Abstract
Aims
EGFR mutations now guide the clinical use of EGFR-targeted therapy in lung cancer. However, standard EGFR mutation analysis requires a minimum amount of tumor tissue, which may not be available in certain situations. In this study, we combined a mass spectrometry genotyping assay (Sequenom) with a mutant-enriched PCR (ME-PCR) to detect EGFR mutations in free plasma DNA from patients with lung cancer.Method
DNAs were extracted from 31 plasma samples from 31 patients and analyzed by both methods for EGFR Exon 19 deletion and EGFR L858R mutation. Results in plasma DNA samples were compared with EGFR mutation status obtained in tumor DNA (18/31 EGFR mutant). The relationship of EGFR mutation status in tumor and/or plasma samples to overall survival was assessed.Results
The EGFR mutation status in plasma DNA was identical to the primary tumor in 61% of patients (19/31). By mass spectrometry genotyping, the plasma samples contained mutant DNA corresponding to 5/14 EGFR Exon 19 deletions and 3/4 EGFR L858R mutations previously diagnosed in the matched tumors. Two samples were positive in plasma DNA but negative in primary tumor tissue. Results were similar for samples studied by ME-PCR. For patients treated with erlotinib, overall survival was correlated with the presence of EGFR mutation in plasma and/or tumor tissue (p=0.002), with the two patients positive only in plasma DNA showing responses and favorable outcomes.Conclusion
The detection of EGFR mutations in plasma DNA samples by mass spectrometry genotyping and ME-PCR is feasible. A positive EGFR result in plasma DNA has a high predictive value for tumor EGFR status and for favorable clinical course on EGFR-targeted therapy and could therefore be useful in guiding clinical decisions in patients with insufficient or unavailable tumor specimens.Free full text

Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors
Abstract
Aims
EGFR mutations now guide the clinical use of EGFR-targeted therapy in lung cancer. However, standard EGFR mutation analysis requires a minimum amount of tumor tissue, which may not be available in certain situations. In this study, we combined a mass spectrometry genotyping assay (Sequenom) with a mutant-enriched PCR (ME-PCR) to detect EGFR mutations in free plasma DNA from patients with lung cancer.
Method
DNAs were extracted from 31 plasma samples from 31 patients and analyzed by both methods for EGFR exon 19 deletion and EGFR L858R mutation. Results in plasma DNA samples were compared with EGFR mutation status obtained in tumor DNA (18/31 EGFR mutant). The relationship of EGFR mutation status in tumor and/or plasma samples to overall survival was assessed.
Results
The EGFR mutation status in plasma DNA was identical to the primary tumor in 61% of patients (19/31). By mass spectrometry genotyping, the plasma samples contained mutant DNA corresponding to 5/14 EGFR exon 19 deletions and 3/4 EGFR L858R mutations previously diagnosed in the matched tumors. Two samples were positive in plasma DNA but negative in primary tumor tissue. Results were similar for ME-PCR. For patients treated with erlotinib, overall survival was correlated with the presence of EGFR mutation in plasma and/or tumor tissue (p=0.002), with the two patients positive only in plasma DNA showing responses and favorable outcomes.
Conclusion
The detection of EGFR mutations in plasma DNA samples by mass spectrometry genotyping and ME-PCR is feasible. A positive EGFR result in plasma DNA has a high predictive value for tumor EGFR status and for favorable clinical course on EGFR-targeted therapy and could therefore be useful in guiding clinical decisions in patients with insufficient or unavailable tumor specimens.
Introduction
The detection of EGFR mutations in lung adenocarcinomas has become a routine molecular test with important implications for patient prognosis and selection of therapy. The presence of an activating mutation predicts response to the EGFR tyrosine kinase inhibitors (TKI) erlotinib or gefitinib, and is prognostically favorable regardless of therapy (1). Unfortunately, in some cases, tumor tissue is either inadequate for molecular testing because of its small quantity or very low tumor content or is not readily available. Therefore, there is a need to develop new techniques for detecting clinically significant EGFR mutations in patients with little or no available tumor DNA.
Plasma samples from patients with lung cancer contain much higher levels of DNA than plasma from cancer-free patients. Most of this excess circulating DNA is believed to be released from the dying lung cancer cells at primary or metastatic sites (2). As such, plasma DNA may therefore provide a noninvasive source of genotypic information which could be used as a substitute for tumor tissue for detecting cancer-specific molecular markers that could be used to predict response and prognosis. Several groups have detected EGFR mutations in DNA isolated from plasma (3–7) or serum samples (8, 9) and show some correlation between mutation status in plasma and tumor tissue (3, 4, 6, 8, 9, 10). Furthermore, EGFR mutation detected in plasma or serum may, by itself, be predictive of response to EGFR TKI (3, 5, 6, 7, 9).
In this study, we report the detection of EGFR L858R mutations and EGFR exon 19 deletions in plasma samples from patients with NSCLC using a novel, mass spectrometry assay. The detection of these mutations in plasma samples is correlated with better survival when patients are treated with TKIs.
Materiel and Methods
Patients characteristics
We studied 31 patients with a biopsy-proven diagnosis of stage III or IV NSCLC and available plasma and tumor tissue. All patients gave informed consent, and the collection and analysis of their health information was approved by the Memorial Sloan-Kettering Cancer Center (MSKCC) Institutional Review Board. The patients were followed for tumor responses and survival outcomes.
Analysis of EGFR mutations in tumors tissues
EGFR Exon 19 deletion assay
Detection of the small in-frame deletions in exon 19 of EGFR was performed by fragment analysis of fluorescently labeled PCR products as previously described (11). Briefly, a 207-bp genomic DNA fragment encompassing the entire exon 19 was amplified using the primers A1 and A2 (Table 1). PCR products were subjected to capillary electrophoresis on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA). This assay can detect an EGFR exon 19 deletion in as little as 5–10% of tumor cells in a given sample (11).
Table 1
Primers listed by assay
| Fragment analysis | |
| A1 | EGFR-Ex19-FW: 5’GCACCATCTCACAATTGCCAGTTA 3’ |
| A2 | EGFR-Ex19-REV: 5’ Fam-AAAAGGTGGGCCTGAGGTTCA 3’ |
| B1 | EGFR-Ex21-FW: 5’CCTCACAGCAGGGTCTTCTCTGT 3’ |
| B2 | EGFR-Ex21-REV: 5’ Fam-TCAGGAAAATGCTGGCTGACCTA 3’ |
| Sequenom Mass Spectrometry assay | |
| C1 | EGFR-Ex19-FW 5’ACGTTGGATGGATCCCAGAAGGTGAGAAAG 3’ |
| C2 | EGFR-Ex19-REV 5’ACGTTGGATGAGCAGAAACTCACATCGAGG 3’ |
| D1 | EGFR-2573-FW 5’ ACGTTGGATGACGTACTGGTGAAAACACCG 3’ |
| D2 | EGFR-2573-REV 5’ ACGTTGGATGTCTTTCTCTTCCGCACCCAG 3’ |
| D3 | EGFR-2573 extension primer-REV: 5’ CACCCAGCAGTTTGGCC 3’ |
| Mass of extension primer: 5131.3 Da | |
| Mass of extension product Call T: 5402.6 Da | |
| Call G: 5378.5 Da | |
| D4 | LNA-EGFR-Ex21-FW: 5’-GGCTGGCCAaactgc/3InvdT/-3’ |
| Mutant-enriched PCR | |
| E1 | EGFR-Ex19-FW: 5’ATCCCAGAAGGTGAGAAAGATAAAATTC 3’ |
| E2 | EGFR-Ex19-REV1: 5’CCTGAGGTTCAGAGCCATGGA 3’ |
| E3 | EGFR-Ex19-REV2: 5’ Fam-CCTGAGGTTCAGAGCCATGGA 3’ |
| F1 | EGFR-Ex21-FW1: 5’CAGCCAGGAACGTACTGGTGA 3’ |
| F2 | EGFR-Ex21-REV1: 5’TCCCTGGTGTCAGGAAAATGCT 3’ |
| F3 | EGFR-Ex21-FW2: 5’CGCAGCATGTCAAGATCACAGAT 3’ |
| F4 | EGFR-Ex21-REV2: 5’ Fam-TCCCTGGTGTCAGGAAAATGCT 3’ |
EGFR Exon 21 L858R assay
This mutation was detected by a PCR-restriction fragment length polymorphism assay (PCRRFLP), based on a Sau961 restriction site created by the mutation 2573T>G as previously described (11). Briefly, a 222-bp genomic fragment encompassing the entire exon 21 was amplified using primers B1 and B2 (Table 1). Digestion of the mutant PCR product with Sau96I enzyme (New England BioLabs) generates a shorter 87 bp fragment. The digested, fluorescently labeled PCR products were analyzed by capillary electrophoresis on an ABI 3730 Genetic analyzer (Applied Biosystems, Foster City, CA). The detection sensitivity of this assay for the EGFR exon 21 L858R mutation is also 5–10% of tumor cells (11).
Blood sample collection and processing
The majority (n=23) of blood samples were collected prior to initiation of therapy, with the rest (n=8) drawn after initiation of therapy (TKI or cytotoxic chemotherapy). Blood samples were collected in tubes containing EDTA, and centrifuged 20 min to separate the plasma. Aliquots of plasma were stored at −80°C until DNA extraction. DNA extraction was performed from 800µL of plasma using the Qiagen DNA Virus extraction kit (without RNAse step, which induces some background in mass spectrometry assays).
Whole genome amplification
For whole genome amplification, plasma DNA was processed by a blunt-end ligation method as described (12, 13). Whole genome amplification was carried out using GenomiPhi V2 DNA amplification kit (GE Healthcare).
Analysis of EGFR mutations by mass spectrometry
Plasma DNA was analyzed by mass spectrometry-based genotyping (Sequenom Inc, San Diego, CA), as described previously (14). Briefly, tumor DNA was subject to a first PCR amplification [total volume: 5µL, 1.25x buffer, 1.625mM MgCl2, 500µM dNTP, 100nM from each primer, 0.5 U HotStar Taq DNA Polymerase (Qiagen)] using primers C1 and C2 (EGFR Exon 19 deletion assay), D1 and D2 (EGFR L858R mutation assay) (Table 1). To increase the sensitivity of the EGFR L858R PCR, we introduced a locked nucleic acid (LNA) oligonucleotide in the PCR mix at the concentration of 0.6 µM (D4, Table 1). Alkaline Phosphatase treatment was performed in a total volume of 2µL (final volume: 7µL), incubated at 37°C for 40min and 85°C for 5 min. A single allele base extension reaction was performed using the indicated primers (Table S1 for EGFR Exon 19 deletions; D3 (Table 1) for EGFR L858R). Two microliters of a solution containing 0.74 µL ddH2O, 0.2 µL Buffer (Sequenom Inc.), 0.1µL Termination Mix (Sequenom Inc.), 0.94 µL of 7–13.67 µM extension primer (depending of the mass) and 0.02 µL of Thermo Sequenase (Sequenom Inc.) were added to the 7 µL SAP (Shrimp Alkaline Phosphatase) treated PCR products. Thermal cycling was performed following the recommendations of Sequenom. A different well was used for each single allele base extension primer. The detection sensitivity of unmodified Sequenom assays for mutant alleles is estimated to be 5% tumor cells (range 1–10%) (15). Using the LNA at 0.6 µM, the sensitivity of the Sequenom EGFR L858R assay increases to 0.1 % (Supplementary figure S1).
Analysis of EGFR mutations by mutant-enriched PCR
EGFR Exon 19 deletion assay
Cases showing discordant results between tumor mutation status and plasma mutation status were further studied using a more sensitive mutant-enriched PCR assay (16). Briefly, a 138-bp genomic fragment of exon 19 was amplified with the primers E1 and E2 (Table 1) for 17 cycles at 60°C. After purification (QIAquick Purification kit, Qiagen), PCR products were subjected to restriction enzyme digestion by the MseI enzyme (New England BioLabs) for 4 hours at 37°C. On an EGFR allele with an exon 19 deletion, the restriction site is absent yielding an undigested PCR product. During a second round of amplification (primers E1 and E3 (Table 1), 40 cycles at 60 °C) only the mutant and the incompletely digested wild type fragments are amplified. The PCR products were analyzed by fragment analysis on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA). This assay has been reported to detect an EGFR exon 19 deletion at a level of 0.05% (16).
EGFR Exon 21 L858R assay
A similar method, described by Asano, et al, was used for the EGFR Exon 21 assay (16). In this assay, the first PCR amplification results in a 130-bp genomic fragment (primers F1 and F2 (Table 1), 17 cycles at 60 °C), After purification (QIAquick Purification kit, Qiagen), the MscI restriction enzyme (New England BioLabs) was used to cut at the TGGCCA sequence of the wild-type allele (37 °C, 4 hours). A second amplification was done using the primers F3 and F4 (Table 1) (40 cycles at 60 °C). After purification, the PCR products were digested with the Sau961 (GGNCC) (New England BioLabs) (2 hours at 37 °C) and analysed by fragment analysis on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA). A positive result corresponds to the presence of a 99-bp fragment. This assay has been reported to detect an EGFR L858R mutation at a level of 0.05% (16).
Statistical analyses
The relationship between EGFR mutation and factors such as sex, age, stage, histologic type and smoking was examined by χ2 test or Fisher’s exact test using GraphPad software (La Jolla, CA). Time from the date of lung cancer diagnosis to death was used to compare overall survival (OS) between the different groups. Survival curves were estimated by the Kaplan-Meier method with log-rank test using MedCalc Software (Mariakerke, Belgium).
Results
Patients and tumor characteristics
Patient characteristics are summarized in Table 2. There were 16 women and 15 men, 30 with lung adenocarcinoma and one patient with poorly differentiated squamous cell carcinoma. Seventeen patients were former or current-smokers and 14 were never smokers. At time of blood collection, all patients were stage IV except one with stage III disease. By the mutation-specific PCR assays used in routine practice at MSKCC to establish EGFR mutation status in tumor tissue (10), 14 tumors had EGFR Exon 19 deletion (15-bp deletion in 12 cases, 18-bp deletion in one case and 9-bp deletion in one case), and 4 tumors had the EGFR L858R mutation. The remaining 13 patients did not have any mutation in the tumor at the time of diagnosis (Table 2).
Table 2
Patient characteristics.
| Variable | N | % | EGFR mutation | EGFR WT | |||
|---|---|---|---|---|---|---|---|
| 31 | 18 | 58.1% | 13 | 41.9% | |||
| Age, years | |||||||
| Mean | 62.2 | 65.22 | 57.85 | ||||
| Range | 38–86 | 41–86 | 38–73 | ||||
| ≥65 years | 16 | 52% | 12 | 67% | 4 | 31% | p=0.07 |
| <65 years | 15 | 48% | 6 | 33% | 9 | 69% | |
| Sex | |||||||
| Male | 15 | 48% | 9 | 50% | 6 | 46% | p=1 |
| Female | 16 | 52% | 9 | 50% | 7 | 54% | |
| Smoking history | |||||||
| Smoker | 17 | 55% | 8 | 44% | 9 | 69% | p=0.2 |
| Never smoker | 14 | 45% | 10 | 56% | 4 | 31% | |
| Histologic subtype | |||||||
| ADC | 30 | 97% | 18 | 100% | 12 | 92% | p=0.4 |
| SCC | 1 | 3% | 0 | 0% | 1 | 8% | |
| Disease stage at blood collection | |||||||
| III | 1 | 3% | 0 | 0% | 1 | 8% | p=0.4 |
| IV | 30 | 97% | 18 | 100% | 12 | 92% | |
| Therapy | |||||||
| Chemotherapy | 24 | 77% | 11 | 61% | 13 | 100% | p=0.08 |
| Erlotinib | 25 | 81% | 18 | 100% | 7 | 54% | |
| Prior treatment | |||||||
| Yes | 8 | 26% | 6 | 33% | 2 | 15% | p=0.4 |
| No | 23 | 74% | 12 | 67% | 11 | 85% | |
Among the 31 patients, 25 patients received the EGFR tyrosine kinase inhibitor (TKI), erlotinib. Among the 18 patients with EGFR mutant tumors, 6 patients had received erlotinib between 10 days and 22 months before the first blood collection while the remaining 12 patients were erlotinib-naïve. Finally, 24 patients received chemotherapy in first line or second line therapy.
EGFR mutations in plasma DNA
Plasma samples were collected before treatment or between 10 days and 22 months after treatment initiation. Plasma samples were first analyzed by Sequenom mass spectrometry assays. The Sequenom EGFR Exon 19 assay was positive in 5/14 plasma samples from patients who had EGFR Exon 19 deletion detected in their tumor (36%). All 5 positive patients showed concordance for the specific subtype (size) of exon 19 deletion detected in the tumor and plasma DNA (Figure 1). The EGFR Exon 19 deletion assay was also positive in 2 patients (WT-1, WT-2) who did not have an EGFR mutation detected in their tumor tissue (Table 3).
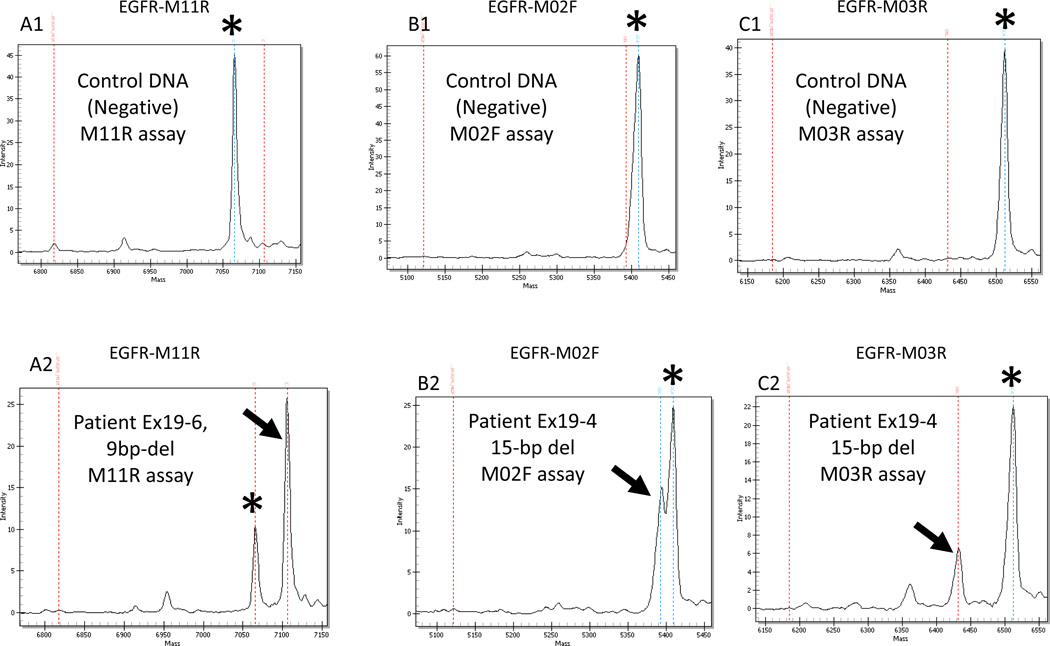
Sequenom assay results for EGFR exon 19 deletion in two plasma samples (patients Ex19-6 and Ex19-4) with the EGFR-M11R (A1 and A2), EGFR-M02F (B1 and B2) and EGFR-M03R (C1 and C2) assays. The control DNA provides a negative control for each assay and only the germline call is visible, indicated by an asterisk (A1, B1, C1). The mutant call is indicated by an arrow. The mass of the product is specific for each assay and is listed in table S1 (Supplementary file). Sample Ex19-6 shows a mutant call for the EGFR-M11R assay (A2) indicating a 9-bp deletion. Sample Ex19-4 shows a mutant call for the EGFR-M02F (B2) and the EGFR-M03R assays (B3) indicating a 15-bp deletion size.
Table 3
Summary of mutations detected in Tumor and Plasma DNA samples.
| Patient | Tumor-RFLP | Plasma-Sequenom | Plasma-ME-PCR | Tumor-Sequenom | Tumor-ME-PCR |
|---|---|---|---|---|---|
| Ex21-1 | L858R | L858R | L858R | NP | NP |
| Ex21-2 | L858R | L858R | - | NP | NP |
| Ex21-3 | L858R | - | - | NP | NP |
| Ex21-4 | L858R | L858R | L858R | NP | NP |
| Ex19-1 | Ex19 del-18bp | Ex19 del-18bp/L858R | Ex19 del-18bp/L858R | unavailable | unavailable |
| Ex19-2 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-3 | Ex19 del-15bp | Ex19 del-15bp | Ex19 del-15bp | NP | NP |
| Ex19-4 | Ex19 del-15bp | Ex19 del-15bp | Ex19 del-15bp | NP | NP |
| Ex19-5 | Ex19 del-15bp | Ex19 del-15bp | Ex19 del-15bp | NP | NP |
| Ex19-6 | Ex19 del-9bp | Ex19 del-9bp | Ex19 del-9bp | NP | NP |
| Ex19-7 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-8 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-9 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-10 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-11 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-12 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-13 | Ex19 del-15bp | - | - | NP | NP |
| Ex19-14 | Ex19 del-15bp | - | - | NP | NP |
| WT-1 | - | Ex19 del-15bp | Ex19 del-15bp | - | - |
| WT-2 | - | Ex19 del-15bp | Ex19 del-15bp | unavailable | unavailable |
| WT-3 | - | - | - | NP | NP |
| WT-4 | - | - | - | NP | NP |
| WT-5 | - | - | - | NP | NP |
| WT-6 | - | - | - | NP | NP |
| WT-7 | - | - | - | NP | NP |
| WT-8 | - | - | - | NP | NP |
| WT-9 | - | - | - | NP | NP |
| WT-10 | - | - | - | NP | NP |
| WT-11 | - | - | - | NP | NP |
| WT-12 | - | - | - | NP | NP |
| WT-13 | - | - | - | NP | NP |
RFLP= Restriction Fragment Length Polymorphism. ME-PCR = Mutant-Enriched PCR. NP=Not performed.
The Sequenom EGFR L858R assay showed the L858R mutation in 3/4 patients with L858R detected in the tumor (75%, Table 3). The EGFR L858R assay was also positive for one patient for which an EGFR Exon 19 deletion, but not a L858R mutation, had been detected in the tumor tissue (patient Ex19-1, Table 3) (Figure 2).

Sequenom assay results for EGFR L858R mutations in two plasma samples (patients Ex21-1 and Ex21-2) with the EGFR-2573 assay. The control DNA provides a negative control and only the germline call is visible, indicated by an asterisk (a T call at 5402.6 Da). The mutant calls are visible at 5378.5 Da (G call) and are indicated by an arrow.
In parallel, all plasma samples were analyzed by ME-PCR for EGFR Exon 19 and Exon 21 mutations. All EGFR Exon 19 deletion positive cases observed in plasma DNA by Sequenom assays were confirmed by ME-PCR (Table 3). The EGFR Exon 19 ME-PCR assay did not detect additional plasma DNA-positive cases among patients with EGFR Exon 19 deletion in their tumor. The ME-PCR technique confirmed the positivity of the two positive plasma samples among patients with tumors thought to be EGFR wild type (WT-1, WT-2) (Table 3). Concerning the EGFR L858R mutation, ME-PCR confirmed the presence of a L858R mutation in the plasma of 3/5 patients (two patients with L858R mutation diagnosed in the tumor and one patient with EGFR L858R mutation diagnosed only in plasma samples) (Table 3).
To verify the discordant EGFR mutations between plasma DNA and primary tumor, we reanalyzed the tumor DNA of one of the 3 cases with discordant positive results between the original analysis of tumor DNA and the Sequenom/ME-PCR assays on plasma DNA (patient WT-1, Table 3). Both Sequenom genotyping and ME-PCR confirmed the absence of EGFR Exon 19 deletion in the tumor of this patient. Tumor DNA was not available for the other 2 patients with discordant positive plasma DNA results (WT-2, Ex19-1) (Table 3). Finally, EGFR status was highly concordant between mutations detected in plasma DNA and tumor DNA, matching in 61% with only 2 patients with EGFR mutation detected only in plasma DNA (positive predictive value = 80%).
Correlation between EGFR mutations in tumor and/or plasma and clinicopathological characteristics
We examined correlations between overall survival (OS), age, sex, and smoking history. Patients with EGFR mutation in the tumor who received erlotinib had a better OS than patients who received erlotinib in the absence of EGFR mutation (p=0.01) (Figure 3A). The correlation was even stronger when patients with EGFR mutation in plasma and/or tumor DNA were compared with patients with EGFR mutations in neither (p=0.002) (Figure 3B). Among the 20 patients treated with erlotinib and presenting EGFR mutations in tumor and/or plasma DNA, the detection of an EGFR mutation in plasma DNA by itself was not a predictor of survival (p=0.4, n=10 in each group). There was no significant correlation between EGFR mutations in plasma DNA and age (p=0.3), sex (p=1), smoking history (p=0.6), or treatment before the blood collection (p=0.3) but the numbers for most of these comparisons were small.

Overall survival curves for the 25 patients treated with erlotinib. Survival curves were evaluated by the Kaplan-Meier method and log-rank test. 3A “Patients with EGFR mutation in tumor DNA (n=18)” versus “Patients without EGFR mutation in tumor DNA (n=7)”, p=0.01. 3B “Patients with EGFR mutation in plasma or tumor DNA (n=20)” versus “Patients without EGFR mutation in either tumor or plasma DNA (n=5)”, p=0.002.
Discussion
The detection of EGFR mutations in tumor tissue has important prognostic and predictive value, and can be used to select therapy for the treatment of lung adenocarcinoma. Many patients with stage IV adenocarcinoma are diagnosed with small biopsies, or by fine needle aspiration of tumors which often yields insufficient DNA for molecular testing. Detection of EGFR mutations in plasma samples would be of value for patients in whom sufficient tumor tissue is not available.
In the present study, we found that Sequenom mass spectrometry genotyping assays on plasma DNA samples show concordance with tumor genotyping in 61% of the cases (19/31). In plasma DNA, we detected 36% (5/14) of EGFR Exon 19 deletion cases, and 75% (3/4) of EGFR L858R mutation cases, compared with the matched primary tumors. Table 4 provides details on the different methods used in previous reports to detect EGFR and KRAS mutations in plasma samples (3–10, 17, 18). Depending of the technique, the concordance between EGFR status in tumor and plasma/serum samples varies from 66 to 100% with the highest correlation index being reported for the denaturing high-performance chromatography (3) and the mutant-enriched PCR (6). It is important to note that cases in which plasma DNA is negative for the mutation found in the tumor cannot be considered as conventional “false-negatives” as the possibility that there is little or tumor DNA being shed in the blood in a given case cannot be excluded.
Table 4
Different methods used in selected previous reports to detect EGFR and KRAS mutations in plasma and serum samples of lung cancer patients.
| Assay design | Gene | n | Sensitivity | Specificity | Sample | |
|---|---|---|---|---|---|---|
| Kimura et al 2006 (8) | PCR + Direct Sequencing | EGFR | 12 | 66% | 63/71 %* | Serum |
| Kimura et al 2007 (9) | Scorpion-amplification refractory mutation system | EGFR | 42 | 85% | 94% | Serum |
| Bai et al 2009 (3) | Denaturing High-Performance liquid chromatography | EGFR | 230 | 82% | 90% | Plasma |
| He C et al 2009 (6) | Mutant-enriched PCR | EGFR | 18 | 100% | 89% | Plasma |
| Mack et al 2009 (5) | Scorpion-amplification refractory mutation system | EGFR | 49 | n.a. | n.a. | Plasma |
| Yung 2009 (4) | Microfluidics digital PCR | EGFR | 35 | 92% | 100% | Plasma |
| Jian 2009 (7) | Lightcycler PCR with Taqman-MGB probes | EGFR | 88 | n.a. | n.a. | Plasma |
| Kuang 2009 (10) | Scorpion-amplification refractory mutation system | EGFR | 30 | 70% | 85% | Plasma |
| Wang 2010 (17) | PCR-restriction fragment length polymorphism | KRAS | 273 | 77% | 95% | Plasma |
| Tsao 2009 (18) | Colorimetric membrane array method | KRAS | 209 | 83% | 96% | Peripheral blood |
| Tsao 2009 (18) | Weighted chemiluminescent membrane array | KRAS | 209 | 93% | 94% | Peripheral blood |
n.a. : Sensitivity and specificity are not available because of a lack of correlation with primary matched tumors.
In the study by Bai et al. (3), 89.6% of the patients received prior therapy before blood collection. Timing of chemotherapy was not reported by He et al (6). In the present study, EGFR mutations were more often detectable in plasma samples from patients who received treatment before the blood collection, 67% (4/6) versus 33% (4/12), although the difference was not statistically significant. That could be explained by an increasing amount of circulating DNA due to tumor cell death in response to treatment. Similar results were observed by Kimura et al (8) who observed the EGFR E746-A750del more frequently in post-treatment samples. In contrast, using a microfluidic digital PCR method, Yung et al described a decreased concentration of the mutant sequences in the plasma after treatment compared with pre-treatment plasma samples (n=5) (4).
In the present study, two mutations detected in plasma samples were not detected in the corresponding tumor samples. One of these 2 cases was a poorly differentiated squamous cell carcinoma. This phenomenon has also been observed in previous studies (3, 5, 6, 8, 9) and could be explained by tumor heterogeneity, or by the presence of a second primary tumor harboring the mutation detected in the plasma DNA. The clinical observation that these 2 patients showed a good survival with EGFR TKIs supports the notion that these plasma DNA mutations were representative of underlying disease rather than being technical false positives.
More surprising was the detection of both EGFR Exon 21 and EGFR Exon 19 mutations in the same plasma DNA sample, while only EGFR Exon 19 deletion was observed in the primary matched tumor. We propose that it is more probable that this patient harbored a second primary tumor rather than both mutations being present in the same clonal tumor. Separate primary tumors with discordant EGFR or KRAS mutations are well described (19).
Mutant-enriched PCR (ME-PCR) has been previously used to detect EGFR L858R mutation and EGFR Exon 19 deletions in plasma samples (6). In the present study, ME-PCR did not detect more EGFR mutations in plasma DNA samples than the mass spectrometry genotyping assays. The Sequenom mass spectrometry platform has the advantage of being able to detect multiple mutations in the same assay, meaning that less sample and less time is needed to screen for different mutations in plasma DNA. For instance, testing for EGFR mutations could be run in parallel with testing for KRAS mutations. Indeed, detection of KRAS mutations in plasma DNA has already been reported (17, 18) using other methods and the detection of KRAS mutations in tumors by Sequenom is also feasible (15).
As observed in previous studies (3, 5, 6, 7, 9), erlotinib was more effective in improving survival in patients with EGFR mutation in the tumor and/or plasma DNA, with the correlation with outcome being enhanced by using either source of DNA to determine the EGFR status.
In conclusion, we show that mass spectrometry genotyping assays on the Sequenom platform can detect EGFR L858R mutations and EGFR Exon 19 deletions in plasma DNA samples from patients with lung cancer. The EGFR mutation status is identical to the primary matched tumor in 61% of the samples but, given the high positive predictive value of the technique (8/10) and the favorable outcome of patients with mutations detected only in plasma DNA, it is likely that most or all patients with a positive result in plasma DNA are correctly assigned to the EGFR mutant group. Thus, patients with TKI-sensitive EGFR mutations detected in plasma DNA could begin EGFR TKI treatment without the need for re-biopsy or delays caused by difficult or unsuccessful retrieval of archival tumor tissue. The recent development and validation of two mutation-specific monoclonal antibodies for the detection of EGFR mutations by immunohistochemistry, generated against the L858R mutant and the exon 19 mutant with the common 15bp/5AA deletion, suggests clinical scenarios in which these could be combined with plasma DNA testing to enhance clinical confidence, for instance in cases where cytologic diagnosis is based on small clusters of cells insufficient for DNA extraction but sufficient for immunohistochemistry (20, 21). Finally, it is important to note that, although we and others have demonstrated that the molecular diagnosis of EGFR status based on plasma DNA samples is feasible, analysis for EGFR mutations in tumor DNA remains the gold standard.
Acknowledgments
Supported by NIH P01 grant CA129243 (ML), the Gretchen and Samuel Feldman Fellowship (MB) and La Fondation de France (MB).
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.lungcan.2010.10.014
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3282180?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.lungcan.2010.10.014
Article citations
A Phase II Study of Osimertinib in Patients with Advanced-Stage Non-Small Cell Lung Cancer following Prior Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor (EGFR TKI) Therapy with EGFR and T790M Mutations Detected in Plasma Circulating Tumour DNA (PLASMA Study).
Cancers (Basel), 15(20):4999, 16 Oct 2023
Cited by: 1 article | PMID: 37894366 | PMCID: PMC10605750
Recent advances in the detection of glioblastoma, from imaging-based methods to proteomics and biosensors: A narrative review.
Cancer Cell Int, 23(1):98, 20 May 2023
Cited by: 3 articles | PMID: 37210528 | PMCID: PMC10199620
Review Free full text in Europe PMC
Liquid biopsy in lung cancer management.
Rom J Morphol Embryol, 63(1):31-38, 01 Jan 2022
Cited by: 1 article | PMID: 36074665 | PMCID: PMC9593132
Review Free full text in Europe PMC
Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS.
Nat Commun, 12(1):3770, 18 Jun 2021
Cited by: 67 articles | PMID: 34145282 | PMCID: PMC8213710
Detection of epidermal growth factor receptor mutations in peripheral blood circulating tumor DNA in patients with advanced non-small cell lung cancer: A PRISMA-compliant meta-analysis and systematic review.
Medicine (Baltimore), 99(40):e21965, 01 Oct 2020
Cited by: 8 articles | PMID: 33019389 | PMCID: PMC7535563
Review Free full text in Europe PMC
Go to all (86) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer.
J Thorac Oncol, 4(12):1466-1472, 01 Dec 2009
Cited by: 41 articles | PMID: 19884861
EGFR Mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler.
Clin Cancer Res, 11(8):2924-2929, 01 Apr 2005
Cited by: 79 articles | PMID: 15837743
Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer.
JAMA Oncol, 2(8):1014-1022, 01 Aug 2016
Cited by: 322 articles | PMID: 27055085 | PMCID: PMC4982795
Picoliter-Droplet Digital Polymerase Chain Reaction-Based Analysis of Cell-Free Plasma DNA to Assess EGFR Mutations in Lung Adenocarcinoma That Confer Resistance to Tyrosine-Kinase Inhibitors.
Oncologist, 21(2):156-164, 14 Jan 2016
Cited by: 36 articles | PMID: 26768482 | PMCID: PMC4746084
Funding
Funders who supported this work.
La Fondation de France
NCI NIH HHS (5)
Grant ID: P01 CA129243-05
Grant ID: T32 CA009207-35
Grant ID: R25 CA020449
Grant ID: T32 CA009207
Grant ID: P01 CA129243
NIH P01 grant (1)
Grant ID: CA129243





