitopes on liposomes or viral membranes (11). Before capturing liposomes or virions, the L1 chip was preconditioned by immobilizing approximately 2000 RU of BSA in each of the four flow cells using amine coupling chemistry to minimize the nonspecific binding of analyte antibody to the blank L1 chip surface. The surface of the L1 chip was further cleaned with a 60 s injection of 40 mM octyl β-D-glucopyranoside at a flow rate of 100 μL/min followed by washes with buffer (PBS, pH 7.4) to remove any traces of detergent, immediately before the injection of virions. Liposomes or virions were immobilized at a previously optimized level of approximately 500 RU. In general, injection of a solution with 5-fold dilution from the frozen virion stocks at a flow rate of 5 μL/min was found to be optimal to achieve an immobilization level of approximately 500 RU despite variations in injection volume among different batches of viruses. Each scFv or mAb at a concentration of 200 μg/mL was then passed over the surface for 2 min at a flow rate of 20 μL/min, followed by a 10 min dissociation phase. The chip was then regenerated by injection of 40 mM octyl β-D-glucopyranoside at a flow rate of 100 μL/min, followed by a 5 s injection of 25 mM NaOH at a flow rate of 50 μL/min. This procedure was repeated until the response level was reduced to that before the injection of liposomes or virions in each flow cell. In the titration experiments performed to measure kinetic parameters, each cycle involved injection of antibodies at different concentrations at a flow rate of 50 μL/min. Using BIAevaluation 4.1 software, the low levels of nonspecific binding of analytes to appropriate controls (an empty flow cell or the level of nonspecific binding of irrelevant antibody) were subtracted to obtain the specific binding and were used to analyze binding of mAbs.
Neutralization Assay and Neutralization-Blocking Assay.
Fifty percent neutralization titers (IC50) were determined by monitoring reductions in luciferase (Luc) reporter-gene expression after a single round of infection by pseudotyped HIV-1 viruses in TZM-bl cells, as described previously (32). The pseudotyped viruses containing Envs derived from HIV-1 isolates, BG1168, and SF162, were preincubated with serial 3-fold dilutions of each of the 4E10 scFv preparations or 4E10 IgG (Polymun) with a starting concentration of 0.5 mg/mL in 96-well plates for 1 h at 37 °C. After mixing with TZM-bl cells and incubating for additional 48 h, IC50 was calculated from the serial luciferase activities recorded by measurements of luminescence using Bright Glo substrate solution as recommended by manufacture (Promega). To analyze the effect of addition of gp41-inter on neutralization by 4E10, serial 3-fold dilutions of 4E10 IgG with a starting concentration of 10 μg/mL were plated and incubated with the BG1168 pseudovirus for 1 h at 37 °C. TZM-bl cells, premixed with either HIV-1 or SIV gp41-inter protein at both 6.25 and 12.5 μg/mL, were then added to the plate. IC50 values were determined, as described above.
Acknowledgments.
We thank the Biological Products Core of the AIDS and Cancer Virus Program, SAIC-Frederick, Inc., National Cancer Institute, for providing chemically inactivated virions. We also acknowledge the technical assistance of Kara Anasti in performing SPR binding assays of 4E10 scFv binding to viral liposomes. This work was supported by a Collaboration for AIDS Vaccine Discovery grant (to B.F.H.) from the Bill and Melinda Gates Foundation and National Institutes of Health Grant R21 AI069972 (to B.C.). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908713106/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0908713106
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2787149?pdf=render
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/106/48/20234
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/106/48/20234.pdf
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/106/48/20234
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0908713106
Article citations
Decoupling HIV-1 antiretroviral drug inhibition from plasma antibody activity to evaluate broadly neutralizing antibody therapeutics and vaccines.
Cell Rep Med, 5(9):101702, 30 Aug 2024
Cited by: 1 article | PMID: 39216479 | PMCID: PMC11524982
Determination of Binding Affinity of Antibodies to HIV-1 Recombinant Envelope Glycoproteins, Pseudoviruses, Infectious Molecular Clones, and Cell-Expressed Trimeric gp160 Using Microscale Thermophoresis.
Cells, 13(1):33, 22 Dec 2023
Cited by: 0 articles | PMID: 38201237 | PMCID: PMC10778174
Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14(1):7218, 08 Nov 2023
Cited by: 0 articles | PMID: 37940661 | PMCID: PMC10632514
Progress with induction of HIV broadly neutralizing antibodies in the Duke Consortia for HIV/AIDS Vaccine Development.
Curr Opin HIV AIDS, 18(6):300-308, 25 Sep 2023
Cited by: 1 article | PMID: 37751363 | PMCID: PMC10552807
Review Free full text in Europe PMC
Parameter estimation and identifiability analysis for a bivalent analyte model of monoclonal antibody-antigen binding.
Anal Biochem, 679:115263, 06 Aug 2023
Cited by: 2 articles | PMID: 37549723 | PMCID: PMC10511885
Go to all (192) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
N-terminal residues of an HIV-1 gp41 membrane-proximal external region antigen influence broadly neutralizing 2F5-like antibodies.
Virol Sin, 30(6):449-456, 21 Dec 2015
Cited by: 4 articles | PMID: 26715302 | PMCID: PMC8200909
Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41.
J Virol, 88(2):1249-1258, 13 Nov 2013
Cited by: 78 articles | PMID: 24227838 | PMCID: PMC3911647
An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10.
J Virol, 81(8):4033-4043, 07 Feb 2007
Cited by: 123 articles | PMID: 17287272 | PMCID: PMC1866125
Antigp41 membrane proximal external region antibodies and the art of using the membrane for neutralization.
Curr Opin HIV AIDS, 12(3):250-256, 01 May 2017
Cited by: 17 articles | PMID: 28422789
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIAID NIH HHS (1)
Grant ID: R21 AI069972





