Abstract
Free full text

PNAS Plus
Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis
Abstract
Helicobacter pylori is a gastric pathogen responsible for a high disease burden worldwide. Deregulated inflammatory responses, possibly involving macrophages, are implicated in H. pylori-induced pathology, and microRNAs, such as miR-155, have recently emerged as crucial regulators of innate immunity and inflammatory responses. miR-155 is regulated by Toll-like receptor (TLR) ligands in monocyte-derived cells and has been shown to be induced in macrophages during H. pylori infection. Here, we investigated the regulation of miR-155 expression in primary murine bone marrow-derived macrophages (BMMs) during H. pylori infection and examined the downstream mRNA targets of this microRNA using microarray analysis. We report TLR2/4- and NOD1/2-independent up-regulation of miR-155, which was found to be dependent on the major H. pylori pathogenicity determinant, the type IV secretion system (T4SS). miR-155 expression was dependent on NF-κB signaling but was independent of CagA. Microarray analysis identified known gene targets of miR-155 in BMMs during H. pylori infection that are proapoptotic. We also identified and validated miR-155 binding sites in the 3′ UTRs of the targets, Tspan14, Lpin1, and Pmaip1. We observed that H. pylori-infected miR-155−/− BMMs were significantly more susceptible to cisplatin DNA damage-induced apoptosis than were wild-type BMMs. Thus, our data suggest a function for the prototypical H. pylori pathogenicity factor, the T4SS, in the up-regulation of miR-155 in BMMs. We propose the antiapoptotic effects of miR-155 could enhance macrophage resistance to apoptosis induced by DNA damage during H. pylori infection.
The Gram-negative bacterium, Helicobacter pylori, is a spiral-shaped, microaerophilic pathogen that is prevalent in ~50% of the world’s population (1). This pathogen causes persistent infections of the gastric mucosa, which can result in both acute and chronic gastric diseases. Although infections frequently are asymptomatic, severe disease outcomes, such as duodenal and gastric ulcers, develop in 5–10% of infected individuals (2). Moreover, early epidemiological studies have shown a convincing association between infections with this pathogen and gastric adenocarcinoma and mucosa-associated lymphatic tissue lymphoma (3, 4). Accordingly, H. pylori was the first bacterial pathogen to be classified as a type I carcinogen by the World Health Organization (5). In recent years the prominent role of the bacterial genomic cag pathogenicity island (cagPAI) in severity of H. pylori disease has become evident. The distribution of cagPAI-positive H. pylori strains is geographically dependent; there are regions of the world where more than 95% of H. pylori strains (e.g., HpEast-Asia) are cagPAI positive, whereas strains in other regions (e.g., HpAfrica2) are cagPAI negative (6). The H. pylori cagPAI encodes the structural components of a type IV secretion system (T4SS) and a unique effector protein, cytotoxin-associated gene A (CagA). Following T4SS-mediated translocation into host cells, CagA is phosphorylated by host-cell kinases whereupon it disrupts epithelial cell function by binding to cellular phosphatases such as SHP-2 (7) and receptors such as c-met (8). Binding of the T4SS via β1-integrins also has been shown to alter signal transduction in host cells (9).
MicroRNAs (miRNAs) are small, noncoding RNAs ~23 nt in size (10). To date, more than 600 miRNAs, which target >60% of the protein-coding genome in mammals, have been identified in humans and mice (11). Many miRNAs are transcribed as primary transcripts by RNA polymerase II. Primary transcripts are cleaved to precursor miRNAs (premiRNAs) by the RNaseIII enzyme Drosha and are exported to the cytoplasm, where further processing via Dicer RNaseIII yields mature miRNAs (12). By binding to the argonaute complex, single miRNAs can regulate a multitude of target genes by binding preferentially to the 3′ UTR of the target gene mRNA, thereby blocking translation of the mRNA and leading to mRNA degradation (13). A crucial feature for miRNA activity is based on complementarities of its seed sequence (10). miRNAs are involved in almost all cellular processes and also play crucial roles in disease, for example, in cancer. One example of a cancer-related miRNA is miR-155, which is overexpressed in different tumors, such as diffuse large B-cell lymphoma and several types of adenocarcinoma (14). This onco-miRNA also is up-regulated during H. pylori infection (15, 16). Targets of miR-155 include, among others, proteins of the toll-like receptor (TLR) pathway, leading, upon miR-155 induction, to attenuation of transcription factor NF-κB activity (17). Interestingly, miR-155 itself is regulated through the TLR pathway via NF-κB (18).
H. pylori can activate the NF-κB pathway by various means; however, published data are inconclusive as to how these mechanisms contribute to NF-κB signaling in general (19). In gastric epithelial cell lines, NF-κB–dependent up-regulation of IL-8 depends largely on T4SS function (20), and H. pylori LPS does not seem to play a crucial role in this setting (21). It has been proposed that T4SS translocation of H. pylori peptidoglycan activates NF-κB via the NOD1 receptor, which belongs to the cytosolic NOD-like receptor family (22). In monocyte-derived cells, such as macrophages and dendritic cells (DC), the TLR family members TLR2, TLR4, and TLR9 clearly are involved in the response to H. pylori infection (23). Nevertheless, discussion is ongoing as to whether H. pylori LPS signals via TLR4 (a common receptor for Gram-negative enterobacterial LPS) or via TLR2 (the main receptor for Gram-positive bacteria lipoteichoic acid), because H. pylori LPS lacks distinct features of the prototypical enterobacterial LPS (24). The RNA receptor RIG-I also has been proposed to be an intracellular receptor for H. pylori-mediated transcriptional activation in DCs (23).
Gram-negative bacteria may act as pro- as well as antiapoptotic factors for macrophages. For example, Salmonella enterica Typhimurium has been described as promoting apoptosis (25), whereas Brucella suis reportedly is antiapoptotic (26). H. pylori encodes both pro- and antiapoptotic effector molecules, such as CagA and VacA (27). Again, which factors drive the pro- or antiapoptotic response remains controversial (28, 29). In human and murine monocytic cell lines such as Raw-264.1 and THP-1, as well as in primary human monocytes, H. pylori was shown to induce apoptosis directly (30, 31). However, primary monocyte-derived cells seemed to be resistant to H. pylori-induced apoptosis (32). The antiapoptotic potential of miR-155 has been described for several cell types, including B lymphocytes (33), and in pancreatic tumors by targeting TP53INP1 (34) and JARID2 (35). Moreover, miR-155 has been reported to promote resistance to specific chemotherapeutics in breast cancer cells by targeting FOXO3A (36), and recent reports suggested proapoptotic potential of miR-155 through targeting SKI2 in human melanoma cells (37) or Kpc1 in murine DCs (38).
Here, we investigated the regulation of miR-155 expression by H. pylori in cell lines and in primary murine bone marrow-derived macrophages (BMMs) isolated from WT and gene-knockout mice. In addition to the known TLR2/TLR4-dependent up-regulation of miR-155, we found that H. pylori also induced miR-155 via a TLR2/TLR4-independent, T4SS-dependent mechanism in BMMs, a potentially unique function of this major pathogenicity factor. Using microarray analysis, we confirmed the miR-155–dependent down-regulation of known miR-155 gene targets during infection and validated miR-155 binding sites in the 3′ UTRs of putative miR-155 targets Tspan14, Lpin1, and Pmaip1. Interestingly, results in both established and putative targets suggested that miR-155 has a role in the regulation of apoptosis in macrophages during infection. Indeed, WT BMMs were more resistant than BMMs lacking miR-155 (miR-155−/− BMMs) to apoptosis upon cisplatin-induced DNA damage during infection, thus demonstrating the antiapoptotic function of miR-155.
Results
Up-Regulation of miR-155 Is Dependent on Multiplicity of Infection and Duration of Infection.
To investigate the regulation of miR-155 in macrophages in response to H. pylori infection, the murine macrophage cell line J774A was infected with the mouse-adapted H. pylori strain Hp76. After 3 h of infection, total RNA was extracted and subjected to Northern blot. We detected up-regulation of the 65-nt premiR-155 and the 23-nt mature miR-155 form (Fig. 1A). Up-regulation of mature miR-155 was validated by quantitative RT-PCR (qRT-PCR) for infections with mouse-adapted strain Hp76 and the human-adapted strain P12 in comparison with mock-infected cells. The murine cell line J774A exhibited an approximately fourfold up-regulation of miR-155 during infection with both H. pylori strains, whereas infection of C57BL/6 WT mice BMMs (WT BMMs) resulted in an ~20-fold up-regulation (Fig. 1B). To determine whether the observed effect was specific and to identify maximal induction of miR-155 expression upon infection, increasing multiplicity of infection (MOI) and an infection time course were analyzed. There was a strong correlation between MOI and the expression of miR-155 in WT BMMs after 3 h of infection with P12. Infection with an MOI of 50 showed the strongest miR-155 up-regulation, which decreased with an MOI of 10 or 1 (Fig. 1C). miR-155 was induced as early as 90 min postinfection (p.i.) and increased up to ~500 fold at 30 h p.i. (Fig. 1D).

Up-regulation of miR-155 by H. pylori in murine macrophages in an MOI- and time-dependent manner. (A) Up-regulation of miR-155 after 3 h of infection with the mouse-adapted strain Hp76 (MOI of 100) was determined by Northern blot in the murine macrophage cell line J774A using U6 snRNA as endogenous control. (B) Regulation of miR-155 by Hp76 and the human-adapted strain P12 in J774A (MOI of 100) and primary murine BMM (MOI of 50; 3 h) observed by qRT-PCR. (C) MOI dependency of up-regulation of miR-155 was observed by qRT-PCR in BMMs after 3 h of infection with H. pylori P12. (D) Time-course experiments reveal an increase in expression of miR-155 over the duration of infection with H. pylori P12 (MOI of 50) observed by qRT-PCR. The values presented here are the ΔΔCt values normalized to RnU6b snRNA as endogenous control. Experiments were performed with at least three biologically independent replicates and are presented as mean + SE.
TLR-Dependent and -Independent Up-Regulation of miR-155.
Previously it has been shown that the expression and regulation of miR-155 in murine macrophages depends on ligands of the TLR family (39). It has been demonstrated that in primary phagocytes H. pylori can signal via TLR2, TLR4, and other members of the TLR family, leading to the release of IL-6 in a MyD88-dependent manner (23). To examine the effectors that are responsible for the observed up-regulation of miR-155, we incubated WT BMMs with TLR-specific ligands and H. pylori strains Hp76 and P12 for 3 h. The TLR4-specific ligand, Escherichia coli LPS (EC LPS), as well as the synthetic TLR2 ligand Pam3CSK4, led to an induction of miR-155 expression (Fig. 2A). A similar result was obtained for purified H. pylori P12 LPS. Heat-inactivated H. pylori P12 (P12 hi) and Hp76 (Hp76 hi) induced miR-155 to levels similar to that induced with viable bacteria. To ascertain whether the induction of miR-155 expression by these factors depended purely on the interaction of the main extracellular TLRs, TLR2 and TLR4, with H. pylori (23), BMMs isolated from C57BL/6 mice lacking receptors for both TLR2 and TLR4 (TLR2/4−/− BMMs) were used. These cells did not up-regulate miR-155 in response to TLR2- and TLR4-specific ligands (EC LPS and Pam3CSK4, respectively) or in response to H. pylori LPS (Fig. 2B). It has been discussed in the literature whether H. pylori LPS signals via TLR2 or TLR4 (40, 41). Here, we found that signaling occurred via both receptors (Fig. S1 A and B) and that signaling was absent when neither TLR2 nor TLR4 was present (Fig. 2B). In contrast to H. pylori LPS, viable H. pylori P12 and Hp76 showed an ~65-fold up-regulation of miR-155 in TLR2/4−/− BMMs. Induction of miR-155 was even higher in TLR2/4−/− BMMs than in WT BMMs (Fig. 2A). This higher induction rate can be explained by a higher endogenous basal level of miR-155 in the mock-infected WT BMMs as compared with TLR2/4−/− BMMs. Heat inactivation of H. pylori significantly reduced the relative expression of miR-155 in TLR2/4−/−BMMs as compared with the nontreated strain, whereas the use of the proteasome inhibitor MG132 (used as an NF-κB inhibitor) completely abolished miR-155 relative expression (Fig. 2B). The expression of miR-155 also was investigated in BMMs lacking the adaptor proteins MyD88 or Trif (Fig. S1 C and D). MyD88 is the adaptor protein for all TLR-dependent NF-κB signaling, with the exception of TLR3- and, in part, TLR4-dependent signaling, which use the adaptor function of Trif (42). Infecting MyD88/Trif−/− BMMs with viable H. pylori led to an ~25-fold induction of miR-155, similar to that observed with WT BMMs (Fig. 2C). Moreover, up-regulation of miR-155 was almost completely absent in MyD88/Trif−/− BMMs infected with heat-inactivated P12 (Fig. 2C). In the literature, H. pylori-derived RNA has been proposed to signal via MyD88-dependent TLRs as well as via Rig-I (23). Therefore, we tested whether transfected P12 RNA induced miR-155 in BMMs. Fig. 2D shows that P12 RNA strongly induced miR-155, by ~60-fold, in TLR2/4−/− BMMs.
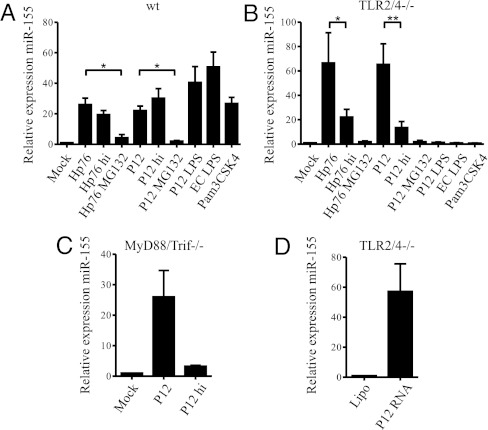
TLR2- and TLR4-dependent and TLR-independent up-regulation of miR-155 by H. pylori. (A) Living and heat-inactivated (hi) H. pylori P12, Hp76 (MOI of 50), P12 LPS (1.5 μg/mL, 2,000 EU/mL), and the specific ligands for TLR2 (Pam3CSK4, 10 ng/mL) and TLR4 (ultra-pure E. coli LPS, 100 ng/mL) up-regulate miR-155 after 3 h of incubation in WT BMMs. (B) In BMMs lacking TLR2 and TLR4 (TLR2/4−/−), a strong reduction of miR-155 expression can be observed using heat-inactivated H. pylori. The proteasome inhibitor MG-132 (10 μM) (used as an NF-κB inhibitor) almost completely abolished the up-regulation of miR-155. (C) The regulation of miR-155 in BMMs lacking both adaptors for TLR signaling (MyD88 and Trif) is similar to WT BMMs when using P12 (MOI of 50; 3 h). Heat-inactivation of P12 (MOI of 50) clearly reduces the up-regulation of miR-155. (D) The effect of transfected P12 RNA on the induction of miR-155 was tested in TLR2/4−/− BMMs. TLR2/4−/− BMMs were used to exclude contamination of P12 LPS. All data were obtained by qRT-PCR using ΔΔCt values with RnU6b or snoR-202 as endogenous control. Experiments were performed with at least three biologically independent replicates and are presented as mean + SE. Statistical significance was analyzed by Student’s t test; *P < 0.05 and **P < 0.01.
Thus, during H. pylori infection, miR-155 expression is induced in BMMs partially via a TLR2/4- and MyD88/Trif-dependent mechanism. However, miR-155 induction in BMMs also has a TLR-independent component that results, at least in part, from the activation of MyD88/Trif-independent pathogen-associated molecular pattern (PAMP) receptors by H. pylori RNA.
H. pylori T4SS Contributes to the Up-Regulation of miR-155.
To probe further the TLR2/4-independent component of miR-155 up-regulation, we analyzed miR-155 expression in TLR2/4−/− BMMs treated with bafilomycin A1 (BafA1) for 1 h before infection. BafA1 blocks the proton pump of the endosome, thereby inhibiting phagosomal maturation (43). As a result, endosomal TLRs or cytosolic receptors cannot detect H. pylori effectors, such as RNA (23, 44, 45). BafA1 treatment did not influence the translocation of CagA (Fig. S2). To ascertain whether BafA1 was functional in this system, up-regulation of IL-6 mRNA was quantified in TLR2/4−/− BMMs after H. pylori infection. The inhibitory effect of BafA1 on IL-6 protein expression in TLR2/4−/− bone marrow-derived DCs (BMDCs) has been reported previously (23) and could be confirmed here at the mRNA level (Fig. S3A). There was almost no expression of IL-6 mRNA in response to P12 infection in BafA1-treated TLR2/4−/− BMMs, whereas there was an ~10,000-fold up-regulation of IL-6 in cells not treated with BafA1. In comparison with the abrogation of IL-6 mRNA expression in the presence of BafA1 during P12 infection, miR-155 expression was reduced by only ~50% in BafA1-treated TLR2/4−/− BMMs (Fig. 3A) (~25-fold up-regulation), compared with nontreated cells (~50-fold up-regulation). Thus, to investigate the origin of the lower sensitivity of miR-155 expression to BafA1, we investigated other bacterial factors that might contribute to the miR-155 up-regulation. H. pylori strains Hp76 and P12ΔcagPAI (an isogenic mutant of P12), both lacking the T4SS and the CagA protein, were used for infections to confirm whether the effect was cagPAI dependent in TLR2/4−/− BMMs. E. coli DH1 (MOI of 10), which does not possess a T4SS, served as control. In the absence of BafA1, all infections of TLR2/4−/− BMMs showed very similar up-regulation of miR-155 (40- to 45-fold) (Fig. 3A). However, the relative expression of miR-155 decreased by ~50% in TLR2/4−/− BMMs during infection with the T4SS-positive strain P12, as previously shown in the presence of BafA1. The effect of BafA1 was significantly larger in strains that lacked the T4SS (Hp76, P12ΔcagPAI, and E. coli DH1): miR-155 expression was reduced by ~80% in the presence of BafA1. The reduced capacity of T4SS-negative strains to induce miR-155 expression in the presence of BafA1 compared with T4SS-positive strains was highly significant (P < 0.001), thus implying that the TLR2/4-independent expression of miR-155 contains a T4SS-dependent component. This effect of BafA1 was not observed in WT BMMs, where the TLR-dependent miR-155 expression likely was dominant (Fig. S3B). This result led us to question whether the observations were dependent only on the presence of the T4SS or also were dependent on T4SS function and CagA. To gain further insights into this process, infections were performed with isogenic mutants of P12 lacking either the CagA protein (P12ΔcagA) or the CagL protein (P12ΔcagL), which is reported to be located at the tip of the T4SS and the absence of which renders the T4SS nonfunctional (46). In all mutants tested, cells not treated with BafA1 did not differ significantly from WT P12 cells in miR-155 up-regulation (40- to 50-fold up-regulation). However, the up-regulation of miR-155 was reduced significantly (P < 0.01) in BafA1-treated TLR2/4−/− BMMs infected with the P12ΔcagL mutant as compared with WT P12; levels were comparable to those observed with the P12ΔcagPAI strain (Fig. 3B). By contrast, the P12ΔcagA mutant behaved similarly to WT P12 (Fig. 3B). These experiments strongly suggested that, although the observed miR-155 expression in BMMs was partially dependent on the T4SS, this process was independent of CagA. Given reports showing that H. pylori peptidoglycan leads to NF-κB translocation in epithelial cells via NOD1 (22), we tested the potential of the NOD1- and NOD2-specific ligands TriDAP and MDP, respectively, to induce miR-155. We found no NOD1 ligand-mediated up-regulation of miR-155 in TLR2/4−/− BMMs (Fig. S4B); however a slight up-regulation of miR-155 via NOD2 could be observed at high concentrations of MDP. Additionally, BMMs lacking Rip2 [thus lacking NOD-dependent NF-κB activation (47)] showed up-regulation of miR-155 upon P12 infection (~60 fold) similar to that in infected TLR2/4−/− BMMs (Fig. S4A). Because the T4SS-mediated effect may be explained by the translocation of peptidoglycan, we generated a mouse mutant lacking MyD88/Trif/Rip2−/− and tested the expression of miR-155 in BMMs derived from these mice. MyD88/Trif/Rip2−/− BMMs did not up-regulate miR-155 upon EC LPS or MDP stimulation, demonstrating that TLR as well as the NOD1/2 signaling truly was abrogated in these cells. However, when we infected MyD88/Trif/Rip2−/− BMMs with H. pylori P12, a strong (45-fold) up-regulation of miR-155 could be observed (Fig. 3C). Pretreatment with BafA1 reduced this up-regulation of miR-155 to ~27-fold, and pretreatment with MG132 further diminished miR-155 induction to approximately fivefold. In H. pylori P12ΔcagPAI without BafA1 pretreatment, miR-155 induction was reduced significantly compared with WT P12, possibly as a direct result of the absent T4SS-dependent activation. Pretreatment with BafA1 further decreased the up-regulation of miR-155 in these BMMs. The P12ΔcagA mutant up-regulated miR-155 expression to levels comparable with WT P12 (in both, an ~50-fold miR-155 induction) during infection of MyD88/Trif/Rip2−/− BMMs. By contrast, P12ΔcagL showed a reduction of miR-155 up-regulation (~50% of that in WT P12 cells). Pretreatment with BafA1 significantly decreased miR-155 up-regulation of P12ΔcagL-infected MyD88/Trif/Rip2−/− BMMs. Thus, NOD1/2 signaling can up-regulate miR-155 expression weakly in BMMs, but the T4SS-dependent regulation of miR-155 expression during H. pylori infection is independent of NOD1/2 signaling.
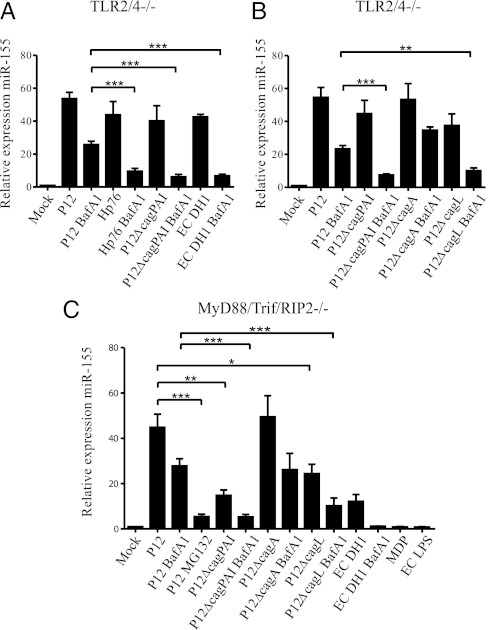
Regulation of miR-155 is dependent on the T4SS. (A) Blocking phagosomal maturation by BafA1 (100 nM) 1 h before infection decreases miR-155 upregulation in TLR2/4−/− BMMs infected (3 h) with P12 (MOI of 50) but not as strongly as in BafA1-treated BMMs infected with Hp76 (MOI of 50) or P12ΔcagPAI (MOI of 50), both of which lack the cagPAI. Similar inhibition can be observed for BafA1-treated cells infected with E. coli DH1 (MOI of 10). (B) BafA1 treatment inhibits the up-regulation of miR-155 significantly more during infection of TLR2/4−/− BMMs with the P12ΔcagL mutant (MOI of 50; 3 h) than with the P12ΔcagA (MOI of 50) mutant. The response to P12ΔcagL is similar to that to P12ΔcagPAI, whereas the response to P12ΔcagA is similar to that to WT P12. (C) MyD88/Trif/Rip2−/− BMMs were pretreated with BafA1 (100 nM) or were left untreated, then were infected for 3 h with P12, P12 P12ΔcagPAI, P12ΔcagA, or P12ΔcagL (all MOI of 50), and were checked for the expression of miR-155. In addition, these cells were infected with E. coli DH1 for 3 h (MOI of 10). As controls, MDP (100 μg/mL) and EC LPS (100 ng/mL) were used. All data presented here are from qRT PCR experiments. Data are ΔΔCt values using snoR-202 as endogenous control and the mock-infected control to normalize. Experiments were performed with at least three biologically independent replicates and are presented as mean + SE. Statistical significance was analyzed by Student’s t test; *P < 0.05; **P < 0.01; ***P < 0.001.
Downstream Effects of miR-155 During H. pylori Infection.
To investigate the role of miR-155 in primary macrophages during infection with H. pylori P12, BMMs from WT mice were infected for 6 h and then cultivated in differentiation medium containing gentamycin for another 24 h to detect targets down-regulated by miR-155 without affecting the survival of the cells. The up-regulation of miR-155 at this time point (30 h p.i.) had been determined previously (Fig. 1D). A microarray analysis of total mRNA was performed with mock-infected WT BMMs and P12-infected WT BMMs (array 1). Additionally, P12-infected WT BMMs were compared with P12-infected BMMs isolated from miR-155−/− mice (miR-155−/− BMMs) completely lacking miR-155 expression (array 2). The numbers of regulated genes are depicted in Fig. 4A. To identify direct targets of miR-155, hits with a cutoff of a 1.5-fold change and a P value <0.00001 that were down-regulated in WT P12 infected BMMs relative to miR-155−/− BMMs were examined for their putative miR-155 binding sites as predicted by TargetScan5.1 (48). Overall, 8.5% (1,234 of 14,506) of genes spotted on the array were predicted putative targets of Mus musculus (mmu)-miR-155 (for genes excluded, see Materials and Methods). Comparison of H. pylori infected WT BMMs with H. pylori infected miR-155−/− BMMs resulted in 17.9% (82/458) of all down-regulated genes with a cutoff of a 1.5-fold change and a P value < 0.00001 that contained a 7-mer or 8-mer complementary to the seed region of mmu-miR-155 (Fig. 4B). Statistical analysis (χ2) showed this gene set to be significantly enriched in targets of mmu-miR-155 (P < 0.0001) and highlighted that miR-155 exerts a strong effect on the overall regulation in response to P12 infection. Direct targets of miR-155 were considered to be down-regulated in WT relative to miR-155−/− BMMs; however, these targets still can be either unregulated or differentially regulated in mock-infected and infected WT BMMs (array 1). We subsequently focused only on the subset of genes that were down-regulated on both arrays to observe the direct biological link between H. pylori infection and the role of miR-155. Overall, 128 genes were down-regulated and overlapping on both arrays with a cutoff of 1.5-fold change and a P value < 0.00001. Table S1 depicts 35 of these mRNAs that have at least one miR-155 binding site in their 3′ UTR as identified by TargetScan5.1 (i.e., 35 genes are down-regulated in an H. pylori- and miR-155–dependent manner as putative direct targets). This result represented a further significant enrichment of miR-155 targets by 27.3% (35/128) (Fig. 4B) as determined by χ2 statistics (P = 0.0422).

mRNA microarray analysis of P12-infected BMMs lacking miR-155 shows enrichment of miR-155 targets. (A) Venn diagram representing differentially regulated genes on array 1 (P12 infected WT BMMs relative to mock-infected WT BMMs) and array 2 (P12 infected WT BMMs relative to miR-155−/− BMMs). Infections were for 30 h using MOI of 50. A total of 6,765 and 1,204 genes were regulated differentially in array 1 and array 2, respectively. Array 1: Groups II, VI, and X contain up-regulated genes; groups I, IX, and VIII contain down-regulated genes. Array 2: Groups III, VI, and IX contain up-regulated genes; groups IV, X, and VIII contain down-regulated genes. Group VIII contains genes down-regulated in both array 1 and array 2. (B) Comparison of all predicted targets of TargetScan (conserved and nonconserved) and all genes on the microarray (genome) with the down-regulated genes in array 2 (IV, X, VIII) reveals a significant bias toward miR-155 target genes when performing χ2 statistics (P < 0.0001). The enrichment is even higher in the overlapping down-regulated genes of arrays 1+ 2 (VIII) (χ2 test, P = 0.0422). (C–F) Validation of regulated mRNAs from the microarray by qRT-PCR. Only in the WT BMMs were targets significantly down-regulated compared with miR-155−/− BMMs. Relative values presented here are ΔΔCt values normalized to β-Actin as an endogenous control and a mock-infected control. Experiments were performed with at least three biologically independent replicates and are presented as mean + SE. Statistical significance was analyzed by Student’s t test; *P < 0.05; ***P < 0.001.
Validation of Direct Targets of miR-155.
Some of the direct miR-155 targets identified by our analysis have been published previously, namely, Bach1, Trp53inp1, Smad5, and Map3k7ip2 (34, 49–51). We additionally identified the putative targets Tspan14, Lpin1, and Pmaip1. To examine further the microarray data and the direct link between miR-155 and H. pylori infection, we validated the putative targets that contained two binding sites for mmu-miR-155 (Tspan14, Lpin1, and Pmaip1) together with Bach1 by qRT-PCR. These putative targets were down-regulated in P12-infected WT BMMs but were largely unregulated in P12-infected miR-155−/− BMMs (Fig. 4 C–F). To identify the direct effect of miR-155 on these newly identified targets, the 3′ UTRs (possessing two mmu-miR-155 binding sites) were cloned into the 3′ region of a Renilla luciferase reporter plasmid containing firefly luciferase as endogenous control (52–56). In addition, the predicted seed regions (7-mer or 8-mer) of the binding sites were mutated from UUA to AAU. For Tspan14 a full-length (Tspan14-2) and a truncated deletion of the 3′ UTR missing the last 83 nt (Tspan14-1) were used. Tspan14-2 contained two mmu-miR-155 binding sites, whereas the truncated version (Tspan14-1) contained only one that subsequently was mutated (Fig. S5). Lpin1 and Pmaip1 both contained two binding sites for mmu-miR-155, both of which were mutated (Fig. S5). These vectors then were transfected into HEK293T cells together with the precursor of mmu-miR-155 (premmu-miR-155) and a nonspecific control, prehsa-miR-198 (human). Cotransfection of premmu-miR-155 with the respective plasmids containing the 3′ UTR of the putative targets Pmaip1, Tspan14, and Lpin1 showed a strong down-regulation of the luciferase activity of the Renilla luciferase normalized to firefly luciferase (Fig. 5 A–C). Mutating the first predicted mmu-miR-155 binding site led to a significant recovery of the Renilla activity of Pmaip1-3′ UTR and Lpin1-3′ UTR, and mutating both binding sites led to almost full recovery of the Renilla luciferase signal; these results indicated that both mmu-miR-155 binding sites needed to be deactivated. For Tspan14 the truncated version (Tspan14-1) exhibited significantly increased Renilla luciferase activity compared with Tspan14-2 when cotransfected with premmumiR-155, whereas mutating the predicted binding site in Tspan14-1 restored the Renilla luciferase signal almost completely. By mutating the mmu-miR-155 putative binding sites of our identified targets, we propose a miR-155–mediated regulation of the genes Tspan14, Pmaip1, and Lpin1.

Validation of direct targets of mmu-miR-155. The 3′ UTRs of the putative targets Pmaip1 (A), Tspan14 (B), and Lpin1(C) were cloned in the 3′ direction to Renilla luciferase in a luciferase reporter plasmid containing Renilla and firefly luciferase as internal control. The putative binding sites subsequently were mutated in the seed region of the miRNA. The UUA motif (nucleotides 2–4) was replaced by AAU. The data are luciferase assays normalized to the untreated plasmid. All experiments were performed with at least three independent experiments and are presented as mean + SE. Statistical significance was analyzed by Student’s t test; *P < 0.05; **P < 0.01.
Cisplatin-Induced Apoptosis Is Inhibited by miR-155.
A functional analysis using Ingenuity Pathway Analysis software was performed with the 35 genes listed in Table S1. Interestingly, 39.4% of those genes were cell death related, whereas, in a control set comprising genes differentially regulated in mock- vs. P12-infected WT BMMs, only 27.4% of the genes were cell death related. Together with this finding, a large proportion of the genes (Table S1) are implicated in DNA damage responses (e.g., the known miR-155 targets Bach1, Trp53inp1, and MAP3K7ip2 and, although less well-established, our identified targets, Pmaip1, and Lpin1). Cisplatin is a well-known drug that induces apoptosis via DNA damage and has been shown previously to up-regulate LPIN1, PMAIP1, and TRP53INP1 (57). Accordingly, cisplatin was tested on P12-infected WT BMMs and miR-155−/− BMMs to elucidate the potential role of miR-155 in apoptosis mediated by DNA damage. As readouts, cleavage of Procaspase-3 to activated apoptosis effector Caspase-3 and the cleavage of the Caspase-3 downstream target poly(ADP ribose) polymerase (PARP) were analyzed by Western blot. BMMs were infected for 6 h and incubated further for another 24 h to emphasize miR-155-dependent effects. Following 18 h of cisplatin treatment, substantial apoptosis was observed in mock-infected cells, whereas the induction of apoptosis mediated by DNA damage was inhibited in WT BMMs infected with H. pylori P12 (Fig. 6A). This result is in contrast to that seen in P12-infected miR-155−/− BMMs, which had PARP/Procaspase-3 cleavage levels comparable to those of mock-infected cells. The quantification of six independent Western blots is shown in Fig. 6 B and C. For further confirmation, Caspase-3/-7 activity was measured using proluminescent Caspase-3/-7 substrate (Fig. 6D). As a control, water-soluble tetrazolium salts (WST-8) assays were performed before cisplatin treatment; no difference between the WT and miR-155−/− cells could be detected (Fig. S6). Finally, during infection, Caspase-3/-7 activity was higher in miR-155−/− than in WT BMMs, further emphasizing the differences in apoptosis and the biological impact of miR-155.
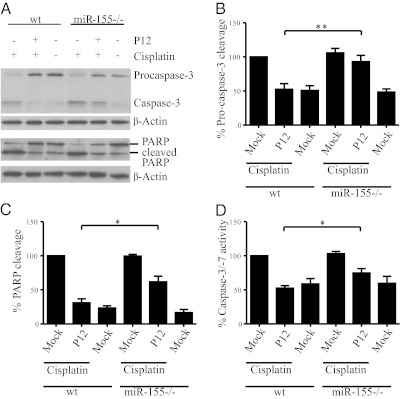
Up-regulation of miR-155 in H. pylori-infected BMMs blocks cisplatin-induced apoptosis. (A) Pro-caspase-3 and PARP cleavage was detected by Western blot after H. pylori infection (MOI of 50) and 18 h of treatment with cisplatin (50 μM). Here, cleaved Procaspase-3 and PARP can be observed in the cisplatin-treated mock-infected BMMs but not in the untreated cells. Procaspase-3 as well as PARP cleavage is inhibited after infection but, as seen in the quantification in B and C, is inhibited significantly less in the miR-155−/− BMMs than in WT BMMs. The blot in A is representative of six biologically independent experiments, and quantification shows the mean + SE of six biologically independent experiments. (D) The activity of caspase-3/-7 was determined by a luciferin-based assay. The activity is less in the P12-infected WT BMMs than in P12-infected miR-155−/− BMMs. Experiments were performed with at least three biologically independent replicates and are presented as mean + SE. Statistical significance was analyzed by Student’s t test; *P < 0.05; **P < 0.01.
Discussion
Here we demonstrate a TLR- and NOD1/2-independent up-regulation of miR-155 in macrophages during H. pylori infection. The TLR-dependent up-regulation of miR-155 via MyD88 and Trif in BMMs has been reported previously (39); here, however, we show there is a TLR-independent up-regulation of miR-155 by H. pylori in macrophages occurring via the NF-κB pathway. The T4SS-dependent up-regulation of miR-155 in macrophages that we report confirms the importance of this microRNA in H. pylori infection and may be of particular relevance in the context of suboptimal TLR activation, resulting in part from reduced MyD88/Trif expression, in gastrointestinal macrophages (58).
We found that the signaling pathway upstream of miR-155 during H. pylori infection appeared to differ from that of other NF-κB–dependent genes, such as IL-6. IL-6 mRNA synthesis was almost completely dependent on MyD88 and abolished by BafA1 treatment of TLR2/4−/− BMMs, consistent with previous data (23). By comparison, miR-155 expression during infection was less sensitive to BafA1 treatment in TLR2/4−/− BMMs and led us to investigate the role of other bacterial factors in the regulation of miR-155 expression during infection. The up-regulation of miR-155 was significantly stronger in T4SS-positive strains than in T4SS-negative strains when extracellular TLR activation/signaling was blocked (TLR2/4−/− BMMs) in conjunction with a block to endosomal maturation (BafA1 treatment) and/or in macrophages devoid of all MyD88/Trif-dependent TLR signaling and NOD1/2 activation (MyD88/Trif/Rip2−/−). Notably, the H. pylori CagA protein itself has been reported to induce NF-κB signaling directly in gastric epithelial cells (59); however, we found that the T4SS-dependent miR-155 up-regulation was independent of this major effector protein. Induction of NF-κB by H. pylori also has been proposed to occur via NOD receptors (22). We have shown here that classical NOD ligands have limited capacity to induce miR-155 in macrophages; however, the T4SS-dependent miR-155 up-regulation during infection was independent of NOD1/2 receptor activity, as demonstrated by the use here of MyD88/Trif/Rip2−/− BMMs. The MyD88/Trif-independent up-regulation of miR-155 observed here displays a phenotype similar to that seen with the induction of TNF-α and type I IFNs or IFN-induced genes, such as CXCL10, demonstrated for intestinal bacteria-infected BMMs (60, 61). IFN-β and TNF-α incubation induced miR-155 expression; however, induction of miR-155 by TLR ligands still occurred in the absence of TNF receptors and IFN-α receptors in BMMs (39). In accordance with our data, a partial SPI1/2 (encoding Salmonella translocated effector proteins and a type III secretion system)-dependent effect in the up-regulation of miR-155 in LPS tolerized macrophages has been demonstrated during Salmonella enterica Typhimurium infection (54). We speculate that the T4SS-dependent pathway could be triggered by an as yet unknown T4SS translocated effector, which is detected by cytosolic receptors in BMMs, or could result from docking of the T4SS to an unidentified plasma membrane receptor(s). Alternatively, the H. pylori T4SS could signal directly via members of the integrin family, e.g., integrin β1 receptors (62). Indeed, downstream effects of this interaction have been described previously (46). To our knowledge the TLR- and NOD1/2-independent regulation of an inflammatory response via NF-κB during bacterial infection has not been reported previously. The T4SS-dependent effect shown here contributes to the ongoing discussion as to whether the bacterial secretion system machinery is involved directly in innate immune detection and responses of professional phagocytes, as shown for Legionella pneumophila and Yersinia pseudotuberculosis (47, 61). Recently, the question of the existence of “vita-PAMPs,” which implicates pattern recognition from live rather than dead bacteria, has been addressed (63). Our study contributes to this discussion in that the functional H. pylori T4SS could resemble such a vita-PAMP. We anticipate that the MyD88/Trif/Rip2−/− generated here will have future applications for investigations into the contribution of bacterial secretion system to the activation of PAMP receptors.
As recently suggested, H. pylori is found not only on the epithelial cell surfaces of the infected gastric mucosa but also in the lamina propria inside mucosal macrophages that are located directly basally of epithelial cells (64). One hypothesis for the mechanism of H. pylori-induced pathology is that during persistent infection an unresolved inflammatory response, possibly involving macrophages, contributes to severity of disease. Depletion of macrophages in vivo has been suggested to confer protection against gastritis in the presence of high bacterial loads (65). We previously reported that miR-155 was up-regulated in gastric biopsies of human volunteers infected with H. pylori for 80 d, and a major infiltration of macrophages/DCs was detected at the RNA level (15). In addition, we recently reported that miR-155 expression is important for T-helper cell 1/17 immunity to H. pylori infection in vivo, because miR-155−/− T cells impaired the ability of mice to control bacterial load; interestingly, miR-155−/− mice also were protected against pathology induced by H. pylori infection (66). We now report that, in addition to its role in T-cell immunity, miR-155 also has a role in resistance to apoptosis caused by DNA damage in macrophages. It is conceivable that increased macrophage survival in the inflammatory environment could contribute to the phenotype of H. pylori infection observed in vivo (66).
We observed remarkable gene-regulatory patterns for miR-155 in BMMs during H. pylori infection and strong enrichment of numerous miR-155 targets, both those identified here and previously published (34, 49–51), demonstrating the importance of miR-155 during infection. Our data show the substantial impact of a single miRNA on the overall mRNA expression profile during infection. Validation of several microarray hits by qRTPCR showed not only that the respective target RNAs were down-regulated during infection with H. pylori in WT BMMs but also that this down-regulation was attenuated almost completely in miR-155−/− BMMs. Overall, there was a significant enrichment in target genes with established roles in determining cell fate (e.g., regulation of apoptosis/DNA damage). In particular, the targets TRP53INP1, BACH1, and Map3k7ip2 (TAB2) (34, 50, 51) were confirmed here as being regulated by miR-155 in murine macrophages and have been reported to be regulated in response to DNA damage (67–69). Three highly regulated genes with two 3′ UTR miR-155 binding sites were selected on the basis of their novelty as miR-155 targets and were validated in depth. These targets, Lpin1 (70), Pmaip1 (57), and Tspan14 (71, 72), have reported roles in cell survival/DNA damage, but these roles are not as well established as those of many other targets of miR-155 that we identified from our screen. Interestingly however, it has been demonstrated that Pmaip1/Noxa−/− BMMs were more resistant than WT BMMs to apoptosis induced by vaccinia virus Ankara (73). Our experiments revealed that H. pylori-infected WT BMMs are substantially more resistant than mock-infected control cells to apoptosis induced by DNA damage. H. pylori-infected miR-155−/− BMMs were almost as sensitive as mock-infected BMMs to apoptosis. Previous reports suggested a direct induction of apoptosis by H. pylori in monocyte-derived cell lines (30, 31). However, in line with our results, others detected no apoptosis in murine BMDCs without sensitization (32), and H. pylori blocked apoptosis induced by DNA damage in Mongolian gerbils (74). In addition, it has been reported that the overall effect of miR-155 may be antiapoptotic/chemoresistant, thereby explaining the high expression of miR-155 in different tumor types (14, 36). Again, this suggestion is in agreement with the results obtained here. It is possible that miR-155 helps stabilize macrophages in the strongly inflammatory environment created during H. pylori infection. Indeed, DNA damage triggered by nitric oxide or reactive oxygen species has been observed in H. pylori-infected patients (75). It has also been reported that in inducible nitric oxide synthase double-knockout mice infected with H. pylori, gastric epithelial cells do not exhibit DNA damage and apoptosis to the same extent as in H. pylori-infected WT mice (76). This and other mechanisms of DNA damage may lead to apoptosis in the surrounding tissue (77), which may contribute to the pathology of H. pylori infections.
Materials and Methods
Cell Culture and Generation of BMMs.
Murine J774A.1 (ACC170; Deutsche Sammlung von Mikroorganismen und Zellkulturen) cells were grown in RPMI medium (Gibco) containing 10% (vol/vol) heat-inactivated FCS. BMMs were generated as described elsewhere (78). In brief, monocytes were isolated from the femur and tibia of C57BL/6 mice and were plated in 10-cm dishes in RPMI medium containing 10% (vol/vol) heat-inactivated FCS and 30% (vol/vol) L929 supernatant as source of macrophage colony-stimulating factor. After 7 d in culture these cells were washed twice with PBS and were replated overnight in RPMI medium containing 10% (vol/vol) heat-inactivated FCS and 10% (vol/vol) L929 supernatant. Purity of the culture was controlled by CD11b+ and F4/80+ antibodies in FACS analysis with the respective isotype control (both from RnD Systems).
Bacterial Culture and Infection Experiments.
H. pylori P12 (strain collection no. 243) and its isogenic mutants P12ΔcagPAI (no. P387), P12ΔcagL (no. P454), and P12ΔcagA (no. P378) were cloned as described previously by replacing the gene with a chloramphenicol or kanamycin cassette (79) and were grown on GC agar plates as described elsewhere (79). Infection was carried out after 3 h of serum starvation in RPMI medium with the respective MOI and time point. Cells were serum starved for the duration of infection, except for the late time points 24 h and 30 h, at which medium was replaced with RPMI, 10% (vol/vol) heat-inactivated FCS, 10 μg/mL gentamicin (Sigma) (at 24 h), and 10% (vol/vol) L929SN (at 30 h). Heat-inactivation of bacteria was performed at 56 °C for 1 h. P12 LPS was purified by the hot phenol-water technique as previously described (80) followed by octyl-Sepharose column, DNaseI, RNaseA, and proteinase K treatment (40). Activity and concentration of P12 LPS was tested by LAL Pyrochrome assay (Pyroquant). P12 RNA was purified with the RNeasy RNA purification kit with on-column DNA digestion (Qiagen). The RNA was transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. E. coli strain DH1 (No. E60) was grown in LB liquid culture and used for experiments in the logarithmic growth phase.
Reagents.
For stimulations, the TLR4 ligand ultrapure E. coli LPS, the NOD1 ligand TriDAP, the NOD2 ligand MDP (all from Invivogen), and the synthetic TLR2 ligand Pam3CSK4 (EMC Microcollections) were used. As inhibitors the proteasome inhibitor MG132 (C2211) and the inhibitor of endosomal maturation Bafilomycin A1 (B1793) (both from Sigma) were used. Cisplatin (Calbiochem), Bafilomycin A1, and MG132 were dissolved in DMSO to final concentrations of 50 μM, 100 nM, and 50 μM, respectively.
RNA Isolation and qRT-PCR.
Total RNA was isolated by the TRIzol (Invitrogen) method following the manufacturer’s protocol. The qRT-PCR for the quantification of miRNAs was performed according to the manufacturer’s protocol (Applied Biosystems) in two steps using specific primers for mmu-miR-155 (Assay ID: 001806), RnU6b (Assay ID: 001093), or snoR-202 (Assay ID: 001232). In total, 10 ng RNA was used for reactions. Relative expression levels were determined by applying the ΔΔCt method (81) using snoR-202 or RnU6b as endogenous controls and normalization to mock-infected cells.
For the detection of mRNAs, the one-step SYBR-green method using the RNA-to-Ct assay was followed in accordance with the manufacturer’s protocol (Applied Biosystems). Ten nanograms of RNA were used for each reaction. Relative expression levels were determined by applying the ΔΔCt method using β-Actin as endogenous control and normalization to the mock-infected cells. Primer sequences are given in Table S2.
Northern blotting was performed according to ref. 54.
Luciferase Assay.
The 3′ UTRs from the genes Lpin1, Tspan14, and Pmaip1 were cloned from murine BMM genomic DNA 3′ to the Renilla luciferase psicheck2 vector (Promega) between the XhoI and NotI restriction sites. This plasmid also contained firefly luciferase as an endogenous control. To identify miR-155 binding sites, mutagenesis experiments were performed using the psicheck2-Lpin1, psicheck2-Pmaip1, and psicheck2-Tspan14-1 vectors as template. The mutagenesis experiments were performed with the Phusion polymerase (Finnzymes) according to the manufacturer’s protocol, followed by a 3-h DpnI digestion (Fermentas) at 37 °C. Selected clones were controlled by plasmid-specific sequencing. Primer sequences are given in Table S2.
These plasmids were transfected into HEK293T cells (P4-8) at a virtual confluency of 70%; plasmids (1 μg/mL) were incubated with Lipofectamine 2000 (Invitrogen) and the respective precursors of miRNAs [(mmu-miR-155) (AM17100 ID: PM13058; Ambion), hsa-miR-198 (AM 17100 ID: 11088; Ambion)] or controls, according to the manufacturer’s protocol. HEK293T cells were incubated for 24 h; then the Dual-Glo Luciferase assay was performed according to the manufacturer’s protocol (Promega).
Microarray Analysis.
Microarray experiments were performed as dual-color hybridizations. To compensate for dye-specific effects, an independent dye-reversal color swap was applied. Quality control and quantification of total RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and a NanoDrop 1000 UV-Vis spectrophotometer (Kisker). RNA labeling was performed with the dual-color Quick-Amp Labeling Kit (Agilent Technologies). In brief, mRNA was reverse transcribed and amplified using an oligo-dT-T7 promoter primer, and resulting cRNA was labeled with Cyanine 3-CTP or Cyanine 5-CTP. After precipitation, purification, and quantification, 1.25 μg of each labeled cRNA was fragmented and hybridized to whole-mouse genome 4 × 44k multipack microarrays (AMADID 014868) according to the supplier’s protocol (Agilent Technologies). Scanning of microarrays was performed with 5-μm resolution using a G2565CA high-resolution laser microarray scanner (Agilent Technologies) with XDR extended range. Raw microarray image data were analyzed with the Image Analysis/Feature Extraction software G2567AA v. A.10.5.1 (Agilent Technologies) using default settings and the GE2_105_Jan09 protocol. The extracted MAGE-ML files were analyzed further with the Rosetta Resolver Biosoftware, Build 7.2.2 SP1.31 (Rosetta Biosoftware). Ratio profiles comprising single hybridizations were combined in an error-weighted fashion to create ratio experiments. A 1.5-fold change expression cutoff for ratio experiments was applied together with anticorrelation of ratio profiles, rendering the microarray analysis highly significant (P < 0.01), robust, and reproducible. Microarray data presented in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession no. GSE29388.
Data Analysis.
To analyze the microarray results further, all regulated genes were compared with the predicted targets of mmu-miR-155 by TargetScan5.1 (48). Because TargetScan5.1 predicts only targets with RefSeq mRNA accession numbers, only genes with a RefSeq accession number were used. In total, the microarray contained 14,506 genes with RefSeq accession numbers. From those genes a set-union of 1,234 genes were matched with conserved and nonconserved mmu-miR-155 predicted targets that also were identified by the microarray analysis. For identification of targets, a cutoff of 1.5-fold change and P < 0.00001 was used. For statistical analysis χ2 statistics were performed. Biological effects of target genes with RefSeq numbers were analyzed subsequently with the Ingenuity Pathway Analysis software package.
Apoptosis Assays.
BMMs were grown in RPMI medium containing 10% (vol/vol) heat inactivated FCS plus 10% (vol/vol) L929 supernatant (standard medium). These cells were infected with H. pylori P12 (MOI of 50) for 6 h in RPMI only; then the medium was replaced by standard medium containing gentamicin, and incubation continued for a further 24 h. After incubation, apoptosis was induced using 50 μM cisplatin for 18 h in RPMI containing 10% (vol/vol) heat-inactivated FCS. Samples were taken for Western blot and for Caspase-Glo-3/-7 assays (Promega). To check for cell viability the WST-8 assay according to the Colorimetric Cell Viability Kit I assay (Promokine) was performed.
Western Blot and Quantification.
For Western blot, Laemmli buffer was added directly to BMMs. Depending on the protein of interest, 5–15% SDS/PAGE gels (CagA: 5%, PARP: 10%, Caspase-3: 15%) were used for blotting onto a PVDF membrane. After blotting, membranes were blocked with TBS buffer containing 0.1% (vol/vol) Tween-20 and 3% (wt/vol) BSA. Antibodies and dilutions were as follows: rabbit anti-Caspase-3, 1:1,000 (9662; Cell Signaling); rabbit anti-PARP, 1:1,000 (sc-7150; Santa Cruz); mouse anti–β-Actin, 1:3,000 (A5441; Sigma); rabbit anti-pY99, 1:1,000 (sc-7020, Santa Cruz); rabbit anti-CagA, 1:1,000 (sc-25766, Santa Cruz); rabbit anti-IQGAP, 1:1,000 (sc-10792; Santa Cruz); sheep anti-mouse HRP, 1:3,000 (NA931V; Amersham); donkey anti-rabbit HRP, 1:3,000 (NA934V’ Amersham). ECL Western blotting substrate either was detected by Hyperfilm ECL (Amersham) or quantified using ImageQuant LAS4000 (GE) (for the apoptosis assay). Signals detected by ImageQuant LAS4000 were maintained below saturation, and images were not contrast adjusted before AIDA Image Analysis (Raytest). All signals were marked with the same pixel size, and densities were determined by AIDA. The 2D region report was analyzed further and normalized to the WT BMM mock-infected cisplatin-treated cells.
Mice.
Mice were bred and maintained according to German and European guidelines for animal care and were kept under specific pathogen-free conditions according to Federation of European Laboratory Animal Science Association recommendations. Mice deficient in TLR2, TLR4, TLR2/4, MyD88, Trif, and MyD88/Trif had a C57BL/6 background and have been described previously (82–85). Mice deficient in Rip2 (B6.129S1-Ripk2tm1Flv/J) and miR-155 (B6.Cg-Mir155tm1.1Rsky/J) were purchased from Jackson Laboratories. Mice deficient in MyD88/Trif/Rip2 were generated by crossing MyD88/Trif−/− mice with Rip2−/− mice. As control, WT BMMs were isolated from C57BL/6J mice.
Statistical Analysis.
Statistical analyses were performed using Student’s t test.
Acknowledgments
We thank Kirstin Hoffmann and Meike Soerensen for excellent technical assistance, and Dr. Daniel Becker and Dr. Kate Holden-Dye for editing the manuscript. MyD88/Trif−/− mice were a gift from the Faculty of Veterinary Medicine, Technical University, Munich. This work was funded by the Sixth Research Framework Programme of the European Union, Project SIROCCO (LSHG-CT-2006-037900) and the Deutsche Forschungsgemeinschaft through SFB633. M.K. was funded by the International Max Planck Research School for Infectious Diseases and Immunology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data presented in this paper have been deposited in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE29388).
See Author Summary on page 7144 (volume 109, number 19).
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1116125109/-/DCSupplemental.
References
Author Summary
Author Summary
The bacterium Helicobacter pylori infects ~50% of the human population worldwide and is a major cause of gastritis, peptic ulcer, and gastric cancer. However, researchers have not yet discovered which factors determine whether infections progress from being relatively benign, and often symptomless, to causing severe disease (in 10–20% of cases). It is evident, however, that bacterial, host, and environmental factors are involved. Here, we report a unique role for the key H. pylori pathogenicity factor, the cagPAI pathogenicity island, a segment in the pathogen’s genome, in modulating an immune response during infection of host immune cells.
Bacterial factors implicated in H. pylori pathogenicity are expressed largely by the bacterial cagPAI island, which encodes the functional type four secretion system (T4SS) and the CagA effector protein. The innate immune responses of the host also are implicated as factors in H. pylori disease; for example, the inflammatory response itself may be potentially damaging to tissues (1). Macrophage immune cells are key mediators of inflammatory responses, but their precise role in H. pylori pathology is undetermined. MicroRNAs (miRNAs) are short sequences of genetic material that can inhibit genetic activity and which may play a role in macrophage-induced pathogenicity.
We wanted to know which bacterial factors are responsible for inducing the expression of miR-155, an miRNA mediator of inflammation that recently was discovered to be up-regulated during H. pylori infection (2). We studied its expression in macrophages and the consequences of this expression during H. pylori infection. To do so, we infected either WT mouse macrophages or macrophages that were mutated to lack Toll-like receptors (TLRs). TLRs are key receptors for bacterial/viral components, such as LPS, that stimulate responses from host immune cells. Macrophages, in particular, are rich in these receptors. For example, when LPS interacts with TLRs in macrophages, these macrophages are stimulated to mount a powerful immune response. Because miR-155 already has been demonstrated to be up-regulated when TLR2 and TLR4 (3) are stimulated in this manner, we used macrophages that lacked TLR2 and TLR4 or key TLR signaling adaptor proteins (MyD88/Trif) to identify other factors that might be responsible for inducing miR-155. We also used different H. pylori strains that either naturally lack some of the bacterial pathogenicity factors or contain antibiotic resistance cassettes in place of some of the pathogenicity factors.
Using quantitative PCR (qPCR) techniques, a method to measure gene expression, we quantified the levels of miR-155 expressed during infection with different H. pylori strains and in the different macrophage cell types. We found that all bacterial strains increased miR-155 expression via activation of TLRs. More interestingly, we observed that strains containing the T4SS up-regulated miR-155 more strongly (when some other signaling pathways were blocked) than strains that lacked the T4SS. Thus, we concluded that, in addition to previous observations implicating TLRs in miR-155 regulation, miR-155 up-regulation also is dependent partly on the direct interaction of the T4SS with macrophages. Moreover, we found that this up-regulation occurred independently of the CagA protein and via the activation of another inflammatory signaling pathway, the NF-κB pathway. Thus, we identified a role for the prototypical H. pylori T4SS pathogenicity factor independent of its major effector protein, both of which are encoded on the bacterial cagPAI island.
MicroRNAs inhibit the expression of their gene targets. We also wanted to identify gene targets of miR-155 in macrophages during H. pylori infection. We used microarray analysis, a method of measuring the expression of large numbers of genes, to screen the gene activity in response to H. pylori in WT macrophages and also macrophages that had been depleted of miR-155. We compared microarray datasets to determine previously unidentified miR-155 gene targets, which were down-regulated during infection. Many of the gene targets identified are known to be associated with cell-death pathways and DNA damage responses. Of particular interest, among the 35 miR-155 down-regulated gene targets with predicted miR-155-binding sites were the genes Pmaip1, Tspan14, and Lpin1, which have been variously linked to cell-death or apoptotic pathways (Fig. P1). We found that H. pylori-infected macrophages lacking miR-155 were more sensitive to cell death induced by the DNA-damaging agent cisplatin. This result suggested that miR-155 is induced during H. pylori infection and protects macrophages against cell death. It is possible that prolonged macrophage survival contributes to H. pylori disease. Certainly, depletion of macrophages protected against H. pylori pathology in a mouse model of gastritis. Furthermore, depletion of miR-155 in T cells, another immune cell, also protected mice against such pathology (4, 5). In both examples, bacterial loads remained high, although pathology was reduced, presenting an intriguing conundrum for H. pylori research.

Model of H. pylori-induced miR-155 expression in macrophage immune cells. Red arrow represents the T4SS-mediated miR-155 regulatory pathway. Macrophages detect the LPS characteristic of H. pylori via TLR2 and TLR4 receptors involving the adaptor proteins MyD88 and Trif, thereby up-regulating miR-155. However, there is an additional T4SS-dependent induction of miR-155 that is independent of the CagA genome segment studied here. miR-155 expression results in the down-regulation of miR-155 target genes, which have a general antiapoptotic, or anti-cell death, function in macrophages during H. pylori infection.
In conclusion, we found that the H. pylori T4SS protein can increase the expression of miR-155 in immune cells independently of the known regulation via TLRs (Fig. P1). Because evidence now suggests that TLR signaling in general is dampened at the gastric surface, our findings may be of particular relevance for persistent H. pylori infections in the human stomach, where it is known that strains containing the T4SS cause more severe disease. Future work is needed to resolve the precise roles of miR-155 and macrophages in H. pylori disease.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1116125109
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/109/19/E1153.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The TLR/MyD88 signalling cascade in inflammation and gastric cancer: the immune regulatory network of Helicobacter pylori.
J Mol Med (Berl), 101(7):767-781, 17 May 2023
Cited by: 4 articles | PMID: 37195446
Review
Saliva microRNA Profile in Children with and without Severe SARS-CoV-2 Infection.
Int J Mol Sci, 24(9):8175, 03 May 2023
Cited by: 1 article | PMID: 37175883 | PMCID: PMC10179619
The Role of miR-155 in Antitumor Immunity.
Cancers (Basel), 14(21):5414, 03 Nov 2022
Cited by: 11 articles | PMID: 36358832 | PMCID: PMC9659277
Review Free full text in Europe PMC
The role of non-coding RNA in the diagnosis and treatment of Helicobacter pylori-related gastric cancer, with a focus on inflammation and immune response.
Front Med (Lausanne), 9:1009021, 13 Oct 2022
Cited by: 1 article | PMID: 36314013 | PMCID: PMC9606473
Review Free full text in Europe PMC
Impact of microRNA Regulated Macrophage Actions on Adipose Tissue Function in Obesity.
Cells, 11(8):1336, 14 Apr 2022
Cited by: 6 articles | PMID: 35456015 | PMCID: PMC9024513
Review Free full text in Europe PMC
Go to all (73) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (3 citations) GEO - GSE29388
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Deficiencies of myeloid differentiation factor 88, Toll-like receptor 2 (TLR2), or TLR4 produce specific defects in macrophage cytokine secretion induced by Helicobacter pylori.
Infect Immun, 75(5):2408-2414, 12 Mar 2007
Cited by: 64 articles | PMID: 17353291 | PMCID: PMC1865764
Induction of TLR-2 and TLR-5 expression by Helicobacter pylori switches cagPAI-dependent signalling leading to the secretion of IL-8 and TNF-α.
PLoS One, 6(5):e19614, 09 May 2011
Cited by: 62 articles | PMID: 21573018 | PMCID: PMC3090411
Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence.
PLoS Pathog, 6(9):e1001120, 23 Sep 2010
Cited by: 84 articles | PMID: 20886104 | PMCID: PMC2944809
Genetic and Epigenetic Regulation of the Innate Immune Response to Gout.
Immunol Invest, 52(3):364-397, 06 Feb 2023
Cited by: 7 articles | PMID: 36745138
Review





