Abstract
Free full text

Therapeutic RNA Manipulation in Liver Disease
Summary
Posttranscriptional regulation of gene expression is increasingly recognized as a model for inherited and acquired disease. Recent work has expanded understanding of the range of mechanisms that regulate several of these distinct steps including mRNA splicing, trafficking, and/or stability. Each of these pathways is implicated in disease pathogenesis and each represent important avenues for therapeutic intervention. This review will summarize important mechanisms controlling mRNA processing and the regulation of mRNA degradation, including the role of miRNAs and RNA binding proteins. These pathways provide important opportunities for therapeutic targeting directed at splicing and degradation in order to attenuate genetic defects in RNA metabolism. We will highlight developments in vector development and validation for therapeutic manipulation of mRNA expression with a focus on potential applications in metabolic and immune-mediated liver disease.
RNA synthesis, processing and post-transcriptional regulation
Messenger RNA synthesis is accompanied by co-transcriptional addition of two major stability elements. These include 7-methyl guanosine (m7G) capping at the 5’ end of the mRNA transcript, which prevents 5’ exonuclease digestion and addition of a variable length poly-A tail to the 3’ end of the mRNA. Splicing of intron-containing transcripts occurs in the nucleus and is mediated by spliceosomal apparati composed of distinctive small nuclear ribonucleoprotein complexes (snRNPs) (1) and amplifies the number of translation products generated from a single genomically templated transcript. Enhancers and repressors of splicing exert additional combinatorial control over other post-transcriptional modifications including RNA editing (1, 2). Following or simultaneous with 5’ and 3’ end modifications and splicing of the nuclear transcript, mRNAs are bound to a complex of nuclear export factors (NXFs) to form a ribonucleoprotein complex (mRNP). The mRNP complex interacts with the nuclear pore complex (NPC) for translocation to the cytoplasm (3). The m7G cap is bound by elongation initiation factor 4E (eIF4E) and other cytoplasmic eIF4 proteins, which then engage via eIF3 directly to ribosomes to facilitate mRNA translational initiation (4). Similarly, poly-A binding protein (PABP) also accelerates mRNA translation via activation of the helicase activity of the eIF4 complex (5).
Mechanisms and pathways for RNA degradation
Eukaryotic mRNA half-lives range from minutes to months, the predominant mechanism limiting mRNA longevity being transcript deadenylation. mRNA transcripts earmarked for destruction undergo initial removal of the 5’ m7G cap by “decapping” enzymes (6) or alternatively the 3’ poly-A tail may be targeted by deadenylases (7). Poly-A tails are deadenylated at differing rates, mRNAs containing long poly-A tails (> 55 adenosines) being generally more efficiently translated (8). During cellular stress (such as glucose deprivation) (9, 10), or when the mRNA decay machinery is overloaded, mRNA transcripts may be shuttled to cytoplasmic processing bodies (P-bodies) where they are sequestered for degradation or later translation, providing an additional layer of regulation of gene expression and a defense against translating aberrant proteins (11).
In addition to the 5’ cap and 3’ poly-A tail, key mRNA stability elements are found within the 3’ untranslated region (UTR). The 3’ UTR of transcripts manifesting rapid turnover or tight regulation commonly contain AU-rich elements (AREs). Most AREs contain the pentanucleotide AUUUA consensus sequence and are classified as class I or II, reflecting the number and context of pentanucleotides, although non-AUUUA AU-rich elements may also direct mRNA decay (12). ARE domains are targeted by ARE-binding proteins including Hu antigen R (HuR, ELAVL1), tristetraprolin (TTP, ZFP36), apolipoprotein B mRNA editing enzyme, catalytic polypolypeptide 1 (APOBEC1) that modulate mRNA stability (13, 14). CUG triplet repeat, RNA binding protein 1 (CUGBP1) is an ARE-binding protein that modulates mRNA splicing as well as stability and directly recruits the deadenylase poly(A)-specific ribonuclease (PARN), linking AU-rich elements and deadenylation (15). ARE-binding protein mediated mRNA stabilization is incompletely understood, but likely involves competition for binding sites with destabilizing ARE-binding proteins or stabilizing interactions between the poly-A tail and PABP (11). Modulation of ARE-binding protein activity occurs through combinatorial interactions with other signaling pathways including p38 mitogen activated protein kinase (MAPK) and Wnt/β-catenin (16). For example, activation of p38 MAP kinase results in phosphorylation of the ARE-binding protein KH-type splicing regulatory protein (KSRP) in developing myocytes, decreasing the affinity of KSRP for ARE containing mRNAs and thus favoring transcript stabilization (17).
Stability elements within mRNA transcripts may also include microRNA (miRNA)-binding sites (Figure 1A). MicroRNAs are small single stranded non-coding RNAs that are processed and incorporated into the RNA-induced silencing complex (RISC). MicroRNAs are transcribed as mono or polycistronic transcripts to form pri-microRNAs, which are cleaved in the nucleus by the RNAse III endonuclease Drosha to 60–90 nucleotide pre-microRNAs and exported from the nucleus by the karyopherin Exportin 5. Cytoplasmic endonucloeolytic processing by Dicer yields 20–22 nucleotide double-stranded microRNA fragments that bind complementary target mRNAs leading variably (discussed below) to translational suppression, trafficking to P-bodies or degradation (18).
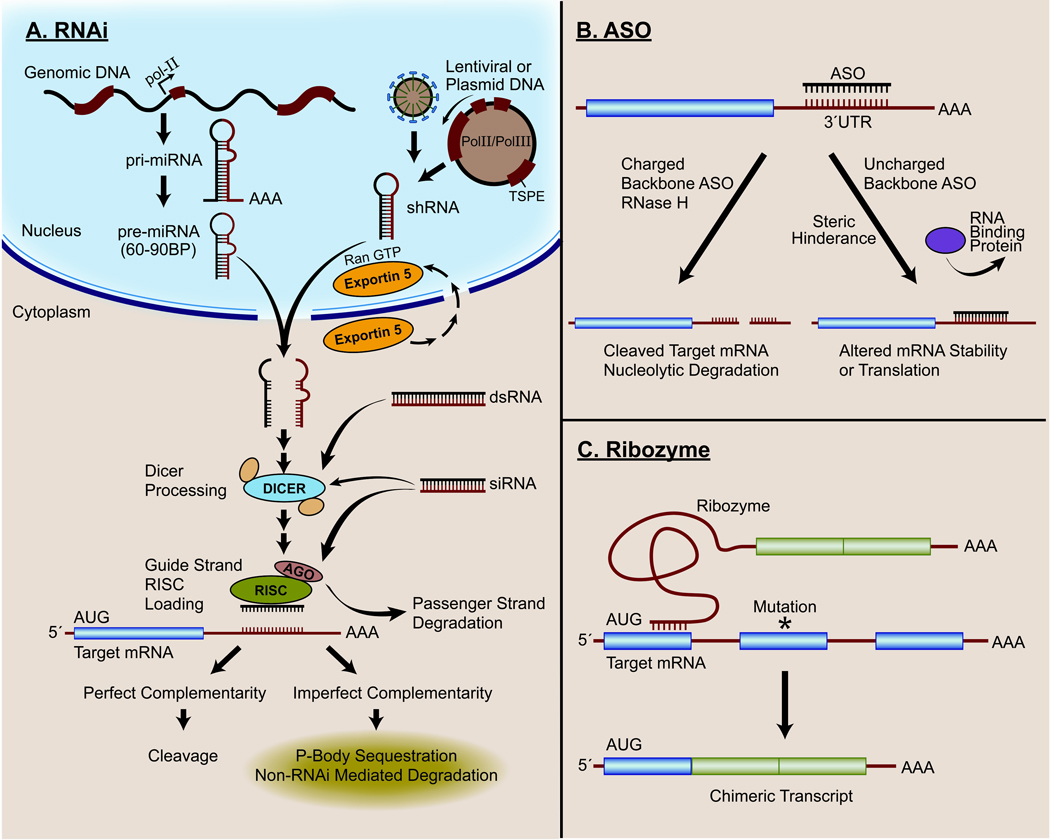
(A) RNA Interference (RNAi). Cellular RNAi machinery may be coopted by a variety of mRNA targeting strategies. Genomically templated pri-miRNA is transcribed in an RNA Polymerase-II (Pol II) dependent fashion and cleaved in the nucleus to pre-miRNA and transported via Exportin 5 to the cytoplasm. Exportin 5 shuttles back to the nucleus, while the pre-miRNA undergoes further processing in the cytoplasm by Dicer. The guide strand is incorporated into the RNA-induced silencing complex (RISC) that includes proteins such as Argonaute (AGO) involved in targeting mRNA and leading to passenger strand degradation. Lentiviral or plasmid encoded short hairpin RNA (shRNA) transcripts may be generated in the nucleus using either Pol II or Pol III driven by a tissue specific promoter/enhancer (TSPE) and are similarly processed to activate RNAi. Exogenously delivered cytoplasmic dsRNA or siRNA may be processed by Dicer and incorporated into the RISC complex and generally require less processing than miRNA or shRNA. Perfect or near perfect complementarity between the guide sequence and target mRNA typically leads to target cleavage. Imperfect complementarity typically results in trafficking of the mRNA to cytoplasmic P-bodies where it is degraded or sequestered for later translation. (B). Antisense oligodeoxynucleotides (ASOs). ASOs may base pair with the target mRNA transcript at almost any location, including the 3’ untranslated region (UTR) which is shown but may also be used to target the 5’ region of the target gene and the initiation AUG codon. Depending on the target location and chemical modification of the ASO, the target mRNA may be cleaved in an RNase H dependent fashion. Alternatively, the ASO may result in steric hindrance of RNA-binding proteins, preventing translation, modifying splicing, or altering mRNA stability. (C) Ribozyme mediated trans-splicing. The guide sequence of the targeting transcript base pairs with the target mRNA upstream of the non-functional or pathologic sequence or mutational site (*). The ribozyme domain of the targeting construct catalyzes splicing of the target transcript to the replacement exons, generating a functional, non-pathogenic mRNA sequence.
Imperfect complementarity between the miRNA and target sequence may result in translational repression. This may occur via alterations in m7G cap-dependent recruitment of the 80S ribosome to mRNA (“cap-dependent repression”) and interference with post-initiation translation (“cap-independent repression”) (reviewed in (19)) or alternatively, targeted transcripts may be trafficked to P-bodies for sequestration (20). In contrast, perfect or near-perfect complementarity generally results in transcript destabilization, which may occur either by direct endonucleolytic cleavage within the target sequence (21) or by accelerated deadenylation, 5’ decapping and 5’ exonuclease activity (19). There is still uncertainty however regarding the fate of the target transcript and the role of target-miR complementarity (reviewed in (22)). For example, near-perfect miRNA-target mRNA interactions may lead to translational repression and not cleavage (23). miRNA-mediated translational repression is also reversible as evidenced by the cationic amino acid transporter-1 (CAT-1) which is repressed in human hepatoma cells by miR-122, and reversed by stress (amino acid deprivation or induction of the unfolded protein response). This response is mediated by competition with HuR for binding sites within the 3’-UTR shared with miR-122. These results suggest that miRNA binding and ARE-binding proteins may interact to modulate overall mRNA stability (24).
Mechanisms of Therapeutic Post-Transcriptional RNA Regulation
Antisense oligonucleotides (ASOs)
ASOs were discovered 30 years ago (25, 26) as single-stranded DNA molecules (13–25 nucleotides) that complement target RNA via sequence-specific base pairing. ASOs with negatively charged backbones recruit RNase H to the temporary duplex causing endonucleolytic mRNA cleavage (27) (Figure 1B). Alternatively, ASOs targeting the 3’-UTR, 5’ region or AUG initiation codon and those with uncharged backbones may suppress mRNA expression by steric mechanisms, leading to decreased or aberrant splicing, translation, or mRNA transport (28, 29). Although ASO-mediated mRNA targeting is susceptible to sequence-specific off-target effects as well as non-specific effects resulting from enzymatic processing (30), the major limitiations are in ASO delivery and short half-life. Chemically modified derivatives, including phosphorothioates and peptide linked morpholino derivatives, are currently in development with targets including bcl-2 and protein kinase C-α (PKC-α, PRKCA) (31, 32). To date, fomivirsen sodium (Vitravene, Isis, Carlsbad, Ca), a phosphorothioate derivative targeting cytomegalovirus mRNA in CMV retinitis, is the only FDA approved ASO therapy.
Targeting of mRNA expression through ASO mediated manipulation of mRNA splicing holds promise for treatment of diseases caused by a variety of hereditary mutations and may offer opportunities for therapeutic targeting in other settings. Morpholino-modified ASOs targeting mutant intronic splice sites in β-globin pre-mRNA from thalassemic patients (33) corrected the splicing defects and restored hemoglobin synthesis. ASOs could thus theoretically be used to target disease-causing splicing mutations, such as in Wilsons disease (34), familial intrahepatic cholestasis, hepatocellular carcinoma (35), or alternative splice variants of hepatitis B (36) and C. Therapeutic manipulation of Apo-B mRNA splicing offers an experimentally testable approach to reduce hepatic Apo B production (37, 38). In this situation, ASOs targeted to the physiological 5’ and 3’ splice sites within exon 27 of APOB induce exon skipping leading to a frame shift within exon 28 leading to production of a truncated protein that is less efficient at exporting lipid (discussed further below). Mutations in the dystropin gene cause Duchenne muscular dystrophy (DMD) as a result of a prematurely truncated and unstable transcript. Less severe forms of DMD result from mutations that maintain the reading frame by skipping the defective exon resulting in a partially functional protein. ASO-mediated rescue produced an mRNA transcript lacking exon 23 with validated cellular localization of the dystrophin gene product in mice with a mutant exon 23 that caused premature termination (39). Accordingly modification of mRNA splicing may provide a strategy to increase the abundance and stability of an otherwise rare or unstable transcript.
Ribozymes
Catalytic RNAs (ribozymes) act in a sequence-specific fashion in cis or trans to cleave mRNA in the absence of proteins (reviewed in (28)) and may direct trans-splicing reactions. Ribozyme-mediated trans-splicing therapeutic constructs contain a targeting domain, the ribozyme sequence and a replacement coding region (Figure 1C). Base pairing of the guide sequence with the target transcript results in a trans-splice reaction and a chimeric transcript (29) in which the disease causing mRNA segment is replaced by a functional coding exon. A shortened version of the Tetrahymena thermophila self-splicing intron coupled to a portion of the γ-globin transcript was used to target the mutant βs-globin mRNA transcript just upstream of the sickle-cell mutation. The ribozyme-directed cleavage and ligation generated a chimeric transcript encoding a non-sickling but functional globin protein (40). Non-ribozyme mediated trans-splicing reactions utilizing the cellular spliceosomal apparatus are also under development (29).
RNAi
RNAi encompasses two gene-silencing processes, siRNA and miRNA (Figure 1A). miRNAs are endogenously transcribed single stranded RNAs that form hairpin structures, which are processed via a series of nucleolytic cleavages to form mature miRNAs. In contrast, siRNAs are endogenously or exogenously derived double stranded non-coding RNAs that are also processed to their mature 21–23 nucleotide form. In both cases, the final “guide” strand is incorporated into the RNA-induced silencing complex (RISC) directing endonucleolytic cleavage or translational repression of target RNAs (28, 41). Both mechanisms may be co-opted for therapeutic purposes and offer advantages over other sequence-specific targeting strategies. These include low effective concentration, limiting toxicity and flexibility in targeting sites within a given mRNA. RNAi guide sequences can be delivered as pre-formed double stranded RNA oligonucleotides (siRNA) or generated within cells using promoter-based expression systems (shRNA). Each approach has advantages and disadvantages (42). siRNA is attractive in that the delivered sequence activates RNAi directly with minimal cellular metabolism being required. The downside is that the effect is transient and administration must be repeated. In contrast, shRNA-based delivery involves delivering a DNA template to the target tissue from which an shRNA is transcribed (Figure 1A). This shRNA undergoes Exportin-5 mediated nuclear export and processing by Dicer to yield more durable siRNA production. Saturation of the Exportin-5 pathway, however, by adenoviral mediated shRNA is hepatotoxic (43), an effect not seen with in-vivo delivery of siRNA (44).
Specific targeting of endogenous miRNAs may also hold therapeutic benefit. Administration of a cholesterol-conjugated, 2’-OMe modified RNA sequence (antagomir) targeting miR-122 produced potent suppression of endogenous hepatic miR-122 levels in mice and upregulation of mRNAs enriched in miR-122 3’-UTR target sites (discussed below) (45).
Hepatotropism of RNA targeting
Early work with modified encapsulated ASOs demonstrated effective hepatotropism after intravenous administration (46) with >80% detectable in the liver at one hour and 26% by four hours. Further experiments demonstrated that subcutaneously injected ASO targeting Fas accumulated in the liver of mice for approximately 10 days (47). Naked siRNAs injected systemically concentrate predominantly in the kidney (48), but modifications to the siRNA backbone increase plasma stability and improve hepatic delivery. Synthetic duplex siRNAs containing one phosphodiester strand and one phosphothioate strand were stable up to 72 hours in 50% mouse serum compared with unmodified single stranded RNA which was degraded in seconds (49). siRNA-lipid complexes including cholesterol, cationic lipid or lipidoid nanoparticles increase transfection efficiency (50) and have been used to optimize in-vivo targeting of apolipoprotein B (APOB) and proprotein convertase subtilisin/kexin type 9 (PCSK9) (discussed below) (51–53).
Potential for off-target effects and toxicity
siRNAs activate the immune system (54) and double stranded RNA can activate the intracellular receptors dsRNA-dependent protein kinase R (PKR, EIF2AK2) or retinoic acid inducible gene-I (RIG-I, DDX58). Activation of PKR results in global suppression of protein synthesis, ultimately leading to apoptosis. Activation of either PKR or RIG-I leads to interferon production and other antiviral responses, including Toll-like receptors TLR7 and TLR8 which are activated by siRNA in a sequence and delivery-specific fashion. U- and G- rich siRNAs, longer dsRNA sequences, single stranded siRNAs as well as cationic lipid complexed siRNAs result in the most potent activation of immunostimulatory signaling pathways. Conversely, shorter siRNA sequences, low G/U content, double stranded, naked and cholesterol-complexed siRNAs appear to be least immunostimulatory (54). In-vivo trials of siRNA therapy have utilized chemical modifications such as 2 nucleotide 3’ overhangs and 2’-O-methyl modifications that diminish immunostimulatory effects of siRNAs as evidenced by attenuated production of interferon-α (IFNA2), interleukin-6 (IL6), and tumor necrosis factor-α (TNF) (51, 55).
RNA therapy mediated off-target effects can also occur via sequence homology to unintended transcripts. ASOs targeting protein kinase C-α RNA suppressed protein kinase C-ζ (PKC-ζ, PRKCZ) mRNA even though these isoforms share only 11/20 base pair homology (56). Additionally, while siRNA binding to target mRNA with perfect complementarity results in mRNA cleavage, imperfect complementarity suppresses translation, similar to that discussed above with miRNA (57).
Experience with RNAi therapy using adeno-associated virus (AAV) has revealed potential hepatotoxicity. AAV mediated shRNA expression targeting transgenically expressed human α-1 antitrypsin (AAT) mRNA (43) suppressed h-AAT mRNA, but was complicated by lethal hepatotoxicity, likely due to oversaturation of Exportin-5 as alluded to above (Figure 1A). Accordingly, successful delivery of an RNAi effector must be accompanied by careful modulation of intracellular concentrations in order to avoid toxicity.
Therapeutic Applications of RNA Modulation in Metabolic Liver Disease
Efforts are underway to utilize RNA-targeting therapy in metabolic conditions involving the liver, in particular the management of hypercholesterolemia, where despite the success of statin-based therapies, only 2/3 of patients achieve their target low density lipoprotein (LDL) goal (58). Patients with hereditary mutations in either the LDL receptor (familial hypercholesterolemia, FH) or in the LDL receptor binding region of ApoB100 fail to clear ApoB100-containing lipoproteins from the serum, leading to accumulation of lipid particles and hypercholesterolemia. ASO targeting of ApoB-100 has been used in proof-of-principle experiments in ApoE-deficient and LDLR-deficient mice, (59), where cholesterol reduction ranged from 25–55%, with LDL-C reductions as high as 88% without hepatic steatosis. A double blind, placebo controlled, randomized trial of mipomersen (ApoB ASOs) in 36 volunteers with mild sporadic dyslipidemia demonstrated decreased serum LDL and ApoB levels of 35% and 50% respectively. There were no serious adverse events with the most common side effect being mild injection site erythema (60). More recently, phase 3 studies with mipomersen in FH subjects revealed a 25% reduction of LDL cholesterol levels after 26 weeks of treatment (32, 61). Further studies are directed toward assessing whether this approach places patients at risk for hepatic steatosis as seen in patients with some naturally occurring ApoB mutations or with familial hypobetalipoproteinemia (62).
Targeting the 3’ end of ApoB mRNA with cholesterol-conjugated siRNA in mice lowered serum ApoB levels and decreased serum LDL (52) and stable nucleic acid lipid particles (SNALP)-formulated siRNA targeting the 5’ end of ApoB mRNA (53) in cynomolgus monkeys suppressed ApoB mRNA by 90% at 48 hours after a single dose, an effect that persisted for 11 days, accompanied by >60% reductions in serum cholesterol.
ASO-mediated therapeutic ApoB RNA splicing inferference was used to induce skipping of exon 27 (38) to generate a truncated ApoB isoform predicted to decrease LDL-cholesterol. The “skip 27” mRNA translated product (ApoB87) was exported from the cells at decreased efficiency compared to ApoB100, an effect reminiscent of that associated with familial hypobetalipoproteinemia (FHBL) patients expressing an APOB truncating mutation (63). Differential processing of the truncated ApoB mRNA and alterated posttranslational lipidation of the protein product might account for the therapeutic efficacy of this targeting strategy.
A second target of hypolipidemic therapy is proprotein convertase subtilisin/kexin type 9 (PCSK9), which regulates hepatic LDL-receptor expression via altered protein processing (64). Gain-of-function mutations result in increased degradation of the LDL-receptor and increased lipid levels. Conversely, loss-of-function mutations result in increased expression of LDL-receptors and decreased serum cholesterol (65). Epidemiologic studies link mutations in PCSK9 to low LDL cholesterol levels in approximately 2% of African Americans (66), with an approximately 40% reduction in LDL-cholesterol and reduction in coronary artery disease (64), making PCSK9 an attractive hypolipidemic target. In-vivo targeting of PCSK9 using ASOs in hyperlipidemic mice fed a high fat diet increased hepatic LDL receptor expression and decreased serum cholesterol (67). siRNA targeting of PCSK9 in rodents and non-human primates using intravenous lipidoid nanoparticles also decreased serum cholesterol (51).
ASOs have also been developed (antagomirs) to target miRNAs involved in hepatic lipid metabolism, including miR-122. In-vivo silencing of miR-122 in mice using a miR-122 antagomir, decreased 3-hydroxy-3-methylglutaryl-Coenzyme A reductase (HMG-CoA reductase, HMGCR) and acyl-Co A synthetase 2 (ACSS1) among other targets, accompanied by a >40% decrease in plasma cholesterol and without adverse effects on hepatic function or elevated transaminase levels (45). Analysis of miRNA expression patterns in patients with non-alcoholic steatohepatitis (NASH) revealed reduced expression of miR-122 with corresponding alterations in several target genes that were replicated in HepG2 cells following either miR-122 knockdown or overexpression (68). These findings and the implications for NASH pathogenesis need to be interpreted with caution however, since miR-122 is known to modulate the expression of ~200 target genes (69) and the precise mechanisms by which alterations in miR-122 expression influence hepatic lipid metabolism in human subjects remains to be defined.
Immune-Mediated Liver Disease
Hepatocytes are susceptible to Fas-mediated apoptosis and disruption of this pathway protects hepatocytes from acute liver injury. Mice treated with ASOs targeting Fas (FAS) were protected from Fas antibody-induced fulminant liver failure and from a sub-lethal dose of acetaminophen (47). Similar experiments using RNAi confirmed that interference with Fas signaling protects from fulminant hepatitis (70). Taken together, these data strongly suggest that RNA-directed therapy may have clinical utility in treating disorders of immune-mediated liver injury as well as fulminant liver failure with an immune-mediated component.
Future Directions and Prospects
The last several years have witnessed great advances in understanding of the physiologic and pathologic roles of regulated mRNA stability. Among the challenges moving forward will be to define how aberrant post-transcritional mRNA processing including mRNA trafficking and stability functions in disease pathogenesis and to define optimal mRNA targets for specific clinical conditions. From the standpoint of drug development, refining potency, specificity, minimization of side effects and tissue delivery will be crucial goals in coming years.
Acknowledgements
We apologize to colleagues whose work we were unable to cite due to space limitations. Work cited in this review was supported by grants HL-38180, DK-52574 and DK-52560 (to NOD) and by a Fellow to Faculty transition award from the Foundation for Digestive Health and Nutrition (to TAK).
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1002/hep.23344
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2964871?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/hep.23344
Article citations
The advent of RNA-based therapeutics for metabolic syndrome and associated conditions: a comprehensive review of the literature.
Mol Biol Rep, 51(1):493, 05 Apr 2024
Cited by: 0 articles | PMID: 38580818
Review
Rosa26-LSL-dCas9-VPR: a versatile mouse model for tissue specific and simultaneous activation of multiple genes for drug discovery.
Sci Rep, 12(1):19268, 10 Nov 2022
Cited by: 0 articles | PMID: 36357523 | PMCID: PMC9649745
Rational Design of an Activatable Reporter for Quantitative Imaging of RNA Aberrant Splicing In Vivo.
Mol Ther Methods Clin Dev, 17:904-911, 18 Apr 2020
Cited by: 3 articles | PMID: 32405512 | PMCID: PMC7210378
Application of Locked Nucleic Acid Oligonucleotides for siRNA Preclinical Bioanalytics.
Sci Rep, 9(1):3566, 05 Mar 2019
Cited by: 12 articles | PMID: 30837588 | PMCID: PMC6401054
MicroRNAs and Malaria - A Dynamic Interaction Still Incompletely Understood.
J Neuroinfect Dis, 6(1):165, 01 Mar 2015
Cited by: 5 articles | PMID: 26005686 | PMCID: PMC4441219
Go to all (14) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MicroRNAs challenge the status quo of therapeutic targeting.
J Cardiovasc Transl Res, 2(1):100-107, 09 Sep 2008
Cited by: 5 articles | PMID: 20559973
Review
miR-122--a key factor and therapeutic target in liver disease.
J Hepatol, 62(2):448-457, 13 Oct 2014
Cited by: 341 articles | PMID: 25308172
Review
Role of identified RNA N6-methyladenosine methylation in liver.
Anal Biochem, 578:45-50, 07 May 2019
Cited by: 15 articles | PMID: 31075230
Review
Reprogramming RNA processing: an emerging therapeutic landscape.
Trends Pharmacol Sci, 43(5):437-454, 21 Mar 2022
Cited by: 5 articles | PMID: 35331569
Review
Funding
Funders who supported this work.
NHLBI NIH HHS (4)
Grant ID: R37 HL038180-24
Grant ID: HL-38180
Grant ID: R37 HL038180
Grant ID: R01 HL038180
NIDDK NIH HHS (5)
Grant ID: R01 DK056260-11
Grant ID: DK-52560
Grant ID: DK-52574
Grant ID: R01 DK056260
Grant ID: P30 DK052574




