Abstract
Free full text

Genome-wide Association Study of N370S Homozygous Gaucher Disease Reveals the Candidacy of CLN8 gene as a Genetic Modifier Contributing to Extreme Phenotypic Variation
Abstract
Mutations in GBA1 gene result in defective acid β-glucosidase and the complex phenotype of Gaucher disease (GD) related to the accumulation of glucosylceramide-laden macrophages. The phenotype is highly variable even among patients harboring identical GBA1 mutations. We hypothesized that modifier gene(s) underlie phenotypic diversity in GD and performed a GWAS study in Ashkenazi Jewish patients with type 1 GD (GD1), homozygous for N370S mutation. Patients were assigned to mild, moderate or severe disease category using composite disease severity scoring systems. Whole-genome genotyping for >500,000 SNPs was performed to search for associations using OQLS algorithm in 139 eligible patients. Several SNPs in linkage disequilibrium within the CLN8 gene locus were associated with the GD1 severity: SNP rs11986414 was associated with GD1 severity at p value 1.26 × 10−6. Compared to mild disease, risk allele A at rs11986414 conferred an odds ratio of 3.72 for moderate/severe disease. Loss of function mutations in CLN8 causes neuronal ceroid-lipofuscinosis but our results indicate that its increased expression may protect against severe GD1. In cultured skin fibroblasts, the relative expression of CLN8 was higher in mild GD compared to severely affected patients in whom CLN8 risk alleles were over-represented. In an in vitro cell model of GD, CLN8 expression was increased which was further enhanced in the presence of bioactive substrate, glucosylsphingosine. Taken together, CLN8 is a candidate modifier gene for GD1 that may function as a protective sphingolipid sensor and/or in glycosphingolipid trafficking. Future studies should explore the role of CLN8 in pathophysiology of GD.
Introduction
In Gaucher disease (GD), the defective activity of acid β-glucosidase due to mutations in the GBA1 gene leads to progressive accumulation of the substrate, glucocerebroside in the lysosomes of mononuclear phagocytes [1]. The result is widespread accumulation of glucocerebroside-laden macrophages, systemic macrophage activation and a complex phenotype involving the liver, the spleen, the bones and the bone marrow [1]. Non-neuronopathic Type 1 Gaucher disease (GD1) represents the most common form accounting for >90% of currently known patients. Acute and chronic neuronopathic forms, type 2 and type 3 GD respectively, manifest neurodegenerative disease in addition to the systemic features [2]. Among lysosomal storage diseases, GD is the most common, with overall frequency of ~ 1 in 40,000 individuals but in the Ashkenazi Jews, as many as ~ 1 in 500 individuals are affected [1]. GD1 has served as an excellent prototype for genotype/phenotype delineation and therapeutic innovation for lysosomal storage diseases [3]. However, the pathways from lysosomal accumulation of glucocerebrosides to clinical manifestations of GD are not known and this has hindered the delineation of basis of phenotypic variation [4-6].
The gene encoding acid β-glucosidase (GBA1) and its highly homologous pseudogene (96% identity) are closely linked on human chromosome 1q21 [7]. More than 200 mutations in the active GBA1 gene have been catalogued, but only 4 account for >90% of disease alleles in patients of Ashkenazi Jewish ancestry and ~ 50% of disease alleles in non-Jewish patients [1, 7]. In studies from North America and Europe, N370S comprise up to 70% of all disease alleles; N370S mutation occurs in homozygous form typically in patients of Ashkenazi Jewish ancestry and it is estimated ~1 in 15 individuals in this population are carriers of this mutation [8]. A disease mutation comprising 55bp deletion in exon 9 spans the site of N370S mutation, which results in erroneous genotype assignment (N370S homozygous in N370S/55bp deletion) when allele-specific PCR assays are employed [7, 9].
There is some genotype/phenotype correlation so that the presence of at least one N370S allele absolutely precludes the development of neuronopathic disease [1]. As a group, N370S homozygous patients tend to have less severe disease compared to compound heterozygous N370S/L444P or N370S/84GG [7, 8, 10, 11]. However, within each GBA1 genotype category and even among affected siblings there is extreme variation of disease severity [11, 15,16]. In fact, it has been suggested that homozygosity for N370S mutation results in such mild disease that a significant number never come to medical attention; yet others develop incapacitating disease at young age [7, 12].
The basis of such extraordinary phenotypic variation in this single gene/single enzyme disorder and widely regarded as involving a single cell type (the mononuclear phagocyte), is not understood. Studies have demonstrated lack of correlation between the residual acid β-glucosidase activity and disease severity, however these studies have been questioned due to failure to measure in situ lysosomal enzyme activity in intact cells [13, 14]. Sib-pairs with GD have been reported to exhibit different pattern of disease manifestations as well differ in overall disease severity [15, 16]. Therefore, modifier gene(s) have been proposed to underlie phenotypic variation [13]. It should be kept in mind, that two pairs of identical twins have been reported with divergent phenotypes, suggesting that the basis of phenotypic diversity may be more complex [17,18].
A number of modulators of residual enzyme activity have been proposed as determinants of disease severity, i.e. lysosomal pH [19], saposin [20] and intracellular trafficking [21]. A priori, the determinants of phenotype variation in GD among patients with the same GBA1 genotype could arise from environmental factors or through role of genetic modifier(s). Anecdotal evidence suggests that environmental factors through viral infections, i.e., EBV, may accelerate disease course [22]. With respect to potential genetic modifier(s) in the gene dense GBA1 locus there are eleven widely distributed polymorphic sites that are in linkage disequilibrium. All N370S chromosomes appear to harbor an identical haplotype, regardless of geographic origin of the patient [7]. Therefore variation in disease severity cannot be explained on the basis of other mutations or polymorphic variations in the GBA1 gene locus, the promoter or enhancer sequences—at least in this ethnic group [7, 23]. Studies have explored SNPs in genes encoding glucosylceramide synthase gene, GBA2, GBA3, vitamin D receptor, IL6 and TNF alpha, but there was no consistent correlation with disease severity [24-28]. No other plausible candidate genes have emerged based on current understanding of pathophysiology to explain phenotypic variation in GD…
We conducted a hypothesis-generating GWAS through unbiased exploration for association of SNPs with GD1 severity. The study was focused on N370S homozygous GD1 patients of Ashkenazi Jewish ancestry who exhibit extreme phenotypic variation with the rationale of conducting the study in genetically homogenous population to minimize population stratification.
Methods
Determination of Disease Severity
Patients were enrolled at the Gaucher Disease Treatment Centers of Yale School of Medicine and NYU School of Medicine. The study was approved by the Human Investigation Committee of the Yale University of Medicine and the Institutional Review Board of NYU School of Medicine.
Eligible patients represented consecutive N370S homozygous GD patients in whom comprehensive phenotype data prior to enzyme replacement therapy (ERT) was available. Patients had confirmed diagnosis of GD1 based on low (i.e., <10% of normal) acid β-glucosidase activity in peripheral blood leucocytes and GBA1 mutation analysis. GBA1 genotyping was performed as described previously to ensure absence of 55bp deletion in exon 9 [9]. Database included longitudinal phenotype data regarding bone, hematology, visceral involvement, age of onset, severity score index and whether prior splenectomy had been performed. Liver and spleen volumes were determined by volumetric MRI and depicted as fold-enlargement compared to normal (x N); normal liver volume is 2.5% body weight and spleen volume 0.2% body weight as described previously [10]. Hemoglobin, platelet count, MRI, Hermann Score [10], DXA scan, Bone Marrow Burden (BMB) score [29] were determined prior to ERT. Overall disease severity was determined according to Severity Score Index (SSI) [30] and the recently validated DS3 score [31]. Age of onset of symptoms, age at splenectomy and age at initiation of ERT were also recorded in the database. We aimed to assess association of SNPs based on ordinal ranking of disease severity, and therefore, the patients were assigned categories of mild, moderate and severe disease (Table I). After initial scoring via DS3 and SSI, patients were ranked by disease severity from most mild to severe. Additionally, experts in Gaucher disease (PKM and GMP) who have followed patients for >10 yrs assigned a global assessment of disease severity of the patient. Using the DS3, SSI score and global assessments, patients were assigned to severity categories of mild, moderate and severe. Illustrative natural history of mildly affected GD1 patient compared to severely affected patient is depicted in Supplementary Figure 1. In addition to variation in overall disease severity, GD is recognized to exhibit different patterns of disease among affected patients, i.e., visceral vs skeletal disease. Therefore, in addition to overall disease severity, we assessed severity of individual disease compartments. A category designation was created for bone disease severity as well as severity of visceral disease. For bone disease, severity was based on pre-ERT Hermann score as described previously [10] with scores 1-2 classified as mild, 3 as moderate, and 4-5 as severe. Patients were also assessed based on visceral disease with spleen size: 1-5 × N as 0 point, 5-10 × N as 1 point, 10-15 × N as 2, and greater than 15 or splenectomy as 3 points. Severity scoring for hepatomegaly was as follows: less than 1.25 × N as 0 point, 1.25-1.5 × N as 1 point, 1.5-1.75 as 2 point and greater than 1.75 as 3 points; presence of cirrhosis/portal hypertension or hepatopulmonary syndrome as 4 points. Aggregate cores for liver and spleen involvement assigned severity of visceral disease: 0-1 classified as mild; 2-3 points, moderate and ≥4 points, severe.
Table I
| Severity | Age | SSI | DS3 | Liver Size | Spleen | % Female | % Splenectomy |
|---|---|---|---|---|---|---|---|
| Mild | 47.7±1.9 (n=80) | 4.2±0.2 (n=80) | 1.7±0.1 (n=80) | 1.1±0.0 (n=80) | 4.9±0.3 (n=80) | 55% | 0% |
| Moderate | 51.8±3.3 (n=35) | 7.4±0.3 (n=35) | 4.3±0.1 (n=35) | 1.3±0.1 (n=35) | 7.7±1.1 (n=32) | 55% | 9.6% |
| Severe | 57.5±3.5 (n=24) | 12.1±0.4 (n=24) | 7.0±0.3 (n=24) | 1.6±0.2 (n=24) | 13.5±2.4 (n=17) | 50% | 29.2% |
| Total | 50.4±1.5 (n=139) | 6.4±0.3 (n=139) | 3.2±0.2 (n=139) | 1.2±0.0 (n=139) | 6.8±0.5 (n=129) | 55% | 7.2% |
Demographic information of the study population is depicted in Table I.
Genotyping and Patient Exclusion Criteria
A total of 186 GD1 patients confirmed to be homozygous for N370S mutation were genotyped on Illumina 650 SNP array, which contains 620,901 SNPs. The GenTrain 2.0 calling algorithm implemented in GenomeStudio from Illumina was used to infer the genotype. Quality control measures included: excluding samples with <95% call rate, excluding SNPs with call rates <94%, MAF less than 0.5% and HWE p-value <1×10−5. The above quality control measurements were implemented in PLINK package [32].
Additionally, we excluded patients under age 18 yrs who were diagnosed through genetic screening because these patients tend to be asymptomatic and exhibit minimal or even no manifestations of the disease. Patients were also excluded if there were insufficient records for determination of SSI and DS3 scores prior to ERT. After these exclusions, we were left with 139 patients for the final analysis of SNP association with disease severity.
Family Structure and Association Analysis
The Kinship program [33] was used to identify cryptic family relationship directly from genotypes. It estimates the IBD (identical by descent) coefficients and calculates kinship coefficients. These coefficients were used to infer duplicated samples, siblings and parent-offspring relationships.
After family relationship among individuals was established, a newly developed method, OQLS was used to compare the allele frequency of SNPs among different phenotype categories while accounting for the ordinal nature of trait information. OQLS (Wang, Z, unpublished) is an extension of the single-marker testing method for a binary trait, MQLS [34]. The advantage of OQLS/MQLS is that it is applicable to completely general combinations of family and case-control designs. OQLS/MQLS explicitly incorporates the phenotyped relatives which provide additional information on the enrichment of predisposing variants, and uses the dependence structure to derive optimal weights to increase the power of association analysis.
For top association regions, LD pruning was performed as described in the PLINK package. SNPs were pruned if their r2 were larger than 0.8. The HaploView [35] was used to calculate and visualize the fine scale LD structure and infer haplotype blocks. The SNAP tool [36] was used to plot the regional association result and LD values. The eQTL browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) was used to search for SNPs associated with the expression of CLN8 [37].
Skin Fibroblast Cell Lines
Two millimeter punch skin biopsy was obtained from 6 patients (3 with severe GD and 3 with mild GD), and 2 healthy controls using the AcuPunch kit (Acuderm Inc, Lauderdale, FL). A sterile 12mm circular cover glass was place on each of the skin biopsy fragments and 6ml of primary growth culture (DMEM supplemented with 15% fetal calf serum, 1% penicillin/streptomycin/gentamicin, 1% NEAA) was added. A fibroblast cell line was established once the outgrowth of fibroblasts skin biopsy covered 75% - 90% of the dish. The cells were digested with 0.05% typsin/EDTA and split at a ratio of 1:3 to 1:4. Cell lines between 4 - 7 generations were used for the experiments.
In vitro studies with fibroblast cell lines
The inhibitor of glucocerebrosidase, conduritol B epoxide (CBE) was purchased from Calbiochem (La Jolla, CA); Glucocerebroside (GL1) and Glucosylsphingosine (Lyso GL1) were purchased from Matreya (Pleasant Gap, PA). Stock solutions of the drug were prepared according to the manufacturer's instructions. Final concentrations of 50μM of CBE, 40μM of GL1, and 20μM of Lyso GL1 were used to treat the cells as described previously [37a]. CBE alone, CBE with GL1 or Lyso GL1 were separately added to the two normal (healthy) cell lines with the same amount of DMSO as control. After incubation for the specified periods, RNA was extracted for real time PCR.
Total RNA from each cell line was prepared using the TRIzol Plus RNA Purification kit (Invitrogen, Carlsbad CA). Briefly, adherent fibroblast cells were scratched and concentrated to an Eppendorf micro-centrifuge tube to which 1ml of TRIzol reagent was added. Following phase separation, the solutions containing RNA were transferred to a spin cartridge. After binding and washing, RNA was eluted into a fresh microtube and RNA concentration was determined.
First strand cDNA synthesis was performed using the AffinityScript Multiple Temperature cDNA Synthesis kit (Agilent Tech, Santa Clara, CA) according to the manufacturer's protocols. Oligo dT was used as primer for the reverse transcription. RNA-Oligo dT mixture was preheated at 70°C for 5min before gradual cooling at room temperature for 10min, then followed by addition of the reaction mixture on ice and and at 42°C for 5min and 50°C for 55min.
Quantitative real-time PCR was performed using a 7500 Real-time PCR system (Applied Biosystems, Carlebad, CA). Primer and probe mixtures for CLN8 and Actin-β (used as internal reference control) were ordered from the inventory of Applied Biosystems and pre-labeled with the reporter dye 6-carboxyfluorescein (6′-FAM) at the 5′ end and with the quencher dye 6-carboxytetramethylrodamine (TAMRA) at the 3′ end. Quantitative PCR was performed with 2μl of 1:3 diluted cDNA, 10μl of the FastStart universal probe master (Roche, Mannheim, Germany), 0.5μl of 900nM primers, and 0.5μl of 250nM probe in a final volume of 20μl. Each reaction was set up in triplicate and the mean values were used for analysis.
The study was approved by the Human Investigation Committee of the Yale University of Medicine and IRB of NYU School of Medicine.
Results
Genotype Quality Control and Family Structure
Five out of 186 samples had genotyping call rates 95% and these samples were removed from further analysis. Among the remaining 181 samples, 2 samples had call rates between 96 and 97%. The remaining 179 samples had call rates > 99%. After SNP cleaning using the quality control criteria described in methods section, 554,340 SNPs were left. The total genotyping rate in post-QC samples is 99.8%. There were 540,902 autosomal SNPs retained for final association analysis.
From pair-wise estimated distribution of IBD coefficients and kinship coefficients, we inferred 14 nuclear families. Among these 14 families, 3 were parent-offspring, 10 were sibling pairs and one family had 3 siblings (supplementary table IB). We found two categories of kinship coefficients, 31 pairs near 0.25 representing parental-child or sibling relationship (supplementary table IA). For a parent offspring pair, they are anticipated to share 1 allele IBD, whereas for a sib pair, the probabilities of sharing 2, 1 or 0 alleles IBD are 0.25, 0.5 and 0.25, respectively. Other sample pairs had kinship coefficients less than 0.05, and therefore less likely to be related with each other.
Association Results
After applying the OQLS algorithm to our ordinal valued phenotype in a combined sample of family and unrelated cases/controls, the genetic association of 540,902 autosomal SNPs was identified in the three categories of disease severity (overall disease severity, severity of visceral disease and severity of bone disease). Figure 1 shows the Manhattan plot for the genome wide association results. No markers achieved genome wide significance based on the stringent Bonferroni correction (p ≤ 9 × 10−8). However, the Bonferroni correction assumes all markers behave independently. In practice, a significant proportion of markers do not segregate independently because they are in LD with each other. Therefore, the top signals displayed in the Manhattan plot are worthy of further investigation. Consequently, we analyzed SNPs with lowest P values for associations with disease severity. In addition, cluster of top associations displayed in the Manhattan plot (spikes) were examined in detail.


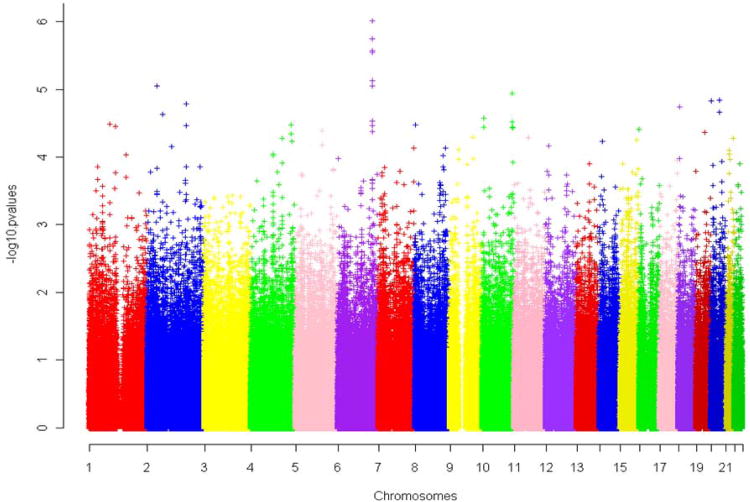
The x-axis shows the chromosomes arranged according to their genome location. The y-axis shows the –log p value of the association strength. Each chromosome is in different colors. A. Overall disease severity. B. Severity of visceral disease. C. Severity of skeletal disease.
Supplementary Table II lists the top associated markers with each of the three disease severity categories. If not within the known gene region, their nearest gene neighborhoods and distances were displayed. The distance to the start of the gene (disstart) was calculated by the gene start position minus the SNP location; the distance to the end of the gene (distend) was calculated by the gene end position minus the SNP location.
When assessing SNP association with overall disease severity, the region harboring CLN8 gene achieved the strongest association signal. In addition, the CLN8 gene region also demonstrated strong association with the severity of visceral disease (Supplementary table II). Association signals for SNPs within CLN8 and its 100 kb flanking regions with the overall disease severity are depicted in Figure 2a. LD structure within 100 kb flanking region is displayed in Figure 2b. Closer resolution of LD structure and haplotype block within the 25 kb flanking region is displayed in Figure 2c. Several SNPs within or near CLN8 regions are in strong LD with each other and majority of these SNPS are strongly associated with the overall disease severity. Supplementary table S III shows the detailed association results for each SNP within the CLN8 region for the overall disease category. Interestingly, among these SNPs two intronic SNPs (rs11136424 and rs4875958, pvalues 1.59E-05 and 2.69E-05) are strongly associated with CLN8 expression as depicted in the eQTL browser. SNP rs11136424 is associated with CLN8 expression in lymphoblastoid cell lines and SNP rs4875958 is associated with CLN8 expression in liver tissue (Supplementary Figure 2). Both eQTL data are derived from people of European ancestry [37].

A. Regional Manhattan plot and LD structure with the top SNP rs11986414. The x-axis shows the chromosome position, the left y-axis shows the –log (p) for the association strength, and the right y-axis shows the recombination rate calculated from Hapmap3 CEU reference population. The color for each marker represents the LD r2 value with the topmost snp rs11986414. B. LD structure. The pair-wise LD pattern is shown for all markers within the 100kb flanking region. The chromosomal location is shown above, the red, blue and white colors represent different LD measured by D′. The blocks bordered with thick black box represent haplotype blocks inferred. C. LD structure and haplotype block for the 25kb flanking region. The value inside each cell displays the LD measurement r2, the color scheme is based on D′. The inferred haplotype block and haplotype frequency are shown too.
Among SNPs within the CLN8 region, the topmost SNP rs11986414 achieved a p value of 1.26 × 10−6. The two alleles of this SNP are A and G; A is the risk allele associated with more severe disease. As shown in Supplementary Table IVa, there was over-representation of A allele in patients with moderate and severe disease compared to the patients with mild disease. Compared to patients with mild disease, A allele conferred odds ratio (OR) of 3.62 (95% CI 1.96 - 6.70) for moderate disease and 3.85 (95% CI 1.96 - 7.55) for severe disease (Table S IVb). The OR between moderate and severe disease categories is 1.06 (95% CI 0.51 - 2.22). Therefore genetic risk conferred by rs11986414 appears to be similar for both moderate and severe disease. The OR with combined severe and moderate vs mild disease category is 3.72 (95% CI 2.19 - 6.31). To understand the genetic inheritance model, the genotypic odds ratios for combined severe and moderate category vs mild disease were further evaluated. Supplementary table IVc shows the OR for AA vs AG, AG vs GG and AA vs GG: 2.38 (95% CI 0.76 -9.04), 4.52 (95% CI 2.05 – 9.97) and 11.84 (95% CI 3.33 - 42.0), respectively. The large confidence interval for odds ratios with genotype AA are due to the fact that there are only 4 patients with genotype AA in the mild category. These results are consistent with the notion that there is a gene dosage effect, where each addition of allele A increases the risk of severe disease.
For SNPs association with the severity of bone disease, the top SNP is rs510933 on chromosome 6 (p value 7.87E-07) (Supplementary table IIc). There are 4 other SNPs from this region. However, this region is devoid of genes within 200kb. The most proximal genes near this region are around 500kb away and include STXBP5, SAMD5, SASH1. A noteworthy candidate modifier gene for severity of bone disease is indicated by SNP rs7920091, a SNP in ADAM12 (p=1.11E-05). ADAM12 is member of the ADAM (a disintegrin and metalloprotease) protein family. Polymorphisms in this gene have been associated with osteoarthritis and its progression [38]. The ADAM family is also thought to regulate human osteoclast differentiation and activity [39]. Other noteworthy associations were with SNPs rs6129532 and rs6124225 on chromosome 20 linked with HSPE pseudogene [HSPE1P1] (p value 1.39E-05 and 2.12E-05 respectively) and rs11087800, intronic SNP in ANGPT4 (p value 1.45E-05). ANGPT4 is involved in angiogenesis. This may be relevant since avascular osteonecrosis in GD is considered a primarily vascular complication reflecting endothelial cell involvement in GD and may involve impaired angiogenic support due to osteoblastic dysfunction [6, 40, 41].
Expression of CLN8 in skin fibroblasts and macrophages
Using Real Time PCR, CLN8 relative expression was higher in GD patients compared to controls (Figure 3b). There was also a trend of GD patients with mild disease having higher expression of CLN8 compared to GD patients with severe disease. To assess the relationship between relative expression of CLN8 and the number of risk alleles in CLN8 gene region, two SNPs (rs4595147 and rs11986414) with the lowest LD were selected from the associated SNPs within the CLN8 region. Of the 6 samples analyzed (3 severe and 3 mild patients), the total number of risk alleles appear to be marginally correlated with relative expression (correlation coefficient -0.796, p value 0.058). In control fibroblasts cultured with CBE, an in vitro chemically-induced cellular model of GD, there was slight increase of CLN8 expression compared to untreated cells (Figure 3a). There was more striking increase (2 - 2.5 fold) of CLN8 expression in CBE-treated cells exposed to potent bioactive lipid substrate of acid β-glucosidase, Lyso GL1. Of the two lipid substrates of acid β-glucosidase, Lyso-GL1 was the more potent inducer of CLN8 expression compared to GL1 (Figure 3a). We performed RT PCR for CLN8 expression in primary monocyte-derived macrophages of GD1 patients, and the results were similar to the findings in skin fibroblasts (data not shown). To further assess the expression pattern of CLN8 in different tissues, we interrogated Novartis expression atlas (www.biogps.org) to determine whether it is expressed preferentially in immune cells [42, 43]. Supplementary Figure 3 depicts the tissue specific expression pattern of CLN8. Interestingly, highest expression is observed in immune cells including macrophages and monocytes. Curiously, lowest expression was observed in brain tissues, which is most vulnerable to loss of function mutations in this gene.

A. CBE alone, CBE with GL1 or Lyso GL1 were separately added to the two normal (healthy) cell lines with replacement of medium and drugs every 5-7days. RNA was then extracted and real time PCR was performed for CLN8 expression. Actin-β was used as internal reference control. Concentration of culture solutions were: 50 μM CBE, 40 μM GL1, 20 μM Lyso GL1. B. CLN8 expression at baseline in mild and severe GD patient fibroblasts compared to control cells by real time PCR. Results are expressed as times control. The cycle threshold for actin was 19.8 and the average control cycle threshold was 27.8. Each patient's risk alleles are displayed on top for the 2 SNPs (rs11986414 and rs4875960) associated with CLN8. Allele C for the first SNP and allele A for second SNP are associated with a higher risk of disease severity.
Discussion
The basis of remarkable variation of severity of GD1 and its heterogeneity among patients with identical GBA1 gene mutations is not understood [1]. Therefore, it is not possible to identify patients who will benefit from pre-emptive intervention with expensive ERT or small molecule therapy before onset of incapacitating disease manifestations. Identification of genetic modifier(s) will enhance the ability to accurately predict natural history in individual patients in order to develop a personalized monitoring and therapeutic strategy as well as refine genetic counseling.
There are significant inherent obstacles to conduct robust GWAS studies for discovery of modifier gene(s) in a Mendelian disorder such as GD. The chronicity and the heterogeneity of the disease as well as the lack of sufficiently well-characterized patients limit the ability to conduct such studies. The greatest hurdle is that the new generation of patients tends to be treated early before the full natural history unfolds [52,53]. Therefore the accuracy of ascertainment of true disease phenotype with respect to severity is severely compromised. Moreover, the instruments to depict disease severity have limitations to adequately capture the full scope of heterogeneity of disease manifestations. The latter is exemplified most strikingly by some GD patients who predominantly exhibit bone manifestations with only mild visceral or hematological disease and vice versa; these patterns are in contrast to occasional patients who manifest uniformly severe disease in all disease compartments [10]. These patterns of disease, may in fact be driven by modifier gene(s) that underlie monocyte trafficking and/or macrophage-mediated determinants of individual organ injury. Therefore, in our study, we elected to depict disease severity by multiple approaches, including SSI, validated DS3 as well as global physician assessment by two investigators with extensive experience of GD. Moreover, we also examined individual disease compartments, including the viscera and the bone. Thus the final dataset of mild, moderate and severe disease are remarkably consistent with these classifications in the context of overall disease severity as well as severity of individual disease compartments (Table I).
Our study revealed several candidate modifier genes, not previously considered on the basis of current understanding of GD pathophysiology: CLN8, ADAM12 and ANGPT4. The lead candidate modifier gene was CLN8. The physiological function of CLN8 is not known but a loss of function mutation is responsible for a subtype of neuronal ceroid-lipofuscinosis (NCL), the Northern epilepsy mental retardation (EPMR) syndrome, characterized by seizures in childhood and progressive mental retardation [44]. The CLN8 gene encodes a predicted 286–amino acid transmembrane protein containing a C-terminal ER-retrieval signal (KKRP) [45] Consistent with this structure, CLN8 protein, a nonglycosylated membrane protein, is located in the ER and shuttles between the ER and the ER-Golgi intermediate compartment. Highest expression of CLN8 is surprisingly in the liver and the spleen (compared to the brain) and it has been suggested that it plays an important role as a sensor of sphingolipids and sphingolipid trafficking in all cell types [46]. CLN8 is a member of a family of homologues that includes ER-resident proteins: yeast Lag1p and human TRAM (translocating chain-associating membrane protein) [47]. A C-terminal motif of 200 amino acid residues designated the Tram- Lag-CLN8 (TLC) domain in these proteins has been proposed to play a role in ceramide synthesis and glycosphingolipid trafficking [47].
In an induced in vitro cellular model of GD, we found there was an induction of CLN8 expression and the level of expression was further increased in the presence of potent bioactive lipid substrate of acid β-glucosidase, Lyso-GL1. In cells derived from, GD1 patients, there was increased basal expression of CLN8 relative to control cells; moreover there was a trend towards increased expression in patients with mild disease compared to patients with moderate/severe disease. We found the SNP alleles in CLN8 region associated with severe GD correlated with relatively lower expression of CLN8 compared to the lower risk alleles (Figure 3). Interestingly, CLN8 and CLN6, two integral ER trans-membrane proteins and cathepsin D, are increased in expression (3-4-fold) in the spleens of mice with conditional deletion of GBA1[6]; loss of function mutations in these three genes result in NCLs [48]. Moreover, MYC, a regulator of CLN8 was marginally associated with GD1 severity (top SNP p value 4.96 × 10−5). Finally interrogation of Novartis gene expression pattern highlighted prominent relative expression of CLN8 in immune cells compared to other cell types and organs. Taken together, CLN8 is a plausible candidate modifier gene of GD and its induction likely protects cells from the damaging effect of accumulating lipids in GD1.
Interestingly loss of function mutation in CLN8 results in lysosomal accumulation of autofluorescent material but with lower levels of plasmalogen [47] and the latter has been suggested to promote lipid peroxidation. In fact, in GD1, serum plasmalogen levels are reduced, the baseline levels correlate negatively with chitotriosidase and furthermore increase after ERT [50]. This observation is also consistent with our notion that induction of CLN8 may be protective towards damaging cellular effects of glycosphingolpid accumulation in GD1.
CLN8 and the other potential candidate modifier genes (ADAM12 and ANGPT4) identified in our GWAS do not achieve the threshold for genome-wide significance after stringent Bonferroni correction [51]. Given the potential impediments to conduct a replication study, an alternative approach to define the potential role of CLN8 as a modifier of GD will derive from studies in transgenic mice since CLN8 KO as well as conditional KO mouse models of GD are available [44, 54].
In conclusion, CLN8 is a plausible candidate modifier gene for Gaucher disease based on our GWAS results and in vitro studies in chemically-induced cellular model of GD. Further studies are warranted to define the physiologic role of CLN8 in health and its role in GD pathophysiology. The availability of mouse models should facilitate these studies.
Supplementary Material
Supp Fig S1-S3
Supplementary Figure 1 Timelines of N370S Homozygous GD patients with severe disease compared to patient with mild disease. The presentation in Gaucher disease is very variable. This timeline shows significant events in the natural history of two N370S homozygous patients enrolled in the current GWAS study. These contrasting patterns illustrate mild vs. severe disease in our patient population. Patient A with severe disease had early onset osteomyelitis/AVN. He had severe splenomegaly and hepatomegaly at an early age and went on to have a splenic rupture and splenectomy. Patient B with mild disease was diagnosed based on maternal diagnosis and has been asymptomatic. She has virtually no disease manifestations and does not require ERT at 41 years of age.
Supplementary Figure 2: QTL association results in CLN8 region. This screenshot from the eQTL browser displays eQTLs near the CLN8 region. The chromosome 8 and CLN8/C8orf61 genes are displayed. And each SNP associated with CLN8 or other genes are displayed below, together with related references.
Supplementary Figure 3: Tissue specific expression profile for CLN8 This figure displays the human tissue specific expression profile from U133plus2 Affymetrics microarray (CLN8, probe ID 219340_s_at). The expression value is a z-score derived from the barcode function of the R package "frma". A z-score > 5 suggests that the gene is expressed in that tissue. Color codes highlight neuronal tissues (blue), immune cells/organs (red), viscera (purple) and miscellaneous (orange).
Supp Table S1-S4
Supplementary Table I-IV Table S I Pairwise IBD coefficients and inferred family relationships. A. Top 50 sample pairs with largest kinship coefficients. Columns ID1 and 2 are IDs from two individuals, D7, D8 and D9 are the last three IBD coefficients inferred assuming no inbreeding, KinshipCoef is the kinship coefficients calculated from 9 IBD coefficients. B. Reconstructed samples potentially within a family, 17 families were inferred.
Table S II Top associated SNPs for the three traits, and their nearest gene neighborhood and distance to gene start and gene end. SNPs are listed in the order of their genomic location. The Symbol column shows the nearest gene neighborhood. The distance to the start of the gene (diststart) is calculated by the gene start bp minus the snp location; the distance to the end of the gene (distend) is calculated by the gene end bp minus the snp location. The chromosome location is from NCBI genome assembly 36.3. A. Overall top SNPs. B. Visceral top SNPs. C. Bone top SNPs
Table S III Association of SNPs in CLN8 region with overall disease severity. In the annotation column, flanking refers to the flanking region of CLN8. The eight strongly associated SNPs within the region are colored in red.
Table S IV. The genotype and allelic configuration for SNP rs11986414 in CLN8 region, allelic pairwise odds ratio and genotypic odds ratio. The genotype configuration for the top SNP rs11986414 associated with CLN8 with a p value of 1.26 × 10-6 for overall disease severity is depicted. Allele A is the risk allele associated with more severe phenotype. The A allele is observed more frequently in moderate and severe disease categories compared to the mild category.
A. genotype and allelic configuration for SNP rs11986414.
B. Allelic pairwise odds ratio.
C. Genotypic odds ratio for combined severe and moderate phenotype vs mild phenotype.
Acknowledgments
We thank the patients at Yale and NYU Gaucher Disease Treatment Centers for participation in this study. The study was supported by the National Gaucher Foundation and by Lysosomal Fellowship Training Grant from Genzyme-Sanofi to PS. PKM thanks Dr Jeffrey Gruen and Karen K. Weisbord for advice.
Contract grant sponsor: PKM is supported by NIDDK K24DK066306 mid-career clinical investigator award, the National Gaucher Foundation and a grant from Genzyme Corporation for investigator-initiated study on biomarker discovery and validation. PS was supported by a Lysosomal Storage Disease Fellowship Training Grant from Genzyme-Sanofi.
References
Full text links
Read article at publisher's site: https://doi.org/10.1002/ajh.23118
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3684025?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/ajh.23118
Article citations
Gaucher disease provides a unique window into Parkinson disease pathogenesis.
Nat Rev Neurol, 20(9):526-540, 06 Aug 2024
Cited by: 0 articles | PMID: 39107435
Review
Adult type I Gaucher disease with splenectomy caused by a compound heterozygous GBA1 mutation in a Chinese patient: a case report.
Ann Hematol, 103(5):1765-1774, 20 Mar 2024
Cited by: 0 articles | PMID: 38509388
Multi-Omics Data Analysis Identifies Prognostic Biomarkers across Cancers.
Med Sci (Basel), 11(3):44, 27 Jun 2023
Cited by: 4 articles | PMID: 37489460 | PMCID: PMC10366886
Neuronopathic Gaucher disease models reveal defects in cell growth promoted by Hippo pathway activation.
Commun Biol, 6(1):431, 19 Apr 2023
Cited by: 2 articles | PMID: 37076591 | PMCID: PMC10115838
A rare homozygous p.Arg87Trp variant of the GBA gene in Gaucher disease: A case report.
Clin Case Rep, 10(5):e05846, 12 May 2022
Cited by: 1 article | PMID: 35592045 | PMCID: PMC9097371
Go to all (37) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 10 of 10)
- (10 citations) dbSNP - rs11986414
- (2 citations) dbSNP - rs11136424
- (2 citations) dbSNP - rs4875958
- (1 citation) dbSNP - rs4875960
- (1 citation) dbSNP - rs11087800
- (1 citation) dbSNP - rs6124225
- (1 citation) dbSNP - rs6129532
- (1 citation) dbSNP - rs7920091
- (1 citation) dbSNP - rs4595147
- (1 citation) dbSNP - rs510933
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Functional and genetic characterization of the non-lysosomal glucosylceramidase 2 as a modifier for Gaucher disease.
Orphanet J Rare Dis, 8:151, 26 Sep 2013
Cited by: 19 articles | PMID: 24070122 | PMCID: PMC3850879
RecTL: a complex allele of the glucocerebrosidase gene associated with a mild clinical course of Gaucher disease.
Am J Med Genet, 50(1):74-78, 01 Mar 1994
Cited by: 14 articles | PMID: 8160756
The underrecognized progressive nature of N370S Gaucher disease and assessment of cancer risk in 403 patients.
Am J Hematol, 84(4):208-214, 01 Apr 2009
Cited by: 102 articles | PMID: 19260119 | PMCID: PMC3008404
Gaucher disease: gene frequencies and genotype/phenotype correlations.
Genet Test, 1(1):5-12, 01 Jan 1997
Cited by: 87 articles | PMID: 10464619
Review
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: M01 RR000071
NIDDK NIH HHS (2)
Grant ID: K24 DK066306
Grant ID: K24DK066306





