Abstract
Free full text

A Kinetochore-Independent Mechanism Drives Anaphase Chromosome Separation During Acentrosomal Meiosis
Abstract
The self-organized assembly of acentrosomal meiotic spindles has been extensively studied1 but little is known about how chromosomes segregate on these spindles. Here, we investigate two chromosome-microtubule interaction mechanisms—kinetochores and chromokinesins—during meiosis in fertilized C. elegans oocytes. We show that the conserved kinetochore protein KNL-1 directs assembly of meiotic kinetochores that orient chromosomes on the acentrosomal spindles. However, in contrast to mitosis, chromosome separation during meiotic anaphase was kinetochore-independent. The chromokinesin KLP-19 did not contribute to chromosome orientation or anaphase, but stabilized late anaphase spindles. Prior to anaphase separation, meiotic kinetochores and spindle poles disassembled along with microtubules on the poleward side of the chromosomes; during anaphase, microtubules were formed between the separating chromosomes. Functional analysis implicated a set of proteins that localize to a ring-shaped domain between the kinetochores in pre-anaphase spindle assembly and anaphase separation. Ring domain proteins are localized by the chromosomal passenger complex (CPC), whose local enrichment is patterned by recombination to control step-wise loss of meiotic cohesion2–4. Thus, meiotic segregation in C. elegans is a two-stage process where kinetochores orient chromosomes but are dispensable for their separation. We suggest that separation is instead controlled by a meiosis-specific chromosomal domain to coordinate step-wise dissolution of cohesion with chromosome segregation.
To study chromosome segregation on acentrosomal meiotic spindles, we used fertilized C. elegans oocytes because both female meiotic divisions and the first embryonic division can be monitored ex-utero. Assembly on a chromatin base containing the histone H3 variant CENP-A (CENtromeric Protein-A) is a universal feature of mitotic kinetochores that is conserved in C. elegans, despite the fact that it has holocentric chromosomes with kinetochores that run along along the length of each chromatid5. In contrast to CENP-A-directed mitotic assembly, a CENP-A-independent mechanism recruits kinetochore components to the chromosome surface during female meiosis in C. elegans6; the assembly of these meiotic kinetochores and their contribution to chromosome segregation has not been addressed. A recent study analyzing acentrosomal spindles in fertilized oocytes arrested in meiosis I implicated KLP-19 (the only C. elegans chromokinesin required for mitotic chromosome segregation7) in chromosome alignment8; however, the role of KLP-19 in chromosome segregation was not addressed. Here, we analyze the contribution of the kinetochore and KLP-19 to meiotic chromosome segregation in normally progressing fertilized oocytes.
In C. elegans, fertilization is followed by two rounds of meiotic chromosome segregation. During both meiotic divisions, the 6 chromosomes adopt a compact oval shape and kinetochore components accumulate on their surface in two opposing cup-like structures (Fig. 1a) separated by a gap. The gap, which represents the mid-bivalent region in meiosis I (MI), and the sister chromatid axis in meiosis II (MII), is enriched for proteins including the chromosomal passenger complex (CPC; Fig. 2b) that control the step-wise dissolution of meiotic cohesion3,4. During meiosis I, but not meiosis II, kinetochore proteins also form rod-shaped structures (similar to structures observed in Drosophila meiosis I oocytes10) prevalent in the spindle region (Fig. S1; S5a) and on the cell cortex6,9.
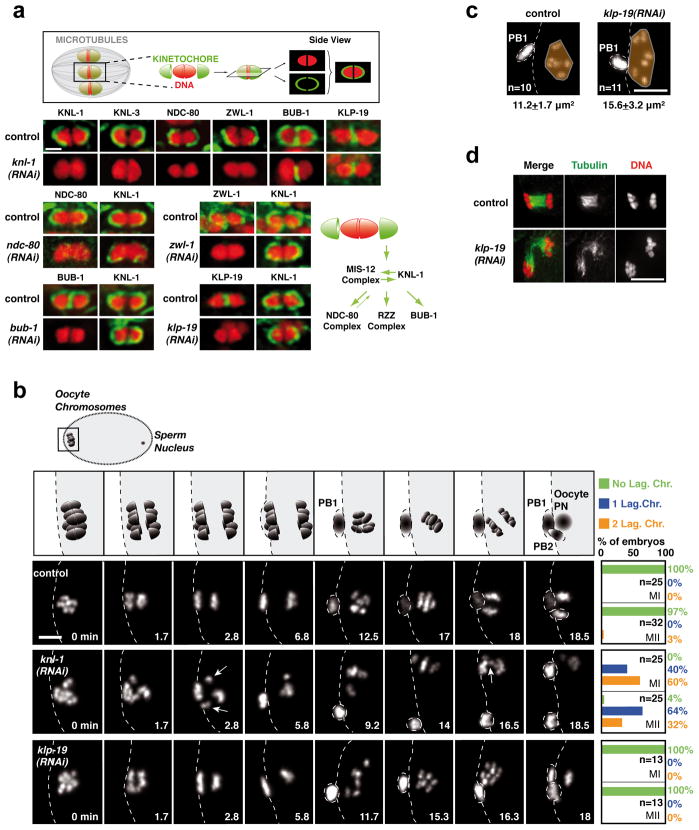
(a) Top: Schematic of cup-shaped meiotic kinetochores. Bottom. Localization of 6 kinetochore components and the chromokinesin KLP-19 on individual meiosis I bivalent chromosomes (see also Fig. S1). Schematic summarizes assembly dependencies for the meiotic kinetochore. Scale bar, 1 μm. (b) Top row: Schematic of chromosome segregation during meiosis I and II. (PB1: First Polar Body; PB2: Second Polar Body; PN: Pronucleus). 3 Bottom rows: stills from movies of fertilized oocytes expressing GFP-histone-H2b. Time relative to Metaphase I is in the lower right corner of each panel. White arrows indicate lagging chromosomes during anaphase I and II in the KNL-1 depletion. Quantification of lagging chromosomes during anaphase I (MI) and anaphase II (MII) is on the right. Scale bar, 5 μm. (c) KLP-19 depletion causes increased chromosome dispersal in late anaphase that is correlated with spindle instability at this stage. (d) Area occupied by the chromosomes (orange) was measured at a similar time after anaphase I onset (Control, 11.4±1.2 min; KLP-19-depleted, 11.9±0.6 min); spindle instability was observed in 8/8 KLP-19-depleted fixed oocytes at this specific cell cycle stage. Scale bars, 5 μm.
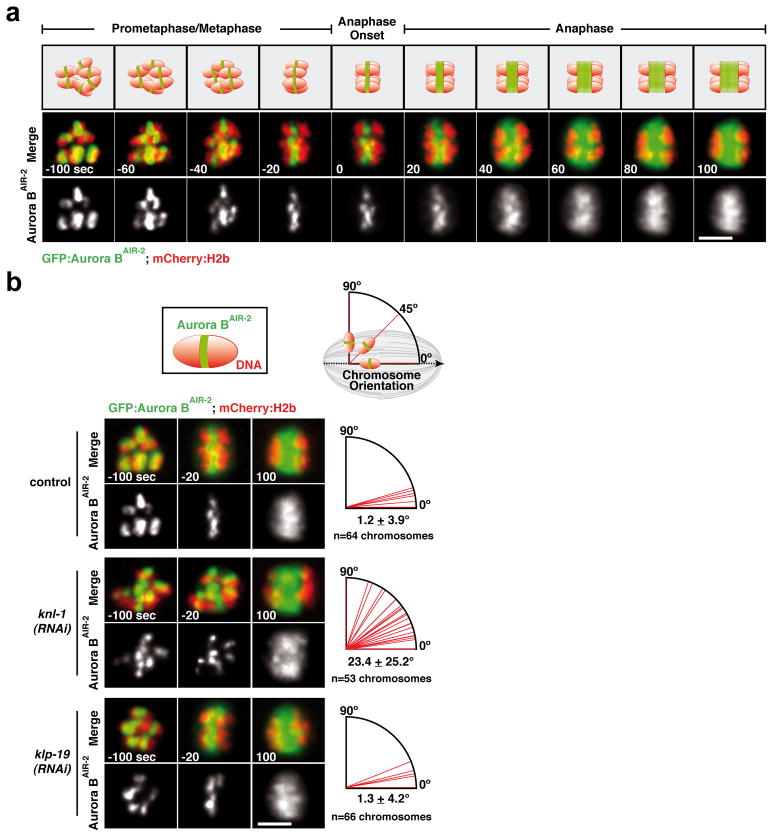
(a) GFP-Aurora BAIR-2 and mCherry-histone-H2b dynamics in control embryos. Time is in seconds relative to anaphase I onset. Scale bar, 5 μm. (b) Top: Schematics of Aurora BAIR-2 signal on an individual chromosome and measurement of chromosome orientation angle using this signal; the spindle axis is defined by anaphase chromosome separation. Bottom: Stills from movies of the indicated conditions; quantification of individual chromosome angles relative to the axis of the meiosis I spindle, measured 20 sec before anaphase onset (n=12 embryos per condition), is presented on the right. Scale bars, 5 μm.
To identify a means to perturb meiotic kinetochore assembly, we analyzed the localization interdependencies between 5 conserved components that localize to the kinetochore cups during both meiotic divisions (KNL-1, the MIS-12 complex, the NDC-80 complex, the RZZ complex, and BUB-111). BUB-1 additionally localizes to the mid-bivalent region (Fig. 1a; Fig. S1; Fig. S2)6. Similar to mitosis12, KNL-1 and the MIS-12 complex (represented here by KNL-3 and KBP-1) are at the top of the meiotic kinetochore assembly pathway (Fig. 1a; Fig. S1; Fig. S2a). In KNL-1 depleted oocytes, the MIS-12 complex still targets to the chromosome surface but not in a stable manner (Fig. S2b,c). Thus KNL-1 is a central scaffold protein required to assemble cup-like meiotic kinetochores. Localization of KNL-1 and the chromokinesin KLP-19, which accumulates at the mid-bivalent region8, were mutually independent (Fig. 1a). Thus, a comparison of control, KNL-1-depleted, and KLP-19-depleted fertilized oocytes should reveal the respective contributions of kinetochores and chromokinesin to chromosome segregation on acentrosomal meiotic spindles.
We analyzed chromosome segregation during both meiotic divisions in dissected fertilized oocytes expressing GFP-histone-H2b (Fig. 1b; Movie S1). Imaging was typically initiated in late prometaphase I. Control embryos took ~16 min to progress from metaphase I to metaphase II, and ~19 min to progress from metaphase II to pronuclear meeting in the one-cell embryo. Chromosomes aligned on a tight metaphase plate near the oocyte cortex during metaphase I (Fig. 1b: 0min). During both anaphase I and II, segregating chromosomes were tightly grouped (Fig. 1b); individual chromosomes were visible at prometaphase II (Fig. 1b: 12.5min) before they aligned on the metaphase II plate.
KNL-1 depletion led to visible segregation defects during both meiotic divisions—tight metaphase plates failed to form and one or more lagging chromosomes were evident during anaphase (Fig. 1b; Movie S1). Chromosome counting revealed significant aneuploidy in meiosis II embryos (Control,6.0; n=23 oocytes; KNL-1-depleted,5.6±1.1; n=39 oocytes). KNL-1 depletion did not affect the timing of meiotic divisions (Fig. S3c), polar body extrusion (Fig. S3B), chromosome number during meiosis I (Control,6.0±0.1; n=68 oocytes; KNL-1-depleted,6.0±0.2; n=61 oocytes), or chromosome structure. Spindle structure also appeared normal, indicating that a gross spindle defect is not the cause of the observed segregation defects (Fig. S4a). In contrast to KNL-1 depletion, KLP-19 depletion did not cause significant defects in meiotic segregation (Fig. 1b; Movie S1), even though the same embryos showed mitotic defects (Fig. S3a) and lacked detectable KLP-19 (Fig. 1a; S1b). The primary meiotic phenotype in KLP-19 depleted embryos was excessive chromosome dispersion following anaphase I (Fig. 1c), which correlated with instability of the late anaphase spindle (Fig. 1d). Thus, direct imaging indicated that kinetochores provide a critical activity for chromosome alignment and accurate segregation on acentriolar meiotic spindles.
The meiotic spindles in the C. elegans oocyte contain numerous thick microtubule bundles that run along the sides of the meiotic chromosomes and are proposed to orient chromosomes relative to the spindle axis8. In KNL-1-depleted embryos undergoing meiosis I and II, chromosomes were improperly oriented (Fig. S1a), despite the persistence of these microtubule bundles (Fig. S4b). This observation suggested that meiotic kinetochores provide the activity that orients chromosomes relative to the spindle axis. To test this hypothesis, we quantified chromosome orientation during meiosis I using a strain expressing GFP-Aurora BAIR-2. GFP-Aurora BAIR-2 marks the short axis of each chromosome (Fig. 2a; Movie S2), enabling measurement of the angle between the spindle axis and the long axis of each chromosome. In control and KLP-19-depleted embryos, the long axis of all six chromosomes became aligned with the spindle axis prior to anaphase I (Fig. 1a; Fig. 2b,-20s; see also3,4). By contrast, in KNL-1 depleted embryos, chromosomes failed to orient relative to the spindle axis, and instead maintained an early prometaphase-like configuration (Fig. 2b; middle panel). Both meiotic divisions exhibited a similar orientation defect; however, the topology of meiosis II, where anaphase frequently occurs perpendicular to the imaging plane, prevented quantification. Thus, meiotic kinetochores are required to orient chromosomes along the spindle axis and ensure their proper segregation during both meiotic divisions.
Despite the severe orientation defects in KNL-1-depleted embryos, (Fig. 1b; Fig. 2b), chromosome masses always separated during anaphase (Fig. 3a; Fig. 1b). This was surprising, as anaphase chromosome separation during mitosis requires kinetochore activity to power poleward chromosome movement in anaphase A and to link chromosomes to spindle poles during anaphase B spindle elongation (in C. elegans mitosis, anaphase B predominates13). These observations suggested that, in contrast to mitosis, anaphase segregation on acentrosomal meiotic spindles is independent of kinetochores. Consistent with this, kymographs of anaphase I (Fig. 3b) revealed that chromosome masses in KNL-1 depleted embryos separated at a speed comparable to controls (Fig. 3c). KLP-19 depletion also had no effect on separation velocity, except in late anaphase when spindle instability led to chromosome dispersal. Thus, despite their central role in aligning chromosomes prior to anaphase onset, kinetochores appear to be dispensable for anaphase chromosome separation during female meiosis in C. elegans.
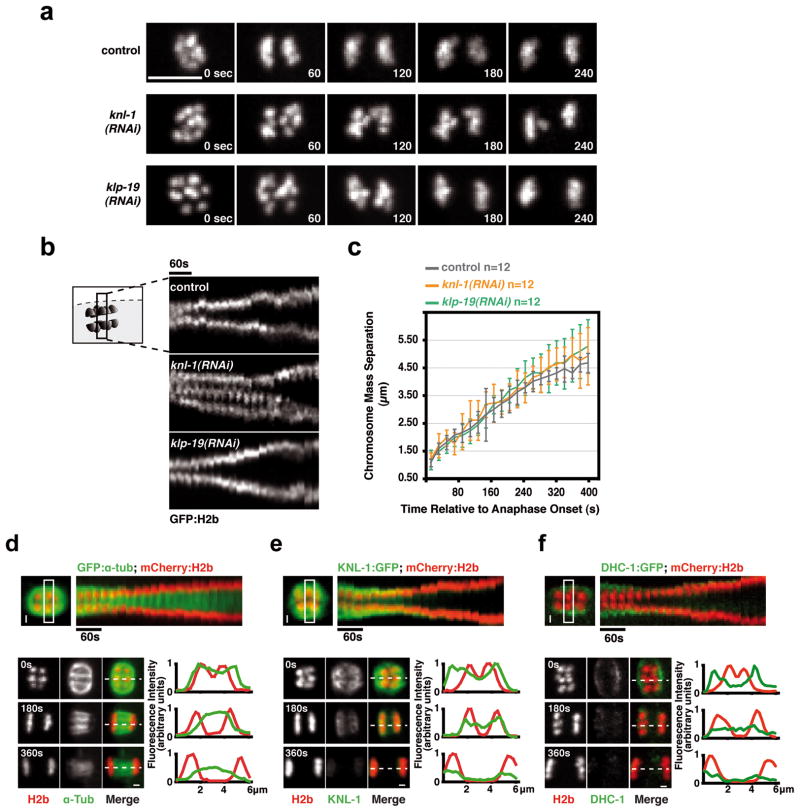
(a) Anaphase I in control, KNL-1-depletion and KLP-19 depletion. Scale bar, 5 μm. (b) Kymographs initiated at anaphase I onset; in the KNL-1 depletion the signal in the middle is a lagging chromosome. The time interval between consecutive strips is 20 sec. (c) Graph plotting average chromosome mass separation versus time, aligned with respect to the onset of anaphase I. The average separation speed is ~0.5 μm/min for all 3 conditions. Error bars represent the standard deviation. (d) & (e) & (f) Top: Kymographs from timelapse sequences of control embryos co-expressing mCherry-histone-H2b and GFP-α-tubulin (d) or KNL-1-GFP (e) or DHC-1-GFP (f). Bottom: Stills from timelapse sequences of the three strains aligned relative to anaphase onset. Normalized fluorescence intensity along a 1-pixel-wide linescan (dashed line) is plotted on the right; position 0 corresponds to the left edge of the linescan. Scale bars, 1 μm.
Consistent with kinetochore-independent anaphase segregation, significant metaphase spindle shrinkage was observed during both meiotic divisions in unperturbed embryos14, so that by anaphase onset, spindle poles were almost directly apposed to the poleward chromosomal surface (Fig. 3d; Movie S3). Shortly after anaphase onset, the meiotic kinetochores and microtubules on the poleward sides of the chromosomes disassembled; during chromosome separation, microtubules were observed primarily between the separating chromosomes (Fig. 3d,e; Movie S3). Consistent with the disassembly of poleward microtubules, the spindle pole marker, GFP-DHC-1 (dynein heavy chain) moved inwards relative to the chromosomes over the 60s immediately preceding anaphase until it was immediately adjacent to the external surface of the chromosomes, and then dissipated coincident with loss of chromosome-associated KNL-1 (Fig. 3f; Movie S3). These observations in unperturbed oocytes, together with the analysis of KNL-1-depleted oocytes, suggest the existence of a kinetochore-independent mechanism that drives anaphase chromosome separation on acentriolar C. elegans meiotic spindles.
To investigate how microtubule assembly occurs predominantly between the separating meiotic chromosomes, we focused on 4 components that initially localize to the mid-bivalent: the CPC, BUB-1, the CENP-F-like proteins HCP-1 and 2, and the Clasp ortholog CLS-2 (Fig. 4a; Fig S5). BUB-1, HCP-1/2 and ClaspCLS-2 also localize to kinetochore cups in a KNL-1-dependent fashion (Fig. 1a; S5)6. Immunofluorescence revealed that the mid-bivalent, which appears as a line in side-view projections, is a tri-laminar ring composed of three concentric layers. The CPC localized to the inner-most layer, BUB-1 and KLP-19 to the middle layer, and HCP-1/2 and ClaspCLS-2 to the outer layer (Fig. 4b; S5b). The CPC was required to recruit BUB-1, which was in turn required to target the other ring components (Fig. 4a,b). Notably, HCP-1/2, ClaspCLS-2 and BUB-1 disassembled from the kinetochore but not from the ring-like structures soon after anaphase onset (Fig. 4d and not shown). After the onset of chromosome segregation the rings began to elongate (Fig. 4d; white arrow) to form bar-shaped linker structures between the separating chromosomes. KNL-1 depletion did not affect localization of ring components (Fig. 1a; ;4c;4c; S2a) or formation of the bar-shaped structures during anaphase (Fig. 4e).
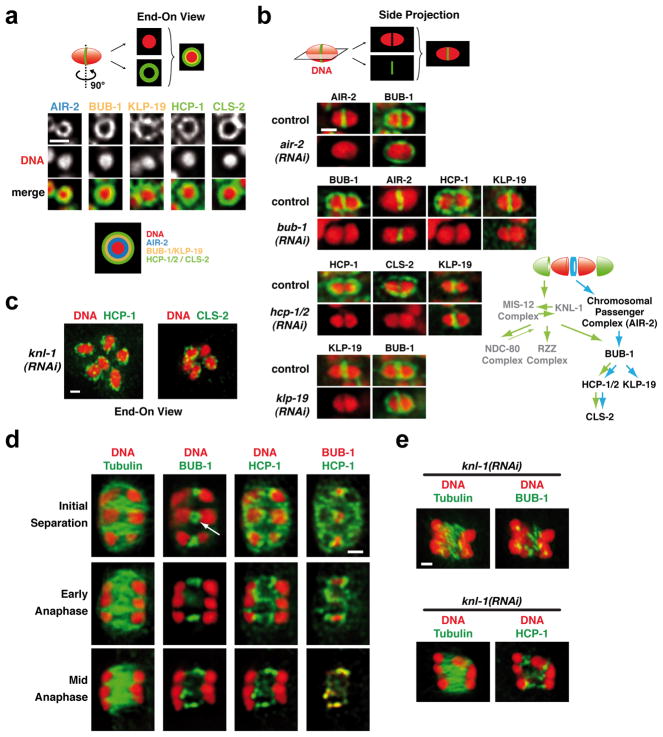
(a) End-on views illustrating the localization of the indicated components to a ring-shaped domain between the two kinetochores; see schematic for summary of the layered composition. See also Fig. S5b. (b) Analysis of the targeting dependencies of the ring protein network and summary of the results. (c) End-on views illustrating that the ring domain localization of HCP-1 and CLS-2 persists in KNL-1-depleted embryos. (d) Localization of HCP-1 and BUB-1 to linker structures between the separating chromosomes during anaphase of meiosis I. Panels are from a quadruple-labeling experiment (DNA, Tubulin, BUB-1, HCP-1); the white arrow shows the persistence of a BUB-1 ring after anaphase onset. (e) BUB-1 and HCP-1 staining during early anaphase I in KNL-1-depleted embryos. Scale bars, 1 μm.
To determine whether the ring protein network contributes to anaphase segregation, we assessed the effect of depleting BUB-1, HCP-1/2 or ClaspCLS-2, in embryos co-expressing mCherry-histone-H2b and GFP-α-tubulin (Fig. 5a; Movie S4). Unlike the CPC, these three proteins do not participate in removal of meiotic cohesion. Parallel comparison revealed a phenotypic series with cls-2(RNAi) being more severe that hcp-1/2(RNAi), which in turn was more severe than bub-1(RNAi). In cls-2(RNAi), no chromosome mass separation or assembly of anaphase microtubule bundles was observed during MI or MII (Fig. 5a; Movie S4); consistent with the severe anaphase I defect, no polar body was formed and 12 univalent chromosomes were present during MII. BUB-1 depletion resulted in a less severe phenotype where aberrant separation of chromosome masses was evident (Fig. 5a; Movie S4). HCP-1/2 depletion led to an intermediate phenotype (Fig 5a; Movie S4). While anaphase bundles are present in bub-1(RNAi) and hcp-1/2(RNAi), they are disorganized relative to controls and homolog segregation is highly aberrant. All three depletions, including that of BUB-1, also led to severe pre-anaphase spindle defects (Fig. 5a; S1; S5; Movie S4). The comparison of the three depletions suggests that ClaspCLS-2 provides a general anaphase assembly activity essential for building the microtubule bundles that separate the chromosome masses. This conclusion is consistent with abrupt loss of spindle microtubules following anaphase onset in Clasp-inhibited MII Xenopus extracts15. The significant pre-anaphase spindle defects in all three depletions indicate that the complete lack of anaphase separation in cls-2(RNAi) cannot solely be explained by pre-anaphase defects. As neither BUB-1 nor HCP-1/2 are direct regulators of microtubule dynamics, we suggest that they promote microtubule assembly between the separating chromosomes by localizing and/or regulating ClaspCLS-2 activity.
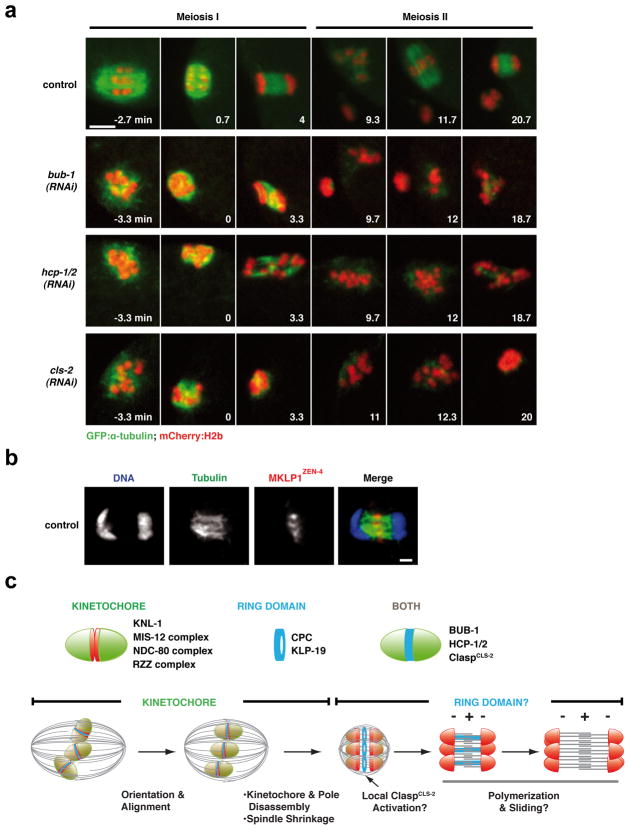
(a) Stills from movies of control, BUB-1-depleted, HCP-1/2-depleted or ClaspCLS-2-depleted fertilized oocytes expressing GFP-α-tubulin and mCherry-histone-H2b. Time, relative to anaphase I onset, is in the lower right corner of each panel. Scale bar, 5 μm. (b) DNA (blue), tubulin (green) and MKLP1ZEN-4 (red) labeling in a control meiotic embryo. Scale bar, 1 μm. (c) (Top) Summary of the composition of the cup-like kinetochores and of the ring domain located between the kinetochores of meiotic chromosomes. (Bottom) A model for chromosome orientation and segregation on acentrosomal meiotic spindles. See text for further details.
Taken together, our results suggest that meiotic chromosomes are aligned and segregated via a two-stage mechanism in C. elegans oocytes. In the first stage, chromosomes are oriented and aligned at the metaphase plate via interactions between kinetochores and spindle microtubules, potentially by a mechanism analogous to mitosis. Chromosome alignment may be facilitated by the spindle shrinkage that occurs during late metaphase, which has the potential to collect dispersed chromosomes and provide an unusual congression mechanism. We did not detect a significant contribution of the chromokinesin KLP-19 during chromosome orientation/alignment, but a subtle contribution cannot be excluded. It is possible that chromokinesins are essential to maintain congressed chromosomes when the cell spends an extended period in a prometaphase/metaphase-like stage—meiosis I mouse oocytes, where this stage lasts 7 hrs, or meiosis I-arrested C. elegans embryos8,16—but that in normally progressing cells (each meiotic division lasts about 20 min in C. elegans), their contribution is limited. After alignment, the kinetochores, the microtubules on the poleward side of the chromosomes, and the spindle poles disassemble, at which time we suggest there is a hand-off to the ring protein network, concentrated between the two chromosome halves (Fig. 5c). We speculate that the ring protein network separates chromosomes by promoting ClaspCLS-2-dependent microtubule assembly. Testing this proposal will require tightly controlled temporal inactivation, because the phenotypes associated with inhibition of ring network components are not exclusive to anaphase.
Outward pushing forces generated by microtubules between the separating chromosomes are the primary candidate for driving chromosome separation. Clustering of chromosomes during separation may be promoted by interstitial structures, analogous to those comprised of the chromokinesin Kid in mouse oocytes17, or direct physical coupling of the inside surface of the chromosomes to microtubule bundles. Microtubule orientation within the bundles, the nature of the pushing force, and the mechanism coupling microtubule assembly to chromosome separation are important issues that remain to be addressed. Photobleaching of GFP-α-tubulin (Fig. S6b; Movie S5) and imaging of GFP-EBP-1 (not shown) failed to reveal a restricted region of plus end polymerization within these bundles. However, MKLP1ZEN-4, a kinesin enriched at the microtubule overlap region of the mitotic anaphase spindle midzone, also localized to the center of the meiotic anaphase bundles (Fig. 5b), suggesting that the meiotic anaphase bundles resemble those in the mitotic midzone (Fig. 5c). Inhibition of MKLP1ZEN-4 and/or the bundling protein PRC1SPD-1 did not prevent formation of microtubule bundles or severely disrupt anaphase chromosome separation (Fig. S6a). This observation suggests that microtubule assembly/bundling during meiotic segregation is likely to primarily be under control of the CPC, and involve the activity of ClaspCLS-2.
Analysis of the ring protein network highlights the importance in anaphase chromosome separation of the mid-bivalent region, which is patterned by the site of recombination at an earlier stage of meiotic prophase18. Asymmetric disassembly of the synaptonemal complex in diakinesis is linked to asymmetric enrichment of the CPC and definition of the mid-bivalent2. This specification plays two critical functions: (1) it restricts cohesion loss to the short axis of the bivalent in MI3,4,18,19, and (2) it directs assembly of the ring protein network, which we suggest contributes to physical separation. Thus, in C. elegans, the specialized architecture of meiotic chromosomes, established following the critical event of recombination, may couple the two essential functions of anaphase: restricted cohesion dissolution and chromosome separation.
Kinetochore-dependent chromosome orientation followed by kinetochore-independent separation may be generally relevant to segregation on acentriolar meiotic spindles or may represent an adaptation specific for holocentric chromosomes. For segregation of recombined homologous chromosomes during meiosis I, monocentricity is thought to be crucial for preventing recombined sister chromatids from being simultaneously pulled toward both spindle poles20–22. Uncoupling kinetochore activity from anaphase could help avert this fate in holocentric organisms. Kinetochore-independent anaphase separation may also represent a general adaptation required for chromosome segregation on acentrosomal meiotic spindles. DNA beads/chromatin injected into mouse oocytes separate into distinct masses without functional kinetochores24 and mid-late anaphase chromosome separation in mouse oocytes resembles that in C. elegans, with robust microtubule bundles present between the chromosome masses23,24. As oocyte spindles are acentrosomal in most species, it will be interesting to test whether the kinetochore-independent segregation mechanism is conserved, especially in mammals that are particularly prone to chromosome segregation errors during oocyte meiosis25,26.
METHODS
Worm strains and antibodies
The genotypes of all strains used are listed in Table S1. The strain expressing a C-terminal GFP fusion with KNL-1 (OD248) was generated by microparticle bombardment27 of DP38 worms ([unc-119(ed3)]) using a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). The genomic sequence of knl-1 was cloned into pASM89. Affinity purified antibody against the C-terminal region (exons 6–7) of klp-19 was generated as previously described28. Other antibodies used in this study were previously described in12,13,28–30.
Live imaging
Worms were dissected in 4 μl L-15 blastomere culture31 on a 24×60 mm coverslip mounted on a homemade metal holder. A ring of petroleum jelly was deposited around the drop of medium to serve as a spacer and prevent compression of the embryos and a 18×18 mm coverslip was placed on top to seal the chamber and prevent evaporation during filming. Live imaging was conducted at 20°C using a spinning-disc confocal head (CSU-10; Yokogawa; Japan) mounted on a Nikon TE2000-E inverted microscope equipped with a krypton-argon 2.5W water-cooled laser (Spectra-Physics, Mountain View, CA) and a charge-coupled device camera (Orca-ER; Hamamatsu, Japan). Acquisition parameters, shutters, and focus were controlled by MetaMorph software (Molecular Devices, Downington, PA). Alternatively, an Andor Revolution spinning disc confocal system controlled by the Andor iQ software (Andor Technology, Belfast, Ireland) was used. In all cases a 60×, 1.4 NA PlanApochromat lens with 2×2 binning was used. For live imaging of chromosome segregation in GFP:histone-H2b embryos, 9 z-sections were collected at 1μm intervals every 10s. For embryos expressing kinetochore components fused to GFP (OD7, OD9, OD11, OD29, OD86, OD87, OD248) or co-expressing GFP-Aurora BAIR-2 and mCherry-histone-H2b (OD224) or GFP-α-tubulin and mCherry-histone-H2b (OD57) or GFP-DHC-1 and mCherry-histone-H2b (OD203) a z-series consisting of 4 planes at 2 μm intervals was collected every 20s. A single informative z-section was presented for OD9, OD11, OD29, OD86, OD87 and OD248. A maximum projection of each time-point was generated using MetaMorph for OD224, OD57 and OD203. All the kymographs were generated using NIH-imageJ (http://rsb.info.nih.gov/ij/).
Analysis of chromosome orientation
The graphs in Figure 2b were generated by measuring the angle between the axis of the meiotic spindle, defined by the direction followed by the two sets of separating chromosomes after anaphase onset, and the line-shaped Aurora BAIR-2 signal on each chromosome, 20s before anaphase onset. Anaphase onset was defined as the first time-point where Aurora BAIR2 started to leave chromosomes and transitioned onto microtubules. The measured angle was converted into the angle between the axis of each chromosome, which is perpendicular to the line-shaped Aurora BAIR-2 signal, and the spindle axis.
RNA-mediated interference
dsRNAs were prepared as described13, using the primers listed in Table S2 to amplify regions from N2 genomic DNA or N2 total cDNA or specific cDNAs. L4 hermaphrodites were injected with dsRNA and incubated at 20°C for 48 hours before dissection.
Immunofluorescence
Immunofluorescence was performed as described previously13 using a 20 minutes cold methanol fixation. All antibodies used were directly labeled with fluorescent dyes (Cy3 or Cy5; Amersham Biosciences) and used at a concentration of 1 μg/ml. Immunofluorescence against tubulin was performed with a FITC labeled α-tubulin antibody diluted 1/100 (DM1-α; Abcam, Cambridge, MA). Images were acquired using a 100X, 1.35 NA Olympus U-Planapo oil objective and a CoolSnap CCD camera (Roper Scientific) mounted on a DeltaVision system (Applied Precision). All fixed images are projections of widefield z-planes acquired every 0.2 μm that were computationally deconvolved using Softworx software (Applied Precision).
FRAP experiment
The FRAP experiments of the anaphase spindle presented in Figure S6b were performed in a strain co-expressing GFP-α-tubulin and mCherry-histone-H2b (OD57) using an Andor Revolution spinning disc confocal system controlled by the Andor iQ software (Andor Technology, Belfast, Ireland) and a 60×, 1.4 NA PlanApochromat lens with 2×2 binning. Stacks of 3 z-sections with a spacing of 2 μm were acquired every 2 seconds in the GFP channel before and after a single FRAP event of the entire surface of the anaphase spindle. The FRAP event was performed during anaphase of meiosis I when the distance between the separating chromosomes (monitored by the mCherry-histone-H2b signal) was ~3 μm. A maximum projection of the 3 z-sections is presented for each time point. 15 individual spindles were photobleached and the average fluorescence was measured in a box around the anaphase spindle (Fspin), in a box away from the spindle in the cytoplasm (Fcyt) and in a box outside of the embryo (Fout). Normalization and correction of the measured fluorescence intensities were performed using the Microsoft Excel software. The fitting was made using the KaleidaGraph software (Synergy Software). To correct for the background due to the noise of the CCD camera, Fout was substracted to Fspin at each timepoint. The background-corrected data curves were then corrected for the photobleaching occuring during acquisition by multiplying each timepoint by Fcyt(0)/Fcyt(t). In order to be able to compare different experiments, the last prebleach time point was normalized to 1 (FcorNormalized(t)=Fcor(t)/FcorPB). The mean value of FcorNormalized was then calculated for 15 individual embryos at each timepoint. The corresponding plot was fitted to a mono-exponential function and the half-time for recovery was extracted.
Supplementary Material
Movie1
Movie2
Movie3
Movie4
Movie5
Suppl_Fig1
Suppl_Fig2
Suppl_Fig3
Suppl_Fig4
Suppl_Fig5
Suppl_Fig6
Suppl_Text
Acknowledgments
We are grateful to Julie Canman and Rebecca Green for critical reading of the manuscript. This work was supported by an EMBO long-term post-doctoral fellowship (to J.D.), grants from Human Frontiers Science Program (RGY0084) and the NIH (GM074215) to A.D., and funding from the Ludwig Institute for Cancer Research to A.D. and K.O.
Footnotes
AUTHOR CONTRIBUTIONS
All experimental data were generated by J.D., who also had primary responsibility for experimental design and data analysis. A.D. and K.O. contributed to experimental design and data analysis. J.D., A.D. and K.O. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb2093
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3052858?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes.
Mol Biol Cell, 35(8):ar105, 12 Jun 2024
Cited by: 1 article | PMID: 38865189 | PMCID: PMC11321039
Phospho-KNL-1 recognition by a TPR domain targets the BUB-1-BUB-3 complex to C. elegans kinetochores.
J Cell Biol, 223(7):e202402036, 05 Apr 2024
Cited by: 0 articles | PMID: 38578284 | PMCID: PMC10996584
Redundant microtubule crosslinkers prevent meiotic spindle bending to ensure diploid offspring in C. elegans.
PLoS Genet, 19(12):e1011090, 27 Dec 2023
Cited by: 4 articles | PMID: 38150489 | PMCID: PMC10775986
Chromosomal evolution in Cryptangieae Benth. (Cyperaceae): Evidence of holocentrism and pseudomonads.
Protoplasma, 261(3):527-541, 21 Dec 2023
Cited by: 1 article | PMID: 38123818
Microtubules oppose cortical actomyosin-driven membrane ingression during C. elegans meiosis I polar body extrusion.
PLoS Genet, 19(10):e1010984, 02 Oct 2023
Cited by: 1 article | PMID: 37782660 | PMCID: PMC10569601
Go to all (126) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Meiotic kinetochores get pushed aside by a CLS act.
Nat Cell Biol, 12(9):849-851, 22 Aug 2010
Cited by: 2 articles | PMID: 20729839
Kinetochore-independent chromosome poleward movement during anaphase of meiosis II in mouse eggs.
PLoS One, 4(4):e5249, 13 Apr 2009
Cited by: 19 articles | PMID: 19365562 | PMCID: PMC2664963
The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis.
Curr Biol, 12(10):798-812, 01 May 2002
Cited by: 163 articles | PMID: 12015116
Chromosome dynamics: new light on Aurora B kinase function.
Curr Biol, 12(13):R458-60, 01 Jul 2002
Cited by: 53 articles | PMID: 12121637
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (3)
Grant ID: GM074215
Grant ID: R01 GM074215-06
Grant ID: R01 GM074215





