Abstract
Free full text

DNA Microarray Profiling of a Diverse Collection of Nosocomial Methicillin-Resistant Staphylococcus aureus Isolates Assigns the Majority to the Correct Sequence Type and Staphylococcal Cassette Chromosome mec (SCCmec) Type and Results in the Subsequent Identification and Characterization of Novel SCCmec-SCCM1 Composite Islands
Associated Data
Abstract
One hundred seventy-five isolates representative of methicillin-resistant Staphylococcus aureus (MRSA) clones that predominated in Irish hospitals between 1971 and 2004 and that previously underwent multilocus sequence typing (MLST) and staphylococcal cassette chromosome mec (SCCmec) typing were characterized by spa typing (175 isolates) and DNA microarray profiling (107 isolates). The isolates belonged to 26 sequence type (ST)-SCCmec types and subtypes and 35 spa types. The array assigned all isolates to the correct MLST clonal complex (CC), and 94% (100/107) were assigned an ST, with 98% (98/100) correlating with MLST. The array assigned all isolates to the correct SCCmec type, but subtyping of only some SCCmec elements was possible. Additional SCCmec/SCC genes or DNA sequence variation not detected by SCCmec typing was detected by array profiling, including the SCC-fusidic acid resistance determinant Q6GD50/fusC. Novel SCCmec/SCC composite islands (CIs) were detected among CC8 isolates and comprised SCCmec IIA-IIE, IVE, IVF, or IVg and a ccrAB4-SCC element with 99% DNA sequence identity to SCCM1 from ST8/t024-MRSA, SCCmec VIII, and SCC-CI in Staphylococcus epidermidis. The array showed that the majority of isolates harbored one or more superantigen (94%; 100/107) and immune evasion cluster (91%; 97/107) genes. Apart from fusidic acid and trimethoprim resistance, the correlation between isolate antimicrobial resistance phenotype and the presence of specific resistance genes was ≥97%. Array profiling allowed high-throughput, accurate assignment of MRSA to CCs/STs and SCCmec types and provided further evidence of the diversity of SCCmec/SCC. In most cases, array profiling can accurately predict the resistance phenotype of an isolate.
INTRODUCTION
Staphylococcus aureus is a versatile pathogen responsible for a wide range of infections, reflecting its ability to express an extensive array of virulence factors and antimicrobial resistance genes. Methicillin-resistant S. aureus (MRSA) was first reported in 1961 in the United Kingdom, and subsequently a variety of different MRSA clones that exhibited resistance to numerous antimicrobial agents emerged (6, 22). Different epidemic and pandemic clones have spread worldwide since the 1970s (6). Today, MRSA strains are a major nosocomial problem worldwide and have also emerged as a significant cause of infections among otherwise healthy individuals in the community and among animals (10, 55).
While many virulence-associated genes in S. aureus are part of the core genome (such as those encoding microbial surface components recognizing adhesive matrix molecules [MSCRAMMs] and the polysaccharide intercellular adhesion [ica] locus), many are encoded on mobile genetic elements (MGEs) that can be horizontally transferred between S. aureus isolates and from coagulase-negative staphylococci (CoNS) (29). Many different types of MGEs encoding virulence genes have been identified in S. aureus, including bacteriophages, pathogenicity islands, plasmids, staphylococcal cassette chromosome (SCC) and SCC-like elements, and genomic islands (3, 32).
Antimicrobial resistance in S. aureus can be due to spontaneous chromosomal mutations, but resistance is more commonly due to the acquisition of MGEs, including plasmids, transposons, and SCC elements harboring antimicrobial resistance genes (32). SCC elements include SCCmec, which harbors the methicillin resistance gene mecA. Two divergent mecA alleles have been described in MRSA to date (20, 46). A search of GenBank revealed an additional 32 variants that differ at the nucleotide sequence level and 28 protein variants. Eleven different SCCmec types, as well as numerous subtypes, have been identified in MRSA, and each type has a unique combination of cassette chromosome recombinase (ccr) genes which are involved in the integration and excision of the element, as well as mec complex genes, including mecA and, when present, the mec regulatory genes mecI and mecR1 (15, 20, 28, 46). Each different SCCmec type has been assigned a Roman-numeral name in the order in which it was identified (20). SCCmec subtypes differ from SCCmec types mainly in the joining or “J” regions, i.e., the regions outside the ccr and mec gene complexes (20). Subtypes are designated by the use of either lower- or uppercase alphabetic suffixes after the relevant Roman-numeral SCCmec-type name (20). Lowercase alphabetic suffixes (e.g., IVa, IVb, or IVc) are used to indicate DNA sequence variation in the J regions of SCCmec subtypes, while uppercase alphabetic suffixes (e.g., IVA, IIA, IA) indicate variation due to the presence or absence of mobile genetic elements (20). SCC elements without mecA but with ccr genes and sometimes with additional antimicrobial resistance or virulence-associated determinants, as well as composite islands (CIs) consisting of a combination of two or more SCC, SCC-like, or SCCmec elements, have also been described in S. aureus and CoNS (2, 11, 18, 20, 21, 25, 30, 37, 47). Evidence suggests that CoNS are reservoirs for SCCmec in S. aureus. Due to the diversity and frequency of identification of novel SCC and SCCmec types and subtypes, some reported novel elements do not follow any standardized or internationally agreed rules of nomenclature. While it is now recommended that nomenclature of novel SCCmec types and SCC types and subtypes should be undertaken in conjunction with the International Working Group on the Classification of SCC elements, this does not always happen, and there are ambiguities in the classification of SCC and SCCmec elements in the published literature. A binary system is currently in development to simplify this confusing SCCmec subtype nomenclature (20).
Effective and informative molecular typing plays an important role in monitoring the emergence, spread, and evolution of different MRSA clones. Multilocus sequence typing (MLST) is commonly used for MRSA typing, and while it has provided invaluable insights into its evolution, it lacks the discriminatory ability required for local or outbreak investigations and is relatively expensive and labor-intensive, as it involves amplification and sequencing of internal fragments of seven housekeeping genes (40). In contrast, spa typing involves DNA sequencing of a variable number tandem repeat (VNTR) region of just one gene (the S. aureus protein A spa gene), provides greater discrimination than MLST, and is widely used internationally for local and global epidemiological studies (16). Accurate identification of the type and subtype of the SCCmec element harbored by a MRSA isolate enhances understanding of the genetic relatedness of MRSA isolates. SCCmec typing commonly involves the use of several multiplex PCR assays to identify the ccr and mec complex genes which are used to define the SCCmec type and to identify the main characteristics of the J regions for SCCmec subtype determination and which can harbor integrated transposons and/or plasmids with additional antimicrobial resistance determinants. In the case of novel SCCmec elements, complete nucleotide sequencing is commonly carried out. The combination of MLST and SCCmec typing data is used to describe MRSA clones (14), and for the purpose of the present study, clones are defined as MRSA isolates with the same sequence type (ST) and the same or very closely related SCCmec type. Strains are defined as isolates not just with the same ST and SCCmec type but also with additional shared traits, such as specific combinations of virulence or antimicrobial resistance genes.
In recent years, major advances in high-throughput whole-genome sequencing technologies have resulted in the development of more in-depth typing methods, including single-nucleotide polymorphism (SNP) analysis of whole-genome sequences (17) and different microarray platforms (29). A recent study demonstrated the potential of SNP analysis of whole-genome sequences for investigating evolutionary relatedness and for epidemiological tracking of MRSA (17). However, this approach is currently expensive and time-consuming, and data analysis is too complicated for routine typing. DNA microarray systems have been developed based on the whole-genome sequences that are available for S. aureus and allow simultaneous hybridization of an isolate's genome against the entire gene content of multiple S. aureus genomes. While these DNA microarray systems provide a large amount of information about the isolate's genome, their use is restricted mainly to specialized research laboratories, as data analysis is complicated (29, 50, 52, 56). In contrast, several groups have recently developed more focused DNA microarrays (12, 35, 36, 43), with one system in particular offering simultaneous high-throughput genotyping of S. aureus isolates by assigning isolates to an MLST clonal complex (CC) or ST and to an SCCmec type and detection of species-specific markers, accessory gene regulator (agr) alleles, capsule types, MSCRAMMs, and a range of clinically relevant antimicrobial resistance and virulence-associated genes (34–36).
Like in many other countries, MRSA has been a major problem in Irish hospitals since the 1970s (4, 7, 19, 45). Previous studies of nosocomial MRSA isolates recovered in Irish hospitals over a 33-year period using MLST and detailed SCCmec typing revealed that the predominant MRSA clone was replaced after a certain period of years (45). Dominant clones identified included ST250-MRSA-I/I-pls in the 1970s and the early 1980s, the ST239-MRSA-IIIHg/IIIHg-p1258/Tn554 clone in the 1980s, and the ST8-MRSA-IIA-IIE, ST36-MRSA-II, and ST22-MRSA-IV clones in the 1990s and early 2000s. The ST22-MRSA-IV clone has continued to predominate in Irish hospitals to the present day (45).
The purpose of the current study was to perform an in-depth molecular characterization of the predominant MRSA clones recovered from patients in Irish hospitals between 1971 and 2004 using spa typing and a DNA microarray system. The DNA microarray was investigated to (i) determine its value for genotyping MRSA isolates representative of major pandemic clones that have been previously investigated by MLST and detailed SCCmec typing, (ii) identify the range of virulence and antimicrobial resistance genes harbored by these MRSA clones, and (iii) study the correlation between antimicrobial resistance phenotype and the presence of antimicrobial resistance genes.
MATERIALS AND METHODS
MRSA isolates.
A total of 175 MRSA isolates, representative of the most prevalent antibiogram-resistogram (AR) and pulsed-field gel electrophoresis (PFGE) types of MRSA isolates recovered among patients in Irish hospitals at different time periods between 1971 and 2004, were investigated (45).
Molecular typing.
SCCmec typing of all isolates was undertaken previously using individual PCR assays to detect mec complexes A and B and ccr complexes 1, 2, and 3 and analysis of the joining (J) regions by a previously described multiplex PCR method (38, 45). For all ST8 isolates previously identified as harboring novel SCCmec II and IV variant elements, in-depth molecular characterization of the SCCmec elements was performed using long-range PCR amplification and sequencing (45). As part of the present study, all isolates harboring SCCmec IV were subtyped using a multiplex PCR method (33) which detects the SCCmec IV subtypes IVA, IVa, IVb, IVc, IVd, IVg, and IVh. Previously described S. aureus reference strains were used as positive controls for SCCmec IV subtyping (48). One hundred thirty of the 175 isolates were previously typed by multilocus sequence typing (MLST) (45). MLST was performed as described previously on the remaining 45 isolates as part of the present study (13, 49). All 175 isolates underwent spa typing as described previously (49).
DNA microarray analysis.
One hundred seven of the 175 MRSA isolates investigated underwent DNA microarray analysis. These included (where possible) three isolates representative of each AR type, sequence type (ST), and SCCmec type/subtype combination previously identified among the 175 isolates during different time periods between 1971 and 2004 (45). The StaphyType kit (Alere Technologies, Jena, Germany) was used for DNA microarray analysis according to the manufacturer's instructions, which have been described in detail elsewhere (35, 36). The StaphyType kit consists of a DNA microarray chip adhered to each well of a microtiter strip; each chip consists of 334 S. aureus target sequences, including species-specific, antimicrobial resistance, and virulence-associated genes, genes involved in attachment, adhesion, and biofilm formation, as well as markers used for typing. Data generated by StaphyType arrays were analyzed for the presence or absence of these genes using Arraymate software (Alere Technologies), which can assign S. aureus isolates to STs and/or clonal complexes (CCs) by comparing each isolate's DNA microarray results to those of a diverse reference collection of previously characterized strains in the Arraymate database (36).
PCR to confirm the absence of ccrAB2 genes.
For five isolates that were previously identified as harboring ccrAB2 and a class B mec complex by SCCmec typing PCR, the ccrAB2 genes were not detected using the DNA microarray. PCR detection of ccrAB2 in these isolates was repeated using a previously described multiplex ccr PCR assay (26) and previously described reference strains (49).
PCR amplification of the S. epidermidis ccrAB4 gene.
The presence of S. epidermidis ccrAB4 was investigated by PCR in all isolates that yielded positive or ambiguous signals for ccrA4 and/or ccrB4 genes using the DNA microarray (which has primers and probes specific for S. aureus ccrA4 and ccrB4) using previously described conditions and primers specific for the S. epidermidis ccrAB4 genes (49). Since the majority of isolates that were positive for these ccrAB4 genes exhibited the ST8-MRSA-IIA-IIE or IVE/IVF and spa type t190 genotype, all isolates belonging to these genotypes that underwent spa typing but not DNA microarray analysis were also investigated by PCR for the presence of the S. epidermidis ccrAB4 genes.
Whole-genome sequencing.
The whole genome of one MRSA isolate (AR13.1/3330.2) representative of the most frequently occurring ST-SCCmec type identified as harboring the S. epidermidis ccrAB4 genes was sequenced to further investigate the location and adjacent sequences of these genes. High-throughput de novo sequencing was undertaken commercially by Geneservice (Source BioScience plc, Nottingham, United Kingdom) using the Illumina Genome Analyzer system (Illumina HiSeq 2000 platform; Illumina, Essex, United Kingdom). The average coverage across the genome was 40×. The reads were assembled into contigs using a Velvet de novo genome assembler (version 1.0.15; Illumina). Contigs were analyzed using the Artemis DNA sequence viewer and annotation tool (42) and BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (1). Contigs identified as containing SCC-associated DNA sequences were aligned using the BioNumerics (version 5.1) (Applied Maths, Ghent, Belgium) and DNA Strider (version 1.3f11) (CEA Saclay, Gif-sur-Yvette, France) software packages. Any gaps identified between the SCC-related contigs in the whole-genome sequence of AR13.1/3330.2 were closed by primer walking using PCR with primers based on the surrounding contigs and the Expand long-template PCR system (Roche Diagnostics Ltd., West Sussex, United Kingdom) followed by amplimer sequencing by Geneservice (SourceBioscience, Guinness Enterprise Centre, Dublin). Data were analyzed, and overlapping sequences were assembled using the BLAST, Bionumerics, and DNA Strider software packages.
Confirmation of the genetic organization and location of ccrAB4.
Having determined the location and genetic organization of the ccrAB4 region of an SCC element adjacent to SCCmec in AR13.1/3330.2 using whole-genome sequencing, the genetic organization of this element was confirmed by PCR and amplimer sequencing using four overlapping primer pairs to amplify from the SCCmec element to the end of the ccrAB4-carrying element of AR13.1/3330.2. The primers used are listed in Table S1 in the supplemental material. These PCR assays were performed using chromosomal template DNA with GoTaq DNA polymerase (Promega Corporation, Madison, WI). PCR products were visualized by agarose gel electrophoresis, and the sizes of amplicons obtained were compared to the expected sizes of amplicons based on the whole-genome sequence (see Table S1).
The presence of the ccrAB4-carrying SCC element and its location adjacent to SCCmec were investigated in one isolate representative of each ST and SCCmec type combination identified as harboring the S. epidermidis ccrAB4 genes, including ST8-MRSA-IIA (AR13/0132), ST8-MRSA-IIB (AR05/1345), ST8-MRSA-IIC (AR14/0246), ST8-MRSA-IID (AR13/3698), ST8-MRSA-IVE (AR43/3330.1), ST8-MRSA-IVF (AR43/3246), and ST94-MRSA-IVg (M03/0169.2), using GoTaq DNA polymerase (Promega). The primers described above for AR13.1/3330.2 were used for these PCR amplifications, except that primers J1_IVc_F2, J1_IVb_F2, and J1_IVg_F1 were used instead of primer J1_IIE_F3 for isolates harboring SCCmec types IVE, IVF, and IVg, respectively (see Table S1 in the supplemental material). PCR products were visualized by agarose gel electrophoresis, and the sizes of amplicons were compared to those obtained with template DNA from AR13.1/3330.2 and to the expected size of amplicons based on the whole-genome sequence of this isolate.
PCR detection of antimicrobial resistance determinants.
Phenotypic resistance to selected antimicrobial agents where corresponding antimicrobial resistance genes were included on the DNA microarray was determined by disk diffusion susceptibility testing as described previously (41). Isolates that exhibited phenotypic resistance to particular antimicrobial agents for which associated resistance genes were not detected by the DNA microarray were further investigated using PCR assays and previously described primers and thermal cycling conditions specific for additional resistance genes. These investigations included PCR detection of the fusidic acid resistance gene fusD (5), the trimethoprim resistance genes dfrD (9), dfrG (44), and dfrK (24), the tetracycline resistance gene tet(L) and tet(O) (53), and PCR DNA amplification and sequencing to detect further mutations in fusA encoding elongation factor G (EF-G) (5). Escherichia coli pBS2187dfrK (23) and S. aureus CM.S2 (44) were used as positive controls for dfrK and dfrG, respectively. Positive controls for fusD and dfrD were not available for use in the present study. Following PCR amplification of the fusA gene, PCR products were purified using the Genelute PCR cleanup kit (Sigma-Aldrich, Tallaght, Dublin, Ireland), sequenced commercially by Geneservice, and analyzed using DNA Strider and Bionumerics software using the fusA gene consensus sequence of MRSA reference strain N315 (GenBank accession number NC_002745) as a reference sequence.
Nucleotide sequence accession number.
The nucleotide sequence of the ccrAB4-carrying element from MRSA isolate AR13.1/3330.2 was deposited in the GenBank database under accession number HE858191.
RESULTS
spa typing.
The 175 MRSA isolates recovered in Irish hospitals between 1971 and 2004 were previously assigned to 26 ST-SCCmec types and subtypes (Table 1) (45, 49). In the present study, these 175 isolates were assigned to 35 spa types (Table 1). The spa type t190 predominated (41%; 73/175) and accounted for 99% (73/74) of ST8-MRSA-IIA-IIE and ST8-MRSA-IVE/IVF isolates (Table 1). Single spa types also accounted for the majority of isolates within four other clones: t008 in ST250-MRSA-I/I-pls (95%; 18/19); t045 in ST5-MRSA-II (100%; 17/17); t037 in ST239-MRSA-III/III-p1258/Tn554 (92%; 12/13), and t018 in ST36-MRSA-II (75%; 9/12) (Table 1). In contrast, the 27 ST22-MRSA-IV isolates exhibited 16 spa types, with spa type t032 predominating (33%; 9/27), followed by t005 (11%, 3/27) and t022 (7%, 2/27), with all other ST22 spa types being represented by single isolates (Table 1). In no instance was the same spa type identified among isolates belonging to different CCs, while only one spa type (t012) was detected among isolates belonging to different STs, ST36 and ST30, but to the same CC, CC30 (Table 1).
Table 1
MLST CCs, STs, SCCmec types, and spa types of 175 MRSA isolates representative of different antibiogram-resistogram, ST, and SCCmec type combinations recovered from patients in Irish hospitals between 1971 and 2004 and the corresponding CCs/STs and SCCmec types deduced from DNA microarray profiles of 107 representative isolates
| CC | MLST and SCCmec typing assignment for 175 isolates testedb | spa types for 175 isolates testedc | DNA microarraya | ||
|---|---|---|---|---|---|
| CC/ST-SCCmec type assignment by microarray for 107 isolates testedd | SCCmec-associated genes detected by microarraye | SCCmec type/subtype assignment following manual inspection of microarray profiles | |||
| 8 | ST250-MRSA-I (10) | t008 (10) | ST250-MRSA-I (4) | mecA, ugpQ, ΔmecR1, ccrAB1, dcs, pls | I |
| 8 | ST250-MRSA-I-pls (9) | t008 (8), t121 (1) | ST250-MRSA-I (6) | mecA, ugpQ, ΔmecR1, ccrAB1, dcs | I-pls |
| 8 | ST239-MRSA-III and SCCHg (9) | t037 (8), t138 (1) | ST239-MRSA-III (8) | mecA, ugpQ, ΔmecR1(5 ΔmecR1 negative), mecR1, mecI, xylR, ccrAB3, ccrC, merA/B, erm(A), tet(K) (1 negative) | III and SCCHg |
| 8 | ST239-MRSA- III and SCCHg -p1258/Tn554 (4) | t037 (4) | ST239-MRSA-III (4) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB3, ccrC, merA/B, erm(A), tet(K) | III and SCCHg |
| 8 | ST247-MRSA-Ia (3) | t3503 (2), t052 (1) | ST247-MRSA-I (3) | mecA, ugpQ, ΔmecR1, ccrAB1, dcs, pls, aadD | Ia |
| 8 | ST8-MRSA-IIA and ccrAB4 (9) | t190 (9) | ST8-MRSA-II and CI (4) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, ccrB4, dcs (1 dcs negative), aadD, erm(A) | IIA or IIB and ccrAB4 |
| 8 | ST8-MRSA-IIB and ccrAB4 (1) | t190 (1) | ST8-MRSA-II and CI (1) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, ccrB4, dcs, aadD, erm(A) | IIA or IIB and ccrAB4 |
| 8 | ST8-MRSA-IIC and ccrAB4 (9) | t190 (9) | ST8-MRSA-II and CI (6) | mecA, ugpQ, ΔmecR1, mecR1, ccrAB2, ccrB4, dcs (1 dcs negative), aadD, erm(A) | IIC and ccrB4 |
| 8 | ST8-MRSA-IIC (1) | t190 (1) | ST8-MRSA-II (1) | mecA, ugpQ, ΔmecR1, mecR1, ccrAB2, dcs, aadD, erm(A) | IIC |
| 8 | ST8-MRSA-IID and ccrAB4 (24) | t190 (23), t2196 (1) | ST8-MRSA-II and CI (12) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, ccrB4, dcs (4 dcs negative), erm(A) | IID and ccrAB4 |
| 8 | ST8-MRSA-IID (3) | t190 (3) | ST8-MRSA-II (3) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, dcs, erm(A) | IID |
| 8 | ST8-MRSA-IIE and ccrAB4 (6) | t190 (6) | ST8-MRSA-II and CI (5) | mecA, ugpQ, ΔmecR1, mecR1, ccrAB2, ccrB4, dcs, erm(A) | IIE and ccrAB4 |
| 8 | ST8-MRSA-IIE (1) | t190 (1) | ST8-MRSA-II and CI (1) | mecA, ugpQ, ΔmecR1, mecR1, ccrAB2, erm(A) | IIE |
| 8 | ST8-MRSA-IVE and ccrAB4 (17) | t190 (17) | ST8-MRSA-IV and CI (2) | mecA, ugpQ, ΔmecR1, ccrAB2, ccrAB4 (1 ccrA4 negative) | IVE or IVF and ccrAB4 |
| CC8-MRSA-VI (1) | mecA, ugpQ, ΔmecR1, ccrAB4 | VI | |||
| 8 | ST8-MRSA-IVF and ccrAB4 (3) | t190 (3) | CC8-MRSA-VI (3) | mecA, ugpQ, ΔmecR1, ccrAB4 (1 ccrA4 negative) | VI |
| 8 | ST94-MRSA-IVg and ccrAB4 (1) | t4691 (1) | ST8-MRSA-IV and CI (1) | mecA, ugpQ, ΔmecR1, ccrAB2, ccrB4, dcs | IV and ccrAB4 |
| 8 | ST609-MRSA-IVA (1) | t064 (1) | CC8-MRSA-IV (1) | mecA, ugpQ, ΔmecR1, ccrAB2, dcs, aadD | IVA |
| 22 | ST22-MRSA-IVh (22) | t032 (9), t022 (2), t3505 (1), t1467 (1), t531 (1), t3506 (1), t3504 (1), t790 (1), t515 (1), t3501 (1), t1802 (1), t2945 (1), t4253 (1) | ST22-MRSA-IV (11) | mecA, ugpQ, ΔmecR1, ccrAB2, dcs | IV |
| 22 | ST22-MRSA-IVa (5) | t005 (3), t902 (1), t4902 (1) | ST22-MRSA-IV (2) | mecA, ugpQ, ΔmecR1, ccrAB2, dcs | IV |
| 5 | ST5-MRSA-II (17) | t045 (17) | ST5-MRSA-II (14) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, dcs (2 dcs negative), kdp, aadD, erm(A) | II |
| 5 | ST5-MRSA-IV nonsubtypeable (1) | t001 (1) | CC5-MRSA-atypical SCCmec element (1) | mecA, ugpQ, ΔmecR1, dcs | Class B mec and dcs, no ccr genes detected |
| 5 | ST496-MRSA-II (1) | t002 (1) | ST5-MRSA-II (1) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, dcs, kdp, aadD, erm(A) | II |
| 30 | ST36-MRSA-II (12) | t018 (9), t012 (3) | ST36/39-MRSA-II (6) | mecA, ugpQ, ΔmecR1, mecR1, mecI, xylR, ccrAB2, dcs, kdp, aadD, erm(A) | II |
| 30 | ST30-MRSA-IV nonsubtypeable (4) | t5093 (1), t012 (2), t021 (1) | ST30-MRSA-IV (4) | mecA, ugpQ, ΔmecR1, ccrAB2, dcs (2 dcs negative), Q6GD50 (fusC) | IV and Q6GD50/fusC |
| 45 | ST45-MRSA-IVa (1) | t727 (1) | ST45-MRSA-IV (1) | mecA, ugpQ, ΔmecR1, ccrAB2, dcs | IV |
| 12 | ST12-MRSA-IVc (1) | t160 (1) | CC12-MRSA-IV (1) | mecA, ugpQ, ΔmecR1, ccrAB2, dcs | IV |
Analysis of the 35 spa types represented among the study isolates using the Based Upon Repeat Pattern (BURP) algorithm assigned 26 of the 35 spa types to four clusters (Fig. 1). Cluster 1 (spa-CC 032/022) consisted of 11 of the 16 spa types identified among ST22-MRSA-IV isolates. Cluster 2 (spa-CC 012) consisted of all spa types exhibited by CC30 isolates and the CC8/ST239-MRSA-III spa types t037 and t138. This was not unexpected, because the ST239 genotype arose as a result of a large chromosomal replacement between ST8 and ST30 isolates encompassing the spa locus (39). Cluster 3 (spa-CC 008) consisted of all except one of the remaining spa types identified among the CC8 isolates, while cluster 4 consisted of two of three spa types found among CC5 isolates. The remaining spa types were not assigned to any cluster, because they consisted of four repeats only, which is considered to be too few data to deduce evolutionary history, or they differed from all other spa types by a cost of ≥5 (where cost indicates the steps of evolution between two spa types).
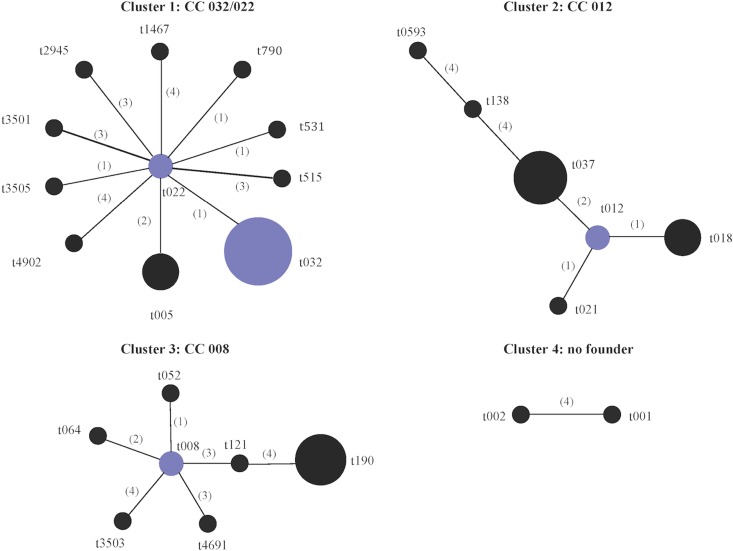
Based Upon Repeat Pattern (BURP) analysis of the spa types identified among 175 MRSA isolates representative of the different antibiogram-resistogram, sequence type, and SCCmec type combinations recovered from patients in Irish hospitals between 1971 and 2004. Twenty-six of the 35 spa types were grouped into four cluster groups using the BURP algorithm. spa types were clustered together if they contained five or more spa repeats and if they had a cost value of ≤4, where cost accounts for the number of steps of evolution between spa types. A low cost value indicates close evolutionary relatedness between two spa types. The following six spa types were defined as singletons by BURP (t045, t160, t902, t3504, t3506, and t4253), as they could not be clustered with any other spa type, i.e., cost value of ≥5. The spa types t727, t1802, and t2196 were excluded, as they consisted of four repeat units only. Cost values are shown in parentheses. Group founders and cofounders (spa types with the second-highest group founder score) are shown in blue and are determined based on the spa type that shares the highest sequence identity with the greatest number of spa types within that cluster as determined by the cost values. No founder was assigned to cluster 4.
Correlation between DNA microarray analysis and MLST.
The DNA microarray assigned the majority of the MRSA isolates investigated to the correct ST (92%; 98/107) (Table 1). Seven isolates were not assigned to any ST but were assigned to the correct CC (7%; 7/107), while two isolates were assigned to incorrect STs (2%; 2/107); but these incorrect STs were within the correct CCs. One isolate with ST496 (which is in CC5) was assigned to ST5, and one isolate belonging to ST94 (which is in CC8) was assigned to ST8 (Table 1). ST496 is a double-locus variant of ST5, while ST94 is a single-locus variant of ST8.
Correlation between SCCmec typing and SCCmec types deduced by DNA microarray analysis.
The majority of MRSA isolates subjected to DNA microarray analysis were assigned to the correct SCCmec type (i.e., to SCCmec types I, II, III, or IV) (95%; 102/107) compared to results obtained previously by conventional SCCmec typing (Table 1). Table S2 in the supplemental material provides an overview of the main characteristics of the SCCmec types and subtypes investigated in the present study. Four ST8/t190 isolates that were previously assigned by SCCmec typing to SCCmec types IVE or IVF (J1 regions IVc and IVb, respectively, as well as class B mec and ccrAB2 and lacking dcs) with an adjacent ccrAB4 were assigned to SCCmec VI by the DNA microarray (Table 1). In these isolates, the class B mec complex and ccrAB4 genes were detected using the DNA microarray, but ccrAB2 was not detected (Table 1). One MRSA isolate belonging to CC5 with spa type t001, which was previously found to harbor SCCmec IV by SCCmec typing, was designated as an atypical SCCmec element by the DNA microarray (Table 1). Manual inspection of the SCCmec-associated DNA microarray signals of this isolate revealed the presence of a class B mec complex and dcs but the absence of ccrAB2. These five isolates with anomalous SCCmec types, as deduced from their DNA microarray profiles because they lacked ccrAB2, were retyped using conventional SCCmec typing by PCR, which confirmed the absence of ccrAB2 and suggested that these isolates may have lost the ccr genes. Lastly, manual inspection of the DNA microarray profiles indicated that while the eight ST239-MRSA-IIIHg isolates yielded signals for mecR1, the probe for the truncated mecR1 gene (ΔmecR1) did not yield a signal for five of these eight isolates (Table 1).
The DNA microarray did not automatically assign SCCmec subtypes to isolates, but manual inspection of the DNA microarray profiles allowed subtyping of SCCmec types I, II, and IIIHg (Table 1). The differentiation of SCCmec subtypes Ia and I-pls is based on the presence of plasmid pUB110 carrying the aminoglycoside resistance gene aadD in SCCmec Ia and the absence of pls in SCCmec I-pls (Table 1). The SCCmec II subtypes IIA, IIB, IIC, IID, and IIE all lack kdp, which is normally associated with SCCmec II elements (Table 1). While the SCCmec subtypes IIA and IIB cannot be further differentiated using the array, SCCmec IIC lacks xylR and mecI due to the presence of a variant mec complex termed class A.3 mec. SCCmec subtype IID is characterized by the absence of aadD, while xylR, mecI, and aadD are absent in SCCmec IIE (Table 1). SCCmec type IIIHg was identified by the presence of the mercury resistance genes merA and merB and by ccrC (Table 1). While the DNA microarray was unable to subtype the majority of SCCmec IV elements, recognition of SCCmec IVA was possible due to the presence of the pUB110-encoding gene aadD (Table 1). The array detected the fusC gene (designated Q6GD50 on the array) indicative of the presence of the SCCfus element (18) in the four ST30 isolates, but additional ccr genes (ccrAB1) normally associated with the presence of this SCC element were not detected. The dcs gene, located between mecA and orfX and normally found in SCCmec II and IV elements, was not detected in eight isolates harboring SCCmec II elements (ST8 and ST5) and eight isolates with SCCmec IV elements (ST8 and ST30), although this has been reported previously for the ST8 SCCmec IVE and IVF elements (45).
The DNA microarray also allowed the identification of the presence of S. epidermidis ccrAB4 in ST8-MRSA isolates (Table 1). The presence of S. epidermidis ccrAB4 has been reported previously in ST8 isolates (49). The array yielded positive or ambiguous signals for ccrA4 and/or ccrB4 in 36 CC8-MRSA isolates, including ST8-MRSA-IIA-IIE isolates (29/33), ST8-MRSA-IVE-IVF isolates (6/6), and the single ST94-MRSA-IVg isolate investigated using the array. The presence of S. epidermidis ccrAB4 was confirmed in these isolates by PCR, indicating the possible presence of an SCC element in addition to SCCmec in these isolates. The presence of S. epidermidis ccrAB4 was also investigated by PCR in the ST8-MRSA-IIA-IIE (n = 21) and ST8-MRSA-IVE-IVF (n = 14) isolates from the present study that did not undergo DNA microarray analysis; the presence of this gene was confirmed in 95% (20/21) and 100% (14/14) of these isolates, respectively.
Detection of a novel SCCmec-SCC composite island in ST8-MRSA-IIE isolate AR13.1/3330.2.
Whole-genome sequencing was performed on one isolate (AR13.1/3330.2) representative of isolates harboring the S. epidermidis ccrAB4 gene to determine the genetic organization of the region surrounding ccrAB4. Whole-genome sequencing of AR13.1/3330.2 yielded 344 contigs ranging in size from ca. 113 bp to 137 kb. The SCC-associated DNA sequences were identified in three contigs (13 kb, 11 kb, and 764 bp in size), and the gaps between these contigs were closed using long-range PCR amplification and sequencing with primers based on the surrounding contigs. The order of the contigs was confirmed by PCR. This analysis revealed the presence of an approximately 14-kb SCC element downstream and immediately adjacent to SCCmec IIE in AR13.1/3330.2 (Fig. 2a). This SCC element exhibited 99% DNA sequence identity to SCCM1 (Fig. 2b) previously identified in the Danish ST8-MRSA isolate M1 (spa type t024) (2).
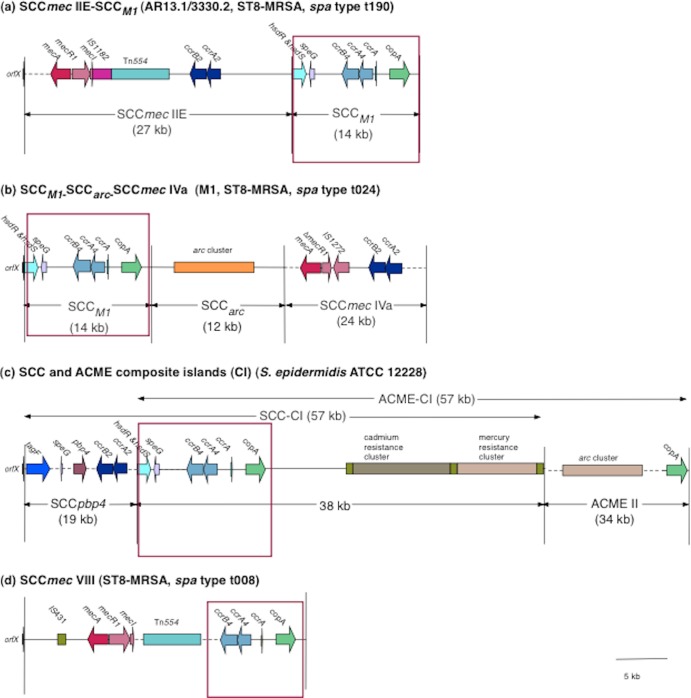
Schematic diagram showing the genomic organization of the novel SCCmec-SCC composite island (CI) SCCmec IIE-SCCM1 identified in the ST8-MRSA spa type t190 isolate AR13.1/3330.2 (GenBank accession number HE858191) (a), the SCCM1-SCCarc-SCCmec element previously reported in ST8-MRSA spa type t024 isolate M1 (HM030720) (b), the ACME- and SCC-CIs previously reported in S. epidermidis ATCC 12228 (NC004461) (c), and SCCmec VIII previously described in ST8-MRSA spa type t008 isolate C10682 (FJ390057) (d). The structure of the novel SCCmec IIE-SCCM1 element was determined by high-throughput whole-genome sequencing of AR13.1/3330.2 and was confirmed using primers spanning the SCCmec/SCC region. The areas surrounded by the red square indicate the regions of high similarity between SCCM1 identified in isolate AR13.1/3330.2 and the other SCCmec/SCC elements.
In MRSA M1, the SCCM1 element exists as part of a composite island and is located directly adjacent to orfX, upstream of a truncated arginine catabolic mobile element (ACME) and SCCmec IVa (Fig. 2b). In contrast, in AR13/3330.2, the SCCmec IIE element was located adjacent to orfX, whereas SCCM1 was located downstream of SCCmec IIE (Fig. 2a). No ACME-arc genes were detected in AR13.1/3330.2, either in the whole-genome sequence or by DNA microarray analysis. The SCCM1 element comprises 18 ORFs that have been described previously (2), most significantly those encoding ccrAB4, a truncated copy of ccrA and a gene encoding a copper-transporting ATPase (copA) (Fig. 2a and andb).b). SCCM1 is similar in its genetic organization to the part of SCC-CI element in S. epidermidis ATCC 12228 outside SCCpbp (99% DNA sequence identity) but significantly smaller in size and lacking the mercury and cadmium resistance genes (Fig. 2c). SCCM1 also shows 99% DNA sequence identity to a region downstream of mecA in SCCmec VIII (Fig. 2d). The composite island (CI) detected in AR13.1/3330.2 was designated SCCmec IIE-SCCM1 (Fig. 2a).
Confirmation of the presence of SCCM1 in representative CC8 isolates harboring the S. epidermidis ccrAB4 genes.
The presence of the SCCM1 element and its location downstream of SCCmec was investigated by PCR in seven additional isolates representative of other ST and SCCmec type combinations identified as harboring the S. epidermidis ccrAB4. Five of the seven isolates, including those representative of ST8-MRSA-IIA, ST8-MRSA-IIB, ST8-MRSA-IIC, ST8-MRSA-IID, and ST94-MRSA-IVg, yielded amplicons of the expected size for SCCM1, and its location was downstream of SCCmec, as found in isolate AR13.1/3330.2 (Fig. 2a). These SCCmec-SCC CIs were assigned the following designations: SCCmec IIA-SCCM1, SCCmec IIB-SCCM1, SCCmec IIC-SCCM1, SCCmec IID-SCCM1, and SCCmec IVg-SCCM1. For the remaining two isolates (ST8-MRSA-IVE and ST8-MRSA-IVF), the primer pairs located within the SCCM1 element (primer pairs 2, 3, and 4; see Table S1 in the supplemental material) yielded amplicons of the expected size, but those amplifying from the respective SCCmec elements to within the SCCM1 element (primer pairs 5 and 6; see Table S1 in the supplemental material) failed to yield any amplicons. These findings indicate that while the SCCM1 element is present in these ST8-MRSA isolates harboring SCCmec IVE and IVF, further investigations are required to determine the genetic organization of the SCCmec and SCCM1 elements in these isolates.
Additional typing markers and antimicrobial resistance, virulence-associated, and adhesion genes detected using the DNA microarray.
The agr capsule and immune evasion cluster (IEC) types, as well as the antimicrobial resistance, virulence-associated, MSCRAMM, adhesion, and biofilm formation genes identified among the 107 MRSA isolates representative of the different ST and SCCmec type combinations investigated by DNA microarray analysis in the present study, are shown in Table 2. The majority of isolates belonged to agr type I (75%; 80/107) and included all CC8 and CC22 isolates (n = 66 and 13, respectively). All CC5 (n = 16) and the single CC12 isolate belonged to agr type II, while isolates within agr III were CC30 (n = 10). The single CC45 isolate belonged to agr type IV. Most isolates belonged to capsule type 5 (78%; 83/107), with the remainder belonging to capsule type 8 (22%; 24/107). Various combinations of IEC genes were detected in the majority of isolates (91%; 97/107), with the most frequently occurring IEC type being type D (39%; 42/107). IEC type D was detected in the CC8 lineages ST239-MRSA-III, ST8-MRSA-IIA–IIE, and ST609-MRSA-IVA and consisted of the enterotoxin A gene sea and the genes encoding staphylokinase (sak) and the staphylococcal complement inhibitor (scn). Other IEC types represented included IEC types A (n = 8 isolates), B (n = 28 isolates), E (n = 5 isolates), F (n = 12 isolates), and G (n = 1 isolate).
Table 2
agr, capsule and IEC types, antimicrobial resistance, virulence-associated, MSCRAMM, adhesion, and biofilm formation genes identified using the DNA microarray among isolates representative of each antibiogram-resistogram, ST, and SCCmec type combination recovered from patients in Irish hospitals between 1971 and 2004a
| CC/ST-SCCmec type (n) | Additional typing marker | Antimicrobial resistance genes | Virulence-associated genes | MSCRAMM, adhesion, and biofilm genes | |||||
|---|---|---|---|---|---|---|---|---|---|
| agr | cap | IEC (n)b | Always present | Variably present (% isolates positive) | Always present | Variably present (% isolates positive) | Always present | Variably present (% isolates positive) | |
| CC8/ST250-MRSA-I/I-pls (10) | I | 5 | B (10) | blaZ, merA/B, erm(A), sdrM, qacA, fosB | tet(K) (70), aacA-aphD (90), aphA3 and sat (40), dfrS1 (40) | sak, chp, scn | seb (90), sek and seq (90) | bbp, clfA, clfb, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | ..... |
| CC8/ST239-MRSA-III and SCCHg/III and SCCHg-p1258/Tn554 (12) | I | 8 | D (12) | blaZ, merA/B, erm(A), tet(M), sdrM, fosB | tet(K) (92), qacA (58), qacC (8), aacA-aphD (83), aadD (8), aphA3 and sat (50), dfrS1 (25), mupA (8), cat-pC194 (pMC524) (8) | sea, sak, scn | sek and seq (83) | clfA, clfB, cna, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | bbp (58) |
| CC8/ST247-MRSA-Ia (3) | I | 5 | A (1), B (2) | blaZ, merA/B tet(M), sdrM, qacA, aacA-aphD, aadD, fosB | erm(A) (67) | sak, chp, scn | sea (33) | clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | ..... |
| CC8/ST8-MRSA-IIA-IIE (33) | I | 5 | D (29), NT (1),c Neg (3) | blaZ, erm(A), aacA-aphD, sdrM, fosB, merA/B | erm(C) (6), tet(K) (3), qacA (97), qacC (3), aadD (36), aphA3 and sat (85), dfrS1 (3), far1/fusB (3), mupA (12) | ..... | sea (91), sak (91), scn (88) | bbp, clfA, clfB, ebh, ebpS, eno, fnbA, fnbB, sdrD, vwb, icaA, icaC, icaD | fib (97), map (97), sdrC (3) |
| CC8/ST8-MRSA-IVE/F (6) | I | 5 | E (5), Neg (1) | blaZ, erm(A), sdrM, aphA3 and sat, fosB | tet(M) (33), qacA (17) | ..... | sak (83), scn (83) | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, vwb, sasG, icaA, icaC, icaD | sdrD (83) |
| CC8/ST94-MRSA-IVg (1) | I | 5 | Neg (1) | blaZ, tet(M), sdrM, aacA-aphD, aphA3 and sat, fosB | ..... | ..... | ..... | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | ..... |
| CC8/ST609-MRSA-IVA (1) | I | 5 | D (1) | blaZ, erm(C), tet(M), sdrM, aacA-aphD, aadD, dfrS1, fosB | ..... | sea, seb, sek and seq, sak, scn | ..... | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC icaD | ..... |
| CC22/ST22-MRSA-IVh/IVa (13) | I | 5 | B (8) Neg (5) | blaZ | erm(C) (46), qacC (8) | egc | sec and sel (85), sak (62), chp (62), scn (62) | clfA, clfB, cna, ebpS, eno, fib, fnbA, sdrC, vwb, sasG, icaA, icaD | fnbB (15), icaC (85), bbp (85), ebh (15), map (92), sdrD (85) |
| CC5/ST5-MRSA-II (14) | II | 5 | F (12) B (1) A (1) | erm(A), sdrM, aadD, fosB | blaZ (86), qacA (21), merA/B (21) | tst, egc,c sak, chp, scn | sea (7), sep (86), sed, j and r (71) | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | ..... |
| CC5/ST5-MRSA-IV nonsubtypeable (1) | II | 5 | B (1) | merA and B, blaZ, erm(A), sdrM, fosB | ..... | egc, sak, chp, scn | ..... | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, map, sdrC, vwb, sasG, icaA, icaC, icaD | ..... |
| CC5/ST496-MRSA-II (1) | II | 5 | B (1) | erm(A), erm(C), aadD, sdrM, fosB | ..... | egc, sak, chp, scn | ..... | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | |
| CC30/ST36-MRSA-II (6) | III | 8 | A (6) | blaZ, erm(A), sdrM, aadD, fosB | ..... | sea, egc, sak, chp, scn | tst (67) | bbp, clfA, clfB, cna, ebh, ebpS, eno, fnbA, fib, map, sdrC, sdrD, vwb, icaA, icaC, icaD | ..... |
| CC30/ST30-MRSA-IV nonsubtypeable (4) | III | 8 | B (4) | blaZ, Q6GD50/fusC, fosB | sdrM (75) | tst, egc, sak, chp, scn | ..... | clfA, clfB, cna, ebh, ebpS, eno, fib, fnbA, map, sdrC, sdrD, vwb, icaA, icaC, icaD | bbp (75) |
| CC45/ST45-MRSA-IVa (1) | IV | 8 | B (1) | blaZ, merA and B, erm(C), far1/fusB, sdrM | ..... | egc, sak, chp, scn | ..... | bbp, clfA, clfB, cna, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, sdrD, vwb, sasG, icaA, icaC, icaD | ..... |
| CC12/ST12-MRSA-IVc (1) | II | 8 | G (1) | blaZ, sdrM, fosB | ..... | sep, seb, sec (M14), sak, scn | ..... | clfA, clfB, cna, ebh, ebpS, eno, fib, fnbA, fnbB, map, sdrC, vwb, icaA, icaC, icaD | ..... |
The main virulence-associated genes identified among the MRSA isolates representative of the different STs and CCs are shown in Table 2. Ninety-four percent of isolates carried one or more superantigen genes with enterotoxin A (sea) (48%; 51/107) predominating, followed by egc (37%; 40/107), tst (21%; 22/107), and sek and seq (19%; 20/107). All isolates lacked the PVL locus genes lukF-PV and lukS-PV; the exfoliative toxin genes etA, etB, and etD; and ACME. All isolates harbored the MSCRAMM, adhesion, and biofilm genes fnbA (fibronectin binding protein A), clfA and clfB (clumping factor A and B), ebpS (elastin binding protein), eno (laminin binding protein), icaA (intracellular adhesion protein A), icaD (intracellular adhesion protein A), and vwb (van Willebrand factor binding protein). The majority of isolates also harbored fib (fibrinogen binding protein) (98%; 105/107) and map (major histocompatibility compound class II analog protein) (97%; 104/107).
The most common antimicrobial resistance genes (apart from mecA, which was detected in all isolates tested) were blaZ (beta-lactamase resistance gene) (97%; 104/107), sdrM (encoding a multidrug efflux pump conferring resistance to norofloxacin, acriflavine, and possibly ethidium bromide [57]) (87%; 93/107), fosB (fosfomycin resistance gene) (87%; 93/107), erm(A) (resistance to macrolides, lincosamides, and streptogramins) (79%; 85/107), merA and merB (mercury resistance genes) (59%; 63/107), qacA (resistance to quaternary ammonium compounds, ethidium bromide) (52%; 56/107), and aacA-aphD (resistance to amikacin, gentamicin, kanamycin, and tobramycin) (53%; 57/107). CC8 isolates harbored the highest number of resistance determinants, with ST8-MRSA-IIA-IIE and ST239-MRSA-III carrying genes encoding resistance to 10 classes of antimicrobials, whereas ST22-MRSA-IV carried resistance to three classes of antimicrobials.
Correlation between antimicrobial resistance phenotype and genotype.
The correlation between antimicrobial agent resistance phenotype (determined by disk diffusion) and genotype for the 107 MRSA isolates (determined by DNA microarray profiling) are shown in Table 3 for antimicrobial resistance genes yielding positive results only, i.e., all isolates yielded negative results for the genes encoding resistance to antimicrobials in the following antimicrobial classes, and results are not included in Table 3: chloramphenicol (cfr and fexA, encoding chloramphenical and florfenicol resistance); glycopeptides (vanA, vanB, and vanZ, encoding vancomycin and/or teicoplanin resistance); macrolides, lincosamides, or streptogramins (MLS) [lnu(A), mef(A), mph(C), msr(A), vat(A), vat(B), vga(A), vgb(A), encoding resistance to a range of MLS antimicrobial compounds].
Table 3
Correlation between antimicrobial resistance gene detection by array profiling and phenotypic detection of resistance by disk diffusionf
| Gene(s) | Conferring resistance to: | No. of isolates with gene profile | Antimicrobial(s) tested (resistance profile obtained)h | No. of isolates resistant | Correlation between genotype and phenotype | |
|---|---|---|---|---|---|---|
| No. of discrepancies | % correlationg | |||||
| Aminoglycoside resistance genes | AMI, GEN, KAN, NEO, TOB | 83 | 1 | 99 (106/107) | ||
    aacA-aphD aacA-aphD | AMI, GEN, KAN, TOB | 14 | AMI, GEN, KAN, ....., TOB | 5 | 0 | |
| ....., GEN, KAN, ....., TOB | 9a | 0a | ||||
    aadD aadD | AMI, KAN, NEO, TOB | 20 | AMI, ....., KAN, NEO, TOB | 18 | 0 | |
| ....., ....., KAN, NEO, TOB | 2a | 0a | ||||
    aphA3 aphA3 | KAN, NEO | 6 | ....., ....., KAN, NEO, ..... | 6 | 0 | |
    aacA-aphD and aadD and aphA3 aacA-aphD and aadD and aphA3 | AMI, GEN, KAN, NEO, TOB | 13 | AMI, GEN, KAN, NEO, TOB | 12 | 0 | |
| ....., ....., KAN, NEO, TOB | 1 | 1 | ||||
    aacA-aphD and aphA3 aacA-aphD and aphA3 | AMI, GEN, KAN, NEO, TOB | 26 | AMI, GEN, KAN, NEO, TOB | 11 | 0 | |
| ....., GEN, KAN, NEO, TOB | 15a | 0a | ||||
    aacA-aphD and aadD aacA-aphD and aadD | AMI, GEN, KAN, NEO, TOB | 4 | AMI, GEN, KAN, NEO, TOB | 4 | 0 | |
    None None | 24 | Susceptible to all (n = 24) | 0 | 0 | ||
| Antiseptics, disinfectants, intercalating dye resistance genes | Ethidium bromide | 58 | 0 | 100 (107/107) | ||
    qacA qacA | QACs,b ethidium bromide, chlorhexidine | 55 | Ethidium bromide | 55 | 0 | |
    qacC qacC | QACs,b ethidium bromide | 2 | Ethidium bromide | 2 | 0 | |
    qacA and qacC qacA and qacC | QACs, ethidium bromide, chlorhexidine | 1 | Ethidium bromide | 1 | 0 | |
    None None | 49 | Susceptible (n = 49) | 0 | 0 | ||
| Beta-lactam (excluding methicillin) resistance gene | Ampicillin | 107 | 3c | 97 (104/107) | ||
    blaZ blaZ | Beta-lactams (not methicillin) | 104 | Ampicillin | 104 | 0 | |
    None None | 3 | Susceptible (n = 0) | 3c | 3c | ||
| Chloramphenicol resistance gene | Chloramphenicol | 1 | 0 | 100 (107/107) | ||
    cat-pC194 (pMC524) cat-pC194 (pMC524) | Chloramphenicol | 1 | Chloramphenicol | 1 | 0 | |
    None None | 106 | Susceptible (n = 106) | 0 | 0 | ||
| Fusidic acid resistance genes | Fusidic acid | 21 | 15 | 86 (92/107) | ||
    far1 (fusB) far1 (fusB) | Fusidic acid | 2 | Fusidic acid | 2 | 0 | |
    Q6GD50 (fusC) Q6GD50 (fusC) | Fusidic acid | 4 | Fusidic acid | 4 | 0 | |
    None None | 101 | Susceptible (n = 86) | 15 | 15 | ||
| Heavy metal ions resistance genes | MER, PMA | 63 | 2 | 98 (105/107) | ||
    merA merA | Mercury ions | 0 | Mercuric chloride | 0 | 0 | |
    merB merB | Organomercurial ions | 0 | Phenylmercuric acetate | 0 | 0 | |
    merA and merB merA and merB | Mercury and organomercurial ions | 63 | MER, PMA | 62 | 1 | |
    None None | 44 | Susceptible (n = 43) | 1 | 1 | ||
| Macrolides, lincoamides, streptogramins (MLS) resistance genes | Erythromycin, (lincomycin) | 93 (47)d | 0 | 100 (107/107) | ||
    erm(A) erm(A) | Erythromycin, Clindamycin | 82 | Erythromycin (lincomycin) | 82 (46)d | 0 | |
    erm(C) erm(C) | Erythromycin, Clindamycin | 8 | Erythromycin (lincomycin) | 8 (1)d | 0 | |
    erm(A) and erm(C) erm(A) and erm(C) | 3 | Erythromycin (lincomycin) | 3 (0)d | 0 | ||
    None None | 14 | Susceptible (n = 14) | 0 | 0 | ||
| Mupirocin resistance gene | Mupirocin (high level) | 5 | 0 | 100 (107/107) | ||
    mupA-ileS2 mupA-ileS2 | Mupirocin (high level) | 5 | Mupirocin (high level) | 5 | 0 | |
    None None | 102 | Susceptible (n = 102) | 0 | 0 | ||
| Tetracycline resistance genes | Tetracycline | 29e | 2e | 98 (105/107)e | ||
    tet(K) tet(K) | Tetracycline | 8 | Tetracycline | 8 | 0 | |
    tet(M) tet(M) | Tetracycline | 8 | Tetracycline | 8 | 0 | |
    tet(K) and tet(M) tet(K) and tet(M) | Tetracycline | 11 | Tetracycline | 11 | 0 | |
    None None | 80 | Susceptible (n = 78e) | 2e | 2e | ||
| Trimethoprim resistance gene | Trimethoprim | 26 | 17 | 84 (90/107) | ||
    dfrS1 dfrS1 | Trimethoprim | 9 | Trimethoprim | 9 | 0 | |
    None None | 98 | Susceptible (n = 81) | 17 | 17 | ||
Eighty-three of the 107 MRSA isolates exhibited both phenotypic and genotypic resistance to aminoglycosides. The three aminoglycoside resistance genes detected by the DNA microarray are aacA-aphD, encoding resistance to amikacin, gentamicin, kanamycin, and tobramycin; aadD, encoding resistance to amikacin, kanamycin, neomycin, and tobramycin; and aphA3, encoding resistance to kanamycin and neomycin. The level of resistance to amikacin encoded by aacA-aphD is low (27) and may be difficult to detect by disk diffusion. Hence, many isolates carrying aacA-aphD or aadD exhibited resistance in the intermediate category or were phenotypically susceptible to amikacin (Table 3). The relative frequencies of occurrence of the various aminoglycoside resistance gene combinations was as follows: aacA-aphD and aphA3, 31% (26/83); aadD, 24% (20/83); aacA-aphD, 17% (14/83); aacA-aphD, aadD, and aphA3, 16% (13/83); and aphA3, 7% (6/83). The phenotypic resistance profiles for each isolate harboring these gene combinations are shown in Table 3. In general (aside from the problem of amikacin resistance with isolates carrying aacA-aphD and/or aadD), correlation between phenotypic and genotypic expression of resistance was excellent (99%), with just one isolate carrying aacA-aphD, aadD, and aphA3 exhibiting phenotypic susceptibility to gentamicin (Table 3).
For all other antimicrobials tested, apart from fusidic acid and trimethoprim, the correlation between phenotype and genotype was ≥97%. A 100% correlation between the antimicrobial resistance phenotype and the presence of a particular gene or genes was observed for high-level mupirocin resistance and mupA, ethidium bromide resistance and qacA and/or qacC, erythromycin resistance and erm(A) and/or erm(C), and chloramphenicol resistance and cat (Table 3). All beta-lactam-resistant isolates harbored mecA, while 97% (104/107) also harbored the beta-lactam resistance gene blaZ. One isolate that lacked blaZ exhibited intermediate resistance to ampicillin. All isolates harboring tet(K), tet(M), or both of these genes exhibited tetracycline resistance. Two isolates exhibited intermediate resistance to tetracycline, lacked tet(K) and tet(M) in their DNA microarray profile, and tested negative for tet(L) and tet(O) when investigated using conventional PCR assays. The mercury resistance genes merA and merB were detected in all but one isolate exhibiting resistance to mercuric chloride, but one isolate exhibiting susceptibility to mercuric chloride harbored merA and merB.
There were major discrepancies with fusidic acid and trimethoprim resistance. Only 29% of fusidic acid-resistant isolates (6/21) and 35% of trimethoprim-resistant isolates (9/26) were DNA microarray gene positive for genes encoding resistance to these agents (Table 3).
The fusidic acid resistance genes far1/fusB were detected in one ST45-MRSA-IV and one ST8-MRSA-IIB isolate, while the Q6GD50/fusC gene was detected in four ST30-MRSA-IV isolates. None of the remaining 15 fusidic acid-resistant isolates were found to harbor the alternative fusidic acid resistance gene fusD by fusD-specific PCRs (fusD is not detected by the DNA microarray). The fusA gene in each of these 15 fusidic acid-resistant isolates was amplified by PCR followed by amplimer sequencing, as mutations in fusA have previously been shown to be associated with fusidic acid resistance in S. aureus. Mutations in fusA were identified in all 15 isolates. Three different combinations of mutations were identified in these isolates, including (i) an amino acid substitution from leucine to lysine at position 461 in the protein sequence (L461K) in eight isolates (seven CC8 isolates: one ST8-MRSA-IID, one ST250-MRSA-I-pls, and five ST250-MRSA-I/I-pls isolates; one ST22-MRSA-IVh isolate), (ii) an amino acid substitution from leucine to serine at position 461 in the protein sequence (L461S) in five isolates (one ST5-MRSA-II, one ST239-MRSA-III, and three ST250-MRSA-I-pls isolates), and (iii) an amino acid substitution from alanine to valine and phenylalanine to leucine at positions 70 (A70V) and 406 (F406K), respectively, in the protein sequence of two isolates (two ST8-MRSA-IIC isolates).
Twenty-six isolates exhibited phenotypic resistance to trimethoprim, and only 35% (9/26) of these harbored the dfrS1 gene, as determined by DNA microarray profiling (all were CC8 isolates as follows: ST609-MRSA-IVA [n = 1], ST8-MRSA-IIE [n = 1], ST239-MRSA-III [n = 3], and ST250-MRSA-I/I-pls [n = 4]). The dfrS1 gene is the only trimethoprim resistance gene detected by the microarray. The remaining 17 trimethoprim-resistant isolates were investigated by a variety of specific PCR assays designed to detect other genes encoding trimethoprim resistance, including dfrD, dfrG, and dfrK. None of the 17 trimethoprim-resistant isolates harbored dfrD or dfrK, whereas three isolates harbored dfrG (all ST239-MRSA-III). The 14 isolates in which no dfr genes were identified belonged to ST8-MRSA-IIC (n = 4), ST8-MRSA-IIE (n = 2), ST8-MRSA-IVE (n = 2), ST8-MRSA-IVF (n = 3), ST5-MRSA-IV (n = 1), and ST239-MRSA-III (n = 2).
DISCUSSION
The application of spa typing and DNA microarray profiling for in-depth molecular characterization of a diverse collection of nosocomial MRSA isolates representative of clones that predominated in Irish hospitals between 1971 and 2004 revealed extensive genetic diversity among the 175 isolates investigated. A total of 35 spa types were identified among these 175 isolates that had previously been assigned to 13 STs representing six CCs by MLST and to 26 ST-SCCmec type combinations representative of pandemic and epidemic MRSA lineages by MLST and conventional SCCmec typing. Almost half of all spa types (46%; 16/35) were identified among isolates of the ST22-MRSA-IV clonal lineage, which represented only 15% of isolates investigated (27/175). In contrast, within the majority of other clonal lineages, isolates were assigned to one or two spa types only. In particular, the majority of ST8 isolates (42% of total isolates investigated) were assigned to a single spa type, suggesting that the spa types of ST8 isolates may be more stable over time than those among ST22-MRSA-IV isolates. In support of this suggestion, Strommenger et al. previously reported that isolates within the different CC8 clones recovered at different time periods and from different geographic locations also exhibited the same spa types (51). A possible explanation for the greater variation in spa types among ST22-MRSA-IV isolates may lie in the higher number of repeat sequences in the VNTR region of the spa gene in this clone. The average number of repeats in the 16 spa types identified among ST22-MRSA-IV isolates in the present study was 13 (range, 4 to 17), with the most frequently occurring spa type (t032) carrying 16 repeats. In contrast, the spa type t190, to which all the ST8-MRSA-IIA-IIE and ST8-MRSA-IVE/IVF isolates were assigned, consists of only seven repeats.
DNA microarray profiling was found to be effective for genotyping MRSA isolates by assigning them to the correct CCs and/or STs and SCCmec types, indicating that this approach has the potential to replace MLST and SCCmec typing for genotyping and for investigating the evolutionary relatedness of MRSA. The advantages of using the DNA microarray for MLST and SCCmec typing include the fact that the DNA microarray also detects clinical relevant virulence and antimicrobial resistance genes and requires only a single multiplex PCR and a 2- to 3-h test procedure per isolate to generate data that require multiple multiplex PCRs and agarose gel electrophoresis to assign an SCCmec type/subtype and at least seven PCRs and subsequent sequencing to assign an ST by conventional methods. In a hospital setting, and particularly in an outbreak investigation, greater discrimination than that provided by ST and SCCmec type assignment is required to effectively type and track MRSA. While each clone included in the present study was found to have a characteristic combination of virulence and antimicrobial resistance genes, there were also differences observed in the carriage of these genes within the clones, e.g., the carriages of sec, sel, erm(C), and IEC were variable in ST22-MRSA-IV. Whether these differences could be used to provide enhanced discrimination and tracking of MRSA in hospitals either using the array as a standalone typing tool or in combination with other methods, such as spa typing, requires further investigation.
It is important to note that the array had difficulty differentiating between some sporadically occurring STs that are single-locus variants of more common STs and with subtyping of some SCCmec elements. While identification of some SCCmec I, II, and IV subtypes was possible following manual inspection of array profiles, in some cases this identification relied on the detection of the presence of a particular resistance determinant associated with a particular SCCmec type, such as aadD located on plasmid pUB110 in SCCmec types IA, IIA, IIB, IIC, and IVA. However, while this may aid in the identification of a particular SCCmec subtype, it is not possible to definitively localize this or any of the other possibly SCCmec-associated resistance determinants, i.e., erm(A), tet(K), and mer, which can be located on Tn554 in SCCmec II and III, pT181 in SCCmec III, and p1258 in SCCmec III with SCCHg, respectively, to within the SCCmec element using the array, and results need to be confirmed by PCR. Array profiling facilitated the identification of possible DNA sequence variation in mecR1 in some SCCmec III elements and in the dcs gene in some SCCmec II and IV elements that were not detected using conventional SCCmec typing PCR assays. In addition, the fusidic acid resistance determinant Q6GD50/fusC was detected in four ST30-MRSA-IV isolates using the array, indicating the possible presence of an SCCfus element in these isolates. Array profiling also indicated that five isolates harboring SCCmec IV may have lost ccrAB2, which was subsequently confirmed by PCR. Four of these isolates were assigned to ST8-MRSA-IVE/F, which also harbored a second set of ccr genes (ccrAB4) outside SCCmec, suggesting that these isolates may have lost ccrAB2 genes because they were not essential for excision or integration of the SCCmec element due to the presence of alternative ccr genes. The ability to accurately identify SCCmec types is essential to the use of this method for genotyping MRSA, and isolates such as these that may have lost ccr genes may be wrongly classified using SCCmec typing because of the presence of the second set of ccr genes outside the SCCmec element.
Several novel SCCmec-SCC-CIs were identified in the present study, each consisting of an SCCmec II or IV element (IIA-IIE, IVE, IVF, or IVg) and SCCM1 harboring ccrAB4. In S. aureus, the DNA sequence corresponding to the SCCM1 element has been identified only in MRSA belonging to CC8 and mainly to ST8 (2, 8, 58). CC8 is one of the largest MRSA lineages, consisting of a substantial number of MRSA clones with extensive diversity in SCCmec (14, 34, 45), and evidence suggests that the diversity in the SCCmec elements of MRSA isolates is due to horizontal transfer and recombination events between SCCmec and SCC elements from S. aureus isolates and CoNS. DNA sequence identity between SCCM1 in ST8-MRSA and the SCC-CI in S. epidermidis ATCC 12228 led Bartels et al. to speculate that the former may have evolved from the latter following recombination events between SCCmec and SCC-CI elements in MRSA and S. epidermidis (2). We previously reported that DNA sequence analysis of the SCCmec elements identified in ST8-MRSA isolates recovered in Ireland revealed the presence of SCCmec elements that may have been generated following recombination and genetic rearrangements between different SCCmec elements (45). The finding of the additional SCCM1 element in these ST8 isolates in the present study suggests that they have acquired the SCCM1 element either from another S. aureus or from CoNS. Only 8% (6/75) of the ST8/t190 or t2196 and ST94/t4691 isolates investigated in the present study did not carry the ccrAB4 element, and while they may have lost SCCM1, it is also possible that they never harbored it. Thus, these ST8 isolates carrying the SCCmec region only may be precursors of isolates harboring the SCCmec-SCCM1 elements.
Whether the acquisition of the SCCM1 element within the ST8 isolates occurred on one or multiple occasions remains to be determined. However, the presence of SCCM1 in association with three distinct groups of SCCmec elements (SCCmec IIA-IIE, SCCmec IVE/IVF, and SCCmec IVg) suggests at least three possible independent acquisitions of SCCM1. Interestingly, the ST8-MRSA-IIA–IIE isolates which harbored the SCCmec-SCCM1 element predominated in Irish hospitals during the 1990s but since 2002 have been recovered only sporadically. This may be due in part to a fitness cost associated with harboring such a large genetic element. While the presence of an SCC element in addition to an SCCmec element in MRSA can complicate SCCmec typing, the presence of SCC elements, such as SCCM1, in MSSA could also compromise the accuracy of rapid MRSA detection assays, such as the GeneXpert real-time PCR assay (49).
DNA microarray profiling allowed the detection of characteristic combinations of antimicrobial resistance and virulence genes among major MRSA clones. Only nine percent of isolates investigated lacked the bacteriophage-encoded IEC genes, and half of these belonged to ST22-MRSA-IV. Ninety-four percent of isolates carried one or more superantigen gene. The ST22-MRSA-IV isolates were the only isolates found to harbor sec and sel, which have been reported previously to be colocated on a pathogenicity island (32). The toxic shock toxin tst gene, which has also been found to be located on various pathogenicity islands (32), was identified in CC30 and CC5 isolates only. The enterotoxin genes sek and seq were identified only in the CC8 lineages ST239 and ST250 and have been reported to be colocated on various pathogenicity islands and bacteriophages, while seb, which has also been found to be located either on a pathogenicity island with sek and seq or on a plasmid, was also detected in ST250 isolates (32). Isolates belonging to ST22-MRSA-IV harbored the fewest antimicrobial resistance genes, with approximately half of the isolates harboring blaZ only. In contrast, isolates belonging to CC8 lineages (ST8-MRSA-IIA-E and ST239-MRSA-III/III-pI258/Tn554) harbored the greatest number of antimicrobial resistance genes (up to 17 in each case).
The present study revealed that, in most cases, the presence of a particular antimicrobial resistance gene correlated with phenotypic resistance to a specific type of antimicrobial agent(s), but absence of the gene was not always indicative of susceptibility. The latter was due in most cases either to the absence of additional genes encoding a particular resistance phenotype on the array (e.g., some trimethoprim-resistant isolates did not harbor the trimethoprim resistance gene dfrS1 that can be detected using the DNA microarray) or to the fact that resistance was due to the presence of mutations in a specific gene which the DNA microarray does not detect (e.g., fusA gene mutations resulting in fusidic acid resistance). This highlights the fact that array profiling cannot replace antimicrobial susceptibility testing but can be used in conjunction with it to help identify the mechanism of resistance. The inclusion of alternative antimicrobial resistance genes on the array, such as additional tetracycline [tet(L) and tet(O)] and trimethoprim (dfrG and dfrK) resistance genes, would enhance the ability of the array to predict a resistance phenotype.
The presence of an aminoglycoside resistance gene(s) detected by array profiling could be used to accurately predict an isolate's phenotypic aminoglycoside resistance pattern. However, the phenotypic expression of aminoglycoside resistance could not always be used to infer which aminoglycoside resistance gene(s) was present. This was partly because of the fact that different aminoglycoside resistance genes encode resistance to the same antimicrobial agents and also because approximately half of the aminoglycoside-resistant isolates (52%; 43/83) carried two or more aminoglycoside resistance genes. For example, all isolates found to harbor a single aminoglycoside resistance gene exhibited resistance to the expected combinations of aminoglycosides, and all isolates exhibiting these phenotypes harbored the corresponding resistance genes. However, for isolates harboring two or three aminoglycoside resistance genes, the phenotype could be predicted from the aminoglycoside resistance genes detected, but the presence of a gene could not be reliably inferred from the phenotype. An advantage of the use of the array was confirmation of reduced susceptibility to amikacin in isolates carrying the aacA-aphD gene (27). This gene encodes low-level amikacin resistance that may not be detected using the standard 30-μg susceptibility testing disk. Among 14 isolates carrying aacA-aphD only, nine showed reduced amikacin susceptibility; the remainder were phenotypically susceptible. Inclusion on the array of genes encoding resistance to the aminocyclitol spectinomycin, including spc (carried on Tn554) and aadA, would be helpful to further differentiate among this MRSA population (31).
In conclusion, the present study showed that DNA microarray profiling can accurately assign isolates representative of major pandemic MRSA clones to the correct MLST and SCCmec types. Microarray profiling allows rapid, high-throughput genotyping and detection of clinically relevant staphylococcal virulence and antimicrobial resistance genes, and can, in most cases, be used to predict the antimicrobial agent resistance phenotype of an isolate. The inclusion of additional antimicrobial resistance genes, including the recently described novel mecA gene (46), in future updated versions of the array would further enhance the usefulness of this system. The present study also provided further evidence of the diversity of SCCmec and SCC elements and the need for complete nucleotide sequencing to determine the genetic organization of these complex SCCmec elements and composite islands.
ACKNOWLEDGMENTS
This study was supported by the Microbiology Research Unit, Dublin Dental University Hospital.
We thank Grainne Brennan and the staff of the Irish National MRSA Reference Laboratory (NMRSARL) for antimicrobial susceptibility testing.
S. Monecke and R. Ehricht are both employees of Alere, the company that manufactures the StaphyType array kit.
Footnotes
Published ahead of print 6 August 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
Articles from Antimicrobial Agents and Chemotherapy are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aac.01247-12
Read article for free, from open access legal sources, via Unpaywall:
https://aac.asm.org/content/aac/56/10/5340.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/aac.01247-12
Article citations
Diversity of clonal complex 22 methicillin-resistant Staphylococcus aureus isolates in Kuwait hospitals.
Front Microbiol, 13:970924, 04 Aug 2022
Cited by: 4 articles | PMID: 35992657 | PMCID: PMC9386227
Characterizing a Novel Staphylococcal Cassette Chromosome mec with a Composite Structure from a Clinical Strain of Staphylococcus hominis, C34847.
Antimicrob Agents Chemother, 65(11):e0077721, 09 Aug 2021
Cited by: 2 articles | PMID: 34370581 | PMCID: PMC8522780
The global prevalence of fusidic acid resistance in clinical isolates of Staphylococcus aureus: a systematic review and meta-analysis.
Antimicrob Resist Infect Control, 10(1):75, 01 May 2021
Cited by: 20 articles | PMID: 33933162 | PMCID: PMC8088720
Review Free full text in Europe PMC
Epidemiological typing of methicillin resistant Staphylococcus aureus recovered from patients attending a maternity hospital in Ireland 2014-2019.
Infect Prev Pract, 3(1):100124, 24 Jan 2021
Cited by: 5 articles | PMID: 34368740 | PMCID: PMC8336322
Updating Molecular Diagnostics for Detecting Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Isolates in Blood Culture Bottles.
J Clin Microbiol, 57(11):e01195-19, 23 Oct 2019
Cited by: 18 articles | PMID: 31484703 | PMCID: PMC6813022
Go to all (21) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (4)
- (2 citations) ENA - HE858191
- (1 citation) ENA - C10682
- (1 citation) ENA - FJ390057
- (1 citation) ENA - HM030720
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NC_002745
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA.
Antimicrob Agents Chemother, 52(12):4407-4419, 13 Oct 2008
Cited by: 48 articles | PMID: 18852274 | PMCID: PMC2592854
Extensive genetic diversity identified among sporadic methicillin-resistant Staphylococcus aureus isolates recovered in Irish hospitals between 2000 and 2012.
Antimicrob Agents Chemother, 58(4):1907-1917, 06 Jan 2014
Cited by: 22 articles | PMID: 24395241 | PMCID: PMC4023797
Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals.
Antimicrob Agents Chemother, 57(1):524-531, 12 Nov 2012
Cited by: 42 articles | PMID: 23147725 | PMCID: PMC3535981
Comparative Genotypes, Staphylococcal Cassette Chromosome mec (SCCmec) Genes and Antimicrobial Resistance amongst Staphylococcus epidermidis and Staphylococcus haemolyticus Isolates from Infections in Humans and Companion Animals.
PLoS One, 10(9):e0138079, 17 Sep 2015
Cited by: 28 articles | PMID: 26379051 | PMCID: PMC4574763
 a,c
a,c 



