Abstract
Free full text

Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1
Associated Data
Abstract
Hepatitis C virus (HCV) infects more than 2% of the global population and is a leading cause of liver cirrhosis, hepatocellular carcinoma, and end-stage liver diseases. Circulating HCV is genetically diverse, and therefore a broadly effective vaccine must target conserved T- and B-cell epitopes of the virus. Human mAb HCV1 has broad neutralizing activity against HCV isolates from at least four major genotypes and protects in the chimpanzee model from primary HCV challenge. The antibody targets a conserved antigenic site (residues 412–423) on the virus E2 envelope glycoprotein. Two crystal structures of HCV1 Fab in complex with an epitope peptide at 1.8-Å resolution reveal that the epitope is a β-hairpin displaying a hydrophilic face and a hydrophobic face on opposing sides of the hairpin. The antibody predominantly interacts with E2 residues Leu413 and Trp420 on the hydrophobic face of the epitope, thus providing an explanation for how HCV isolates bearing mutations at Asn415 on the same binding face escape neutralization by this antibody. The results provide structural information for a neutralizing epitope on the HCV E2 glycoprotein and should help guide rational design of HCV immunogens to elicit similar broadly neutralizing antibodies through vaccination.
Hepatitis C virus (HCV) infects >2% of the world population, with an estimated >500,000 new infections annually in the highest endemic country, Egypt (1, 2). In the United States, the rate of symptomatic HCV infection declined over the last decade and began to level out at ~4 million cases around 2005 (3). Alarmingly, however, in developed countries, new cases are often associated with the younger age group (15-24 y) because of illegal injection drug use (4). Although some HCV-infected individuals can resolve infection without drug treatment, ~70% develop chronic hepatitis and, over a period of 20–30 y, 20–30% will develop liver cirrhosis and 1–5% hepatocellular carcinoma (5). Furthermore, HCV infection is associated with several extrahepatic manifestations, neuropathy, and autoimmune diseases including mixed cryoglobulinemia and Sjögren’s syndrome (6). The standard-of-care treatment for HCV infection uses a combination of pegylated IFN-α and ribavirin, which is effective in approximately 50% of treated patients but has many side effects. Two direct-acting antiviral drugs targeting the virus protease NS3 have recently been approved in the United States for triple therapy with IFN-α and ribavirin to improve success rates and to shorten treatment (7). To solve the global HCV problem and to eradicate the virus, more effective, tolerable, and affordable drugs against HCV, as well as a vaccine, are needed. Potent direct-acting antiviral drugs against additional viral targets are currently under development and show promise in IFN-free treatments (8). In the past few years, progress has also been made in vaccine development for prophylaxis and therapeutic purposes (9, 10).
A major challenge in vaccine design against HCV is the extreme diversity of the virus. HCV is highly heterogeneous, with 6 major genotypes (>30% overall nucleotide sequence difference) and more than 50 subtypes (10–25% difference in nucleotide sequence) (11). Genotypes 1 and 3 are the most widely distributed in the world. Genotype 2 is also found worldwide, with genotype 4 predominantly in Egypt, genotype 5 in South Africa, and genotype 6 in Southeast Asia. This great diversity of HCV is fueled by the poor fidelity of its RNA polymerase and rapid turnover of the virus, as evidenced by the estimate that an individual produces as many as 1012 virions per day (12). Consequently, any given vaccine or drug effective against one isolate will not necessarily be useful against more divergent isolates. To overcome the challenge of viral diversity, a broadly effective vaccine must target conserved immune epitopes of the virus.
We, and others, reported previously that antibodies to the CD81-receptor binding site (CD81bs) on E2 mediate cross-neutralization of diverse HCV isolates. These antibodies include the mouse mAb AP33 (13), rat mAb 3/11 (14), and human mAb HCV1 (15), which block the interaction of E2 to CD81 by binding to linear epitopes located within the highly conserved E2 antigenic site, residues 412–423 (the standardized genome numbering of the HCV prototypic strain H77 will be used throughout). Other mAbs recognize discontinuous E2 epitopes overlapping with the CD81bs on E2 involving residues 395–424, 425–447, and/or 523–540 (16–19). In addition, human polyclonal antibodies purified from the sera of HCV patients using peptides spanning E2 residues 412–419 or 412–423 were also found to neutralize HCV (20, 21). The fact that cross-neutralizing antibodies to the E2 antigenic site 412–423 have been isolated in multiple laboratories suggests that this conserved site is a prime target for HCV vaccine design. However, it has been reported recently that only ~2% of chronic HCV patients are able to produce an antibody response to this antigenic site (21, 22), indicating that it is only weakly immunogenic when presented on native virions.
In this study, we determined two crystal structures of human mAb HCV1 Fab in complex with the E2 peptide 412–423 at 1.8-Å resolution. The mAb HCV1 was isolated by MassBiologics (15) and has been shown to protect the chimpanzee model from infection by an HCV-infected human serum inoculum (23). The mAb is currently under evaluation in a phase II clinical trial to prevent recurrent HCV after liver transplantation in HCV patients (ClinicalTrials.gov identifier NCT01121185). The mAb HCV1 cross-neutralizes HCV isolates from genotypes 1a, 1b, 2b, 3a, and 4a by binding to its highly conserved linear epitope with nanomolar affinity. The residues critical for antibody binding have previously been mapped to Leu413 and Trp420 by alanine scanning mutagenesis (15). The present work reveals the structural determinants of this interaction, allowing the precise design of immunogens as potential vaccine candidates to properly present this epitope.
Results
Expression and Characterization of Recombinant mAb HCV1.
The variable domains of heavy and light chains of mAb HCV1 (15) were inserted into a human IgG1 expression vector (19) to generate full-length antibody. The biological activities of the recombinant mAb were verified by a series of assays. First, mAb HCV1 binds both native and reduced E1E2 antigens in ELISA, suggesting that it recognizes a continuous or linear epitope (Fig. 1A). Second, HCV1 specifically recognizes a peptide containing the mapped epitope (Fig. 1B). Third, HCV1 bound E1E2 with an apparent affinity of ~3 nM, consistent with the affinity reported previously for this mAb (15) (Fig. S1). Finally, as expected, HCV1 was able to cross-neutralize HCV genotypes 1a and 1b (Fig. 1C).

Biological activities of recombinant mAb HCV1. (A) Binding of mAb HCV1 to E1E2 in ELISA. E1E2 antigens expressed in 293T cells, at their native (closed symbols) or reduced (open symbols) form, were captured onto microwells by lectin. The control mAbs A4 and AR3A recognize a continuous E1 epitope (46) and a discontinuous E2 epitope (19), respectively. (B) HCV1 epitope. The specificity of mAb HCV1 was verified by binding to overlapping peptides (250 ng per well) containing the sequence of the epitope (in bold type). The mAb did not bind peptides with truncated epitope sequences. Note that the amount of captured E1E2 is not equivalent to the directly coated peptides. (C) Neutralization of HCV by mAb HCV1. The recombinant mAb neutralized HCV pseudotype virus particles (HCVpp) displaying the genotype 1a or 1b E1E2, but not the control envelope glycoprotein G from vesicular stomatitis virus (VSVpp). The mAbs AR3A and AR4A are control neutralizing mAbs to E2 and the E1E2 complex, respectively (39). (D) Peptide candidates for crystallography. The binding of mAb HCV1 to three candidate peptides (R12-mer, 15-mer, and 18-mer) was first evaluated by direct ELISA (Left). The mAb bound poorly to the 15-mer and R12-mer when the peptides were coated directly onto microwells. However, when in solution, all three peptides blocked the mAb binding to E1E2 at equivalent levels (Right). The epitope sequence is highlighted in bold in the above peptides.
To determine the crystal structure of mAb HCV1 in complex with the epitope, we produced the Fab HCV1 and investigated peptides for complex formation. The Fab fragment of mAb HCV1 was generated by deleting the hinge, CH2, and CH3 sequences in the pIgG1 expression vector, and then expressed and purified. Three peptides (18-mer, 15-mer, and R12-mer) were evaluated for complex formation. In the R12-mer, an arginine was added to the N terminus of the 12-aa epitope to improve peptide solubility. Interestingly, the antibody bound poorly to the shorter 15-mer and R12-mer compared with the 18-mer in ELISA when the peptides were coated directly onto the microwells (Fig. 1D, Left). Nevertheless, in competition ELISA all three peptides (in solution) blocked mAb HCV1 binding to the E1E2 antigen equally well (Fig. 1D, Right). The shorter and more soluble R12 peptide (R-412-QLINTNGSWHIN-423) was selected for complex formation with HCV1 Fab, and that complex was submitted for crystallization screening.
Crystal Structures of Fab HCV1 in Complex with Its Peptide Epitope.
Crystal trials were conducted on purified protein complex samples concentrated to 10 mg/mL at both 4 °C and 20 °C as described in Materials and Methods. The complex crystallized under several conditions containing PEG and two crystal forms (space groups C2 and P21) diffracted to 1.8-Å resolution (Table S1). Electron density for the peptide was well defined in the initial difference maps in the antibody combining regions of both structures. In the P21 form, an initial concern was the possibility that the peptide structure might be affected by crystal contacts, because a symmetry-mate crowds the peptide-binding region (Fig. S2A). However, in the C2 form, the peptide-binding region is exposed with no neighboring contacts, and the peptide structure is very similar to that in the P21 form, with an rmsd of 0.38 Å for all peptide backbone atoms (excluding the added terminal arginine) after aligning all peptide backbone atoms in common (Fig. S2B). The engineered arginine is ordered only in the P21 form because of crystal contacts. Therefore, the peptide–Fab interaction presented in these structures likely reflects how the antibody recognizes the peptide in solution and the epitope in the intact E2 protein.
The peptide forms an extensive β-hairpin loop with a type I’ β-turn at residues 416–419 (TNGS). The hairpin is straddled between the mAb CDR2 and CDR3 loops of the heavy chain and the CDR3 loop of the light chain (Fig. 2A). Although the antiparallel β-hairpin of the peptide is mostly in contact with VH, the β-turn region interacts with VL. The low crystallographic B-values of the side chains on the peptide facing the antibody (36.0 Å2 in the C2 structure and 19.1 Å2 in the P21 structure) compared with those facing toward solvent (55.5 Å2 in the C2 structure and 32.8 Å2 in the P21 structure), and the correspondingly low B-values of Fab residues contacting the peptide compared with surrounding residues, indicate a highly ordered and stable interaction (Fig. 2B). The interaction is primarily hydrophobic, as shown by the electrostatic surface potential map of the epitope and the paratope (Fig. 2C), as well as at least 88 van der Waals interactions (Table S2). As expected, the side chains of residues on the peptide facing away from the interaction site are more flexible, as observed when the peptides from each structure are superimposed on each other (Fig. 2D). The Fab-facing residues of the peptide (Leu413 in the first β-strand, and Trp420 and Ile422 in the second β-strand) are predominantly hydrophobic and fit into a deep hydrophobic depression (Table S2). This depression is formed by aliphatic and aromatic residues from the base of CDR H2 and CDR H3, VH framework regions (FRs) 2 and 3, CDR L3, and VL FR3 (Fig. 3A). Hydrogen bonds also stabilize the interaction: one at the tip of the turn and two on the edges of the sheet for the P21 structure (Fig. 3B), with three more hydrogen bonds to the edge in the C2 structure (Table S3). Altogether, the interactions are mainly focused on four residues of the peptide: Leu413, Asn415, and Trp420 on the β-strands and Gly418 at the tip of the β-turn.

Structure of mAb HCV1 in complex with its HCV E2 peptide epitope. Two X-ray structures of a broadly neutralizing antibody in complex with its epitope from two crystals forms, P21 and C2, were solved and refined to1.8-Å resolution. (A) The structure from the P21 crystal is shown in cartoon representation. The peptide epitope (red) is inserted between the heavy chain (blue) CDR2 and CDR3 loops and makes contact with the light chain (green) CDR3 loop. Arg1 in the peptide, which was introduced to enhance the solubility of the peptide and is therefore not part of the HCV antigenic region, is colored gray. (B) B-values of the crystal structure were mapped onto the molecular surface of the paratope (Left) and a stick representation of the peptide (Right) by temperature gradient coloring from 9.8 Å2 (deep blue) to 96.5 Å2 (red). The paratope is shown in gray to highlight the peptide at Right. (C) The adaptive Poisson-Boltzmann solver was used to calculate the surface potential across the solvent-accessible surfaces of both the paratope and the peptide [−3 kT/e (red) to 3 kT/e (blue)]. For the peptide, the surface potential is shown looking from above the antibody (Upper) and from the paratope toward the binding surface, which is a 180° rotation (Lower). The engineered arginine was included for the calculation and is shown here boxed in gray. (D) A comparison of the peptides between the two crystal structures. The peptides are superimposed on each other, with the P21 structure in red and the C2 structure in blue. The two strands of the peptide are separated and reoriented to better visualize the differences in side chains between the structures. The dots indicate where the two strands meet at the turn. Overall, the side chains pointing away from the antibody have greater differences between the structures than those pointing into the binding site. The main chains mostly overlap each other. (E) The peptide forms a β-hairpin, and the backbone hydrogen bonding that stabilizes this structure is indicated.

Molecular details of antibody binding to the HCV peptide. (A) The Leu413and Trp420 residues on the peptide (red) are shown buried in a hydrophobic depression formed by LC CDR3 residues (green) and by heavy chain FR2, CDR2, and CDR3 residues (blue). (B) Three hydrogen bonds between peptide and antibody also stabilize the interaction, as depicted in wall-eye stereo. Bonds and distances are labeled in black. (C) To further analyze the peptide binding, HCV sequence conservation across 2,161 isolates for this region, crystallographic B-value, rmsd between the two structures, and surface burial by antibody on the peptide are shown. Sequence conservation was taken from the ViPR database (Table S4), whereas the B-values were extracted from the structure and the binding data from ref. 15. The rmsd was calculated on a residue-by-residue basis in PyMOL. Surface burial corresponds to the accessible surface area of each residue on the peptide in the bound structure (P21 structure) normalized by the surface area calculated after the Fab is removed.
The peptide hairpin loop itself is stabilized by a number of backbone hydrogen bonds (Fig. 2E), and this structural motif is in good agreement with previously published secondary structure predictions based on the amino acid sequence (24, 25). The two asparagine residues on the peptide facing away from the Fab (Asn417 and Asn423) are parts of N-linked glycosylation sequons that are likely glycosylated in the native protein (26) (Fig. S3). Another interesting feature of the interaction is that the paratope contains residues near the base of the CDR loops and FR2, which are generally more conserved than those at the tips of the CDR loops (Fig. 3 A and B). Some of these residues are highly conserved across human antibodies for structural reasons, such as Trp47 on VH FR2, which is 97.5% conserved, and Tyr58 on CDR H2, which is 45.6% conserved, across 16,946 distinct antibodies (Abysis Database: www.bioinf.org.uk/abysis) (27) (Fig. S4). Although other contacting residues, such as VH Val50, Phe100C and Ile100D, and VL Asn93 are less than 15% conserved, more than 50% of their alternative substitutions have similar chemical properties (hydrophobicity or polarity) (Fig. S4).
Structural Explanation for Broad Neutralization of HCV by mAb HCV1 and Virus Escape.
The basis for the broad neutralizing activity of mAb HCV1 is the high sequence conservation of the target epitope (Fig. 3C and Table S4) and the apparent solvent accessibility of the binding face of the peptide in the intact E2 protein. The peptide residues facing the antibody are highly conserved (Leu413, Asn415, Trp420, and Ile422 in 99.9%, 97.2%, 99.9%, and 96.9%, respectively, of 2,161 HCV E2 sequences in the Virus Pathogen Resource Database, www.viprbrc.org), whereas some of the residues facing away from the binding site are slightly more variable (Gln412, Ile414, and Thr416 in 93.5%, 67.6%, and 85.1% of E2 sequences, respectively). Several residues on the peptide are likely conserved because of structural constraints. For example, residues in the turn are conserved in line with β-turn propensities (28). Another structurally important residue is Asn415 because it makes a hydrogen bond to a backbone amide of Gly418, a residue at the tip of the turn (Fig. S5). Incidentally, this is the only residue facing the hydrophobic depression of the Fab that is not hydrophobic.
Previous alanine scanning mapping experiments using bacterially expressed fusion proteins containing the epitope showed that Leu413 and Trp420 are critical for the binding of mAb HCV1 (15). These results agree well with the crystal structures that show these two residues to be the most stabilized and buried within the binding site (Fig. 3C). However, the same study also showed that alanine substitution of Asn415 or Gly418 did not affect mAb binding. This result is surprising because the crystal structure shows that Asn415 hydrogen bonds to the tip of the β-turn, and Gly418 is a highly preferred amino acid at this position for this type of β-turn. Therefore, we repeated the analysis using alanine-scanning mutants of E1E2 (Fig. 4A). The results demonstrated that replacement of Asn415 or Gly418 by alanine abolished antibody–antigen interaction, suggesting that proper folding of the tip of the β-turn, in addition to the hydrophobic residues Leu413 and Trp420, is important for the antibody recognition of this epitope. Interestingly, alanine substitution of E2 Gln412, Ile414, Asn417, Ser419, and Ile422 reduced binding of the mAb to varying extents, indicating that these substitutions can perturb the optimal conformation of the epitope for antibody binding.
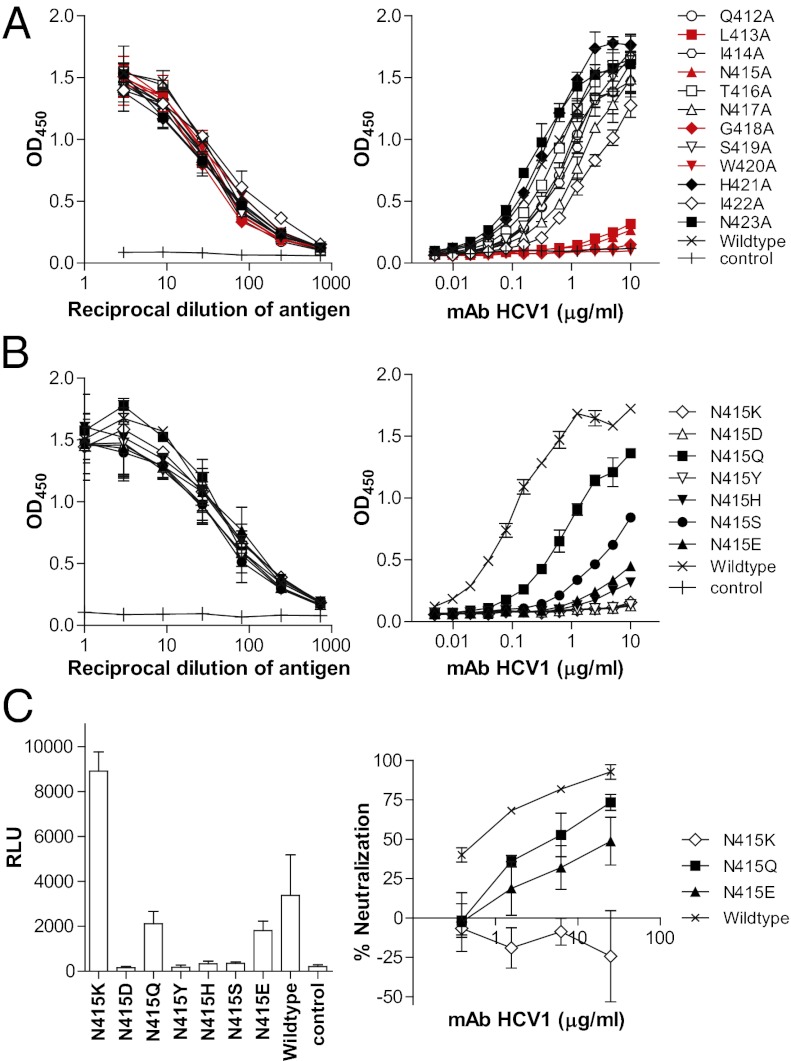
Viral escape through substitutions at E2 residue 415. (A) Alanine scanning mapping of the HCV1 epitope. (B) Binding of mAb HCV1 to E1E2 variants with substitutions at position 415, including naturally occurring substitutions and Gln. (A and B) Left: Expression of the variants confirmed by mAb AR2A (1 μg/mL) (19). Right: Binding of mAb HCV to the variants. (C) Escape of mAb HCV1 by substitution at E2 Asn415. HCVpp bearing the specific substitutions were generated as described previously (19). The infectivity of the variant panel was compared according to the activity of the reporter gene luciferase (relative light unit, RLU) (Left). Only the K, Q, and E variants produced HCVpp with significantly higher signals (>10-fold) than the control pseudotype virus generated without E1E2. The sensitivity of these variants to mAb HCV1 was determined by incubating them with serially diluted mAb (Right). The results shown are the means ± SD of two independent experiments of triplicate measurement.
At a low incidence, some alternative substitutions are found in natural viruses for the antibody facing residues, and these may represent virus variants that could escape neutralization by this particular antibody (Table S4). We were particularly interested in substitutions of Asn415 because this residue may be involved in stabilizing the β-turn and binds within a less tightly packed region of the hydrophobic depression of the mAb. In steric clash analysis of different rotamers modeled into position 415, the hydrophobic depression will not accommodate residues with bulky side chains or side chains with carbons beyond Cγ (e.g., lysine, histidine, tyrosine, glutamate, and glutamine) without significant movement or conformational change in the peptide and/or the antibody paratope (Fig. S5). To evaluate whether this analysis successfully predicted virus escape from the mAb, Asn415 was replaced with these low-frequency variants found in nature or by a glutamine, which is similar in polarity but has a side chain that extends beyond the Cγ position. Although all of the seven mutated E1E2 antigens expressed at a similar level in transient transfection experiments, they all bound mAb HCV1 at a lower level compared with the wild-type E1E2 (Fig. 4B). The loss of antibody binding to the bulky lysine, glutamine, histidine, and tyrosine mutants is consistent with the clash analysis results (Fig. S5). The analysis also predicted that the aspartate and serine mutants could still fit the hydrophobic cavity, but antibody binding was abolished for the aspartate mutant, which would introduce a buried negative charge, and reduced significantly for the serine mutant, which would no longer stabilize the β-turn with a hydrogen bond to the backbone amide of the glycine at the tip. Interestingly, the glutamine mutant bound the antibody at a respectable level despite predicted steric clashes. The results suggest that some minor conformational rearrangements must take place in the binding site to accommodate the larger glutamine residue.
In terms of biological function, substitution of Asn415 with aspartate, histidine, tyrosine, or serine abolished most virus infectivity in the pseudotype virus system (Fig. 4C). The glutamine and glutamate substitutions did not have a significant effect on virus infectivity, whereas the lysine substitution increased virus infectivity by two- to fourfold. These results are surprising because one would expect that viral quasispecies harboring these mutations would be less fit because they are rarely observed in nature (combined <1.5% of 2,161 E2 sequences in the National Institute of Allergy and Infectious Diseases Virus Pathogen Database and Analysis Resource (ViPR) database; Table S4). In neutralization experiments, the N415K substitution enabled the virus to escape mAb HCV1 entirely, whereas the N415Q and N415E substitutions increased virus resistance to the mAb (Fig. 4C). Interestingly, the N415Q substitution is not found in the HCV sequence database, although the mutagenesis data suggest that the virus could accommodate this mutation. Altogether, these results suggest that the lysine, glutamine, and glutamate substitutions may replace asparagine in stabilizing the β-hairpin fold in E2, and to different extents facilitate virus escape of the mAb.
Discussion
A major challenge in the design of an HCV vaccine is to maximize the cross-reactive immune response to protect against the huge diversity of HCV isolates found in nature (>30% genetic difference). In vaccine research, both cellular (29, 30) and humoral immunity (31, 32) have been shown to protect in the chimpanzee model from challenge with homologous or closely related HCV. Choo and colleagues (31, 33) demonstrated the possibility of eliciting sterilizing immunity against low-dose homologous virus challenge by vaccination of chimpanzees with recombinant E1E2 proteins, and protection was associated with high antibody titers to E2 and the ability of the antibodies to inhibit E2 binding to cells expressing HCV receptors. However, when the vaccinated chimpanzees were challenged with a heterologous HCV isolate, none of the animals were protected from acute infection, although the majority (90% vs. 40% in the control group) resolved acute infection and did not progress to the carrier state (9).These experiments support the notion that vaccination with E1E2 proteins may protect infected humans from progressing to chronic infection, which is a major risk factor for cirrhosis and hepatocellular carcinoma. However, the lower level of protection against heterologous challenge is consistent with observations that antibody responses to E1E2 immunization are biased to isolate-specific epitopes and not effective against divergent HCV genotypes. This conclusion is supported by the observations that both passive antibodies against the hypervariable 1 region (HVR1) of E2 (32) and immunization with a mixture of recombinant E1, E2 proteins and multiple HVR1 peptides (34) were unable to protect against heterologous HCV challenge in chimpanzees. These early studies highlight that a broadly effective HCV vaccine must target conserved immune epitopes.
In this study, we determined crystal structures of a B-cell epitope located within the conserved antigenic site 412–423 of the E2 envelope glycoprotein that is also involved in binding to the coreceptor CD81. The mAb HCV1 is one of the antibodies known to recognize this antigenic site and is currently under development to treat recurrent HCV in liver transplant patients (15, 23). The mAb has broad activity against diverse HCV isolates and protected in the chimpanzee model from challenge by an HCV-infected human serum inoculum (15, 23). This antigenic site is poorly immunogenic on virus: only ~2% of chronic HCV patients seem to generate antibodies to this site (21, 22). In patients who make such antibodies, the quantities do not seem to be sufficient to mediate virus neutralization or select for escape virus (22). The structural studies here reveal that the conserved E2 antigenic site 412–423 forms an extended β-hairpin structure, and mAb HCV1 neutralizes HCV by contacting residues on the hydrophobic face of the β-hairpin that are nearly 100% conserved (Fig. 3 and Table S4). Such high conservation of the contacting residues on the virus likely explains the broad activity of this mAb. The antibody–epitope interaction is dependent on inserting residues from the hydrophobic face of the epitope into a hydrophobic depression on the antibody combining site. Replacement with bulky, polar, and charged residues can weaken or abolish the hydrophobic interactions between the antibody and antigen by steric clash or charge burial, thus resulting in virus escape. However, these hydrophobic residues are hardly ever substituted in natural viruses, indicating that these residues are required for the function of the virus. However, the polar Asn415, which also points toward the hydrophobic depression but is not as intimately involved as Leu413 and Trp420, seems to be required for formation of the β-hairpin structure. Consequently, most mutations at Asn415 likely destabilize the β-hairpin, thus perturbing E2 function, but a limited number of residues (lysine, glutamine, or glutamate) can still generate equally or more infectious virus in vitro (35) (Fig. 4C). Interestingly, these mutations are rarely observed in circulating HCV, suggesting that they do not improve viral fitness when other host factors (e.g., adaptive immunity) are present. This suggestion is also supported by another study in which HCV carrying the mutation N415D, T416A, N417S, or I422L was more sensitive to antibodies targeting other neutralizing epitopes (36). Nevertheless, the possibility of virus escape through mutations in this antigenic site should be considered in the design and evaluation of immunogens.
The structure of the HCV peptide that we derived from its complex with a broadly neutralizing antibody has some interesting implications on how the E2 protein is folded in that region. The side of the β-hairpin that binds to the antibody is highly hydrophobic and, therefore, one would predict and expect that it would most likely face the protein core. In fact, when a full-length primary sequence of the E2 protein was submitted to ab initio folding on the Robetta server, only structures where that face of the peptide was covered by protein were predicted (37). However, from the crystal structure, it is clearly accessible to the antibody and, therefore, should be solvent exposed in the native protein on the virus. On the other hand, the nonbinding face of the β-hairpin should also be exposed to accommodate the two putative N-linked glycans in that region (Fig. S3).
Taken together, the results impose several constraints on the structure of this antigenic site in the native context: (i) it is a β-hairpin, and disruption of the hairpin reduces viral fitness; (ii) it is solvent exposed on both sides, despite its hydrophobic character, but perhaps masked to some extent by the hydrophilic N-linked glycans at Asn417and Asn423 (26); and (iii) it extends away from the rest of the protein because otherwise there would be steric clashes with the antibody to the rest of the protein. This antigenic site has previously been predicted to fold into antiparallel β-strands (24, 25), and Krey et al. (24) proposed that it forms part of “domain I” in class II viral fusion proteins. Within domain I, a succession of β-hairpins forms a β-sandwich, in which the nonpolar sides of the β-hairpins face toward mainly the hydrophobic core of the sandwich, and the opposite sides are exposed to solvent. The crystal structures here show that the hydrophobic side of this hairpin is accessible to the antibody, whereas the N-linked glycan(s) on the opposite side will likely prevent it from participating in the proposed β-sandwich core (displayed in Fig. S3). Thus, our data suggest that this particular hairpin folds into a flap-like structure on E2 and is not part of domain I as predicted previously (24).
In conclusion, the structures of the mAb HCV1 epitope presented here provide a structural basis for how this mAb neutralizes diverse HCV strains. The structures also suggest a limited number of virus mutations that may escape this mAb, but fortunately have not yet been found to any extent in known HCV viruses. Vaccination is the most cost-effective solution to many infectious diseases, but there is no prophylactic or therapeutic vaccine available for HCV. The structure of this conserved HCV-neutralizing epitope can be used as a template for the design of biomimetic antigens to direct the immune system to elicit similar cross-NAbs in vaccination (38). Clearly, the long-term goal is to determine the crystal structures of E2 and E1E2 in complex with other broadly neutralizing mAbs (19, 39) to reveal additional conserved B-cell epitopes, as has been achieved so effectively for HIV-1 and influenza virus (40–44). This will greatly aid the rational design of a broadly effective vaccine to eradicate hepatitis C.
Materials and Methods
Expression of Recombinant mAb HCV1.
The variable domains of heavy and light chains of mAb HCV1 (15) were cloned into the pIgG1 vector (19) for expression as a full-length human IgG1 in stably transfected CHO-K1 cells. Fab fragments for crystallization were prepared by deleting the hinge, CH2, and CH3 sequences of the mAb expression vector and transient transfection of FreeStyle 293-F cells. The purified Fab was concentrated to 10 mg/mL in 20 mM Tris·HCl and 140 mM NaCl buffer (pH 7) and allowed to form a complex with the R12-mer peptide at a 1:10 (protein:peptide) molar ratio overnight at 4 °C.
Protein Crystallization and Structure Determination.
The antibody–antigen complex was screened for crystallization using the International AIDS Vaccine Initiative (IAVI)-Joint Center for Structural Genomics (JCSG)-The Scripps Research Institute (TSRI) CrystalMation robot (Rigaku) (45). Multiple crystals were obtained, and datasets were collected from two crystals at the Advanced Photon Source (APS) beamline 23ID-D. The C2 form crystal was grown in 25% (wt/vol) PEG 4000, 0.2 M ammonium sulfate, and 0.1 M sodium acetate (pH 4.6), and the P21 form crystal in 40 mM potassium dihydrogen phosphate, 20% (vol/vol) glycerol, and 16% (wt/vol) PEG 8000. Both crystals diffracted to 1.8-Å resolution. The structures were determined by the molecular replacement method using an unrelated anti-CD20 Fab structure (Protein Data Bank ID code 3GIZ). All structural visualizations were generated with PyMOL (PyMOL Molecular Graphics System, version 1.2r3pre; Schrödinger, LLC). Electron density for the peptide epitope was clear in both crystal forms (Fig. S6).
Detailed methods and the associated references can be found in SI Materials and Methods.
Acknowledgments
We thank MassBiologics for discovering mAb HCV1 for the HCV field, Jonathan Ball and François-Loïc Cosset for generous sharing of reagents, Travis Nieusma for excellent technical support with antibody production, Henry Tien for setting up crystal screens on the CrystalMation robot, Tinashe Ruwona for comments on the manuscript, and Alex Tarr for valuable discussions on the immunogenicity of the E2 antigenic site. We acknowledge the ViPR database which is funded by the National Institutes of Health (NIH) Contract HHSN272200900041C. This work is supported by NIH Grants AI80916 and AI79031 (to M.L.), AI71084 (to D.R.B), and AI84817 (to I.A.W.), and by The Skaggs Institute (I.A.W.). The GM/CA CAT 23-ID-B beamline has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Sciences (NIGMS) (Y1-GM-1104). Use of the Advanced Photon Source (APS) was supported by the US Department of Energy, Basic Energy Sciences, and Office of Science under contract no. DE-AC02-06CH11357. This is The Scripps Research Institute manuscript number 21617.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PBD ID codes 4DGY and 4DGV).
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1202924109/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1202924109
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/109/24/9499.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633(8030):704-709, 04 Sep 2024
Cited by: 0 articles | PMID: 39232163
Hepatitis C Virus E1E2 Structure, Diversity, and Implications for Vaccine Development.
Viruses, 16(5):803, 18 May 2024
Cited by: 0 articles | PMID: 38793684 | PMCID: PMC11125608
Review Free full text in Europe PMC
HCV E1 influences the fitness landscape of E2 and may enhance escape from E2-specific antibodies.
Virus Evol, 9(2):vead068, 18 Nov 2023
Cited by: 1 article | PMID: 38107333 | PMCID: PMC10722114
Prospects for developing an Hepatitis C virus E1E2-based nanoparticle vaccine.
Rev Med Virol, 33(5):e2474, 11 Aug 2023
Cited by: 3 articles | PMID: 37565536 | PMCID: PMC10626635
Review Free full text in Europe PMC
Current Hepatitis C Vaccine Candidates Based on the Induction of Neutralizing Antibodies.
Viruses, 15(5):1151, 11 May 2023
Cited by: 2 articles | PMID: 37243237 | PMCID: PMC10220683
Review Free full text in Europe PMC
Go to all (106) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Clinical Trials
- (1 citation) ClinicalTrials.gov - NCT01121185
Protein structures in PDBe (3)
-
(1 citation)
PDBe - 4DGVView structure
-
(1 citation)
PDBe - 4DGYView structure
-
(1 citation)
PDBe - 3GIZView structure
Protocols & materials
Related Immune Epitope Information - Immune Epitope Database and Analysis Resource
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody.
J Biol Chem, 290(16):10117-10125, 03 Mar 2015
Cited by: 50 articles | PMID: 25737449 | PMCID: PMC4400327
Structure-Based Design of Hepatitis C Virus Vaccines That Elicit Neutralizing Antibody Responses to a Conserved Epitope.
J Virol, 91(20):e01032-17, 27 Sep 2017
Cited by: 45 articles | PMID: 28794021 | PMCID: PMC5625506
Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes.
J Virol, 92(9):e02066-17, 13 Apr 2018
Cited by: 17 articles | PMID: 29467319 | PMCID: PMC5899191
HCV Glycoprotein Structure and Implications for B-Cell Vaccine Development.
Int J Mol Sci, 21(18):E6781, 16 Sep 2020
Cited by: 7 articles | PMID: 32947858 | PMCID: PMC7555785
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: Y1-CO-1020
NIAID NIH HHS (10)
Grant ID: AI80916
Grant ID: R01 AI071084
Grant ID: R21 AI080916
Grant ID: AI79031
Grant ID: R56 AI084817
Grant ID: AI71084
Grant ID: R01 AI079031
Grant ID: AI84817
Grant ID: R01 AI084817
Grant ID: HHSN272200900041C
NIGMS NIH HHS (1)
Grant ID: Y1-GM-1104





