Abstract
Free full text

Metabolic Brain Networks in Translational Neurology: Concepts and Applications
Abstract
Over the past 2 decades, functional imaging techniques have become commonplace in the study of brain disease. Nevertheless, very few validated analytical methods have been developed specifically to identify and measure systems-level abnormalities in living patients. Network approaches are particularly relevant for translational research in the neurodegenerative disorders, which often involve stereotyped abnormalities in brain organization. In recent years, spatial covariance mapping, a multivariate analytical tool applied mainly to metabolic images acquired in the resting state, has provided a useful means of objectively assessing brain disorders at the network level. By quantifying network activity in individual subjects on a scan-by-scan basis, this technique makes it possible to objectively assess disease progression and the response to treatment on a system-wide basis. To illustrate the utility of network imaging in neurological research, we review recent applications of this approach in the study of Parkinson disease and related movement disorders. Novel uses of the technique are discussed, including the prediction of cognitive responses to dopaminergic therapy, evaluation of the effects of placebo treatment on network activity, assessment of preclinical disease progression, and the use of automated pattern-based algorithms to enhance diagnostic accuracy.
Metabolic Network Mapping in Brain Disease
Functional imaging techniques to measure regional cerebral blood flow (CBF) and metabolism, as well as other indices of energy consumption, have been used for many years to map changes in local neural activity associated with brain disease. Regional abnormalities have typically been detected by whole brain voxel-by-voxel searches to identify significant signal differences in comparisons of patients and healthy subjects. However, this mass-univariate strategy is not designed to capture disease-related changes that occur at the systems level. This is especially relevant in the study of neurodegenerative disorders, which have been found to exhibit stereotyped connectivity changes involving discrete sets of brain regions.1 A number of multivariate strategies have been developed for systems-level analysis of functional imaging data, particularly from the resting state.2,3 Spatial covariance analysis has proven particularly useful in characterizing specific network abnormalities in metabolic imaging data from patients with neurodegenerative disease.4 A major feature of this general approach has been the ability to quantify network activity in individual subjects by a single numerical measure. It is this particular attribute that has allowed for the quantitative assessment of the network-level changes that accompany disease progression and the response to treatment. This review will highlight the recent advances that have been made using spatial covariance analysis in the study of the parkinsonian movement disorders as an example of its broader use in translational neurology. In recent years, this approach has additionally advanced the understanding of the systems-level changes that underlie other neurodegenerative disorders, including Alzheimer disease5,6 and Huntington disease,7,8 as well as brain diseases without localized histopathological changes such as Tourette syndrome9 and the dystonias.10 Furthermore, we will emphasize new insights derived by applying this approach to studies of brain metabolism conducted in the resting state. In this vein, we note that of late a variety of connectivity-based approaches have been used to examine multiregional relationships in patients with these diseases undergoing activation paradigms during brain imaging. 11 Progress in this general field of investigation has been reviewed elsewhere.12
The work that we discuss is based largely on a computational technique known as the Scaled Subprofile Model (SSM).3,4,13 The operational details of this principal component analysis (PCA)-based method have been presented elsewhere.14 (Software is freely available at http://www.fil.ion.ucl.ac.uk/spm/ext/.) This particular approach is noteworthy in that resting imaging data from patients and control subjects are merged and analyzed as a combined group. Under these circumstances, measures of cerebral function often exhibit skewed log-normal distributions, as is commonly encountered in biological systems, wherein individual values are small and non-negative, and possess relatively large variance.15 In this situation, variability in resting brain activity can be modeled as the multiplicative product of a large number of independent spatial elements. After removing the between-subject and between-region variability in the natural log-transformed imaging data, SSM/PCA yields residual values, albeit small, that contain relevant biological signals that are independent of the global mean. In accordance with the log-normal distribution inherent in the model, global brain activity is represented by the geometrical mean of the voxel-wise measurements, and is, therefore, invariant under commonly used ratio scaling methods.14 Importantly, the model does not depend on prior assumptions regarding the functional intercorrelations between brain regions. Indeed, the resulting spatial covariance patterns (ie, the metabolic networks) are entirely data-driven, reflecting the relative contributions of all voxels within the covariance network, and are not limited to isolated clusters (blobs).
An advantage of this analytical approach is that these patterns are invariant in prospective cohorts. Their scalar expression in individual subjects (ie, the subject scores, which quantify network activity in each scan) can be directly utilized to test the model,14 to quantify rates of disease progression,16,17 and to evaluate objectively potential treatment effects.18,19 The algorithm identifies linearly independent (orthogonal) sources of variability in the imaging data. The resulting spatial covariance patterns are considered to be disease-related if the associated subject scores discriminate patients from controls according to stringent prespecified criteria.14 Rigorous cross-validation procedures such as bootstrapping are typically performed to assess the stability of the extracted network topographies. Nonetheless, it is essential to understand that the relevance of a given candidate network is determined ultimately by independent replication in new populations. 4,20,21 The use of a particular covariance pattern as a biomarker can be supported by the presence of consistent correlations between quantifiers of its expression in individual patients and independent clinical, physiological, and/or genotypic descriptors of the disease process. 4,22 Even so, the network activity measure should be demonstrated to have a high degree of stability on test–retest evaluation23 before further use as a reliable disease biomarker. Lastly, although not essential, it is very useful to demonstrate that the candidate network exhibits high sensitivity and specificity for the disease in question, as can be determined by long-term clinical evaluation, postmortem confirmation, or both.24
Abnormal Metabolic Networks in Parkinson Disease
Parkinson Disease-Related Covariance Pattern
Parkinson disease (PD) is characterized by both motor and nonmotor clinical manifestations. Network analysis of resting state images of brain metabolism acquired using [18F]-fluorodeoxyglucose (FDG) positron emission tomography (PET) has led to the identification of several distinct spatial covariance patterns associated with the various clinical manifestations of the disorder. The prominent network abnormality seen in PD is characterized by increased pallidothalamic and pontine metabolic activity, associated with reductions in premotor cortex and in parietal association regions (Fig 1A). This PD-related metabolic pattern (PDRP) has been replicated in 7 previously reported populations of patients and healthy control subjects,4,20 and recently in 3 additional cohorts scanned using contemporary, commercially available PET devices (see Fig 1B). Of note, an abnormal spatial covariance topography homologous to the PDRP has been described in an experimental nonhuman primate model of parkinsonism.25 The spatial features of both the human and primate parkinsonism-related metabolic networks accord well with the distribution of synaptic changes that characterizes this movement disorder.26
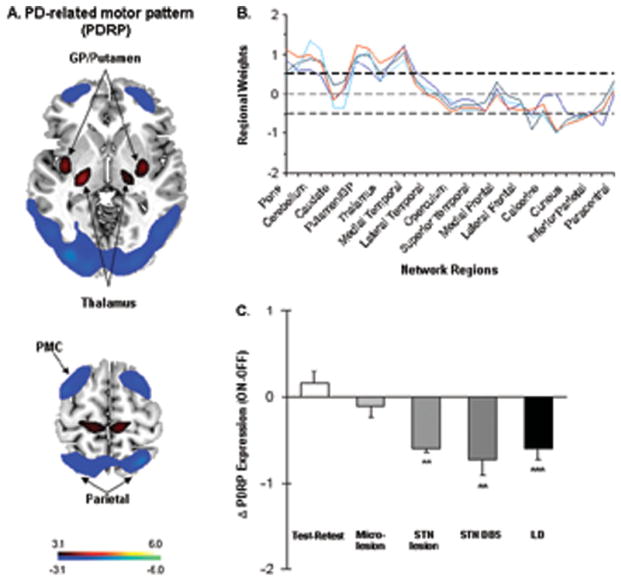
Parkinson disease (PD)-related spatial covariance pattern. (A) PD-related metabolic pattern (PDRP) identified by spatial covariance analysis of metabolic brain images from 20 PD patients and 20 age-matched healthy volunteer subjects scanned with [18F]-fluorodeoxyglucose (FDG) positron emission tomography (PET) using the GE Advance tomograph (4.0mm, full width at half-maximum [FWHM]) at The Feinstein Institute for Medical Research (Manhasset, NY). This pattern was characterized by increased metabolic activity (red) in the globus pallidus (GP)/putamen, thalamus, pons, cerebellum, and sensorimotor cortex, associated with relative reductions (blue) in the lateral premotor cortex (PMC) and parieto-occipital association regions.23 In this combined group, analyzed by principal component analysis, the PDRP was represented by the first principal component pattern (PC1, accounting for 19.5% of the subject × voxel variance), which constituted the largest effect in the data. The display of the voxel weights (ie, the regional loadings) on the resulting pattern were displayed at a reliability threshold of Z = 3, (p < 0.001; bootstrap estimation) and overlaid on T1-weighted magnetic resonance template images. (B) Region weights on PD-related spatial covariance patterns identified in 4 independent cohorts of patient and healthy control subjects scanned with FDG PET. Significant disease-related topographies from the different populations are depicted by colored lines connecting the loadings on 30 standardized regions of interest (ROIs) defined using an automated atlas.21 The PDRP gold standard pattern (A) is represented by a red line. Additional disease-related metabolic patterns were subsequently identified by spatial covariance analysis of data from separate groups of PD patients and control subjects scanned using a Siemens Biograph PET/CT camera (4.5mm FWHM) at Huashan Hospital (Shanghai, China), the GE Discovery PET/CT camera (5.2mm FWHM) at the Institute of Nuclear Medicine and Allied Sciences (New Delhi, India), and the Siemens HR+ PET camera (4.1mm FWHM) at Groningen University Hospital (Groningen, the Netherlands). These patterns are respectively depicted by green, light blue, and dark blue lines. Voxel weights on the PDRP exhibited a close correlation with corresponding regional values on the subsequent disease-related metabolic topographies (r ≥ 0.90, p < 0.001). Spatial covariance analysis was applied separately to combined group FDG PET data from the 3 independent validation samples, which were each comprised of approximately 20 PD patients and 20 age-matched healthy subjects. Disease-related metabolic patterns were identified in each cohort according to prespecified criteria provided elsewhere.14 In all 3 cohorts, the resulting PD covariance topography was represented by the largest effect in the data (PC1, accounting for between 16.0 and 20.9% of the subject × voxel variance). Region weights (y-axis) of absolute value ≥0.5 (dashed lines) denote ROIs in which local glucose metabolism contributed significantly to network activity (p < 0.025). (C) Treatment-mediated changes in PDRP expression (mean ± standard error) following stereotactic surgical interventions (shaded bars) targeting the subthalamic nucleus (STN): microlesion (n = 6),46 subthalamotomy (n = 6),43 and deep brain stimulation (DBS; n = 18).19 Changes in network expression during L-dopa (LD) administration (n = 18; solid bar)47 as well as the test–retest variability of this measure (n = 14; open bar)37 are depicted for comparison. Significant PDRP modulation was evident following subthalamotomy, STN DBS, and L-dopa treatment but not microlesion. **p < 0.01, ***p < 0.001 for the comparison of changes in PDRP expression with each intervention with those observed during test–retest evaluation, repeated measures analysis of variance. Adapted from Asanuma K, Tang C, Ma Y, et al. Network modulation in the treatment of Parkinson’s disease. Brain 2006;129:2667–2678, by permission of Oxford University Press; Pourfar M, Tang CC, Lin T, et al. Assess the microlesion effect of subthalamic deep brain stimulation surgery with FDG PET. J Neurosurg 2009;110:1278–1282, with permission from American Association of Neurological Surgeons; Mattis P, Tang CC, Ma Y, et al. Network correlates of the cognitive response to levodopa in Parkinson’s disease. Neurology 2011;77:858–865, with permission from Elsevier.
In the absence of medication, measurements of PDRP expression made in scans of CBF are closely intercorrelated with corresponding network values computed in scans of cerebral glucose metabolism from the same subjects.27,28 Thus, PDRP expression can be quantified in cerebral perfusion scans acquired using PET,23,27 single photon emission computed tomography,29,30 and more recently with magnetic resonance arterial spin labeling (ASL) techniques.31,32 A significant correlation has been noted between off-medication PDRP scores measured with ASL magnetic resonance imaging (MRI) and FDG PET in the same subjects.31 That said, perfusion imaging techniques may not be optimal for network quantification in the presence of antiparkinsonian medications with substantial vascular effects (see below). To date, there is comparatively limited experience in using resting state functional MRI (fMRI) scans to identify and validate PD-related covariance topographies. The blood oxygen level-dependent signal reflects complex interactions between local neuronal activity, oxygen extraction, and CBF and blood volume, and the interpretation of fMRI-based network topographies may, therefore, not be straightforward. Recent reports have been encouraging, suggesting that abnormal PD-related patterns are expressed in fMRI data, with spatial topographies similar to those described using PET methods.33,34
Clinical and Physiological Correlates of Network Activity
Prospective measurements of PDRP activity have revealed significant correlations with independent clinical severity ratings in multiple patient cohorts.4,30,35–37 In general, PDRP expression has been found to correlate with the akinetic–rigid manifestations of the disorder, rather than tremor.19,38,39 The unique functional concomitants of PDRP expression are highlighted by the association of network activity with measurements of subthalamic nucleus (STN) firing rate recorded intraoperatively during deep brain stimulation (DBS) surgery.22 On the whole, this network can be viewed as a representation of abnormal basal ganglia output, mediated largely through the STN and its efferent projections.40,41 Indeed, substantial reductions in PDRP activity have been noted following effective stereotaxic interventions targeting this structure for motor symptoms (see Fig 1C, shaded bars). Therapeutic STN lesioning (subthalamotomy) is associated with sustained PDRP reductions, and with concurrent normalization of baseline overactivity of the internal globus pallidus (GPi), ventrolateral thalamus, and dorsal pons.42 A similar degree of network suppression has been noted during STN DBS for advanced PD motor symptoms.37,43,44
Clinical outcome in individual patients has been found to relate closely to the degree of PDRP suppression observed during STN stimulation,37,43 and an analogous relationship has been noted following experimental STN adeno-associated viral vector glutamic acid decarboxylase gene therapy.18 Of note, a microlesion created by inserting the DBS electrode into the STN without electrical stimulation has been found to reduce metabolic activity at the target site, and remotely in the GPi and ventrolateral thalamus.45,46 Despite producing significant local metabolic changes in key PDRP regions, the STN microlesion did not substantially alter the activity of the network as a whole46 (see Fig 1C, second bar), nor did it produce lasting clinical benefit. These findings suggest that a critical threshold exists for significant network modulation to occur following treatment. Although isolated effects on regional brain function can be seen below this threshold, symptomatic improvement becomes evident only once a sufficient degree of change has occurred at the circuit level.
L-dopa–Induced Dyskinesia
Treatment-mediated suppression of PDRP activity has also been noted in response to L-dopa treatment (see Fig 1C, black bar).37,47 As with STN stimulation, the response of motor symptoms to L-dopa correlated with the degree of treatment-mediated network modulation that was observed.37,48 That said, side effects such as L-dopa–induced dyskinesia (LID) cannot be readily attributed to excessive suppression of PDRP activity by treatment. 37 Rather, the possibility is emerging that LID is associated with localized microvascular changes in network regions, potentiated in all likelihood by chronic L-dopa exposure.49–51 Consistent with this notion, we reported a marked dissociation between the effects of L-dopa on PDRP expression measured in scans of glucose utilization (reflecting treatment-mediated modulation of synaptic activity) and corresponding changes measured in scans of cerebral perfusion (reflecting hemodynamic responses to treatment) measured concurrently in the same patients.28 At a regional level, this phenomenon was most pronounced in the putamen/GPi and dorsal pontine nodes of the PDRP network. Similar findings have been recently described using quantitative autoradiography in an experimental rat model of LID.52 Importantly, in these regions, flow–metabolism dissociation during L-dopa treatment was substantially greater in PD patients with LID, with larger hemodynamic responses to L-dopa and higher on-medication CBF values than in their nondyskinetic counterparts. It is tempting to speculate that the specific regional changes observed with LID are a consequence of vascular endothelial growth factor upregulation51 giving rise to angiogenesis and abnormal blood–brain barrier permeability in susceptible network regions.28,49,52
Other PD-Related Metabolic Networks
Tremor-Related Covariance Pattern
Resting tremor is a common and important clinical feature of PD. The pathophysiology of tremor is thought to be different than that of bradykinesia or rigidity,53 with involvement of cerebellothalamocortical pathways in addition to corticostriatopallidothalamocortical motor circuits.54 This may explain why PDRP expression is not influenced by the presence or severity of coincident parkinsonian tremor.30,35,38,55 In a recent study, metabolic images from tremulous PD patients were acquired at baseline and following the suppression of tremor by stimulation of the ventral intermediate (Vim) thalamic nucleus.19 A novel spatial covariance mapping approach56 was applied to the paired on/off scan data from each patient to identify a network that was specifically related to parkinsonian tremor.19 The resulting PD tremor-related pattern (PDTP) was characterized by increased metabolic activity in the cerebellum and dorsal pons, primary motor cortex, and putamen (Fig 2A), brain regions known to be interconnected through the Vim thalamic nucleus.57,58 Prospective PDTP computations in an independent group of 41 PD patients revealed this network to be related specifically to the severity of parkinsonian tremor as opposed to akinetic rigidity.19 Tremor ratings correlated with PDTP expression and not with PDRP values measured in the same patients (see Fig 2B, top). Likewise, in this sample, PDTP expression was found to correlate with clinical ratings for tremor and not akinesia–rigidity (see Fig 2B, bottom).
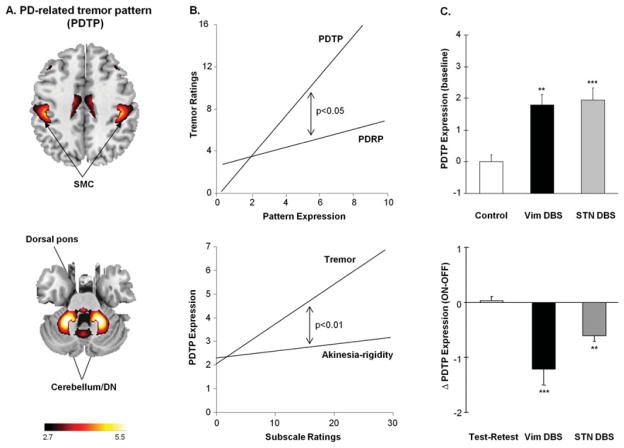
Parkinson disease (PD) tremor-related spatial covariance pattern. (A) PD tremor-related metabolic pattern (PDTP) identified by spatial covariance analysis of [18F]-fluorodeoxyglucose (FDG) positron emission tomography (PET) scans from 9 tremor-dominant PD patients scanned at baseline and during ventral intermediate (Vim) thalamic nucleus deep brain stimulation (DBS).19 This pattern was characterized by increased metabolic activity in the anterior cerebellum/dentate nucleus (DN), dorsal pons, primary sensorimotor cortex (SMC), and the caudate/putamen. The display of the covariance map was thresholded at Z = 2.70, p < 0.01 and overlaid on T1-weighted magnetic resonance template images. (B) Top: Unified Parkinson Disease Rating Scale (UPDRS) tremor ratings correlated with subject scores for the PDTP (r = 0.54, p < 0.001), but not the PD-related metabolic pattern (PDRP; r = 0.25, p = 0.16). The correlation between tremor and network activity was significantly greater for the PDTP values relative to PDRP (p < 0.05, multiple linear regression). Bottom: Although PDTP expression correlated with UPDRS tremor ratings, these values did not correlate with concurrent ratings for akinesia and rigidity from the same patients (r = 0.23, p = 0.15). Moreover, the correlation between PDTP expression and clinical disability was significantly greater for the tremor ratings relative to akinesia/rigidity (p < 0.01, multiple linear regression). (C) Top: PDTP expression (mean ± standard error [SE]) was elevated at baseline (off-stimulation) in PD patients treated with either Vim DBS (n = 9, black) or subthalamic nucleus (STN) DBS (n = 9, gray), compared with corresponding values from healthy control subjects (n = 20, white). There was a significant difference in PDTP expression across groups (p < 0.001, 1-way analysis of variance [ANOVA]), with comparable elevations in pattern expression in the 2 patient cohorts. **p < 0.005, ***p < 0.001, Student t tests relative to the healthy control subjects. Bottom: Treatment-mediated changes in PDTP expression (mean ± SE) in the Vim thalamic DBS (black), STN DBS (gray), and test–retest (white) patient groups. The degree of PDTP modulation differed across the 3 groups (p < 0.001; 1-way ANOVA), with both Vim thalamic and STN stimulation providing significant reductions in network activity (Vim DBS, ***p < 0.001; STN DBS, **p < 0.01).19 Of note, the change in PDTP expression during Vim thalamic DBS was larger than with STN stimulation (p < 0.05, post hoc test). Reprinted from Mure H, Hirano S, Tang CC, et al. Parkinson’s disease tremor-related metabolic network: characterization, progression, and treatment effects. Neuroimage 2011;54:1244–1253, with permission from Elsevier.
The functional difference between these 2 PD-related metabolic networks was further highlighted in a comparison of the effects of Vim thalamic and STN stimulation on network activity during tremor suppression. 19 PDTP expression measured at baseline (off-stimulation) was abnormally elevated in PD patients with implanted Vim thalamic or STN DBS electrodes (see Fig 2C, top). Whereas tremor was effectively abolished by stimulation at either target, akinesia and rigidity responded only to STN stimulation. Accordingly, abnormal baseline PDTP elevations were significantly reduced with either Vim thalamic or STN stimulation (see Fig 2C, bottom), although the magnitude of the network response was notably greater for the former intervention. Baseline PDRP elevations, conversely, were suppressed by STN DBS but not by Vim thalamic stimulation.19 These findings, and the significantly different longitudinal progression rates observed for PDRP and PDTP values,19 support the view that these networks are functionally independent, both as disease biomarkers and as systems-level targets of intervention.
PD Cognition-Related Metabolic Brain Network
PD is clinically defined by its motor features, but nonmotor symptoms are common and may begin early in the disease process.59 Spatial covariance analysis has been particularly useful in providing a network-level understanding of the changes in brain function that underlie the cognitive features of this disorder. Cortical metabolic reductions are often evident in cognitively intact PD patients, even before the initiation of antiparkinsonian therapy.60–62 Network analysis has revealed a specific metabolic topography associated with cognitive functioning in nondemented PD patients. A significant spatial covariance pattern has been identified with reproducible correlations between individual subject expression and performance on tests of learning and memory.47,60,63 This PD-related cognitive pattern (PDCP) is characterized by metabolic reductions in the medial prefrontal, premotor, and parietal association regions, associated with relative increases in the cerebellar vermis and dentate nuclei (Fig 3A). Importantly, the results of both longitudinal and cross-sectional analysis of metabolic data from PD patients indicate that PDCP expression progresses at a slower rate than PDRP,4,16 reaching abnormal levels 4 to 6 years after symptom onset. Indeed, the longitudinal data suggest that significant regional abnormalities occur in frontal and parietal PDCP areas before the appearance of cognition-related network changes in these patients.17,60 These findings accord with the notion discussed above that clinical manifestations are linked to the appearance of significant abnormalities at the network level, as opposed to isolated changes in regional brain function. The role of specific neurotransmitters such as acetylcholine and serotonin in modulating PDCP expression is currently being examined. A number of studies have suggested that cognitive functioning in early PD is influenced by changes in the integrity of dopaminergic afferents to the caudate nucleus, which can vary considerably across patients.64–66 Recent evidence suggests that increases in PDCP expression are associated with loss of dopaminergic input to this structure and not the putamen. 67 That said, the magnitude of this correlation is relatively modest, suggesting that other factors are involved in determining baseline PDCP levels.
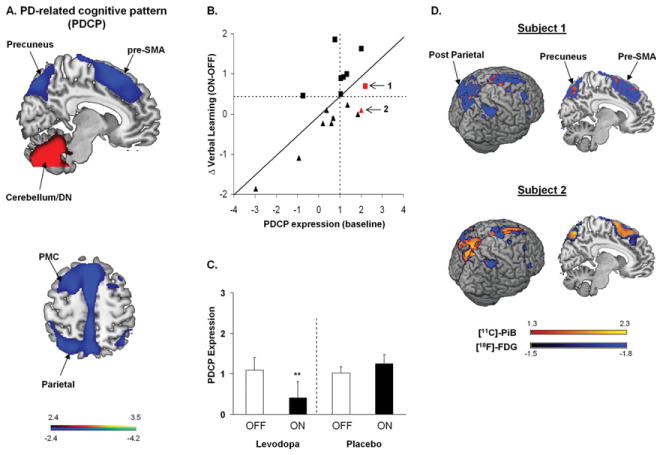
Parkinson disease (PD) related cognitive pattern (PDCP). (A) Identified by spatial covariance analysis of [18F]-fluorodeoxyglucose (FDG) positron emission tomography (PET) scans from 15 nondemented PD patients with mild to moderate motor symptoms. This pattern was characterized by reduced metabolic activity (blue) in the rostral supplementary motor area (pre-SMA), precuneus, and posterior parietal and prefrontal regions, with relative increases (red) in the cerebellar/dentate nucleus (DN).63 The display of the covariance map was thresholded at Z = 2.44, p < 0.01 and overlaid on T1-weighted magnetic resonance template images. PMC = premotor cortex. Reprinted from Huang C, Mattis P, Tang C, et al. Metabolic brain networks associated with cognitive function in Parkinson’s disease. Neuroimage 2007;34:714–723, with permission from Elsevier. (B) Relationship between baseline PDCP expression and L-dopa–mediated changes in verbal learning performance. Higher baseline PDCP scores correlated with greater improvement in cognitive functioning during L-dopa treatment (r = 0.70, p < 0.005; n = 17). Patients with meaningful improvement in verbal learning performance during treatment (defined by an independently determined Reliable Change Index [RCI]82 for this psychometric measure47) are depicted by squares; those without meaningful change are depicted by triangles. The horizontal dashed line represents the RCI cutoff of 0.44 for verbal learning test performance 47; the vertical dashed line represents the estimated minimal baseline network expression value of 1.01 associated with a meaningful cognitive response to medication. The red symbols denote 2 patients who in addition to FDG PET, underwent PET imaging with [N-methyl-11C]2-(4′-methylaminophenyl)-6-hydroxybenzothiazole (Pittsburgh compound B [PiB]) for the assessment of cortical protein aggregates. (C) Treatment-mediated changes in PDCP expression differed for the 8 PD patients (left) who exceeded the RCI cutoff for cognitive response to L-dopa as compared to 7 others (right) who exhibited a cognitive response of similar magnitude to placebo treatment (p = 0.02; 2 × 2 repeated measures analysis of variance).47 Significant PDCP modulation was observed in cognitive responders to L-dopa (**p < 0.01, post hoc test) but not to placebo (p = 0.38). B, C reprinted from Mattis P, Tang CC, Ma Y, et al. Network correlates of the cognitive response to levodopa in Parkinson’s disease. Neurology 2011;77:858–865, with permission from Elsevier. (D) Subject 1, with baseline PDCP expression of 2.19, was a cognitive responder to L-dopa by RCI criteria (B, arrow). This patient exhibited normal levels of [11C]-PiB binding (1.09, specific uptake ratio relative to the cerebellum; normal: 1.09 ± 0.10) in cortical areas with significantly low metabolic activity. By contrast, Subject 2, with baseline PDCP expression of 2.01, was a cognitive nonresponder to treatment (B, arrow). This patient exhibited elevated radioligand binding (1.43) in these hypometabolic regions. Thus, despite baseline PDCP elevations of similar magnitude in the 2 patients, Subject 2 additionally exhibited abnormal levels of protein aggregate binding in key network areas (see text). Maps of [11C]-PiB binding (yellow-red) from the 2 subjects were overlaid on a statistical parametric map of abnormal metabolic reductions (blue) identified by voxel-wise comparison of FDG PET scans from 14 PD patients and 15 age-matched healthy volunteer subjects (p < 0.005, uncorrected). [11C]-PiB binding in hypometabolic cortical areas was quantified in the 2 patients and compared with reference values measured in the corresponding scans from the healthy control cohort.
The clinical relevance of the PDCP is highlighted in a recent study of the cognitive response to L-dopa for the treatment of PD motor symptoms.47 In contrast to the PDRP, which is consistently modulated by effective antiparkinsonian interventions,37 PDCP expression is not altered by the treatment of motor symptoms—at least not at the group level.37,63 That said, the cognitive response to dopaminergic therapy may be influenced by baseline (ie, off-medication) functioning.68 Consistent with this observation, L-dopa–mediated improvement in performance on the California Verbal Learning Test, a neuropsychological test of verbal learning and memory, was found to correlate with baseline PDCP expression (see Fig 3B). During L-dopa treatment, neuropsychological test performance improved in nondemented PD patients with elevated baseline network activity, with consequent normalization of these values (see Fig 3C, left). By contrast, L-dopa did not improve test performance in patients with normal baseline network activity, nor did PDCP expression change during treatment.
Of note, PDCP modulation may not be a universal concomitant of the cognitive treatment response. This is demonstrated by the absence of significant changes in PDCP activity in a group of PD patients with improved cognitive performance in response to placebo (see Fig 3C, right).47 This suggests that brain networks other than the PDCP may be deployed in response to placebo treatment to achieve a cognitive outcome of comparable degree to that observed with dopaminergic pharmacotherapy. Moreover, PET imaging with [N-methyl-11C]2-(4′-methylaminophenyl)-6-hydroxybenzothiazole (Pittsburgh compound B) to measure the deposition of fibrillar protein aggregates in the brain may enhance the accuracy of network-based predictions of the cognitive response to antiparkinsonian therapy. Specifically, preliminary data (D.E., personal communication) suggest that treatment-mediated PDCP modulation, and concomitant improvement in cognitive performance, does not always occur in patients with elevated baseline network expression (see Fig 3B, arrows). In such cases, network modulation during treatment may be limited by strategically positioned deposition of protein aggregates in key network regions (see Fig 3D).
Network Progression in Preclinical PD
As representations of the stereotyped changes in functional connectivity that underlie many of the neurodegenerative disorders, characteristic spatial topographies like that of the PDRP remain essentially unchanged with advancing disease.14 This contrasts with the results of univariate analysis of longitudinal imaging data in which regional differences vary considerably over time.5,16,17 In this regard, the continuous increases in network activity observed in early PD may reflect compensatory responses of the brain at the synaptic level.69,70 Alternatively, these changes may reflect systems-level abnormalities stemming from the neurodegenerative process itself.71 These 2 possibilities, however, are not mutually exclusive, and both can potentially coexist at early disease stages.
A recent analysis of progression data from unilaterally affected (ie, hemiparkinsonian) PD patients has provided some insight into this issue. In that study, network activity was quantified in the cerebral hemispheres ipsilateral and contralateral to the initially involved body side in scans acquired at baseline (within 2 years of diagnosis), and again after 2 and 4 years of follow-up.17 Contrary to expectation, PDRP expression was significantly elevated in both hemispheres at all time points (Fig 4A). Moreover, network activity in the preclinical hemisphere (ie, on the side opposite the initially unaffected presymptomatic body side) was virtually identical in magnitude to that measured in the contralateral side (ie, in the hemisphere opposite the initially affected limbs). Of note, extrapolation of the longitudinal PDRP data supported the existence of a discrete interval of 3 to 5 years over which the network abnormality developed prior to clinical onset.17,71 Although this observation is consistent with a compensatory mechanism, data from cross-sectional patient cohorts suggest that PDRP increases continuously with advancing disease for at least 15 years after symptom onset.4,14,71
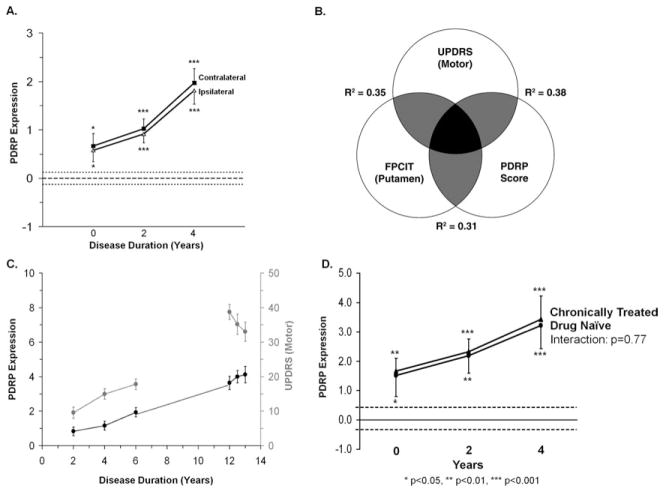
Changes in Parkinson disease (PD)-related metabolic pattern (PDRP) activity with disease progression. (A) Time course of PDRP expression in the contralateral (squares) and ipsilateral (triangles) hemispheres of 15 early stage hemiparkinsonian patients scanned at baseline, and 2 and 4 years.17 On both sides, hemispheric PDRP expression was found to be abnormally elevated at each time point. Network activity increased linearly over time (p < 0.001, repeated measures analysis of variance [RMANOVA]), rising in parallel on both sides. Broken lines denote the mean value (±1 standard error) for PDRP expression measured in 15 age-matched healthy control subjects. *p < 0.05, ***p < 0.001, Student t test comparisons of hemispheric values in patients relative to control subjects. Reprinted from Tang C, Poston K, Dhawan V, et al. Abnormalities in metabolic network activity precede the onset of motor symptoms in Parkinson’s disease. J Neurosci 2010;30:1049–1056, with permission from Society for Neuroscience. (B) Schematic showing significant correlations (p < 0.01) between changes in Unified Parkinson Disease Rating Scale (UPDRS) motor ratings, PDRP network activity, and striatal dopamine transporter binding during the progression of early stage PD. The gray areas indicate overlap between pairs of measures, represented by the strength (R2) of their within-subject correlations.16 The black area indicates the commonality (interaction effect) of the 3 measures. FPCIT=18F-FPCIT PET Reprinted from Eckert T, Tang C, Eidelberg D. Assessment of the progression of Parkinson’s disease: a metabolic network approach. Lancet Neurol 2007;6:926–932, with permission from Elsevier. (C) Time course of PDRP expression in 23 advanced PD patients (black line, right) randomized to sham surgery and followed for 1 year as part of a blinded clinical trial of subthalamic nucleus gene therapy.76 A significant linear increase in whole-brain PDRP expression over time was observed in this group (p < 0.001, RMANOVA), which was in continuity (broken line) with network measurements (black line, left) obtained in a separate longitudinal study of early stage PD patients.16 By contrast, UPDRS motor ratings (gray line, right) declined in the sham-operated patient group (p < 0.001, RMANOVA), compatible with placebo effect, whereas this measure increased in the longitudinal cohort of early PD patients (gray line, left). The left and right y-axes denote PDRP expression and UPDRS motor ratings, respectively. The x-axis denotes disease duration. Adapted from Tang C, Poston K, Dhawan V, et al. Abnormalities in metabolic network activity precede the onset of motor symptoms in Parkinson’s disease. J Neurosci 2010;30:1049–1056, with permission from Society for Neuroscience. (D) Longitudinal changes in PDRP expression measured in 15 early stage PD patients.16 In this study, 8 of the subjects (circles) were drug-naive at baseline but were receiving chronic oral L-dopa/carbidopa by the 2-year time point. The remaining 7 subjects (triangles) were chronically treated with L-dopa/carbidopa for at least 3 months before the initial time point. PDRP expression for the 2 groups was similar at baseline (p = 0.86) and at the subsequent 2 time points (p > 0.85). Importantly, the rate of network progression was similar for de novo and chronically treated subjects, indicating that the estimates were not altered by the introduction of symptomatic therapy midstudy (see text).
The symmetry of the metabolic network changes in early PD contrast with the asymmetric loss of putamen dopaminergic innervations and the lateralized clinical manifestations observed in these patients.17 This may be understood in light of the finding that metabolic activity in the regions with the greatest local contributions to the PDRP topography (ie, those with the highest voxel weights on the pattern: the globus pallidus, ventral thalamus, and dorsal pons) exhibited minimal side-to-side differences during disease progression.16 In other words, PDRP expression is influenced most by symmetrical disease- related changes occurring downstream from the site of the dopaminergic pathology. Although significant correlations have been noted between whole-brain PDRP expression, Unified Parkinson Disease Rating Scale motor ratings, and putamen dopamine transporter binding (see Fig 4B), these relationships are of moderate size, accounting for approximately only a third of the overall variance in each pair of measurements. These data suggest that the PDRP represents a specific feature of the neurodegenerative process that is largely independent of concurrent changes in clinical ratings and imaging measures of nigrostriatal dopaminergic function.
The demonstration that PDRP network progression is a linear process that begins preclinically and spans the period of symptom onset is critical if this measure is to be used as a biomarker in disease modification trials. The data from 2 separate longitudinal PD studies indicate that network progression proceeds at a constant rate for at least a decade after the start of motor symptoms, without a discernible placebo effect (see Fig 4C). Moreover, the effects of symptomatic dopaminergic therapy on network activity are relatively small in comparison with the effects of disease progression. Systematic effects of L-dopa treatment on the estimated PDRP progression rate are minimized by scanning patients 12 hours after the last medication dose (as is routinely done in our laboratory). Under these conditions, we found no difference in the rate of network progression computed in chronically medicated early stage patients relative to their age- and duration-matched counterparts who were initially drug-naive and started L-dopa treatment 1 to 2 years later (see Fig 4D). Based on the longitudinal data from our early stage cohort,16 we estimate that for a placebo-controlled clinical trial of a disease-modifying agent, approximately 56 early stage PD patients would need to be randomized to detect a 20% change in the PDRP progression rate with 80% power (α = 0.05) over 2 years. Analogous calculations can be performed using the PDCP as a potential biomarker in clinical trials directed at the cognitive manifestations of the disease.
Metabolic Brain Networks: Role in Differential Diagnosis
In recent years, there has been a growing interest in optimizing the design of clinical trials of new therapies for PD and other neurodegenerative disorders. This, in turn, has motivated efforts to improve the accuracy of diagnosis in the selection of suitable trial participants. Concerns regarding potential misdiagnosis are particularly relevant in the assessment of new interventions directed at patients with early stage disease, in whom clinical ascertainment may be challenging.72 To determine whether improved accuracy of assignment can be achieved using network methods, we studied 167 patients with parkinsonism who were referred for FDG PET because of uncertain clinical diagnosis.24 After imaging, all patients were followed for an average of 2.6 years before a final clinical diagnosis was made by a movement disorders specialist blinded to the imaging findings. Likelihoods of idiopathic PD, as well as multiple system atrophy (MSA), and progressive supranuclear palsy (PSP), 2 causes of progressive parkinsonism that are frequently misdiagnosed as PD,73 were determined for each case using computed subject scores for the corresponding disease- related patterns (Fig 5A).74,75 Logistical regression analysis was used to classify each scan based upon the computed probabilities of disease conditions.24 The resulting group assignments were compared with the final clinical diagnosis using receiver operating characteristic analysis. Indeed, the pattern-based classifications proved to be very accurate, with excellent positive predictive value (PPV) for discriminating PD from atypical parkinsonism (PPV ≥ 97%) and for differentiating MSA from PSP (PPV ≥ 91%). Importantly, both PPV and diagnostic specificity remained high (≥92%) when assessed in 55 (33%) patients in this cohort with short (≤2 years) symptom duration. The utility of this pattern-based classification approach has recently been substantiated in an independent FDG PET study of 134 patients of clinically indeterminate parkinsonism, of whom 86 (64%) had symptoms for no more than 2 years. Using the same algorithm employed by Tang and colleagues, the investigators (Dr. M. Tripathi, personal communication) accurately discriminated PD from atypical parkinsonism (PPV ≥ 97%, specificity ≥ 97%), even in short-duration cases (see Fig 5B).
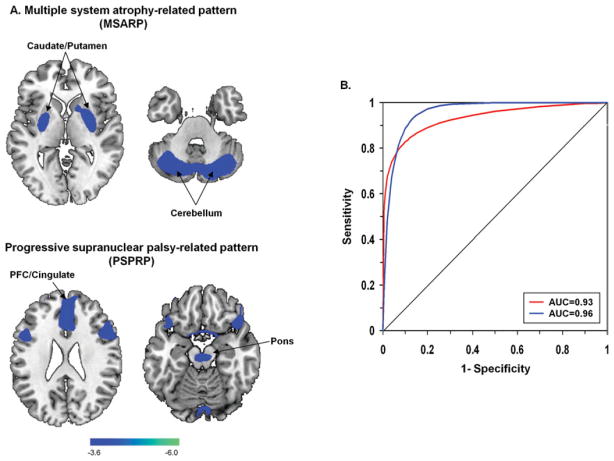
Disease-related spatial covariance patterns for multiple system atrophy (MSA) and progressive supranuclear palsy (PSP). (A) Top: Multiple system atrophy-related metabolic pattern (MSARP) identified by spatial covariance analysis of [18F]-fluorodeoxyglucose (FDG) positron emission tomography (PET) scans from 10 MSA patients and 10 healthy volunteer subjects.74,81 This pattern was characterized by reduced metabolic activity (blue) in the putamen and the cerebellum. Bottom: Progressive supranuclear palsy-related metabolic pattern (PSPRP), identified by spatial covariance analysis of FDG PET scans from 10 PSP patients and 10 healthy volunteer subjects, was characterized by reduced metabolic activity (blue) in the medial prefrontal cortex (PFC), the frontal eye fields, the ventrolateral prefrontal cortex, the caudate nuclei, the medial thalamus, and the upper brainstem.74 The display of the MSARP and PSPRP covariance maps was thresholded at Z = 3.61, p < 0.001 and overlaid on T1-weighted magnetic resonance template images. Reprinted from Eckert T, Tang C, Ma Y, et al. Abnormal metabolic networks in atypical parkinsonism. Mov Disord 2008;23:727–733, with permission from John Wiley & Sons. (B) Receiver operating characteristic (ROC) curves showing accurate network-based classification of FDG PET scans from patients with parkinsonian symptoms of short duration (≤2 years) and indeterminate clinical diagnosis (see text). An ROC curve (red) from a cohort comprised of 55 short-duration patients24 disclosed accurate classification of the individual scans (area under the curve [AUC] = 0.93, p < 0.001; 95% confidence interval [CI], 0.86–0.99). Of these individuals, 30 were subsequently diagnosed clinically as having classical Parkinson disease (PD); the remaining 25 were diagnosed with either MSA (n = 11) or PSP (n = 14). The validity of the approach is supported by findings from a separate group of 86 patients with clinically indeterminate parkinsonism of short duration (≤2 years; M. Tripathi, personal communication). Members of this testing group were individually classified based on their FDG PET scans according to the same diagnostic algorithm in the first group. An ROC curve (blue) based on these data disclosed a similar degree of diagnostic accuracy (AUC = 0.96, p < 0.001; 95% CI, 0.91–0.99) for the second group. Of these patients, 47 were subsequently diagnosed as having PD; the remaining 39 patients were found to have either MSA (n = 18) or PSP (n = 21).
This network-based classification algorithm was recently used to screen participants in a phase II trial of STN gene therapy for advanced PD,76 and in the assessment of patients referred for more routine antiparkinsonian interventions such as STN DBS.77 Similar algorithms may prove to have utility in studies of prodromal parkinsonism. Rapid eye movement behavior disorder (RBD) has been found to presage the development of parkinsonism in approximately 60% of subjects followed for 5 years or longer.78 Indeed, several imaging studies have demonstrated presynaptic nigrostriatal dopaminergic dysfunction in RBD patients.79,80 Although this disorder can be viewed as a prodromal form of PD, it can also evolve into other synucleinopathies such as MSA and diffuse Lewy body disease. Network classification procedures may make it possible to differentiate among these diagnostic possibilities even before the onset of motor symptoms. For example, 2 RBD patients without clinical signs of parkinsonism underwent metabolic imaging, and PDRP and MSA-related metabolic pattern expression values were separately quantified in both cases (Fig 6A, B). Qualitative review of the individual case data indicated that the first RBD patient exhibited higher PDRP expression and lower MSA-related metabolic pattern expression than the second. Graphical display of the subject scores for the 2 patterns in each of these subjects and in the individual data from the independent PD and MSA reference groups81 (see Fig 6C) illustrates the utility of network- based classification in this circumstance. There was a clear difference between the 2 RBD subjects, with the former sorting with PD and the latter with MSA. Needless to say, the validity of image-based assignments in individuals with prodromal disease can only be determined following long-term prospective evaluation of the subjects until distinguishing clinical features emerge.
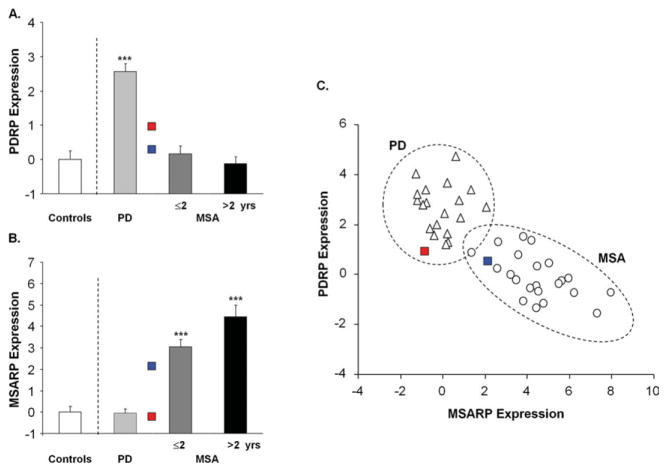
Network activity in presymptomatic patients with rapid eye movement (REM) sleep behavior disorder. (A) Parkinson disease (PD)-related metabolic pattern (PDRP) expression measured in 2 REM-sleep behavior disorder (RBD) patients without clinical signs of parkinsonism. These values were compared with corresponding measurements81 (mean ± standard error [SE]) from 20 PD patients (light gray) and 22 multiple system atrophy (MSA) patients (8 (dark gray) with short (≤2 year) symptom duration and 14 (black) with longer (>2 year) symptom duration). PDRP values were abnormally elevated in the PD patients (***p < 0.001, Student t test comparison with values [white] from 20 age-matched healthy control subjects), but not in their MSA counterparts. PDRP expression was higher in the first RBD subject (red square) than in the second (blue square). (B) MSA-related metabolic pattern (MSARP) expression in the 2 RBD patients compared with corresponding network values (mean ± SE) in the PD, MSA, and healthy control groups (see above). MSARP expression was abnormally elevated in the MSA patients (***p < 0.001, Student t test comparison with healthy control values), but not in their PD counterparts. MSARP expression was higher in the second RBD than in the first. (C) Bivariate scatter plot depicting individual PDRP and MSARP expression values for the PD (triangles) and MSA (circles) subjects; the 2 RBD patients are represented by red and blue squares (see above). Based on subject scores for the 2 patterns, the first RBD patient is seen to cluster with the PD group (dotted circle), whereas the second clustered with the MSA group (dotted ellipse). Of note, both RBD subjects had network values at the low end of the range for their assigned categories, consistent with their presymptomatic status (see text).
Summary
Network analysis methods can be used to identify specific spatial covariance patterns associated with neurodegenerative diseases. These patterns likely reflect interconnected functional changes in multiple brain regions, relating to the underlying pathology, and potentially to compensatory responses. Once identified, disease-related metabolic networks can be quantified on an individual case basis and used as quantitative descriptors of disease severity and progression. Disease-specific networks can also aid in differential diagnosis, an approach that may prove useful in other brain diseases including dementia. Moreover, network activity can be modified by treatment, thereby facilitating the objective evaluation of new therapies for these disorders. Including metabolic network assessments in future clinical trials will define their ultimate role as disease biomarkers.
Acknowledgments
This work was supported by grant number P50NS071675 (Morris K. Udall Center of Excellence in Parkinson’s Disease Research at the Feinstein Institute for Medical Research) from the National Institute of Neurological Disorders and Stroke.
We thank Drs C.C. Tang, Y. Ma, and A. Feigin for their thoughtful comments and for many valuable discussions; T. Fitzpatrick and M. Choi for critical editorial assistance; and Drs K.L. Leenders, L.K. Teune, C. Zuo, and M. Tripathi for generously sharing FDG PET scans from their respective institutions. These data were essential for the comparison of the various disease-related spatial topographies presented in Figure 1B.
Footnotes
Potential Conflicts of Interest
D.E.: board membership, Michael J. Fox Foundation for Parkinson’s Research, Thomas Hartman Foundation for Parkinson’s Research, Bachmann–Strauss Dystonia and Parkinson Foundation; consultancy, Neurologix; employment, Feinstein Institute for Medical Research.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the NIH.
References
Full text links
Read article at publisher's site: https://doi.org/10.1002/ana.23631
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4564117?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Connectivity based on glucose dynamics reveals exaggerated sensorimotor network coupling on subject-level in Parkinson's disease.
Eur J Nucl Med Mol Imaging, 51(12):3630-3642, 17 Jun 2024
Cited by: 0 articles | PMID: 38884774 | PMCID: PMC11445336
Micro-and mesoscale aspects of neurodegeneration in engineered human neural networks carrying the LRRK2 G2019S mutation.
Front Cell Neurosci, 18:1366098, 05 Apr 2024
Cited by: 0 articles | PMID: 38644975 | PMCID: PMC11026646
Investigating the aspect of asymmetry in brain-first versus body-first Parkinson's disease.
NPJ Parkinsons Dis, 10(1):74, 30 Mar 2024
Cited by: 0 articles | PMID: 38555343 | PMCID: PMC10981719
Synaptic loss and its association with symptom severity in Parkinson's disease.
NPJ Parkinsons Dis, 10(1):42, 24 Feb 2024
Cited by: 1 article | PMID: 38402233 | PMCID: PMC10894197
How should we be using biomarkers in trials of disease modification in Parkinson's disease?
Brain, 146(12):4845-4869, 01 Dec 2023
Cited by: 4 articles | PMID: 37536279 | PMCID: PMC10690028
Review Free full text in Europe PMC
Go to all (91) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Assessment of the progression of Parkinson's disease: a metabolic network approach.
Lancet Neurol, 6(10):926-932, 01 Oct 2007
Cited by: 95 articles | PMID: 17884682 | PMCID: PMC2870718
Review Free full text in Europe PMC
Functional brain networks in the evaluation of patients with neurodegenerative disorders.
Nat Rev Neurol, 19(2):73-90, 20 Dec 2022
Cited by: 20 articles | PMID: 36539533
Review
Functional brain networks in Parkinson's disease.
Parkinsonism Relat Disord, 8(2):91-94, 01 Oct 2001
Cited by: 22 articles | PMID: 11489673
Review
[POTENTIAL CLINICAL APPLICATIONS OF RESTING-STATE-FMRI IN NEUROLOGY].
Harefuah, 158(6):378-382, 01 Jun 2019
Cited by: 1 article | PMID: 31215190
Review
Funding
Funders who supported this work.
NINDS NIH HHS (2)
Grant ID: P50 NS071675
Grant ID: P50NS071675




