Abstract
Free full text

Expression Profiling Reveals an Unexpected Growth-Stimulating Effect of Surplus Iron on the Yeast Saccharomyces cerevisiae
Abstract
Iron homeostasis plays a crucial role in growth and division of cells in all kingdoms of life. Although yeast iron metabolism has been extensively studied, little is known about the molecular mechanism of response to surplus iron. In this study, expression profiling of Saccharomyces cerevisiae in the presence of surplus iron revealed a dual effect at 1 and 4 h. A cluster of stress-responsive genes was upregulated via activation of the stress-resistance transcription factor Msn4, which indicated the stress effect of surplus iron on yeast metabolism. Genes involved in aerobic metabolism and several anabolic pathways are also upregulated in iron-surplus conditions, which could significantly accelerate yeast growth. This dual effect suggested that surplus iron might participate in a more complex metabolic network, in addition to serving as a stress inducer. These findings contribute to our understanding of the global response of yeast to the fluctuating availability of iron in the environment.
INTRODUCTION
Iron is an essential micronutrient element found in almost all living organisms. It participates in many fundamental metabolic processes such as cellular respiration, iron-sulfur cluster biosynthesis, and the reactions of iron-containing enzymes (Andrews, 2008; Dunn et al., 2007). The mechanism of iron utilization and homeostasis has been elucidated in Saccharomyces cerevisiae (Kwak and Kosman, 2006). Fe3+ is first reduced to Fe2+ by two plasma membrane proteins, Fre1 and Fre2, then transported by the low-afffinity iron transporter Fet4 (apparent Km of 30 mM) in iron-repleted conditions, or by the multicopper oxidase Fet3, a high afffinity iron system (apparent Km of 0.15 mM) in iron-depleted conditions (De Freitas et al., 2000; Dix et al., 1997). S. cerevisiae uses two iron-responsive transcription factors, Aft1/2, to activate the transcription of a cluster of genes in the iron regulon that is implicated in iron-sulfur biosynthesis, heme utilization, and iron intracellular redistribution (Courel et al., 2005; Rutherford et al., 2003, Shakoury-Elizeh et al., 2004). Almost all studies on iron metabolism have used an iron concentration of 1 mM or lower (Kumanovics et al., 2008; Puig et al., 2008), while the yeast response to 1 mM or higher iron, designated an iron-surplus condition, has not yet been extensively studied. Surplus iron has long been recognized as producing free radicals in the presence of oxygen, and is thus cytotoxic to cells (Felice et al., 2005; Yamaguchi-Iwai et al., 1996). For instance, iron-mediated lipid peroxidation attacks mitochondria. This is responsible for many degenerative diseases, such as Friedreich’s ataxia and myohemoglobinuria (Almeida et al., 2006; Zager, 1996). To counteract unfavorable iron toxicity, yeast cells store excessive iron in specified organelles such as the vacuole. A recent study ascribed iron toxicity at high concentrations in yeast to the accumulation of cytosolic iron and indicated that this could be alleviated by several mitochondrial iron transporters that transport iron from the cytosol into mitochondria (Lin et al., 2011). An in-depth genome-wide analysis of the metabolic mechanism of yeast upon surplus iron has not yet been determined, however.
S. cerevisiae is an ideal eukaryotic organism to study global gene expression in response to a variety of environmental variations including oxidative stress, heavy metal ions and heat shock. Previously, yeast transcriptome analysis under surplus cobalt stress revealed that the iron regulon participates in cobalt tolerance (Stadler and Schweyen, 2002). In contrast, we observed that surplus iron can serve as a growth stimulator to significantly accelerate the growth of yeast cells. To systematically understand the cellular response to iron-surplus conditions, we determined the expression profiling of S. cerevisiae in surplus iron at 1 and 4 h. The data from the yeast genomic expression profile revealed a dual effect that is involved in the expression of stress-responsive and growth-stimulating genes. We further found that the stress-resistance transcription factor Msn4 might participate in the cellular response that counteracts the stress effects of surplus iron. Our study provides novel insights into understanding complex iron metabolism in yeast.
MATERIALS AND METHODS
Yeast strain preparation and determination of growth curve
S. cerevisiae wild-type strain S288C stock (genotype: MATα SUC2 mal mel gal2 CUP1 flo1 flo8-1 hap1) was inoculated in 200 ml yeast extract peptone dextrose Medium (YPD, 20 g tryptone, 10 g yeast extract, and 20 g glucose per liter) at 30°C overnight. The yeast of 500 ml at an OD600 nm of 0.1 was transferred to five flasks, with FeCl3 added to a final concentration of 0, 0.5, 1, 3, 5 mM (the background content of iron in YPD is about 25 μM). The optical density of each culture was monitored at 0, 1, 2, 3, 4, 5, 6, 7 and 8 h, respectively. Then, yeast cells were collected by centrifugation at 2,000 × g for 10 min at room temperature. The cell pellets were flash-frozen in liquid nitrogen and stored in a −80°C refrigerator for preparing RNA.
Extraction of RNA and synthesis of first-strand cDNA
The total RNA was extracted from the frozen cells using the Trizol reagent (Invitrogen, USA) according to the protocol provided by the manufacturer. Deoxyribonuclease I (Invitrogen, USA) was used to remove trace of genomic DNA. The first-strand cDNA was synthesized using M-MLV Reverse Transcriptase following the recommended protocols (Promega, USA). The RNA was subsequently digested using RNase H (MBI Fermentas, Canada).
Microarray construction and hybridization
The yeast genome 70-mer oligonucleotides microarrays were obtained from CapitalBio, and the hybridization was done by the CapitalBio Company (China) (Zhang et al., 2002). The microarrays were monitored on a GenePix scanner (Axon Instrument, USA). For each test and control sample, two hybridizations were performed by using a reverse fluorescence strategy, and 1.5-fold averaged over the two biological replicates and q-value ≤ 1% was set for differentially expressed genes.
Trascriptional analysis of MSN4/MSN2 under different concentration of iron-surplus condition
S. cerevisiae S288C was inoculated in 150 ml YPD medium at 30°C. The yeast at an OD600 nm of 0.1 was transferred into six flasks, with FeCl3 added to a final concentration of 0, 0.5, 1, 2, 5 mM respectively. The yeast cells were collected after being cultured for 4 h. The total RNA was extracted as described above. The amount of RNA was quantified by measuring the absorbance at 260 nm. Semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed as described previously (Frohloff et al., 2001). MSN4 primers used are (forward: 5′-CTCATAACAACAACAATGGTAAGGTTC-3′; reverse: 5′-GATGTTGTGATAAATTGTCACTTCTAC-3′); MSN2 primers used are (forward: 5′-GAAGGAAAGAAGGCCAAGTTACAG-3′; reverse: 5′-GTCTCCATGTTTTTTATGAGTCTTG-3′) and house-keeping gene β-ACTIN primers used are (forward: 5′-AAACCGCTGCTCAATCTTC-3′; reverse: 5′-CATTCTTTCGGCAATACCTG-3′). And in the quantitative RT-PCR (qRT-PCR), immunoQuantArray 96-well plates were prepared by the addition of 0.4 μl of 10 μM forward & reverse primers per well. To each well was then added 10 μl of SYBR Premix Ex Taq, 2 μl of cDNA and 6.8 μl of ddH2O. QRT-PCR data were collected on the ABI Prism 7,000 Sequence Detection System software using the default reaction conditions (1 cycle at 95°C for 5 min, 40 cycles at 95°C for 15 s and at 60ºC for 1 min). The values of threshold cycle (Ct) were exported for Global Pattern Recognition Algorithm (GPR) analysis (Akilesh et al., 2003; Livak et al., 2001), that is, ΔCtgene = Ctgene − Ctnormalizer, ΔΔCt = ΔCtgene − ΔCtcontrol, amount of target gene = 2−ΔΔCt.
Fluorescence microscopy using green fluorescence protein (GFP)-tagging method
The W303-lA strain (genotype: MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11, 15) were transformed with the plasmid pAdh1-Msn4-GFP and grown in SD-Ura medium (lacking uracil). Cells were cultured to the early exponential phase at OD600 nm of 0.6 before being treated with 5 mM Fe3+ for 5 min. Cells were harvested and fixed by adding 1 volume of 4% formaldehyde in phosphate-buffered saline (PBS). After a 15-min incubation at room temperature (RT), cells were washed with cold PBS, kept on ice for 5 min, and washed once more with cold PBS. Nuclei were stained by addition of 2 μg ml−1 of 4, 6-diamidino-2-phenylindole (DAPI) before microscopy observations using a Zeiss LSM710 META laser scanning confocal microscope.
RESULTS AND DISCUSSION
The growth-stimulating effect of surplus iron on S. cerevisiae
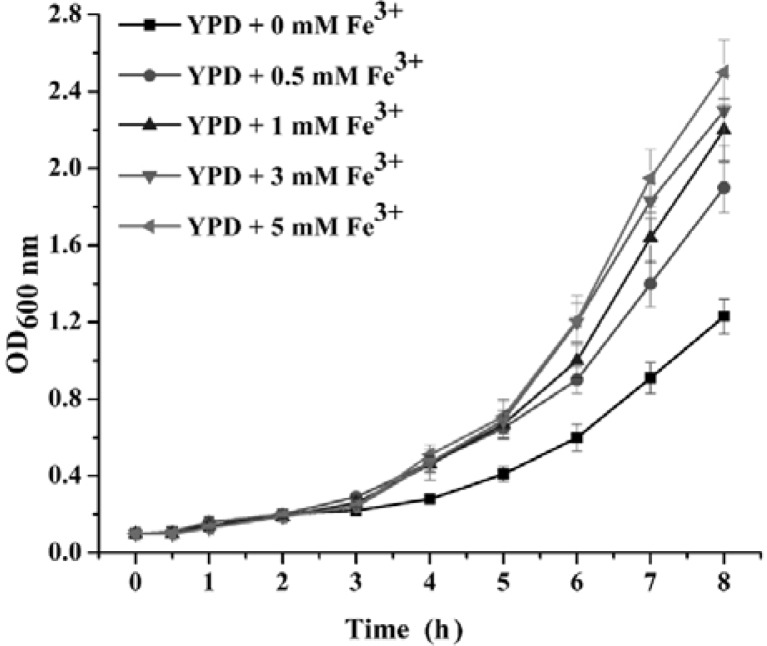
To investigate the effect of surplus iron on yeast metabolism, we first determined a growth curve for the wild-type strain S288C during logarithmic growth in YPD medium supplemented with 0.5, 1, 3, 5 mM Fe3+. Unexpectedly, yeast grew faster in iron-surplus medium than in the normal medium, that is, Fe3+ improved the growth of yeast in a dosage-dependent manner in a specific range of concentrations (Fig. 1). This result implied that surplus iron acted as an enhancer of cell growth, rather than a heavy metal stress. This unexpected finding prompted us to explore the molecular mechanism of the yeast response to surplus iron using DNA microarrays.
Transcriptional profiles of yeast in response to surplus iron
To compare the profiles of the yeast transcriptomic response to surplus iron, early logarithmic yeast S288C grown in liquid YPD medium were exposed to 5 mM Fe3+ for 1 or 4 h. The number of genes that were expressed differentially compared to the control at indicated time points (1 h and 4 h) is in Fig. 2A. Among the 5935 genes covered by the DNA microarray, the expression levels of 48 genes changed more than 2-fold and 135 genes showed a changed between 1.5- and 2-fold at 1 h, whereas 346 genes changed more than 2-fold and 487 genes showed a change between 1.5- and 2-fold at 4 h. The expression profiles indicated that the yeast response to surplus iron is multiphasic. Only 183 stress-related genes responded promptly by 1 h. At 4 h, 833 genes involved in metabolic pathways were expressed. Venn diagrams revealed that two genes were upregulated more than 2-fold, compared to 25 genes that were commonly downregulated by more than 2-fold both at 1 and 4 h (Fig. 2B). These downregulated genes were mainly related to iron transport and homeostasis, and consisted of 17 open reading frames (ORFs) at 1 h and 26 ORFs at 4 h (Supplementary Tables 1 and 2). For instance, the genes FIT1, FIT2, and FIT3 encode cell wall-anchored mannoproteins that can retain siderophore-iron in the cell wall. The high-affinity iron transporter (FET3 and FTR1) and low-affinity iron transporter (FET4) are involved in iron uptake through the plasma membrane. Several downregulated genes were also associated with iron metabolism, such as the iron-sulfur cluster synthesizing gene ISU2 and the ferric reductases encoded by genes FRE6/7. All these genes indicated that surplus iron downregulated the transcription of iron-related genes, thus maintaining the proper intracellular iron concentration.
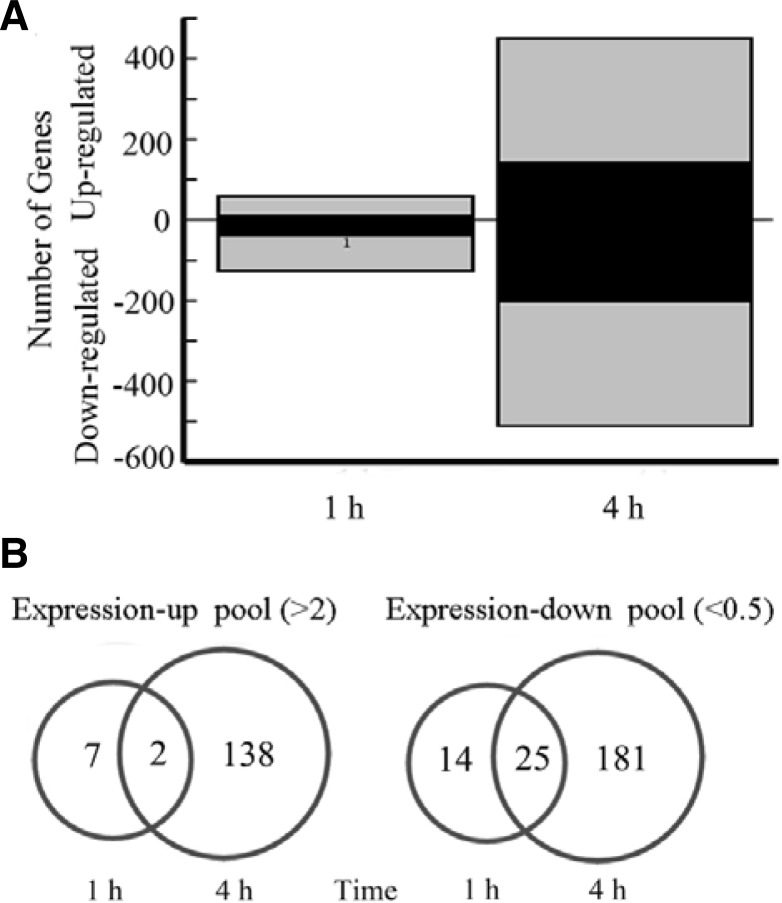
(A) Statistical comparisons of the transcriptomic responses upon 5 mM Fe3+. Black bars indicate genes that strongly responded (≥ 2.0-fold), whereas gray bars indicate genes that weakly responded (1.5~2.0-fold). (B) Venn diagrams of up-regulated genes and down-regulated genes of S. cerevisiae upon surplus iron in 1 h versus 4 h. The genes are shown here whose transcriptional level changed more than 2.0-fold.
When yeast cells were exposed to surplus iron for a considerable time such as 4 h, genes involved in the tricarboxylic acid cycle, electron transport, and oxidative phosphorylation were all upregulated, compared to the downregulated genes of anaerobic respiration. These data showed that surplus iron drove yeast cells to perform aerobic respiration and thus facilitated faster growth, concomitant with the upregulation of genes inamino acid/protein biosynthesis, assembly and modification, sugar/lipid biosynthesis, and cell wall formation (Supplementary Table 2). Furthermore, several stress-responsive genes were also upregulated in surplus iron at 4 h, such as HSP12, HOR7, LSP1, SOD2, GRX4, CCC1, GSY1, and HSP31. In particular, HSP12, HOR7, SOD2, CCC1 and HSP31 contain a stress response element (STRE) consensus sequence in their promoter regions, and all are target genes of the stress-resistant transcription factors Msn2/4 (Gauci et al., 2009; Martinez-Pastor et al., 1996). For instance, Hsp12 is a plasma membrane protein induced by stress conditions that probably initially perceives extracellular surplus iron. The vacuolar iron transporter Ccc1 that is induced by Msn2/4 facilitates the storage of surplus intracellular iron in the vacuole to alleviate high-iron toxicity. In addition to Msn2/4-dependent stress-responsive genes, many upregulated genes in other metabolic pathways are also targets of Msn2/4 (Table 1) (Gauci et al., 2009). Therefore, we further checked changes in the transcription of MSN2/4, with the result that MSN4 increased 1.5-fold and 1.75-fold under 5 mM of Fe3+ at 1 and 4 h, respectively, compared to the stable transcription of MSN2 in iron-surplus conditions.
Table 1.
A cluster of upregulated target genes of the stress-resistant transcription factor Msn4
| Gene | ORF | Induction fold | Number of STREsa | Gene description |
|---|---|---|---|---|
| HSP12 | YFL014W | 4.78 | 7 | Protects membranes from desiccation (response to stress) |
| HOR7 | YMR251W-A | 3.50 | 4 | Glycine dehydrogenase, induced under hyperosmotic stress |
| QCR8 | YJL166W | 3.22 | 1 | Subunit 8 of the ubiquinol cytochrome-c reductase complex |
| COX5B | YIL111W | 3.01 | 2 | Subunit Vb of cytochrome c oxidase |
| FMP10 | YER182W | 2.92 | 2 | Found in mitochondrial proteome |
| YFH1 | YDL120W | 2.74 | 1 | Regulates mitochondrial iron accumulation |
| MDH1 | YKL085W | 2.57 | 1 | Mitochondrial malate dehydrogenase |
| ATP7 | YKL016C | 2.37 | 1 | Subunit d of stator stalk of mitochondrial F1Fo ATP synthase |
| NFU1 | YKL040C | 2.33 | 1 | Protein involved in iron metabolism in mitochondria |
| PBI2 | YNL015W | 2.26 | 1 | Cytosolic inhibitor of vacuolar proteinase B |
| SOD2 | YHR008C | 2.18 | 1 | Superoxide dismutase, protects cells against oxygen toxicity |
| SHM2 | YLR058C | 2.17 | 1 | Cytosolic serine hydroxymethyltransferase |
| CCC1 | YLR220W | 2.03 | 2 | Fe2+/Mn2+ transporter preventing iron accumulation |
| HXK1 | YFR053C | 2.02 | 5 | Hexokinase isoenzyme 1 |
| MSC1 | YML128C | 2.02 | 2 | Mutants are defective in directing meiotic recombination event |
| GPH1 | YPR160W | 1.93 | 3 | Glycogen phosphorylase |
| HSP31 | YDR533C | 1.68 | 1 | Chaperone and cysteine protease |
| STF2 | YGR008C | 1.61 | 2 | Regulation of the mitochondrial F1Fo ATP synthase |
| NCE102 | YPR149W | 1.50 | 6 | Component of the glycolipid-enriched complexes |
To further confirm this finding, RT-PCR assays were performed (Figs. 3A and and3B).3B). The 2−ΔΔCt was calculated to indicate relative changes in gene expression. The results showed that transcription of MSN4 was significantly upregulated upon increased iron concentration in the medium, up by approximately 2-fold at 5 mM of Fe3+, using YPD medium as the background control.
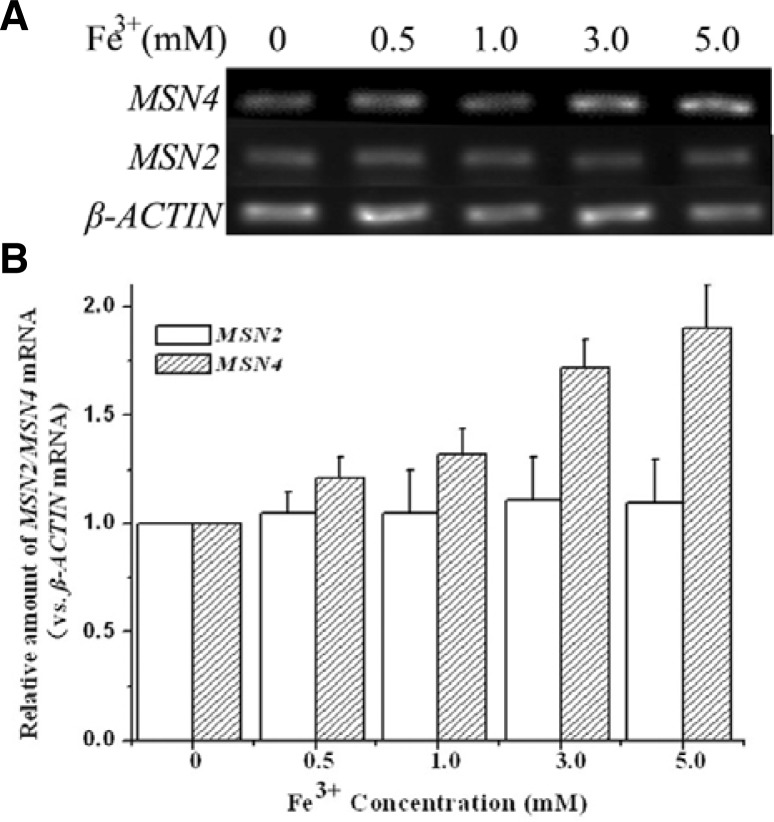
The relative amounts of mRNA of MSN2/MSN4 determined by RT-PCR (A) and qRT-PCR (B) upon different concentration of surplus iron. The transcription of MSN4 was increased dependent on iron concentration, while MSN2 was not. β-ACTIN (house-keeping gene) was as internal control. The results represent three independent assays with error bars.
Surplus iron serves as a stress inducer mediated by Msn4
As a global stress-resistant transcription factor, inactivated Msn4 is ubiquitously distributed in the cytoplasm. Upon stress, Msn4 is hyperphosphorylated and relocalized to the nucleus, then displays a periodic nucleo-cytoplasmic shuttling behavior. DNA binding of Msn4 may be controlled inside the nucleus or by the regulation of nuclear entry (Martinez-Pastor et al., 1996). Therefore, we selected Msn4 fused with a green fluorescent protein (GFP) tag, designated Msn4-GFP, to monitor its dynamic distribution in surplus iron conditions. W303-1A cells with pretransformed Msn4-GFP plasmid were treated with 5 mM Fe3+ and grown to early exponential phase to an OD600 nm of 0.6. After about 5 min, Msn4-GFP was accumulated in the nuclei in almost all cells (Fig. 4A), using DAPI staining as a control (Fig. 4B). The dynamic distribution of the nuclear localization of Msn4-GFP was also observed by real-time capture (Fig. 4C) to illustrate explicitly the stress effect of surplus iron on the yeast cells. Previous studies demonstrated that Msn4 is a transcription factor that is activated only in stress conditions (Martinez-Pastor et al., 1996). Our results further supported that surplus iron is a stress inducer that is probably mediated by Msn4.
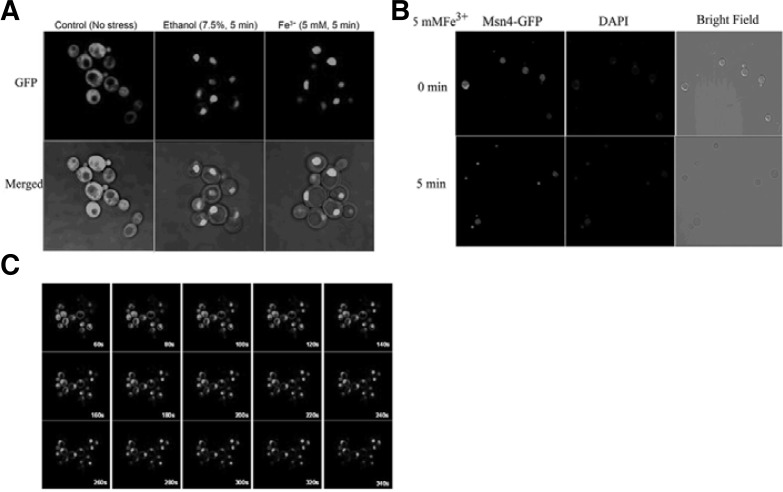
(A) Subcellular localization of Msn4 upon surplus iron. The fluorescence was monitored by GFP that is fused with Msn4. (B) DAPI staining control. (C) The dynamic distribution of Msn4 upon surplus iron captured at an interval of 20 s.
In summary, we used DNA microarrays to determine and analyze expression profiles of S. cerevisiae in surplus iron conditions. Here, we reported the dual effect of surplus iron on S. cerevisiae by yeast transcriptome analysis, and showed that surplus iron acted as a stress inducer (Fig. 5). Surplus iron induced the expression of stress-responsive genes including a considerable number of target genes of the stress-resistant transcription factor Msn4. Surplus iron also downregulated iron utilization related genes. The expression of these genes was an active response of yeast to cope with the stress effect of surplus iron. Surplus iron also induced the aerobic metabolism and several anabolic pathways to stimulate cell growth. However, the faster “cancer cell-like” growth rate compared to the normal growth of yeast could result in undue consumption of intracellular biomolecules and breakdown of metabolic homeostasis, which might be considered an abnormal state. Taken together, our study contributes to our understanding of the intricate iron metabolism of yeast to cope with the fluctuating availability of iron from the environment.
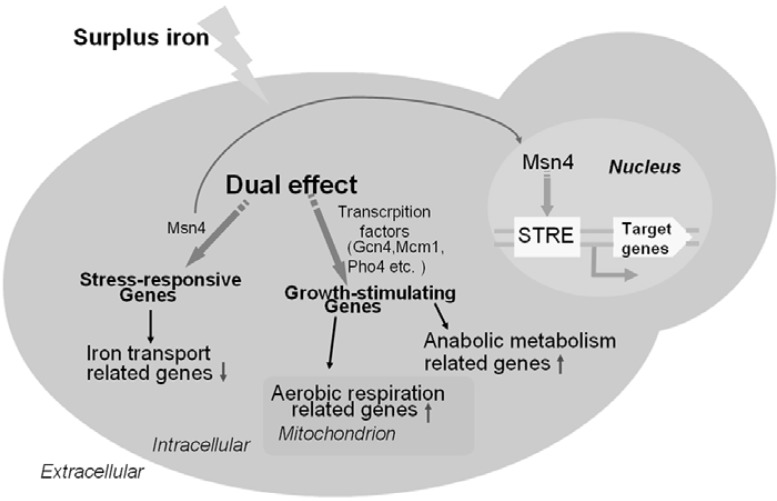
Schematic representation of the dual effect of responses in yeast upon surplus iron. Red down-arrow indicates the downregulated genes and Green up-arrow indicates the upregulated genes. Stress-responsive genes can be mediated by Msn4 and other unknown transcription factors; Growth-stimulating genes can also be mediated by certain transcription factors such as Gcn4 (regulating amino acid biosysthesis gene LEU1), Mcm1 (regulating cell mating gene STE3) and Pip2 (regulating lipid biosysthesis gene FAA1).
Acknowledgments
This work was supported by the 973 project from the Ministry of Science and Technology of China (Project 2012CB911002) and the Chinese National Natural Science Foundation (Grant 30870490). We also thank Prof. Shingo Izawa (Kyoto University) for providing the plasmid pAdh1-Msn4-GFP.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Akilesh S., Shaffer D.J., Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res. 2003;13:1719–1727. [Europe PMC free article] [Abstract] [Google Scholar]
- Almeida A.M., Bertoncini C.R., Borecky J., Souza-Pinto N.C., Vercesi A.E. Mitochondrial DNA damage associated with lipid peroxidation of the mitochondrial membrane induced by Fe2+-citrate. An Acad. Bras. Cienc. 2006;78:505–514. [Abstract] [Google Scholar]
- Andrews N.C. Forging a field: the golden age of iron biology. Blood. 2008;112:219–230. [Europe PMC free article] [Abstract] [Google Scholar]
- Courel M., Lallet S., Camadro J.M., Blaiseau P.L. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol. Cell. Biol. 2005;25:6760–6771. [Europe PMC free article] [Abstract] [Google Scholar]
- De Freitas J.M., Liba A., Meneghini R., Valentine J.S., Gralla E.B. Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. Role of oxidative stress in iron metabolism. J. Biol. Chem. 2000;275:11645–11649. [Abstract] [Google Scholar]
- Dix D., Bridgham J., Broderius M., Eide D. Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J. Biol. Chem. 1997;272:11770–11777. [Abstract] [Google Scholar]
- Dunn L.L., Rahmanto Y.S., Richardson D.R. Iron uptake and metabolism in the new millennium. Trends Cell Biol. 2007;17:93–100. [Abstract] [Google Scholar]
- Felice M.R., De Domenico I., Li L., Ward D.M., Bartok B., Musci G., Kaplan J. Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 2005;280:22181–22190. [Abstract] [Google Scholar]
- Frohloff F., Fichtner L., Jablonowski D., Breunig K.D., Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. [Europe PMC free article] [Abstract] [Google Scholar]
- Gauci V.J., Beckhouse A.G., Lyons V., Beh E.J., Rogers P.J., Dawes I.W., Higgins V.J. Zinc starvation induces a stress response in Saccharomyces cerevisiae that is mediated by the Msn2p and Msn4p transcriptional activators. FEMS Yeast Res. 2009;9:1187–1195. [Abstract] [Google Scholar]
- Kumánovics A., Chen O.S., Li L., Bagley D., Adkins E.M., Lin H., Dingra N.N., Outten C.E., Keller G., Winge D., et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 2008;283:10276–10286. [Europe PMC free article] [Abstract] [Google Scholar]
- Kwak E., Kosman D. Iron in yeast: Mechanisms involved in homeostasis. Top. Curr. Gene. 2006;14:59–99. [Google Scholar]
- Lin H., Li L., Jia X., Ward D.M., Kaplan J. Genetic and biochemical analysis of high iron toxicity in yeast: iron toxity is due to the accumulation of cytosolic iron and occurs under both aeroboc and anaerobic conditions. J. Biol. Chem. 2011;286:3851–3862. [Europe PMC free article] [Abstract] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. [Abstract] [Google Scholar]
- Martinez-Pastor M.T., Marchler G., Schuller C., Marchler-Bauer A., Ruis H., Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [Europe PMC free article] [Abstract] [Google Scholar]
- Puig S., Vergara S.V., Thiele D.J. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7:555–564. [Europe PMC free article] [Abstract] [Google Scholar]
- Rutherford J.C., Jaron S., Winge D.R. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 2003;278:27636–27643. [Abstract] [Google Scholar]
- Shakoury-Elizeh M., Tiedeman J., Rashford J., Ferea T., Demeter J., Garcia E., Rolfes R., Brown P.O., Botstein D., Philpott C.C. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:1233–1243. [Europe PMC free article] [Abstract] [Google Scholar]
- Stadler J.A., Schweyen R.J. The yeast iron regulon is induced upon cobalt stress and crucial for cobalt tolerance. J. Biol. Chem. 2002;277:39649–39654. [Abstract] [Google Scholar]
- Yamaguchi-Iwai Y., Stearman R., Dancis A., Klausner R.D. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [Europe PMC free article] [Abstract] [Google Scholar]
- Zager R.A. Mitochondrial free radical production induces lipid peroxidation during myohemoglobinuria. Kidney Int. 1996;49:741–751. [Abstract] [Google Scholar]
- Zhang L., Zhang Y., Zhou Y., An S., Cheng J. Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother. 2002;49:905–915. [Abstract] [Google Scholar]
Articles from Molecules and Cells are provided here courtesy of Korean Society for Molecular and Cellular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1007/s10059-012-2242-0
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3887821?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1007/s10059-012-2242-0
Article citations
Functional and Evolutionary Integration of a Fungal Gene With a Bacterial Operon.
Mol Biol Evol, 41(4):msae045, 01 Apr 2024
Cited by: 0 articles | PMID: 38415839 | PMCID: PMC11043216
Impact of silver ions and silver nanoparticles on biochemical parameters and antioxidant enzyme modulations in Saccharomyces cerevisiae under co-exposure to static magnetic field: a comparative investigation.
Int Microbiol, 27(4):953-966, 16 Nov 2023
Cited by: 0 articles | PMID: 37971657
Physiological Response of Saccharomyces cerevisiae to Silver Stress.
J Fungi (Basel), 8(5):539, 22 May 2022
Cited by: 0 articles | PMID: 35628793 | PMCID: PMC9145431
Structure and function of the vacuolar Ccc1/VIT1 family of iron transporters and its regulation in fungi.
Comput Struct Biotechnol J, 18:3712-3722, 23 Nov 2020
Cited by: 12 articles | PMID: 33304466 | PMCID: PMC7714665
Review Free full text in Europe PMC
Iron Regulatory Mechanisms in Saccharomyces cerevisiae.
Front Microbiol, 11:582830, 09 Sep 2020
Cited by: 36 articles | PMID: 33013818 | PMCID: PMC7509046
Review Free full text in Europe PMC
Go to all (9) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast.
J Biol Chem, 292(37):15577-15586, 31 Jul 2017
Cited by: 19 articles | PMID: 28760824 | PMCID: PMC5602413
Novel insights into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae.
BMC Genomics, 10:130, 25 Mar 2009
Cited by: 30 articles | PMID: 19321002 | PMCID: PMC2669097
Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae.
BMC Genomics, 11:660, 24 Nov 2010
Cited by: 95 articles | PMID: 21106074 | PMCID: PMC3091778
Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae.
World J Microbiol Biotechnol, 33(4):75, 17 Mar 2017
Cited by: 37 articles | PMID: 28315258
Review