Abstract
Free full text

Genetic variation within the HLA class III influences T1D susceptibility conferred by high risk HLA haplotypes
Abstract
HLA class II DRB1 and DQB1 represent the major type 1 diabetes (T1D) genetic susceptibility loci; however, other genes in the HLA region are also involved in T1D risk. We analyzed 1411 pedigrees (2865 affected individuals) from the type 1 diabetes genetics consortium (T1DGC) genotyped for HLA classical loci and for 12 SNPs in the class III region previously shown to be associated with T1D in a subset of 886 pedigrees. Using the transmission disequilibrium test, we compared the proportion of SNP alleles transmitted from within the high risk DR3 and DR4 haplotypes to affected offspring. Markers rs4151659 (mapping to CFB) and rs7762619 (mapping 5′ of LTA) were the most strongly associated with T1D on DR3 (p=1.2 × 10−9 and p=2 × 10−12 respectively) and DR4 (p=4 × 10−15 and p= 8 × 10−8 respectively) haplotypes. They remained significantly associated after stratifying individuals in analyses for B*1801, A*0101-B*0801, DPB1*0301, DPB1*0202, DPB1*0401 or DPB1*0402. Rs7762619 and rs4151659 are in strong linkage disequilibrium (LD) (r2=0.82) with each other, but a joint analysis showed that the association for each SNP was not solely due to LD. Our data support a role for more than one locus in the class III region contributing to risk of T1D.
Introduction
Type 1 diabetes (T1D) is a form of diabetes mellitus and an autoimmune disease resulting from the immunological destruction of the insulin producing beta cells of the pancreas, which leads to dysregulation of glucose metabolism. T1D has a strong genetic component and approximately 40–50% of the familial clustering of T1D can be attributed to allelic variation in the HLA region (1). The direct involvement of the HLA class II DRB1, DQA1 and DQB1 genes in T1D is well established, with a hierarchy ranging from very predisposing to very protective effects (see 2–3 and references therein). The most highly predisposing DRB1-DQB1 haplotypes are DRB1*0301 DQA1*0501 DQB1*0201 (DR3 for short) and DRB1*0401/2/4/5 DQA1*0301 DQB1*0302 (DR4 for short) with odds ratios > 3.0 for DR3 and > 8.0 for the most predisposing DRB1*04 subtypes (0401 and 0405) (see 3). However, allelic variation at these loci cannot account fully for the pattern of HLA haplotype sharing among affected sib pairs (4). Results from association analyses of the classical HLA loci (class I and DPB1) have shown that other loci are involved in susceptibility to T1D (4–8), with HLA-B demonstrating the strongest association in class I (5–7). Several authors have also reported consistent and reproducible associations of DPB1 with T1D (e.g. 8). Moreover, other polymorphisms within the HLA region yet outside the classical loci have been found to contribute to T1D susceptibility, such as the gene region telomeric to HLA-A associated with T1D after conditioning for DR-DQ, HLA-A, -B and –C (9).
The human MHC class III region is of particular interest. It has been implicated in risk for T1D and the associations observed are not due to linkage disequilibrium (LD) with the DR-DQ genes (10–13). A recent study identified polymorphisms in the AIF-1 gene, mapping to class III, to be significantly associated with T1D risk after conditioning on DRB1, DQB1, HLA-A and HLA-B variation (12). The human MHC class III region comprises a 700 kb sequence located between the centromeric class II (HLA-DRA) and the telomeric class I regions (MICB), and encodes over 60 genes resulting in one of the most gene-dense regions of the human genome (13). These genes include those involved in the activation cascades of the complement system (C2, factor B [BF], C4), hormonal synthesis (steroid 21-hydroxylase, CYP21), inflammation and cell stress (tumour necrosis factor [TNFα], lymphotoxins α and β, 70 kD heat shock proteins (HSPA1A, HSPA1B, HSPA1L), extracellular matrix organization (Tenascin [TNX] as well as immunoglobulin superfamily (Ig-SF) members (1C7, G6f and G6b). The remainder (the majority) of loci are primarily involved in more ‘core’ biological functions with no immediate implication for the immune system. (14–15).
In a previous study of 886 T1D families (17), we applied an overall conditional genotype method to detect associations between variants within the MHC class III region and disease risk after adjusting for the DRB1-DQB1 effect. The null hypothesis of no association assumed that the relative genotype frequencies at marker loci not involved in disease risk are the same in cases and controls on specific DRB1-DQB1 genotypes. This conditional method was applied to the probands from each of the 886 families and the affected family based controls. We tested 356 SNPs within the MHC class III and identified 13 polymorphisms that showed a significant association with T1D risk after adjusting for multiple testing, and conditioning for the DRB1-DQB1 genotype, in both discovery (n=289 families) and replication datasets (n=597 families). Those families represented the set of type 1 diabetes genetics consortium (T1DGC) Caucasian pedigrees with HLA molecular typing available at that time (17).
We hypothesized that if these SNPs were truly implicated in T1D genetic risk, they would affect the risk of the highly predisposing DRB1-DQB1 haplotypes DR3 and DR4 even after matching for DPB1 or for HLA-A and HLA-B class I alleles. The aim of the present study was to assess whether associations we have previously reported in the class III region (17) were reproducible using a different methodological approach and matching for additional classical HLA loci. To test this hypothesis, we analyzed all the complete families from the T1DGC with classical HLA typing. ‘This was possible as high resolution molecular HLA typing became available during 2008 for many more T1DGC samples. The large sample size of this collection (1411 pedigrees, 2865 affected individuals) makes stratification analysis for extended class I-class II haplotypes possible (changed order of words), allowing, in turn, the investigation of additional genetic loci within the MHC class III region that have been suggested by previous studies (10–13).
Results
Using the informative transmissions for the SNPs previously identified by us (15), we assessed whether these polymorphisms significantly affected the risk of predisposing DR3 and DR4 haplotypes to T1D (Figure 1) using both all samples and the subset of samples not previously tested. The strongest association on DR4 haplotypes was observed for rs4151659 (p =4 × 10−15) whereas the strongest association on DR3 haplotypes was observed for rs7762619 (p =4 × 10−12). We decided to concentrate on the role of these two SNPs. First, we explored whether the association was due only to one or a few cohorts, or if there was heterogeneity between cohorts. No evidence for heterogeneity was detected between cohorts with respect to the association of these SNPs on DR3 and DR4 haplotypes (Figure 2). The p-values from meta-analyses are similar to those obtained by pooling all cohorts: p =4 × 10−15 from pooling vs p<5 × 10−18 from meta-analyses and p =4 × 10−12 from pooling vs p= 7 × 10−11 for DR4-rs4151659 and DR3-rs7762619, respectively. Hence, we concluded that pooling data from all cohorts was a valid way of analyzing the association of these SNPs on DR3 and DR4 haplotypes, adjusting for additional classical HLA loci. Alleles within the class II DPB1 gene and the class I HLA-A and HLA-B loci, however, have also been shown to influence risk to T1D (e.g. 4–8), in addition to DR-DQ. We therefore assessed whether the association for rs4151659 or rs7762619 remained present after matching for DPB1, HLA- A or HLA-B alleles on DR3 and DR4 haplotypes. For HLA-A and -B class I genes, we stratified by the following categories when there were at least 20 informative transmissions: A*2401, B*1801, and B*4001 and the highly conserved A*0101-B*0801 on DR3 haplotypes, and A*0201 B*1501 on DR4 haplotypes. For HLA-DPB1, we stratified by the well-known additional effects of: DPB1*0101, *0301, *0201, *0202, *0401, *0402 when there was sufficient sample size. Results show that the presence of minor alleles for both rs4151659 and rs7762619 significantly affect the risk of extended haplotypes including those carrying alleles previously shown to be associated with T1D (Figure 3) (4–8). The increased risk due to the presence of the minor allele of rs4151659 and rs7762619 was not statistically significant on every extended haplotype tested due to the limited number of informative transmissions for some of them, but in every instance the trend was the same. We also included the extended DPB1*0101 DR3 A*0101-B*0801 haplotype. Although the number of transmissions is extremely small (n=10), the association between T1D and both markers was statistically significant (Figure 3). We conclude that the effect of rs4151659 and rs7762619 is not due to these alleles marking extended haplotypes which contain specific alleles at other classical HLA loci. For haplotypes where a large number of informative transmissions were available, e.g. DR4 DPB1*0401, the observed p-values are in the 10−9 range. For less common haplotypes, the p-values are higher, but the same effect is seen consistently, i.e. the minor allele at these two markers is associated with higher risk of T1D among DR3 and DR4 haplotypes regardless of the DPB1 or class I alleles carried.
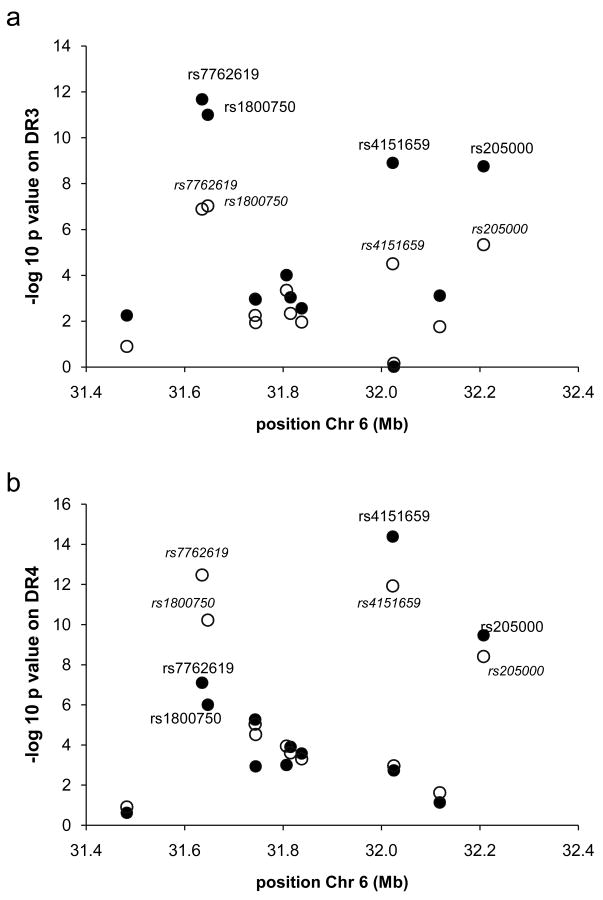
Genetic association between 12 MHC class II SNPs and T1D among (A) DR3 haplotypes (B) DR4 haplotypes. Empty circles are p-values only for samples not included in a previous study (15). Filled circles include all samples. DR3 = DRB1*0301 DQA1*0501 DQB1*0201 DR4= DRB1*0401/2/4/5 DQA1*0301 DQB1*0302, all other DR4 subtypes are excluded.
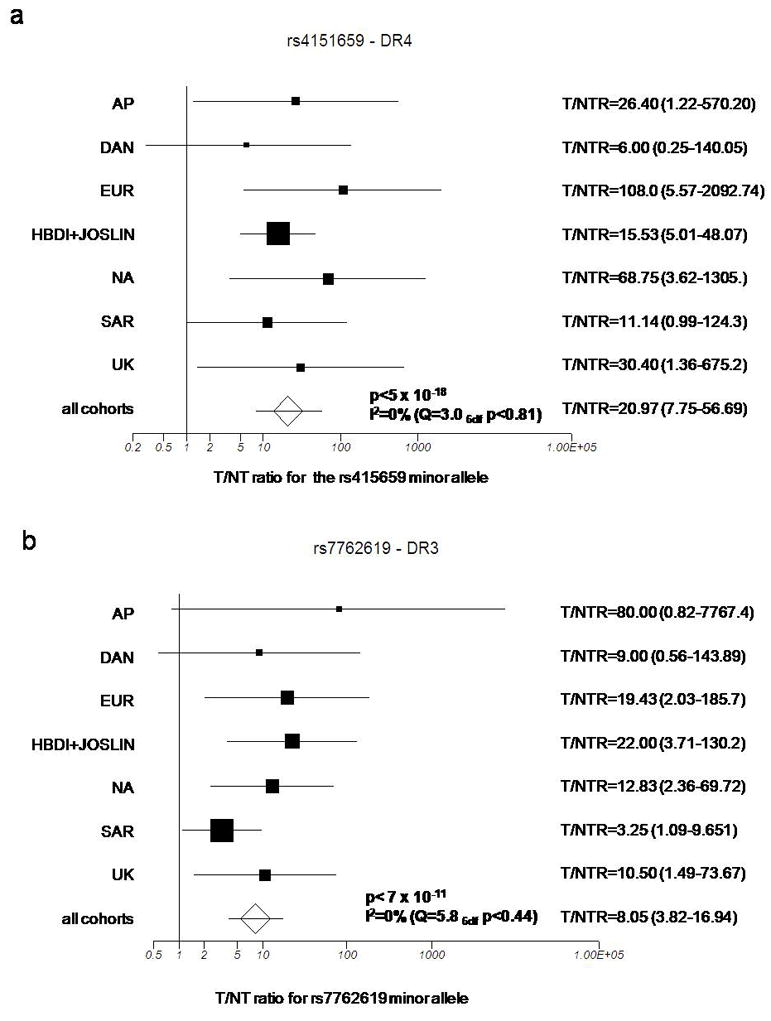
Forest plot of study-specific estimates and fixed effects summary transmitted/non-transmitted ratio (T/NTR) and 95% confidence interval (CI) for the association between the minor allele at (A) rs4151659 on DR4 haplotypes and (B) rs7762619 on DR3 haplotypes in the individual cohorts from the T1DGC collection.
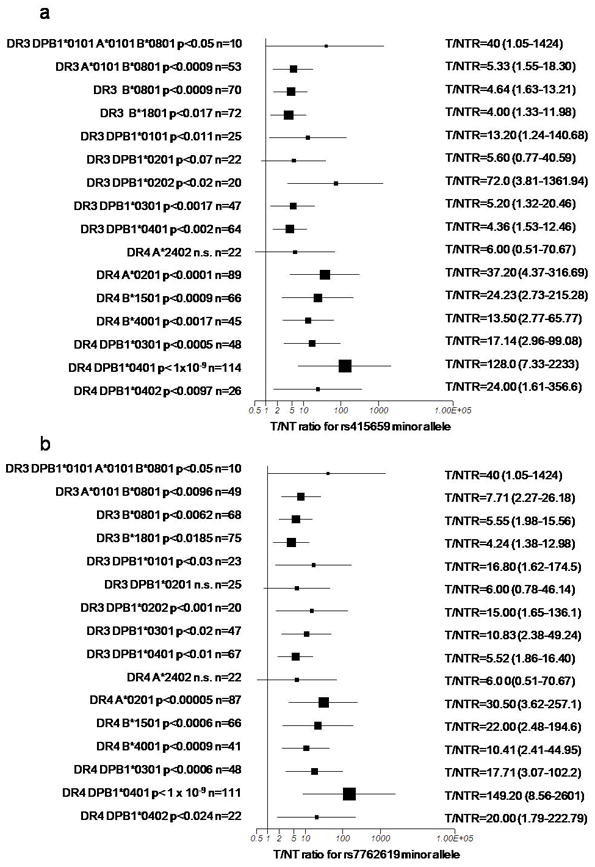
Forest plot showing the effect of (A) rs4151659 on extended DR3 and DR4 haplotypes and (B) rs7762619 on extended DR3 and DR4 haplotypes. The p-value and number of informative transmissions for each comparison are shown.
Rs4151659 and rs7762619 map approximately 400 kilobases apart from each other (Table 2). In spite of this, both SNPs are in high LD, with r2=0.8071 on DR3 haplotypes and r2=0.8397 on DR4 haplotypes. Thus, both SNPs could be marking the same association in spite of the large physical distance between them. To test if this was the case, we carried out a stratified analysis equivalent to that performed above on HLA-B, HLA-A and DPB1. We found that among DR3 haplotypes with the major, low risk T allele at rs7762619, carrying the minor G allele at rs4151659 resulted in significantly higher risk (p<0.004; Table 3). A similar trend that did not reach statistical significance was seen on DR4 (p<0.06). For both DR3 and DR4 haplotypes that carry the major low risk A allele at rs4151659, the presence of the minor allele G at rs7762619 resulted in significantly higher risk of DR4 (p<0.006) and DR3 haplotypes (p<0.02) (Table 3). These data indicate that there are most likely two distinct, albeit correlated, genetic effects on T1D susceptibility marked by rs4151659 and rs7762619.
Table 2
Name and characteristics of MHC region SNPs studied in T1DGC families
| SNP ID | Position on chr 6* | Nearest gene(s) | position in gene | Minor/Major allele | Trans% of minor/major in DR3 haplotypes | Trans % of minor/major in DR4 haplotypes | MAF in present study | In LD (r2 ≥ 0.5) with |
|---|---|---|---|---|---|---|---|---|
| rs1063632 | 31,486,489 | MICA 3′ | Arg87Arg | A/G | 73.5/58.7%(313) | 86.3/78.7%(297) | 10.90% | N/A |
| rs7762619 | 31,639,289 | NFKBIL1, 5′LTA | N/A | G/T | 79.7/31.3%(211) | 95.9/48.4%(288) | 6.10% | rs1800750, rs205000, rs4151664, rs4151659 |
| rs1800750 | 31,647,267 | TNF | 5′ near gene | A/G | 79.1/33.3%(219) | 94.5/53.1%(297) | 6.20% | rs7762619, rs205000, rs4151664, rs4151659 |
| rs2142234 | 31,747,108 | LY6G5B | intron | T/C | 70.1/55.9%(473) | 88.0/68.3%(577) | 13.00% | rs9267532, rs400547, rs707915, rs707936 |
| rs9267532 | 31,744,256 | LY6G5B | Cys176Arg | A/G | 69.8/55.3%(501) | 87.5/70.7%(603) | 12.00% | rs2142234, rs400547, rs707915, rs707936 |
| rs400547 | 31,806,987 | CLIC1 | intron | A/G | 71.9/54.1%(447) | 87.1/69.1%(533) | 12.60% | rs2142234, rs9267532, rs707915, rs707936 |
| rs707915 | 31,815,244 | MSH5 | intron | T/A | 70.4/55.8%(485) | 86.6/69.4%(570) | 12.10% | rs2142234, rs9267532 rs400547, rs707936 |
| rs707936 | 31,837,927 | C6orf27 | intron | T/C | 69.9/56.7%(481) | 86.1/69.7%(568) | 12.10% | rs2142234, rs9267532 |
| rs4151659 | 32,026,443 | CFB | Lys565Glu | G/A | 79.3/38.0%(214) | 96.3/48.4%(281) | 6.40% | rs400547, rs707915, rs1800750, rs205000, rs4151664, rs7762619 |
| rs4151664 | 32,028,852 | RDBP | intron | A/G | 66.8/67.1%(351) | 88.4/68.7%(388) | 8.60% | rs1800750, rs205000, rs4151659, rs7762619 |
| rs17421133 | 32,118,251 | TNXB | Ile2Asn | T/A | 61.3/77.9%(354) | 69.2/82.2%(272) | 14.20% | N/A |
| rs205000 | 32,207,323 | EGFL8 | 3′ UTR | G/C | 77.0/40.2%(263) | 93.7/57.8%(322) | 7.10% | rs1800750, rs7762619, rs4151664 rs4151659 |
Table 3
Effect of rs4151659 on association of DR3-rs7762619 and DR4-rs7762619 haplotypes using informative transmissions for rs4151659 and effect of rs7762619 on association of DR3-rs4151659 and DR4-rs4151659 haplotypes using informative transmissions for rs7762619
| DRB1 | DQB1 | rs7762619 | rs4151659 | Not Trans | Trans. | Not Trans | Trans. | Fishers Exact Test |
|---|---|---|---|---|---|---|---|---|
| 0301 | 0201 | G | A | 0 | 1 | 0.00% | 1.04% | |
| 0301 | 0201 | G | G | 23 | 95 | 100.00% | 98.96% | 1 |
| 0301 | 0201 | T | A | 49 | 29 | 94.23% | 70.73% | |
| 0301 | 0201 | T | G | 3 | 12 | 5.77% | 29.27% | 0.0035 |
| 0401/2/4/5 | 0302 | G | A | 0 | 2 | 0.00% | 1.11% | |
| 0401/2/4/5 | 0302 | G | G | 7 | 178 | 100.00% | 98.89% | 1 |
| 0401/2/4/5 | 0302 | T | A | 47 | 42 | 100.00% | 89.36% | |
| 0401/2/4/5 | 0302 | T | G | 0 | 5 | 0.00% | 10.64% | 0.0559 |
| DRB1 | DQB1 | rs4151659 | rs7762619 | Not Trans | Trans. | Not Trans | Trans. | Fisher’s Exact Test |
| 0301 | 0201 | A | G | 1 | 5 | 1.79% | 16.67% | |
| 0301 | 0201 | A | T | 55 | 25 | 98.21% | 83.33% | 0.0182 |
| 0301 | 0201 | G | G | 23 | 95 | 92.00% | 98.96% | |
| 0301 | 0201 | G | T | 2 | 1 | 8.00% | 1.04% | 0.108 |
| 0401/2/4/5 | 0302 | A | G | 0 | 8 | 0.00% | 16.33% | |
| 0401/2/4/5 | 0302 | A | T | 47 | 41 | 100.00% | 83.67% | 0.0058 |
| 0401/2/4/5 | 0302 | G | G | 8 | 178 | 100.00% | 100.00% | |
| 0401/2/4/5 | 0302 | G | T | 0 | 0 | 0.00% | 0.00% | N/A |
Not transmitted = SNP allele frequency among DR3-SNP or DR4-SNP haplotypes not transmitted to affected offspring from heterozygote parents. Transmitted= allele frequency among DR3 or DR4 haplotypes transmitted to affected offspring. % trans= percent times that a given haplotype was transmitted to affected offspring = Transmitted/(Not Transmitted + Transmitted).
We assessed whether the association of these two markers was confined only to the highly predisposing DR3 and DR4 haplotypes or if similar effects would be seen on other DRB1-DQB1 haplotypes. We only tested haplotypes for which 30 or more informative transmissions were available. We found strong significant associations overall (Figure 4) and no evidence for heterogeneity for the effect of rs4151659 (I2=0% Q(3 df)= 0.52 n.s.) and rs7762619 (I2=0% Q(3 df)= 0.52 n.s.) among the four haplotypes studied. These data indicate that the predisposing effect of the minor alleles at both rs4151659 and rs7762619 are not limited to DR3 and DR4 haplotypes but are seen more generally, including on neutral/protective DRB1-DQB1 haplotypes. In addition, the deviation from the null observed (Figure 4) in the presence of these haplotypes is comparable to that seen for DR3 and DR4 (Figure 2). Furthermore, in spite of the low MAF of these SNPs, there was a sufficient number of informative transmissions of SNP alleles on other haplotypes to obtain highly statistically significant results with p<1 × 10−7 (Figure 4).

Forest plot of study-specific estimates and fixed effects summary transmitted/non-transmitted ratio (T/NTR) and 95% confidence interval (CI) for the association on four different DRB1-DQB1 haplotypes between the minor allele at (A) rs4151659 and (B) rs7762619.
All of the above analyses focused on the difference in transmitted versus not transmitted proportions stratified by classical loci haplotypes. We also investigated the association effect size of rs4151659 and rs7762619 on T1D by conditioning on HLA classical loci haplotypes using the overall conditional haplotype method. The results (Table 4) are similar to those obtained from the previous analyses, although the p-values are in the 10−5–10−8 range. This method also enabled us to compute an estimate of the genetic association effect size which ranged from 2.28 to 3.44.
Table 4
Conditional haplotype method applied to HLA classical loci in haplotypes with rs4151659 and rs7762619. The odds ratio is computed using the observed T1D frequency to the expected frequency given the linkage disequilibrium with classical loci haplotypes.
| SNP | allele | Conditioned on LD with | exp T1D freq | LD | observed T1D freq | chi2 | p-value | OR obs/exp | 95% CI | FPRP | posterior credibility |
|---|---|---|---|---|---|---|---|---|---|---|
| rs7762619 | G | DRB1-DQB1 | 2.43% | 7.60% | 30.45 | 3.42×10−8 | 3.31 | (2.14–5.10) | 0.001 | 0.976 |
| rs7762619 | G | DPB1-DRB1-DQB1 | 2.44% | 7.60% | 30.32 | 3.66×10−8 | 3.29 | (2.13–5.07) | 0.001 | 0.974 |
| rs7762619 | G | DRB1-DQB1-HLA-B | 2.35% | 7.61% | 31.51 | 1.99×10−8 | 3.43 | (2.20–5.32) | 0.001 | 0.985 |
| rs7762619 | G | DRB1-DQB1-HLA-A | 2.34% | 7.60% | 31.74 | 1.76×10−8 | 3.44 | (2.20–5.34) | 0.001 | 0.986 |
| rs4151659 | T | DRB1-DQB1 | 3.03% | 7.85% | 24.96 | 5.87×10−7 | 2.72 | (1.83–4.03) | 0.001 | 0.759 |
| rs4151659 | T | DPB1-DRB1-DQB1 | 2.96% | 7.85% | 25.85 | 3.70×10−7 | 2.80 | (1.87–4.16) | 0.008 | 0.827 |
| rs4151659 | T | DRB1-DQB1-HLA-B | 3.61% | 7.86% | 18.87 | 1.40×10−5 | 2.28 | (1.57–3.28) | 0.173 | 0.148 |
| rs4151659 | T | DRB1-DQB1-HLA-A | 2.76% | 7.85% | 28.37 | 1.00×10−7 | 3.00 | (1.99–4.53) | 0.003 | 0.940 |
Furthermore, using the methods by Wacholder and co-workers (18), and Ioannidis’ (19) Bayesian approach and assuming a conservative prior probability of 1 × 10−4, (average alternative effect OR=2.0) we found that the association of rs7762619 had a false positive report probability (FPRP) of 0.001 and a posterior credibility ranging from 0.974 to 986. For the association of rs4151659, the FPRP ranges from 0.001 to 0.178 and posterior credibility from 0.148 to0.94. Thus, overall, the observed associations appear unlikely to be false positives. However, the credibility of the rs4151659 association when conditioning on DRB1-DQB1-HLA-B is much lower than when conditioning for the presence of other loci.
Finally, we explored whether these two markers are in LD with a SNP mapping to the AIF-1 gene in the class III previously reported to be strongly associated with T1D, rs2259571. We found that although both rs4151659 and rs7762619 were in statistically significant LD with rs2259571, they were not highly correlated with it (r2 =0.028 for both), so they are unlikely to be tagging the AIF-1 associated allele.
Discussion
In this study, we show that two SNP variants within the HLA class III region modify risk for T1D conferred by the high risk DR3 and DR4 haplotypes and appear to also influence the risk conferred by other DRB1-DQB1 haplotypes. Although these two SNPs are highly correlated, we determined that association at these markers may not be exclusively due to LD with each other, suggesting that more than one T1D class III gene is present. Moreover, our results indicate that genetic variation in the class III region contributes to risk of T1D in addition to classical class I and class II variation. These results also demonstrate that utilization of a large, well characterized T1D dataset, such as the one assembled by the T1DGC, makes it possible to dissect various and complex HLA risk components of T1D susceptibility.
One of the markers identified by our study maps 5′ of the lymphotoxin alpha gene (LTA; GeneID:4049) which encodes a highly inducible, secreted, cytokine that mediates a variety of inflammatory, immunostimulatory, and antiviral responses (16). The other marker is a non-synonymous change (K565E) in the complement factor B gene (CFB GeneID:629). Human complement factor B (BF) is an essential component of the alternate complement pathway, and therefore important in innate immune and autoimmune responses (15–16). An association between CFB genetic variation and T1D has, indeed, been reported recently in the North Indian population (20), but unfortunately the genotyping information available in the current study does not allow us to determine which BF allotypes are in LD with rs7762619.
Both markers map to biologically relevant candidates that may be involved in the pathogenesis of T1D. However, because there are other markers in the region that demonstrate very strong LD with one or both of these associated markers (see Table 2 and reference (17)), we cannot exclude the possibility that true causal variants may not be the ones identified by us. An investigation of functional relevance of the genetic associations reported here at both the LTA and CFB and neighboring genes should improve our understanding of the pathogenesis of T1D.
We note several study limitations. The current study focused on DR3 and DR4 haplotypes and only on a limited number of markers which had shown an association, overall, in a previous study. There may be other markers within class III associated with T1D risk. In addition, there may be markers that do not modify the risk of DR3 or DR4, but do modify the risk of e.g. protective haplotypes such as those carrying DQB1*0602 (see e.g. (21)). Furthermore, our study has examined only cases of European descent, and these associations may not generalize to other ethnic groups.
Our findings suggest the involvement of MHC regions in addition to those reported in other studies. An independent study (12), which also used the T1DGC families for replication, identified another class III locus, the AIF-1 gene, as associated with T1D. This could, in part, be due to the different study strategies, i.e., the AIF-1 gene was identified in 434 Norwegian families where the authors tested 53 markers, and subsequently studied the top 3 markers for replication in the T1DGC families. In the current study, we initially examined 356 markers in 889 families, and subsequently studied the top 12 for replication in the whole group of T1DGC families. A large UK study (7) utilizing genotype data from classical HLA loci and SNPs in the MHC region failed to identify any markers in the class III region associated with T1D. This is in contrast with our results and those reported by the Norwegian investigators (12). Differences in marker sets investigated and sample subsets used for discovery may help explain why some variants that were not identified previously, are identified in the current study. There is, in fact, very little overlap among the markers analysed in the current study and those tested by Nejenstev and co-workers (7). Of the class III SNPs tested here, only one - rs1800750 - was included in the large UK study (7). Rs1800750 was found by Nejenstev and co-workers to be nominally associated (p<0.038) with T1D after conditioning for DRB1 and DQB1. These results lend support to our current findings. However, the unconditional association of rs1800750 in the UK study with a considerably smaller sample size compared to the current study has a significance of p<1.09 × 10−7. Using an unconditional TDT for the same SNP in the current set of 1411 pedigrees demonstrates a p<4.88×10−14. The difference in statistical significance between the two studies for the unconditional association results suggests that the conditional effect size of genetic associations at the markers we identified may be smaller in other samples, even if the association is present.
Genetic association results are not always replicated with the same strength in all data sets. It is also possible that different results could be attributed to the use of different methodological approaches. In the present case we have used two different methods and arrived at very similar conclusions. Nevertheless, although failure to find an effect with one analytic approach does not necessarily mean that the effect it is not there, only replications of associations reported here will increase confidence in our findings.
Both our results and those from other research groups (12) strongly suggest that further research on the role of genetic variation in the MHC class III region on MHC susceptibility should be pursued.
Subjects and methods
Study subjects
The T1DGC is a large worldwide collaborative study of new T1D families that have been collected in a highly standardized fashion from various populations, to aid in the search for additional T1D genes within and outside the HLA region (22). High resolution HLA typing has been performed at eight classical MHC loci by four genotyping centers using standardized typing protocols, reagents and quality control procedures (3) and SNP genotyping for over 1800 polymorphisms in the MHC has been carried out in most of those samples (see below).
The samples used in this analysis are derived from extant T1D family collections as well as from the T1DGC regional network collections. The first comprises the following collections; Danish, Sardinian, and two US collections: Human Biological Data Interchange (HBDI) repository (Philadelphia, PA) and Joslin Diabetes Collection (Boston, MA), which we have merged into a single extant US cohort. The T1DGC collection comprises, among others, over one thousand Caucasian T1D families that were recruited for the T1DGC through four regional networks, Asia-Pacific (Australia, New Zealand and Asia), Europe, North America and the United Kingdom. All collections, both new and extant, were HLA-typed using the T1DGC protocols. A detailed description of the collection protocol is published elsewhere (23). The number of individuals and pedigrees per cohort, as well as descriptive statistics for the study subjects are shown in Table 1. Only individuals of Caucasian origin were included in the current analysis.
Table 1
Descriptive characteristics of T1DGC families
| Cohort | Age at T1D onset | SD | Num Pedigrees | Num T1D cases |
|---|---|---|---|---|
| Asia Pacific | 10.29 | 7.42 | 144 | 289 |
| Danish | 11.74 | 8.30 | 95 | 195 |
| Europe | 10.59 | 7.29 | 332 | 661 |
| HBDI | 11.91 | 8.22 | 418 | 881 |
| Joslin | 11.23 | 7.31 | 57 | 122 |
| North America | 8.19 | 5.78 | 220 | 429 |
| Sardinian | 12.21 | 7.88 | 52 | 103 |
| UK | 7.55 | 4.50 | 93 | 185 |
| Total | 1411 | 2865 |
HLA typing
The methods of HLA genotyping and statistical analysis have previously been described (3). Briefly, four digit HLA typing was achieved using oligonucleotide probes that were immobilized to a nylon membrane and hybridized with biotin-labeled locus specific PCR products. Genotyping was performed using the Stripscan and Score proprietary software prior to upload to the coordinating center. Regular quality assurance typing performed on blind samples and between laboratories indicates typing agreement to be greater than 98%. Family based data allowed the assignment of five locus haplotypes (DRB1-DQA1-DQB1-DPA1-DPB1) for statistical analyses. Phased genotypes for HLA classical loci were generated using in-house built macros and double checked manually by two independent individuals. Recombinant cases were excluded if the case could be identified; otherwise the whole family was excluded.
SNP genotyping
High throughput genotyping was carried out by the Wellcome Trust Sanger Institute on the Illumina Golden Gate platform. A detailed description of sample preparation, genotype calling, and quality control procedures is published elsewhere (24). Briefly, the GENCALL score, which reflects the distance of a sample from the centre of its corresponding cluster, was used to flag SNPs or samples with problems. A call rate cut-off of 80% and more than one error per plate were used to flag a SNP as failed. A 50% quantile GENCALL score for each sample indicated sample quality <0.4 and the entire sample was flagged as failed. Mendelian checks were performed using the PEDCHECK software (25), and the family relationship estimation software PREST (26) was used to aid in the determination of family structure problems. Of the 1536 SNPs genotyped, the current analysis refers exclusively to 12 SNPs that map to the MHC class III region and which we reported to be associated with T1D in a two-stage analysis of a smaller subset of samples (17). The SNPs tested in this study are rs1063632, rs7762619, rs1800750, rs2142234, rs9267532, rs400547, rs707915, rs707936, rs4151659, rs4151664, rs17421133, and rs205000. SNP rs2395106 which had been found associated in our previous study (17) was excluded from the present analyses as the call rate when all T1DGC samples, and not merely a subset, were included was <85% The location, minor allele frequency in HapMap (www.hapmap.org) Caucasian samples and in the current sample set, mapping information, and the list of which SNPs of this set are in LD with each other are shown in Table 2.
Statistical Analysis
Under the null hypothesis that within the HLA region no other loci modify the effect on T1D susceptibility of DR3 and DR4 haplotypes, we expect the frequencies of all SNP alleles on these haplotypes to be the same in transmitted and non-transmitted haplotypes (the conditional haplotype method (27–28)); similarly with extension, to include additional predisposition effects from other classical HLA loci: in this case DR-HLA-B, DR-HLA-A or DR-DPB1 haplotypes. If the SNP under study has no effect on T1D risk, the transmission proportions of both alleles should be the same conditioned on the DRB1-DQB1 haplotype. Let DRi-Aj(T) and DRi-Aj(NT) denote the transmitted and non-transmitted counts of haplotype DRi-Aj respectively where Aj denotes allele j at a SNP under investigation. Under the null hypothesis the T/NT ratio= DRi-A1(T) × DRi-A2(NT)/DRi-A2(T) × DRi-A1(NT) should equal 1. This ratio (T/NTR), although not a measure of association in the usual sense, has the same mathematical properties of an odds ratio (OR) so we used it to report the size of the deviation from the null hypothesis.
Both affected sibs from each family were used. Information on the transmission proportion of each allele of each SNP studied within DR3 and DR4 haplotypes and the number of informative transmissions is included in Table 2. Deviations from the random expectation were assessed using a chi-square test where the total sum in the contingency table was ≥200, or a Fisher’s exact test otherwise.
Inter-cohort hetereogeneity
We used the T/NT ratios to test the assumption of heterogeneity for each specified analysis using the method of DerSimonian and Laird based on work first presented by Cochran (29). In the absence of inter-cohort heterogeneity within samples we constructed a Mantel-Haenszel meta-analysis of data from the samples to assess the overall evidence of association. The Mantel-Haenszel chi-squared test and the Mantel-Haenszel estimate of the OR (25) were used to provide a summary test and T/NT ratio. The StatsDirect v 2.7 software (StatsDirect Ltd, Altricham UK) was used.
Overall Conditional Haplotype Method
Control haplotypes (n=967) were determined based on the affected family-based control (AFBAC) method (30). The transmission of overall haplotypes at all four loci (DPA1, DPB1, DRB1, DQA1 DQB1, HLA-B) was used to determine AFBAC haplotypes. Because only the proband from each family is used in the analysis and the AFBAC method uses at most two haplotypes from each pedigree, this approach does not introduce a bias due to the non-independence between sibs.
Adjustment for linkage disequilibrium (LD) with DRB1-DQB1 (HLA-B/HLA-A/DPB1) haplotypes
The expected allele frequencies were computed, given LD and haplotype relative penetrances. Briefly, the null hypothesis (H0) (is that rs4151659 or rs7762619 allele frequencies will differ between patients and controls due to (a) linkage disequilibrium between the SNPs and DRB1-DQB1 (and DRB1-DQB1-HLA-B, DRB1-DQB1-DPB1, DRB1-DQB1-HLA-A) and (b) due to chance (sampling), thus implying that rs4151659 or rs7762619 are neutral relative to disease predisposition.
Under H0 the expected allele frequencies at rs4151659 or rs7762619 alleles can be computed using
the equation derived by Thomson (31):
where Dij denotes the pairwise linkage disequilibrium coefficient between the ith DRB1-DQB1 (HLA-B/HLA-A/DPB1) haplotype and the jth rs4151659 or rs7762619 allele in the control sample, q denotes the allele or haplotype frequency in patients, p denotes the frequency in the affected family based controls and qexp denotes the expected frequency in patients under the assumption of no involvement of the SNP in disease. This method relies on sampling estimates of pairwise linkage disequilibrium between the SNPs and the classical loci haplotypes, and of the proband and control frequencies derived from the samples under study. Thus, there will be a sampling error associated with the computed value for expected SNP alleles. The larger the control sample the smaller this error would be. This has been taken into account in the statistical tests carried out as previously described (6). Given the large number of classical loci haplotypes and the error that very rare haplotypes could introduce, only haplotypes at the susceptibility loci with an average frequency of 0.5% or higher in combined cases and controls were used. Moreover, only HLA+SNP haplotypes where the expected frequency in controls in the absence of LD would be >0.05% were included in the analysis
False positive probability
Two methods were used to estimate the credibility of the genetic associations identified. Both methods rely on the prior probability of a true association of the tested genetic variant with a disease, on the observed P value, and on statistical power to detect the OR of the alternative hypothesis at the given significance level. The first method used was developed by Wacholder and coworkers (18) to compute the probability of no true association between a genetic variant and disease given a statistically significant finding, and is called the false positive report probability (FPRP). The second method used, developed by Ioannidis (19), calculates the Bayes factor under a spike and smear prior using as an alternative, an average genetic effect to estimate a posterior credibility of the associations found.
Acknowledgments
This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. This publication was also supported in part by NIH/NIAID contract number HHSN266200400076C, ABD N01-AI-40076 (GT and LFB). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. AMV is supported by the FP7 large programme grant 200800 TREAT-OA. We thank Evangelis Evangelou for providing us with the excel spreadsheet to compute the credibility of the genetic associations.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/gene.2009.104
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2858242?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Human Leukocyte Antigen (HLA) System: Genetics and Association with Bacterial and Viral Infections.
J Immunol Res, 2022:9710376, 26 May 2022
Cited by: 27 articles | PMID: 35664353 | PMCID: PMC9162874
Review Free full text in Europe PMC
Impact of Human Leukocyte Antigen Loci and Haplotypes on Intestinal Acute Graft-versus-host Disease after Human Leukocyte Antigen-matched Sibling Peripheral Blood Stem Cell Transplantation.
Chin Med J (Engl), 130(11):1290-1295, 01 Jun 2017
Cited by: 0 articles | PMID: 28524827 | PMCID: PMC5455037
Early prediction of autoimmune (type 1) diabetes.
Diabetologia, 60(8):1370-1381, 26 May 2017
Cited by: 88 articles | PMID: 28550517 | PMCID: PMC5491594
Review Free full text in Europe PMC
Amino acid position 11 of HLA-DRβ1 is a major determinant of chromosome 6p association with ulcerative colitis.
Genes Immun, 13(3):245-252, 15 Dec 2011
Cited by: 29 articles | PMID: 22170232 | PMCID: PMC3341846
Genetics of type 1 diabetes.
Cold Spring Harb Perspect Med, 2(1):a007732, 01 Jan 2012
Cited by: 146 articles | PMID: 22315720 | PMCID: PMC3253030
Review Free full text in Europe PMC
Go to all (8) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 14 of 14)
- (27 citations) dbSNP - rs7762619
- (26 citations) dbSNP - rs4151659
- (7 citations) dbSNP - rs1800750
- (6 citations) dbSNP - rs707936
- (6 citations) dbSNP - rs2142234
- (6 citations) dbSNP - rs4151664
- (6 citations) dbSNP - rs205000
- (6 citations) dbSNP - rs400547
- (6 citations) dbSNP - rs707915
- (5 citations) dbSNP - rs9267532
- (2 citations) dbSNP - rs1063632
- (2 citations) dbSNP - rs17421133
- (1 citation) dbSNP - rs2259571
- (1 citation) dbSNP - rs2395106
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Type 1 diabetes risk for human leukocyte antigen (HLA)-DR3 haplotypes depends on genotypic context: association of DPB1 and HLA class I loci among DR3- and DR4-matched Italian patients and controls.
Hum Immunol, 69(4-5):291-300, 26 Mar 2008
Cited by: 17 articles | PMID: 18486765 | PMCID: PMC2505335
The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos.
Tissue Antigens, 59(6):452-469, 01 Jun 2002
Cited by: 44 articles | PMID: 12445315
Several loci in the HLA class III region are associated with T1D risk after adjusting for DRB1-DQB1.
Diabetes Obes Metab, 11 Suppl 1:46-52, 01 Feb 2009
Cited by: 22 articles | PMID: 19143814 | PMCID: PMC2755069
Genetics of the HLA region in the prediction of type 1 diabetes.
Curr Diab Rep, 11(6):533-542, 01 Dec 2011
Cited by: 218 articles | PMID: 21912932 | PMCID: PMC3233362
Review Free full text in Europe PMC
Funding
Funders who supported this work.
European Commission FP7 (1)
Grant ID: FP7_200800
NIAID NIH HHS (2)
Grant ID: N01AI40076
Grant ID: ABD N01-AI-40076
NIDDK NIH HHS (2)
Grant ID: U01 DK062418
Grant ID: U01 DK062418-01
PHS HHS (1)
Grant ID: HHSN266200400076C




