Abstract
Free full text
Regulation of Breast Cancer Stem Cell Activity by Signalling Through the Notch4 Receptor
Abstract
The Notch receptor signalling pathway plays an important role in breast development, regulation of stem cells and differentiation of luminal progenitor cells. The pathway also plays a significant role in breast cancer development and progression. However, which of the Notch receptors that regulate breast cancer stem cells is unknown.
We assessed stem cell activity in breast cancer cell lines and nine primary human tumour samples. In vitro and in vivo breast cancer stem cell activity was enriched using selection of anoikis resistant cells or cells expressing the membrane phenotype ESA+/CD44+/CD24low. We compared the activation of Notch receptors in the breast cancer stem cell-enriched population to luminally differentiated cells and studied the effects of pathway inhibition, both in vitro and in vivo.
We find that Notch4 signalling activity is 8-fold higher in the breast cancer stem cell-enriched cells compared to the differentiated cells while Notch1 activation is 4-fold lower in breast cancer stem cells. Furthermore, pharmacological or genetic Notch inhibition markedly reduced breast cancer stem cell activity in vitro, and significantly reduced tumour formation in vivo. Importantly, cells with Notch4 knock-down using specific shRNA formed fewer mammosphere colonies than Notch1 knock-down cells. In vivo Notch1 knock-down, like pharmacological inhibition, reduced the number and size of tumours but Notch4 knock-down suppressed tumour initiation completely.
Our findings indicate that Notch4-targeted therapies will be more effective than targeting Notch1 in suppressing breast cancer recurrence initiated by breast cancer stem cells.
Introduction
Identification of cell surface markers has allowed the enrichment of cancer stem cells (CSCs) from the total cell population in leukaemia (1) and a number of solid tumours including prostate, colorectal and brain (2-4). In the breast, CSCs are enriched by sorting for ESA+/CD44+/CD24low and this population is tumour initiating in vivo (5).
Comparisons between CD44+ and CD24+ breast cancer cells from patient samples have shown that the breast CD44+ cells are basal-like, similar to normal breast stem cells. In contrast, CD24+ cells express markers of luminal differentiation (6). These two cell types, the basal CD44+ breast cancer stem cells (BCSCs) and the luminally differentiated CD24+ cells, have also been shown to exist in a breast cancer cell lines (7), indicating a similar cellular hierarchy to primary breast cancer tissue.
In addition to cell surface phenotype, suspension mammosphere culture can be used to study normal and cancer stem cells in vitro. This has been successfully used to grow colonies from stem cells in non-adherent culture and to measure the capacity of breast cells to self-renew and produce differentiated progeny, two known characteristics of normal and cancer stem cells (8-10).
The resistance of BCSCs to treatment has been demonstrated by studying breast cancer cells taken from patients before and after neoadjuvant chemotherapy. An increased proportion of CD44+/CD24low breast cancer cells was observed after treatment suggesting that BCSCs were able to preferentially survive treatment compared to the more differentiated cancer cells (11). The Notch pathway has also been linked to radiation resistance in breast cancer cell lines; BCSCs, isolated by non-adherent culture, have increased resistance to radiation compared to the more differentiated non-stem cells and Notch signalling is increased in these cells (12).
The Notch signalling pathway plays an important role in normal breast development, cell fate and stem cell self-renewal (13) and its deregulation has been shown to play a role in cancer. A role for Notch was first identified in mouse mammary tumours which had frequent integration of the mouse mammary tumour virus (MMTV) into the Notch4 receptor resulting in a truncated intracellular form of the receptor which is constitutively activated (14).
Aberrant Notch signalling has been implicated in the development and progression of both pre-invasive ductal carcinoma in situ (DCIS) (10) and invasive breast cancer (15, 16). Over-expression of Notch receptors has been reported in both DCIS (10) and invasive cancer (16). Furthermore, high levels of ligands (17-19), down stream targets (16) as well as down-regulation of Numb (15) have been reported in invasive breast cancer.
Although Notch signalling is clearly important in the development and progression of breast cancer, little is known about its activity in the BCSC sub-population. We show that BCSC activity depends preferentially on Notch4, rather than Notch1, receptor signalling. This improved knowledge of the role of Notch signalling in BCSCs will allow the design of more successful breast cancer treatments.
Materials and Methods
Primary cell isolation
Pleural effusion (PE) samples from patients with metastatic breast cancer (n=7) and primary solid tumour (ST) samples (n=2) were collected with fully informed consent (COREC# 05/Q1402/25 and 05/Q1403/159). For details see Supplemental Table1. PE cells were harvested as previously described (20). ST were cut into ≤1 cm pieces and disaggregated in complete medium with 12 × 1 minute compressions using the Stomacher80 Biomaster, Seward. Remaining leukocytes were removed with CD45-negative magnetic sorting according to manufacturer’s instructions (Miltenyi Biotech, UK).
Monolayer and mammosphere culture
Monolayers of the human breast cancer cell lines were grown as previously described (10, 21). Monolayer cells were enzymatically, 0.125% Trypsin-EDTA (Sigma), and manually, 25 gauge needle, disaggregated to a single cell suspension. Primary cells were resuspended as single cells in PBS. Cells were plated at 500 cells/cm2 in non-adherent culture, flasks coated in 1.2% poly(2-hydroxyethylmethacrylate)/95% ethanol (poly-HEMA [Sigma]). Cells were grown for 7 days in DMEM/F12 containing B27, MEGM SingleQuots (hEGF, Insulin, Hydrocortisone and GA-1000) (Cambrex) and were maintained in a humidified incubator at 370C at an atmospheric pressure in 5% (v/v) carbon dioxide/air. Percentage mammosphere forming units (%MFU) was calculated as number of mammospheres (≥50μm) formed divided by the cell number plated and multiplied by a hundred.
Viable cell count
Annexin/PI staining was carried out according to manufacturer’s instructions (Apoptosis Detection Kit I, BD Bioscience). Staining was assessed using the Becton Dickinson FACS Calibur and levels were analysed using WinMIDI 2.8.
Flow cytometric analysis and sorting
Cells were resuspended at ≤ 1 × 106 in 100μl sorting buffer (PBS containing 0.5% BSA, 2mM EDTA) and incubated with pre-conjugated primary antibodies (dilution); BEREP4-FITC (1:10, Biomeda, USA), CD44-APC (1:20, BD Pharmingen, Oxford, UK) and CD24-PE (1:10, Beckman Coulter, London, UK) for 10 minutes at 40C. The cells were washed in PBS and centrifuged at 800g for 2 minutes. For analysis, cells were resuspended in 500μl of sorting buffer and fluorescence was measured using the FACS Calibur and analysed using WinMIDI 2.8. For sorting, cells were resuspended in 500μl of 1x Hanks Buffered Saline Solution (HBSS, Invitrogen) after incubation with the primary antibodies. Cells were sorted, with HBSS as sheath fluid, at 16PSI using the FACS Aria.
Immunoblot Analysis
Western blotting was carried out as previously described (10, 21). For details of antibodies used see Supplemental Table 2. Densitometry was performed using ImageJ software freely available at http://rsb.info.nih.gov/ij/. Mean band intensity was measured (minus background intensity) and fold change from actin control was calculated.
In vitro inhibition of the Notch pathway
For gamma secretase inhibition (GSI) of signalling, 10μM of the GSI, DAPT (N -[ N -(3,5-difluorophenacetyl-l -alanyl)]-S-phenylglycine t -butyl ester) (Calbiochem, Nottingham, UK) in 0.01% DMSO, was added to monolayer or mammosphere culture at day of plating. 10μM DBZ in DBZ buffer (0.5% Methocel, 0.1% Tween80) was used to treat MCF7 in adherent culture for 3 days for Western blot analysis of signalling inhibition.
Pre-designed siRNA sequences were acquired from Dharmacon to target 4 unique sequences in the Notch1 and Notch4 receptors (For sequence details see Supplemental Table 3). MCF7 cells were transfected according to manufacturer’s instructions.
Production of over-expressing and knock-down cell lines
MDA-MB-231_Numb cell production was previously described in Stylianou et al (2007) (16). To produce inducible N1-ICD cells, MCF7 and MDA-MB-231 cells were transduced with p201-doubleTet lentivirus in presence of 8μg/ml polybrene and selected with 1mg/ml and 1.5mg/ml G418 respectively. DoubleTet lines were then infected with lentivirus (p199-YN1-ICD-iTK-SVzeo) and then selected with 1.5mg/ml Zeocin (Sigma). Doxycycline inducible stable shRNA cell lines were produced using the Clontech pSingle-tTS-shRNA vector for the Notch1 and Notch4 receptors and a scrambled control according to manufacturer’s instructions. Cell lines were grown in DMEM containing 10% tetracycline free foetal bovine serum (Biosera, East Sussex, UK), L-glutamine, antibiotics and 0.5μg/ml G418 (Gibco).
In vivo limiting dilution and Notch inhibition
All procedures were performed in accordance with the Animals (Scientific Procedures) Act 1986 and approved by the UK Home Office. Cell lines were resuspended in 0.2ml PBS and injected subcutaneously into mice treated with oestrogen pellets (0.72mg, Innovative Research of America).
For inducible shRNA, knock out mice were given 2mg/ml Doxycycline (Sigma) and 5% sucrose (Sigma) in their drinking water from the day of cell injection, control mice were given sucrose only. For GSI, 1mg/ml Dibenzazepine (DBZ, a kind gift from Adrian Harris, Oxford) was delivered by intra-peritoneal injection on the day of cell injection and every subsequent 3 days.
Statistical Methods
Throughout the paper data is represented as mean ±SEM taken over a minimum of three independent experiments, unless otherwise stated. Statistical significance was measured using parametric testing, assuming equal variance, in the majority of experiments with standard t-Tests for two paired samples used to assess difference between test and control samples. Analysis of variance (2 way ANOVA) was used to assess difference in tumour formation in vivo.
Results
Identification of breast cancer stem cells (BCSCs) in cell lines and primary samples
We evaluated in vitro mammosphere culture of breast cancer cell lines and invasive primary tumours and malignant pleural effusions from patients. Mammospheres formed in all cell types tested (Figure 1A). Formation rate varied considerably but did not correlate with grade or steroid receptor status (Supplemental Table S1). Importantly, we demonstrated that mammospheres formed from single cells (Supplemental Figure 1A), contained a single label retaining cell in ≥80% of cases (Supplemental Figure 1B and C) and a single mammosphere forming unit (MFU; Supplemental Figure 1D), indicating their clonal origin.
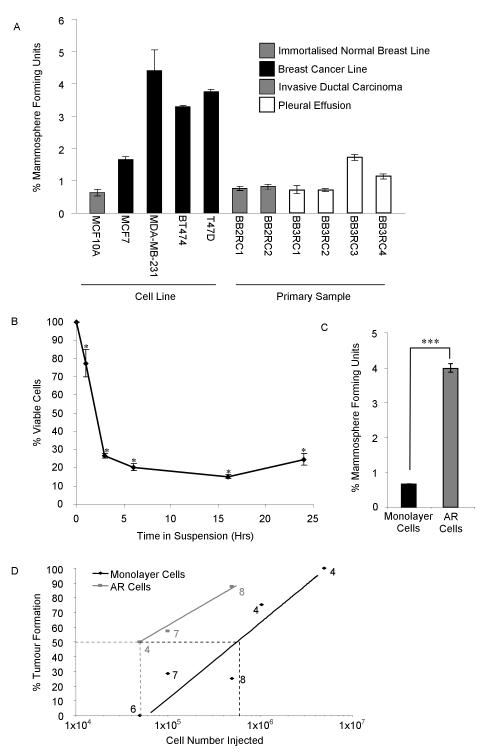
Percentage mammosphere forming units in different cell types (A). Cell viability assessed using Annexin/PI staining and analysed using the Becton Dickinson FACS Calibur after a time course of non-adherent culture (B). Mammosphere forming unit number in anoikis resistant cells, collected at 16 hours (C). Percentage tumour formation of AR and monolayer cells (D, number represents mice in group, 50% formation represented as dotted line).
* P < 0.05 compared to 0 hour control and *** P <0.001, A-C data presented as mean ± SEM.
AR anoikis resistant
At early time points in non-adherent culture, MFU we predicted would be enriched since differentiated cells undergo anoikis in these conditions (8). 85% underwent anoikis within 16 hours of culture (Figure 1B, P<0.05). The anoikis resistant (AR) cells, collected at 16 hours, are significantly enriched for in vitro MFU (5.7-fold P<0.001) (Figure 1C). In vivo, 50% tumour formation required 5×104 AR cells whereas 6×105 monolayer cells are required for the same level of tumour formation suggesting a 12-fold enrichment for tumour initiating cells (Figure 1D). When AR tumours were collected and cells passaged in vivo, all could initiate secondary tumour formation (Data not shown), which demonstrates self-renewal (22). These results show that AR cells are enriched for MFU and tumour initiating cells and suggests, but does not formally prove, that these are over-lapping cell populations which are enriched for self-renewing BCSCs.
Isolation of breast cancer stem cells (BCSCs)
The cell surface phenotype of ESA+/CD44+/CD24low isolates cells that initiate tumours (5) and mammosphere culture enriches for the cells in vitro (9). CD24 is a marker of differentiated luminal cells and cells with low expression of this marker are basal, myoepithelial cells in the normal mammary epithelium (23). Flow cytometric analysis of MCF7 cells demonstrates that they range from the basal CD44+ population to a more differentiated, luminal CD44−/CD24+ population (Figure 2A). AR MCF7 cells are significantly enriched for ESA+/CD44+/CD24low cells with 70% expressing this surface phenotype compared to 6% in monolayer cells (Figure 2A, P<0.02).
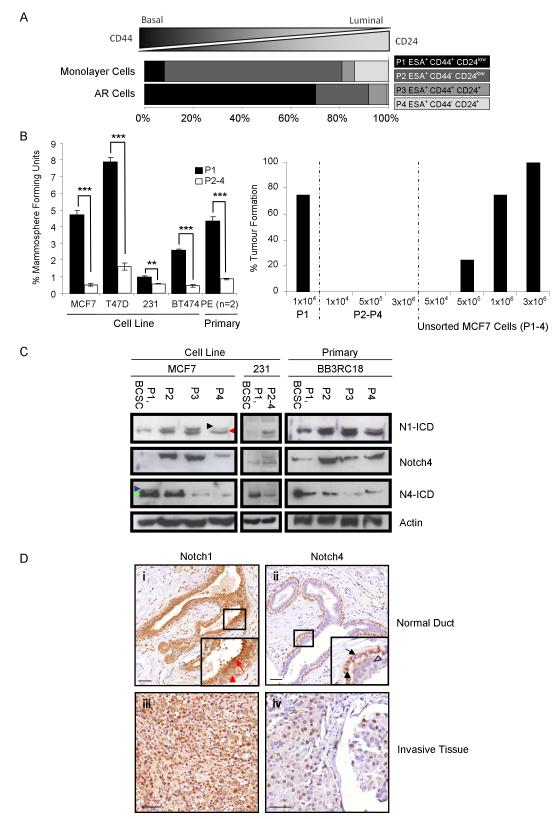
AR and monolayer MCF7 cells were analysed for cell surface phenotypes of breast cancer cells ranging from basal like BCSCs (CD44+/CD24low) to more differentiated, luminal cells (CD44−/CD24+) using the Becton Dickinson FACS Calibur (A). MCF7, T47D, MDA-MB-231, BT474 and primary pleural effusion cells were sorted (P1 BCSC enriched ESA+/CD44+/CD24low, P2 ESA+/CD44−/CD24low, P3 ESA+/CD44+/CD24+, and P4 ESA+/CD44−/CD24+) and plated in mammosphere culture. Percentage mammosphere forming unit number was calculated at day 7 in BSCS enriched cells (P1) and BSCS depleted cells (P2-4) (B). Percentage tumour formation measured over 5 weeks in mice injected with a limiting dilution of sorted and unsorted MCF7 cells (B).
Immuno-blots for full length and cleaved Notch receptors (Notch1-ICD, Notch4 and Notch4-ICD) in sorted cell populations from MCF7, MDA-MB-231 cell lines and a primary pleural effusion sample. Black arrowhead represents Notch1 extracellular truncation (Notch1-EXT), red arrowhead represents Notch1-ICD. Blue and green arrowheads represent Notch4-EXT and Notch4-ICD, respectively (C).
Representative photomicrographs of breast tissue sections stained with antibodies to Notch receptors (D). Cleaved Notch1 (Notch1-ICD) expression was assessed in normal (i, red arrow shows highly positive nucleus in luminal cell, red arrowhead shows weak staining in basal cell nucleus) and invasive tumour tissue (iii). Notch4-ICD staining in normal (ii, black arrow shows inactive cytoplasmic staining, filled black arrowhead shows positive nucleus, hollow black arrowhead indicates cell negative for the stain) and invasive tumour samples (iv). Scale bar equals 100μm.
** P < 0.01 *** P<0.001, B data represented as mean ± SEM).
AR anoikis resistant, ESA epithelial specific antigen, ICD Intracellular domain, N1 Notch1, N4 Notch4
Breast cancer cell lines and primary cells collected from pleural effusion samples were sorted using FACS into four populations (P) based on their cell surface phenotype. Cells were classified as the putatively BCSC-enriched ESA+/CD44+/CD24low population (P1), ESA+/CD44−/CD24low (P2), ESA+/CD44+/CD24+ (P3) or ESA+/CD44−/CD24+ (P4). The BCSC-enriched cells (P1) and BCSC-depleted cells (P2-4) were plated in mammosphere culture to assess MFU enrichment. P1 showed an enrichment for MFU in all cell types tested (MCF7 - 12-fold, T47D - 4-fold, MDA-MB-231 - 2-fold, BT474 - 5-fold and primary breast cancer samples - 5.4-fold) compared to other cell sub-populations (Figure 2B, P<0.01). Mice injected with 1×104 P1 MCF7 cells formed sub-cutaneous tumours in 75% of cases (Figure 2B, n=4) whilst up to 3×106 P2-4 MCF7 cells resulted in no tumour growth which is supportive of previously published work (5). When unsorted MCF7 cells were injected, more than 1×106 were required to form tumours with a 75% take rate (Figure 2B), suggesting a greater than 100-fold enrichment for tumour initiating cells in sorted P1 cells. These data suggest the putative BCSC sub-population is essential for tumour initiation and growth.
Notch receptors are differentially activated in ESA+/CD44+/CD24low cells
Next, we examined Notch activation in BCSCs. Notch receptor and ligand expression are differentially expressed in breast cancer, in particular, Notch1 and Jagged1 (19). Previously published data has studied protein expression in the whole cell population comparing cancer to the normal breast (16). In contrast, we aimed to look specifically at the activity of the Notch signalling pathway in BCSCs.
To do this, we examined the differential activity of Notch1 and Notch4 receptors in sorted MCF7 and MDA-MB-231 cells and primary breast cancer samples (Figure 2C). The antibody to cleaved Notch1 recognised the Notch1 extracellular truncation (Notch1-EXT, upper band) and the intracellular domain (Notch1-ICD, lower band). A significantly lower level of the cleaved Notch1 intracellular domain (Notch1-ICD) was seen in the BCSC enriched MCF7 cells (P1) compared to the other sub-populations. Notch1-ICD levels in P2, P3 and P4 were 3.5±1.2, 4.1±1.3 and 3.4±1.5 fold higher, respectively, than in P1 (Figure 2C, P<0.05), suggesting lower signalling through Notch1 receptor in BCSC. In contrast, significantly higher levels (up to 20-fold) of the active cleaved intracellular domain (Notch4-ICD) are detected in the BCSCs (P1; P<0.05) suggesting strong signalling through this receptor (Figure 2C). A similar pattern of Notch1 and Notch4 activation in differentiated versus stem cell-enriched populations, respectively, was observed in MDA-MB-231 and primary breast cancer samples (Figure 2C).
In order to confirm Notch1 and Notch4 activation patterns in vivo, immunohistochemical analysis of normal and malignant breast samples was performed using antibodies recognising cleaved Notch1-ICD and Notch4-ICD. In normal epithelium, the basal cell layer had higher nuclear expression of Notch4–ICD whereas nuclear Notch1-ICD was detected most strongly in the luminal cell layer (Figure 2D). In invasive breast cancers, Notch1-ICD was present in the majority of cells whereas Notch4-ICD was more infrequent and strongly stained the nuclei of invasive cancer cells (Figure 2D). These observations corroborate the pattern of signalling seen in MCF7 and primary breast cancer cells sorted for basal and luminal cell surface markers.
Notch inhibition reduces BCSC number and activity in vitro and in vivo
Next, we aimed to target Notch signalling and test its effects on BCSCs. One method of Notch inhibition is the use of gamma secretase inhibitors (GSI), such as N - [N -(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (DAPT), Dibenzazepine (DBZ) and MK-0752 which stop Notch-ICD cleavage and its subsequent nuclear translocation. DAPT has previously been shown, in vitro, to decrease the formation of normal and DCIS mammospheres (10, 13) and MK-0752, is currently being trialled in patients (24). We therefore examined the effect of DAPT, DBZ and specific Notch receptor knock-down on BCSC activity in MCF7 and MDA-MB-231 cells and primary breast cancer samples.
In vitro DAPT (10μM) treatment of MCF7 monolayer cells caused a 30% decrease in the proportion of ESA+/CD44+/CD24low cells (Figure 3A, P<0.01). However, Notch4 but not Notch1 siRNA reduced the numbers of ESA+/CD44+/CD24low cells (Figure 3A, P<0.05), suggesting DAPT affects this population through inhibition of other Notch receptors. In MCF7 and MDA-MB-231 cells, primary IDC and pleural effusion (PE) samples, a decrease in MFU was observed (50% decrease in MCF7 and MDA-MB-231 and 30% decrease in PE and IDC samples, Figure 3B, P<0.05). A corresponding decrease in Notch downstream target genes, HES1 and HEY2, was seen in the whole cell population (data not shown). No significant reduction in mammosphere formation was seen in cells treated with DAPT when N1-ICD was activated, suggesting GSI specifically blocked the effects of Notch receptor signalling (Figure 3C).
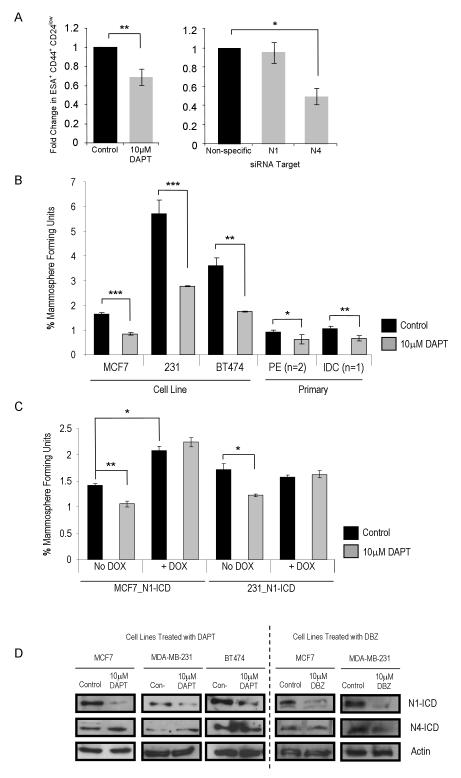
ESA+/CD44+/CD24low cells assessed in vitro following Notch inhibition of MCF7 cells with gamma secretase inhibitor, 10μM DAPT or DMSO control and siRNA to Notch1 and Notch4 (A). Percentage mammosphere forming unit number was assessed in MCF7, MDA-MB-231 and BT474 cells, pleural effusion and primary IDC samples (BB3RC2, BB3RC3 and BB2RC2) after 7 days in non-adherent culture with / without DAPT (B). Cell lines were produced with Doxycycline inducible expression of Notch1-ICD. These lines, MCF7_N1-ICD and MDA-MB-231_N1-ICD, were cultured with / without DAPT either in the absence (control) or presence (activated N1-ICD) of Doxycycline (C). Immunoblot of cleaved Notch receptors (Notch1-ICD and Notch4-ICD) was carried out in MCF7, BT474 and MDA-MB-231 cells with or without DAPT treatment and in MCF7 and MDA-MB-231 with or without DBZ (D).
* P=0.05, ** P<0.01 and *** P<0.001, A-C, E and H data are presented as mean ± SEM.
ICD intracellular domain, N1 Notch1, N4 Notch4, PE pleural effusion, IDC invasive ductal carcinoma, DOX Doxycycline.
We then examined the effects of a GSI DBZ on tumour inhibition in vivo. DBZ significantly decreased MCF7 but not MDA-MB-231 tumours (Table 1, P=0.02) and increased latency compared to control mice (18 to 28 days). DBZ-treated MCF7 tumours that did form had significantly reduced tumour volumes (P=0.03). To validate inhibition of Notch signalling, DAPT and DBZ effects on N1-and N4-ICD were measured in MCF7, BT474 and MDA-MB-231 cells. Both inhibitors caused marked reductions in N1-ICD but had no effect on N4-ICD levels in three cell lines examined (Figure 3D). This suggests a specific or preferential inhibitory effect on the cleavage of the Notch1 receptor at this concentration. This differential effect of GSI on Notch receptors could allow continued activity of Notch4 in the BCSCs, and thus initiation and maintenance of tumours. To assess the effect of inhibition of all Notch receptor activity, MDA-MB-231 cells over-expressing Numb, the Notch inhibitor, were transplanted into mice. Tumour initiation was completely ablated whereas vector control cells grew tumours (Table 1), suggesting inhibition of multiple Notch receptors has a more profound effect than GSI.
Table 1
Table shows cell types injected sub-cutaneously into nude mice. Tumour growth was assessed twice weekly for 30 days. Mice positive for tumour growth / mice in group. Days to growth was calculated as average time until palpable tumour was present in each mouse. Final mean tumour volume was calculated as an average volume of mice positive for growth. Analysis of variance was used to calculate significance of difference.
| Cell Type | MCF7 | MDA-MB-231 | MDA-MB-231_Numb | MCF7scr | MCF7Notch1 | MCF7Nocth4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | Vehicle Control | DBZ | Vehicle Control | DBZ | Vector control | Numb cDNA | −DOX | +DOX | −DOX | +DOX | −DOX | +DOX |
| Positive Tumour Growth / Injection | 4/4 | 2/4 | 3/6 | 3/6 | 7/7 | 0/7 | 8/8 | 9/9 | 4/4 | 2/3 | 3/5 | 0/5 |
| Average Days to Growth | 18 | 28 | 7.3 | 9.3 | 11 | N/A | 12 | 12 | 12 | 22 | 18 | N/A |
| Mean Tumour Volume | 145.4 | 14.9 | 18.7 | 15.1 | 33.7 | N/A | 211.04 | 192.98 | 136.39 | 44.95 | 100.6 | N/A |
| Difference from Control (P Value) | N/A | 0.03 | N/A | 0.78 | N/A | >0.001 | N/A | 0.12 | 0.42 | 0.08 | 0.12 | 0.01 |
Notch4 signalling has a greater effect on BCSC activity than Notch1
Since DAPT and DBZ preferentially affect Notch1 activity, we compared the effect of Notch1 and Notch4 specific knock-down (using RNAi) on mammosphere and tumour formation. Doxycycline (Dox) inducible shRNA MCF7 cell lines targeting each receptor (MCF7Notch1 and MCF7Notch4) were generated, as well as a scrambled control (MCF7scr). In vitro culture was used to assess Dox-induced receptor knock-down and immuno-blot analysis was carried out to measure specificity. In MCF7scr cells, Notch1 and Notch4 receptor expression were not affected after addition of Dox. Knock-down was specific to the targeted receptor with complete knock-down of Notch1 in MCF7Notch1 cells and decreased in expression of Notch4 in MCF7Notch4 cells (Figure 4A). In addition, Notch4-ICD expression is seen to increase in MCF7Notch1 cells, suggesting a switch to signalling through Notch4 when Notch1 is not available. The MCF7scr cells showed no effect on MFU number (Figure 4B) with or without Dox treatment. When compared to the MCF7scr control cells, Notch1 knock-down (MCF7Notch1 plus Dox) resulted in a 58% decrease in MFU whilst Notch4 knock-down (MCF7Notch4 plus Dox) caused a 75% decrease (Figure 4B, P<0.05). Notch4 knock-down caused a significantly greater decrease in MFU than Notch1 (P<0.05).
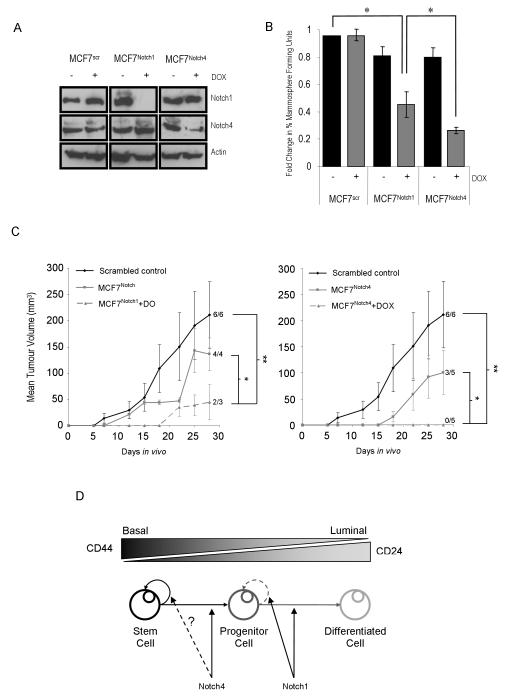
Immunoblot of Notch1 and Notch4 receptors to test the extent of knock-down, and the target specificity of each shRNA cell line (A, Actin as loading control). Mammosphere forming unit number in scrambled control (MCF7scr) and inducible shRNA cell lines (MCF7Notch1 and MCF7Notch4) was measured after 7 days of culture with (grey bars) and without (black bars) Doxycycline (B, * P < 0.05; Small sample t-test, data are presented as mean ± SEM).
Mean tumour volumes over 28 days in vivo growth (C, * P<0.05, ** P<0.01).
A putative model for Notch signalling activity in the cellular hierarchy of breast cancer (D).
MFU mammosphere forming unit, DOX Doxycycline
Next in vivo tumour forming ability for each shRNA cell line was assessed by sub-cutaneous injection into athymic nude mice. Table 1 shows the tumour take and final tumour volume in mice injected with 3×106 MCF7scr, MCF7Notch1 and MCF7Notch4 cells with/without Dox. Tumours formed in all control mice (MCF7scr plus or minus Dox) by day 11, in 100% of the MCF7Notch1 minus Dox mice by day 12 and in 60% of MCF7Notch4 minus Dox by day 18. No significant difference was seen between the tumour volume of these groups (P>0.05).
2 out of 3 Notch1 knock-down mice (MCF7Notch1 plus Dox) formed tumours and tumour volume was not significantly different from control groups (P=0.08). However, there was a significant difference in the tumour growth rate in the Notch1 knock-down mice (Figure 4C, P=0.02). These results were comparable to the inhibition of Notch signalling using DBZ and suggest a role for Notch1 in tumour growth and proliferation. In contrast, no tumours formed by day 28 in Notch4 knock-down mice (MCF7Notch4 plus Dox) (Figure 4D and Table 1). Taken together, these results suggest that Notch4 inhibition has a greater effect on BCSC than Notch1 inhibition and that Notch4 is required for tumour initiation.
Discussion
Our data suggest that BCSCs exist within breast cell lines and primary samples and that these cells are self-renewing, anoikis resistant and tumour initiating. The collection of anoikis resistant cells isolates a BCSC-enriched sub-population with 70% of cells expressing ESA+/CD44+/CD24low. These cells are self-renewing, mammosphere and tumour-initiating as defined by in vitro and in vivo assay techniques.
Aberrant Notch signalling has been shown to play an important role in breast cancer (16, 19). Most work to date has compared cancerous tissue to the normal breast but we aimed to elucidate Notch receptor signalling pathway activity within BCSCs. Our results show that cell lines and primary cells have differential activity of Notch1 and Notch4 receptors in BCSCs In the BCSC-enriched population, the majority of the Notch4 receptor is present in the cleaved/activated form. In normal human breast, Raouf et al showed Notch1 mRNA to be highly expressed in luminal progenitor cells while Notch4 mRNA was at higher levels in basal cells (25). In agreement with these findings, we observed more Notch1-ICD in the luminal cells of normal breast epithelium while more nuclear Notch4 staining was detected in a basal cell population. In addition, we demonstrate that Notch4 but not Notch1 knock-down affects the percentage of ESA+/CD44+/CD24low cells suggesting they are active in different tumour cell populations.
The differential expression of Notch1 versus Notch4 receptors in BCSCs and more differentiated cells suggests different roles for each receptor. The roles of Notch1 and Notch4 are not clearly understood in breast tissue but Notch1 receptor has been reported to be restricted to luminal cells in the normal breast (16) and implicated in cell-fate determination (26, 27). An activating truncation of the Notch1 receptor gene is not sufficient for mammary tumourigenesis in mice. In contrast, MMTV-induced activation of the Notch4 receptor gene has been shown to inhibit mammary epithelial cell differentiation and induce tumour formation in mouse models (14). The constitutively active Notch4 results in the development of poorly differentiated, basal-like tumours.
The Notch pathway has become an attractive drug target for breast cancer treatment and the use of Notch inhibitors is predicted to be effective in reducing tumour growth, perhaps in conjunction with other treatments. Gamma secretase inhibitors (GSI), which were originally used in the treatment of Alzheimer’s disease (28-30), are currently undergoing clinical trials for treatment of leukaemia (31) and a number of solid tumours including those of the intestine (32) and the breast (24). GSIs re-sensitise cancer cells to treatment with trastuzimab, tamoxifen and chemotherapy (16, 33, 34). Our data show that inhibition of Notch signalling using both GSI and shRNA were successful in decreasing BCSC activity. However, inhibiting Notch4 receptor signalling had the greatest effect. GSI treatment inhibited Notch1 and not Notch4 activity in MCF7, BT474 and MDA-MB-231, and reduced BCSC number in vitro by approximately 50%. It has previously been suggested that Notch4 gene transcription is regulated downstream of Notch1 (35) but in the three breast cancer cell lines where we examined Notch4 activity it was unaffected by Notch1 inhibition using GSI, suggesting Notch1-independent regulation of Notch4 (Figure 3D). In contrast, there is a slight increase in Notch4 receptor expression where Notch1 is knocked down (Figure 4A). This may result from differences in the percentage of ESA+/CD44+/CD24low cells following DAPT compared to Notch1 knock-down demonstrated in Figure 3A rather than direct effects on Notch4 activity. We observed similar reductions in tumour growth in vivo using either DBZ or Notch1 shRNA. In comparison to GSIs or Notch1 shRNA, Notch4 shRNA had a greater inhibitory effect on mammospheres and tumour initiation. These findings suggest that a better, more specific target in BCSCs will be the Notch4 receptor. The reason why a GSI has little effect on Notch4 cleavage is unknown. Some evidence exists suggesting that a constitutively active, truncated form of the receptor exist within breast cancer cell lines (36). Another possibility is that the BCSCs are able to efflux the GSI at the dose given and they are, therefore, unaffected by treatment. The ability to efflux drugs is a known characteristic of stem cells (37) and this remains a plausible explanation.
We propose a model (Figure 4D) where Notch4 regulates exit of BCSCs into the proliferating progenitor population whereas Notch1 activity regulates progenitor proliferation and luminal differentiation. This model may explain why DAPT and Notch1 knock-down can partially inhibit MFUs and tumour formation through their effects on progenitor proliferation. The superior effect of Notch4 knock-down on these processes would be explained by preventing the production of progenitors downstream of the stem cell-enriched population (P1). It remains unknown whether Notch4 also has a direct effect on stem cell self-renewal activity.
Overall, our findings indicate that specifically targeting the Notch4 receptor in BCSCs for treatment of breast cancer will be superior to inhibiting gamma secretase or targeting the Notch1 receptor. This is highly topical since gamma secretase inhibitors to target Notch receptors are currently in clinical trials in combination with taxanes, to which BCSCs are known to be resistant (11). Our results indicate that Notch4 receptor plays an important role in the control of BCSC activity in two cell lines and primary samples representative of different tumour types. Further work is required to assess whether this is true in all sub-types of breast cancer using primary tumour samples.
Supplementary Material
1
2
3
4
5
Acknowledgements
We thank patients from The Christie NHS Foundation Trust and the University Hospitals of South Manchester who donated tumour samples for this research. We are grateful to Anthony Howell for reading and commenting on the manuscript, to Rognald Blance and the Paterson Institute Biological Resources Unit for assistance with in vivo experiments and to Ji-Liang Li and Adrian Harris for supplying the DBZ.
Grant support HH was supported by EU NEST 12930, GF and RBC are Breast Cancer Campaign-funded Research Fellows, KRB was supported by the Wellcome Trust and SJH was funded by The Christie.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-09-1681
Read article for free, from open access legal sources, via Unpaywall:
https://aacrjournals.org/cancerres/article-pdf/70/2/709/2642493/709.pdf
Free to read at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/content/abstract/70/2/709
Free after 12 months at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/reprint/70/2/709.pdf
Free after 12 months at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/content/full/70/2/709
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/0008-5472.can-09-1681
Article citations
Interactions between hedgehog signaling pathway and the complex tumor microenvironment in breast cancer: current knowledge and therapeutic promises.
Cell Commun Signal, 22(1):432, 09 Sep 2024
Cited by: 0 articles | PMID: 39252010 | PMCID: PMC11382420
Review Free full text in Europe PMC
Novel Anti-Cancer Stem Cell Compounds: A Comprehensive Review.
Pharmaceutics, 16(8):1024, 01 Aug 2024
Cited by: 0 articles | PMID: 39204369 | PMCID: PMC11360402
Review Free full text in Europe PMC
Lipid metabolism dynamics in cancer stem cells: potential targets for cancers.
Front Pharmacol, 15:1367981, 27 Jun 2024
Cited by: 1 article | PMID: 38994204 | PMCID: PMC11236562
Review Free full text in Europe PMC
C-terminally phosphorylated p27 activates self-renewal driver genes to program cancer stem cell expansion, mammary hyperplasia and cancer.
Nat Commun, 15(1):5152, 17 Jun 2024
Cited by: 0 articles | PMID: 38886396 | PMCID: PMC11183067
Associations of stem cell markers CD44, CD24 and ALDH1A1 with mammographic breast density in women with benign breast biopsies.
Br J Cancer, 131(2):325-333, 07 Jun 2024
Cited by: 0 articles | PMID: 38849477
Go to all (326) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nicastrin and Notch4 drive endocrine therapy resistance and epithelial to mesenchymal transition in MCF7 breast cancer cells.
Breast Cancer Res, 16(3):R62, 11 Jun 2014
Cited by: 47 articles | PMID: 24919951 | PMCID: PMC4095694
Notch4 Signaling Confers Susceptibility to TRAIL-Induced Apoptosis in Breast Cancer Cells.
J Cell Biochem, 116(7):1371-1380, 01 Jul 2015
Cited by: 10 articles | PMID: 25704336
Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity.
Cell Rep, 12(12):1968-1977, 17 Sep 2015
Cited by: 117 articles | PMID: 26387946 | PMCID: PMC4594158
Breast cancer stem cells: something out of notching?
Cancer Res, 70(22):8973-8976, 02 Nov 2010
Cited by: 52 articles | PMID: 21045140
Review
Funding
Funders who supported this work.
Breast Cancer Now (2)
Role of Notch in proliferation and apoptosis of ductal carcinoma in situ
Keith Brennan, University of Manchester
Grant ID: 2006MAYPR19
Survival and self-renewal signalling pathways in human breast cancer stem cells
Professor Clarke, University of Manchester
Grant ID: 2006MAYSF01
Wellcome Trust (1)
Elucidating the role of Notch signalling in mammary gland development.
Keith Brennan, University of Manchester
Grant ID: 065994




