Abstract
Background
The combination of ataxia and hypogonadism was first described more than a century ago, but its genetic basis has remained elusive.Methods
We performed whole-exome sequencing in a patient with ataxia and hypogonadotropic hypogonadism, followed by targeted sequencing of candidate genes in similarly affected patients. Neurologic and reproductive endocrine phenotypes were characterized in detail. The effects of sequence variants and the presence of an epistatic interaction were tested in a zebrafish model.Results
Digenic homozygous mutations in RNF216 and OTUD4, which encode a ubiquitin E3 ligase and a deubiquitinase, respectively, were found in three affected siblings in a consanguineous family. Additional screening identified compound heterozygous truncating mutations in RNF216 in an unrelated patient and single heterozygous deleterious mutations in four other patients. Knockdown of rnf216 or otud4 in zebrafish embryos induced defects in the eye, optic tectum, and cerebellum; combinatorial suppression of both genes exacerbated these phenotypes, which were rescued by nonmutant, but not mutant, human RNF216 or OTUD4 messenger RNA. All patients had progressive ataxia and dementia. Neuronal loss was observed in cerebellar pathways and the hippocampus; surviving hippocampal neurons contained ubiquitin-immunoreactive intranuclear inclusions. Defects were detected at the hypothalamic and pituitary levels of the reproductive endocrine axis.Conclusions
The syndrome of hypogonadotropic hypogonadism, ataxia, and dementia can be caused by inactivating mutations in RNF216 or by the combination of mutations in RNF216 and OTUD4. These findings link disordered ubiquitination to neurodegeneration and reproductive dysfunction and highlight the power of whole-exome sequencing in combination with functional studies to unveil genetic interactions that cause disease. (Funded by the National Institutes of Health and others.).Free full text

Ataxia, Dementia, and Hypogonadotropism Caused by Disordered Ubiquitination
Associated Data
Abstract
BACKGROUND
The combination of ataxia and hypogonadism was first described more than a century ago, but its genetic basis has remained elusive.
METHODS
We performed whole-exome sequencing in a patient with ataxia and hypogonadotropic hypogonadism, followed by targeted sequencing of candidate genes in similarly affected patients. Neurologic and reproductive endocrine phenotypes were characterized in detail. The effects of sequence variants and the presence of an epistatic interaction were tested in a zebrafish model.
RESULTS
Digenic homozygous mutations in RNF216 and OTUD4, which encode a ubiquitin E3 ligase and a deubiquitinase, respectively, were found in three affected siblings in a consanguineous family. Additional screening identified compound heterozygous truncating mutations in RNF216 in an unrelated patient and single heterozygous deleterious mutations in four other patients. Knockdown of rnf216 or otud4 in zebrafish embryos induced defects in the eye, optic tectum, and cerebellum; combinatorial suppression of both genes exacerbated these phenotypes, which were rescued by nonmutant, but not mutant, human RNF216 or OTUD4 messenger RNA. All patients had progressive ataxia and dementia. Neuronal loss was observed in cerebellar pathways and the hippocampus; surviving hippocampal neurons contained ubiquitin-immunoreactive intranuclear inclusions. Defects were detected at the hypothalamic and pituitary levels of the reproductive endocrine axis.
CONCLUSIONS
The syndrome of hypogonadotropic hypogonadism, ataxia, and dementia can be caused by inactivating mutations in RNF216 or by the combination of mutations in RNF216 and OTUD4. These findings link disordered ubiquitination to neurodegeneration and reproductive dysfunction and highlight the power of whole-exome sequencing in combination with functional studies to unveil genetic interactions that cause disease. (Funded by the National Institutes of Health and others.)
In recent years, we have seen great advances in the elucidation of genetic causes of cerebellar ataxia, with newly identified genes regulating a wide spectrum of cellular functions, including intracellular signaling, tau regulation, and mitochondrial function.1 However, a genetic defect cannot be found in approximately 40% of patients with ataxia,1 including those in whom ataxia is associated with reproductive endocrine failure, a syndrome first reported by Gordon Holmes in 1908.2 Most patients with this syndrome have a hypogonadotropic condition, with defective secretion of gonadotropins by the pituitary gland.3–12 Strikingly, genes associated with ataxia have little functional overlap with genes associated with hypogonadotropic hypogonadism, which encode proteins involved in the biologic function of the neurons that secrete gonadotropin-releasing hormone (GnRH).13
A decade ago, we described a consanguineous family with a syndrome of cerebellar ataxia, dementia, and hypogonadotropic hypogonadism.12 Here we report the results of whole-exome and targeted sequencing performed to identify mutations that underlie the syndrome in this kindred and in unrelated patients.
METHODS
STUDY PATIENTS
Our study included 12 patients with ataxia and hypogonadotropic hypogonadism from eight families. The pedigrees of the index family and four of the other seven families are shown in Figure 1. The patients were referred to the Massachusetts General Hospital for clinical or genetic evaluation between 2000 and 2010. The study was approved by the hospital’s human research committee, and written informed consent for all participants was provided by the participant or an authorized representative.
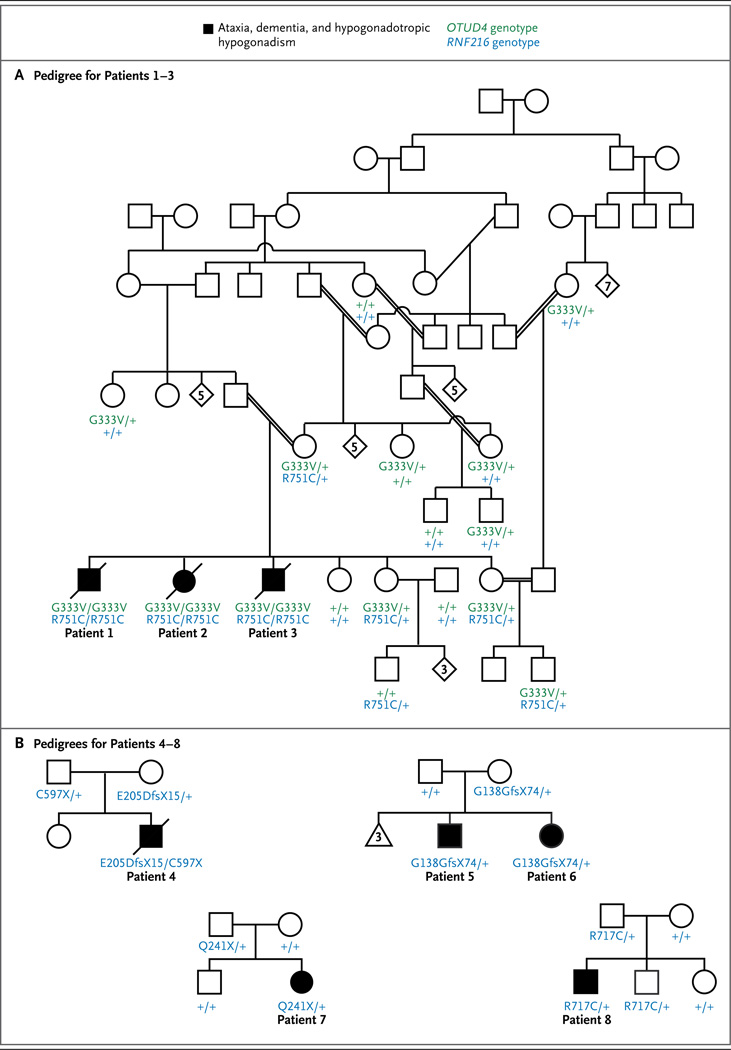
The seven-generation pedigree shown in Panel A includes Patients 1, 2, and 3, all of whom presented with ataxia, dementia, and hypogonadotropic hypogonadism and were homozygous for both RNF216 p.R751C and OTUD4 p.G333V. Double lines indicate consanguineous unions. Genotyped, unaffected family members are shown to be either homozygous for the nonmutated alleles (denoted with a + symbol) or heterozygous for one or both changes. The pedigrees shown in Panel B are for the families of additional RNF216 mutation-positive patients (Patients 4 through 8), all of whom presented with ataxia and hypogonadotropic hypogonadism. Squares denote male family members, circles female family members, solid symbols affected family members, slashes deceased family members, diamonds siblings of either sex, the triangle miscarriages, and Arabic numbers the number of siblings or miscarriages.
GENETIC ANALYSIS
We performed exome sequencing with DNA from Patient 3 in the index family. The data sets used for exome analysis in this study were obtained from dbGaP at www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000475. v1.p1. Candidate genes were sequenced in family members and in unrelated affected persons. Computer algorithms were used to predict the pathogenicity of variants and to identify interactions between candidate genes and genes known to be associated with ataxia or hypogonadotropic hypogonadism. Allele-specific reverse-transcriptase–polymerase-chain-reaction (RT-PCR) assays were performed with RNA from Patients 5, 6, and 7 (see the Methods section in the Supplementary Appendix, available with the full text of this article at NEJM.org).
NEUROPATHOLOGICAL AND ENDOCRINE EVALUATION
The brain of Patient 2 was obtained within 6 hours after death. Immunohistochemical analysis was performed with the use of antibodies against ubiquitin, tau, and α-synuclein. Electron microscopy was performed according to standard procedures. Detailed reproductive endocrine pheno-typing was performed in 5 patients, as described in our previous report12 and in the Methods section in the Supplementary Appendix.
ZEBRAFISH INVESTIGATIONS
Morpholino oligonucleotides (MO) for the silencing of zebrafish rnf216 and otud4 were injected either alone or with nonmutant or mutant human messenger RNA (mRNA) encoding RNF216, mRNA encoding OTUD4, or both (see the Methods section in the Supplementary Appendix).
RESULTS
GENETIC STUDIES
The consanguineous pedigree of the index family (a self-reported Palestinian family) includes three siblings (Patients 1, 2, and 3) with ataxia and hypogonadotropic hypogonadism. Exome sequencing performed with DNA from Patient 3 identified 13 homozygous variants that were rare and predicted to be deleterious (Table S1 in the Supplementary Appendix), 2 of which were also homozygous in the two other affected siblings: RNF216 (NM_207111.3) c.2251C→T, p.R751C and OTUD4 (NM_001102653.1) c.998G→T, p.G333V; these variants were not identified or were heterozygous in the unaffected family members (Fig. 1). RNF216 encodes an E3 ubiquitin-protein ligase. The R751 residue of RNF216 resides within the second of two domains called “really interesting new gene” (RING) finger domains and is conserved across vertebrates (Fig. S1 in the Supplementary Appendix). The R751C variant is predicted to be deleterious by four prediction programs and is absent in 13,006 control chromosomes from the National Heart, Lung, and Blood Institute’s Exome Sequencing Project (ESP) and in 672 chromosomes from Middle Eastern persons (including 36 chromosomes from Palestinian persons). OTUD4 encodes a deubiquitinase (Fig. S1 in the Supplementary Appendix). The G333V variant is predicted to be deleterious by three of four prediction programs and is found in 2 of the 13,006 chromosomes from the ESP and in none of the 672 chromosomes from Middle Eastern persons.
Both OTUD4 and RNF216 were sequenced in nine affected persons from seven unrelated families. No rare variants were identified in OTUD4, but mutations in RNF216 were identified in four probands (Fig. 1, and Fig. S1 in the Supplementary Appendix). Patient 4 had compound heterozygous frameshift and nonsense mutations ([c.615_616delGA; p.E205DfsX15] and [c.1791T→A; p.C597X]). Three additional heterozygous mutations were identified: c.414delG, p.G138GfsX74 in Patients 5 and 6 (siblings); c.721C→T, p.Q241X in Patient 7; and c.2149C→T, p.R717C in Patient 8; the missense mutation in Patient 8 was predicted to be deleterious by four prediction programs.
None of these RNF216 variants were present in the ESP, which did identify five other variants considered to be overtly deleterious and nine considered likely to be deleterious. Given this background level of genetic variation, the presence of deleterious mutations in five of eight probands in this study exceeded what would be expected by chance (P<1×10−13 by Fisher’s exact test). Grail,14 DAPPLE,15 Endeavour,16 InWeb scored network,17 and CNVconnect18 were used to identify potential connections among RNF216, OTUD4, and genes known to be associated with ataxia or hypogonadotropic hypogonadism; no such connections were found. Allele-specific RT-PCR assays failed to identify occult mutations affecting transcription or mRNA stability in the nonmutant alleles of Patients 5, 6, and 7 (Fig. S2 in the Supplementary Appendix).
FUNCTIONAL TESTING
Zebrafish were used to interrogate the functional consequences and suggestive epistatic interactions of the RNF216 and OTUD4 mutations.19,20 The injection of a MO that disrupted the splicing of zebrafish rnf216 (Fig. S3 in the Supplementary Appendix) caused a reduction in the size of the eye cup and optic tecta and disorganization of the cerebellum, as well as a slight reduction in overall head size (Fig. 2 and Fig. 3, and Fig. S4 in the Supplementary Appendix). The tectal phenotype was rescued by the coinjection of human RNF216 mRNA, but the injection of human RNF216 mRNA encoding R751C failed to rescue the phenotype (Fig. 2), suggesting that the mutation affects the function of the encoded protein. The injection of a splice-blocking MO against zebrafish otud4 (Fig. S3 in the Supplementary Appendix) also induced a reduction in size of the optic tecta and cerebellum (Fig. 3).

Panels A through D show dorsal views of control zebrafish embryos (Panel A) and embryos injected with rnf216 morpholino oligonucleotides (MO) (Panel B), rnf216 MO plus nonmutant human RNF216 (Panel C), and rnf216 MO plus mutant human RNF216 (with RNF216 carrying the p.R751C mutation identified in the index pedigree) (Panel D) at 3 days after fertilization (staining with an antibody against α acetylated tubulin). The circles outline the area of the optic tectum, the structure on which all measurements were based. The bar graph in Panel E shows the relative size of the optic tectum in control embryos and the embryos injected with rnf216 MO, rnf216 MO plus nonmutant human RNF216, and rnf216 MO plus mutant human RNF216. P values are based on two-tailed t-tests. I bars indicate standard errors. AU denotes arbitrary units.
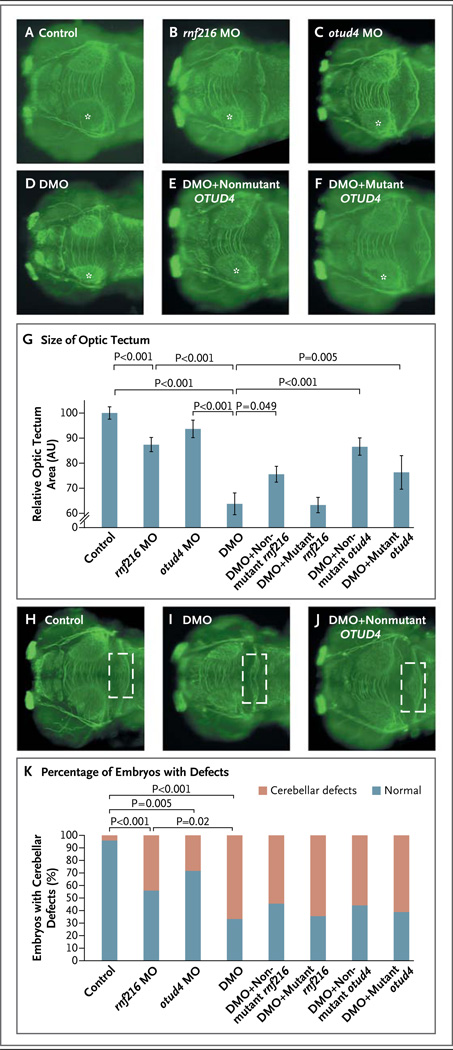
Panels A through F show dorsal views of control zebrafish embryos (Panel A) and embryos injected with rnf216 MO (morpholino oligonucleotides) (Panel B), otud4 MO (Panel C), double MO (DMO, rnf216 MO plus otud4 MO) (Panel D), double MO plus nonmutant human OTUD4 (Panel E), and double (DMO) plus mutant human OTUD4 (OTUD4 carrying the p.G333V mutation identified in the index pedigree) (Panel F) at 3 days after fertilization (anti-α acetylated tubulin stain). The asterisks indicate the optic tecta that were measured to assess the differences between the conditions being evaluated. The bar graph in Panel G shows the mean relative size of the optic tecta in control embryos and the five groups of injected embryos. I bars indicate standard errors. P values are based on two-tailed t-tests. Panels H, I, and J show dorsal views of control embryos (Panel H) and embryos injected with DMO (Panel I) and DMO plus nonmutant human OTUD4 (Panel J) at 3 days after fertilization (anti-α acetylated tubulin stain). The rectangles outline the cerebellar area; maximum disorganization is observed in embryos injected only with DMO (Panel I). The bar graph in Panel K shows the percentage of embryos with cerebellar defects under the conditions being evaluated (as shown in Panels A through F and Panels H, I, and J).
Coinjection of both rnf216 and otud4 MOs induced a significant reduction in the size of the optic tecta as compared with the injection of the rnf216 MO alone (P<0.001) (Fig. 3). Double-MO injection also caused marked disorganization of the cerebellum in more than 60% of embryos (Fig. 3) and the development of a severe cerebellar phenotype with complete loss of structural integrity in approximately 30% of embryos, as compared with only 5 to 10% of embryos injected with either the rnf216 or the otud4 MO alone (data not shown). Furthermore, double-MO injection resulted in marked microphthalmia as compared with modest microphthalmia when either MO injection was used alone (Fig. S4 in the Supplementary Appendix). These phenotypes were specific, since coinjection of nonmutant human RNF216 or OTUD4 mRNA rescued all phenotypes (Fig. 3, and Fig. S4 in the Supplementary Appendix). RNF216 mRNA encoding R751C and OTUD4 mRNA encoding G333V were less effective in rescuing the phenotypes induced by double-MO injection (Fig. 3, and Fig. S4 in the Supplementary Appendix), suggesting not only that these mutant alleles encode functionally deficient proteins but also that epistatic interactions between these mutations contribute to the disease phenotype in the index pedigree.
CLINICAL CHARACTERISTICS OF THE STUDY PATIENTS
Patients 1 through 8, who carried variants in RNF216, had similar clinical histories (Table 1). They presented in adolescence or early adulthood with hypogonadotropic hypogonadism but no other pituitary abnormalities. Dysarthria was the initial neurologic symptom in some patients, but ataxia developed in all patients, leading to wheelchair dependency and to bed confinement for some patients. Dementia was also prominent, with personality changes and memory loss occurring at the onset of the disease and mutism and uncoordinated, purposeless movements during the end stages. Nystagmus was absent. The presentation of Patients 9 through 12, who did not have variants in RNF216, was quite different from that of Patients 1 through 8 (Table 1). Extensive evaluation did not reveal any known causes of ataxia in any of the patients; mitochondrial abnormalities were identified in Patients 7 and 8 (Table S2 in the Supplementary Appendix).
Table 1
Clinical Phenc itypes and RNF216 and OTUD4 Genotypes.*
| Patient and Race or Ethnic Group | Sex | Clinical Features | Imaging Findings | RNF216 genotype | OTUD4 Genotype |
|---|---|---|---|---|---|
| Family 1, Palestinian | |||||
| Patient 1 | Male | No spontaneous puberty; at 22 yr, dysarthria, followed by progressive ataxia and dementia; at 43 yr, death (aspiration pneumonia) | At 30 yr, CT revealed prominent cerebellar and mild cortical atrophy, with hypodensities in cerebral white matter | R751C+R751C | G333V + G333V |
| Patient 2 | Female | At 16 yr, menarche, followed by secondary amenorrhea; at 20 yr, personality change; at 30 yr, dysarthria, followed by progressive ataxia and dementia; at 41 yr, death (aspiration pneumonia) | At 30 yr, CT revealed prominent cerebellar and mild cortical atrophy, with hypodensities in cerebral white matter | R751C+ R751C | G333V + G333V |
| Patient 3 | Male | Normal puberty; at 20 yr, erectile dysfunction; at 29 yr, dysarthria, followed by progressive ataxia and de- mentia; at 47 yr, death (possibly from pulmonary embolism) | At 35 yr, MRI revealed diffuse parenchymal volume loss in cerebellum and cerebral cortex, with multiple punctate and confluent areas of hyperintensity on T2-weighted and FLAIR imaging | R751C+R751C | G333V + G333V |
| Family 2, white | |||||
| Patient 4 | Male | No spontaneous puberty; at 22 yr, dysarthria, ataxia, and dementia; at 30 yr, prominent chorea; at 36 yr, death | At 23 yr, MRI revealed cerebellar atrophy and widespread foci of hyperintensity in cerebral white matter and thalami | C597X + E205DfsX15 | Nonmutant + Nonmutant |
| Family 3, white | |||||
| Patient 5 | Male | Normal puberty; at 36 yr, hypogonadotropism and chorea followed by progressive ataxia and dementia | At 42 yr, MRI revealed global atrophy, with diffusely scattered periventricular foci of hyperintensity on T2-weighted and FLAIR imaging | G138GfsX74 + Nonmutant | Nonmutant + Nonmutant |
| Patient 6 | Female | Normal puberty; at 27 yr, oligomenorrhea, followed by amenorrhea; memory problems; at 31 yr, chorea, followed by progressive ataxia and dementia | At 31 yr, MRI revealed mild-to-moderate cerebellar atrophy and mild prominence of ventricles and sulci, with multiple small foci of hyperintensity on T2-weighted imaging | G138GfsX74 + Nonmutant | Nonmutant+ Nonmutant |
| Family 4, white | |||||
| Patient 7 | Female | Primary amenorrhea; at 27 yr, ataxia and dysarthria, followed by progressive ataxia and dementia | At 18 yr, MRI revealed a normal cerebellum and multiple foci of hyperintensity in subcortical white matter on T2-weighted imaging; at 35 yr, MRI revealed marked cerebellar atrophy, with an increased number of foci of T2-weighted hyperintensity | Q241X + Nonmutant | Nonmutant+ Nonmutant |
| Family 5, white | |||||
| Patient 8 | Male | No spontaneous puberty; at 19–21 yr, partial response to treatment with exogenous pulsatile GnRH; at 21 yr, slurred speech and imbalance, followed by progressive ataxia, mood changes, and memory impairment | At 17 yr, MRI revealed slight prominence of fissures in cerebellum; at 23 yr, M Rl revealed severe cerebellar and mild cerebral atrophy, multiple foci of T2-weighted and FLAIR hyperintensity | R717C + Nonmutant | Nonmutant+ Nonmutant |
| Family 6, white | |||||
| Patient 9 | Male | No spontaneous puberty; at 5 yr, ataxia, nystagmus; at 34 yr, still able to walk | Nonmutant + Nonmutant | Nonmutant+ Nonmutant | |
| Patient 10 | Male | No spontaneous puberty; at 5 yr, ataxia, nystagmus; at 32 yr, still able to walk | Nonmutant + Nonmutant | Nonmutant+ Nonmutant | |
| Family 7, white | |||||
| Patient 11 | Male | No spontaneous puberty; at 4 yr, ataxia; at 28 yr, ataxia stable | At 4 yr, CT revealed normal cerebellum; at 17 yr, MRI revealed cerebellopontine atrophy, global white- matter abnormality in basal ganglia and cortex | Nonmutant+ Nonmutant | Nonmutant+ Nonmutant |
| Family 8, Asian | |||||
| Patient 12 | Female | Normal puberty; at 17 yr, behavior problems, followed by tremor, dysarthria, and ataxia; at 19 yr, internuclear ophthalmoplegia; at 24 yr, hypogonadotropism | At 21 yr, MRI revealed mild atrophy, T2-weighted and FLAIR imaging revealed hyperintensity in brain stem, thalami, internal capsules, insulae, basal ganglia, and periventricular regions | Nonmutant+ Nonmutant | Nonmutant + Nonmutant |
Neuroimaging performed in Patients 1 through 8 revealed striking similarities, with cerebellar and cortical atrophy but no abnormalities of the pituitary gland. The subcortical white matter contained patchy areas of hyperintensity on T2-weighted imaging and fluid-attenuated inversion recovery (FLAIR) imaging (Table 1 and Fig. 4). In Patient 7, these areas of hyperintensity were present approximately 9 years before the onset of neurologic symptoms; the cerebellum appeared normal at that earlier point in time.

Panel A shows a sagittal T2-weighted magnetic resonance imaging scan of the brain in Patient 3. Diffuse cerebellar atrophy (arrow) and cortical atrophy can be seen. Panel B shows a transverse image obtained with fluid-attenuated inversion recovery imaging, revealing multiple distinct and confluent foci of hyperintensity in the white matter. In Panel C, immunohistochemical analysis of a hippocampal brain section from Patient 2 shows a neuronal intranuclear inclusion with immunoreactivity (brown) to an antibody against ubiquitin, counterstained with hematoxylin and eosin. An electron micrograph of the hippocampal neurons, in Panel D, also shows an intranuclear inclusion, which consists of aggregates of granular material and fine filaments, 10 to 15 nm in diameter (arrow), that are for the most part randomly oriented. The scale bar corresponds to 1 µm.
NEUROPATHOLOGICAL STUDIES
The formalin-fixed brain of Patient 2 weighed 940 g (normal weight, 1300 g). The cerebellum and inferior olives were atrophic. Histopathological analysis revealed gliosis and virtually complete loss of inferior olivary neurons, cerebellar Purkinje’s cells, and neurons in hippocampal regions CA3 and CA4, whereas neurons were well preserved in regions CA1 and CA2. Ubiquitin-immunoreactive nuclear inclusions were present in 1 to 5% of the pyramidal neurons in hippocampal regions CA1 and CA2 (Fig. 4) and were also found in granule-cell neurons in the dentate gyrus; these inclusions were not immunoreactive to antibodies against tau or α-synuclein (not shown). On electron microscopy, the intranuclear inclusions appeared as aggregates of fine filaments and granular material (Fig. 4).
REPRODUCTIVE ENDOCRINE STUDIES
When Patient 6 reached 32 years of age, 1 year after the development of neurologic symptoms, low-amplitude pulses of luteinizing hormone were detected, indicating that GnRH secretion, although present, was diminished (Fig. 5). The administration of pulsatile GnRH for 7 days induced robust increases in levels of gonadotropins and estradiol (Fig. 5) as well as the growth of a dominant ovarian follicle, observed on ultraso-nography (not shown). Although the secretion of luteinizing hormone increased in response to the administration of GnRH, the typical peaked pattern of luteinizing hormone pulses21 was not seen, which suggested a degree of pituitary dysfunction. Indeed, the patient’s pituitary responsiveness waned over time, with a diminished response to GnRH on day 1 of treatment 15 months after the initial endocrine study (Fig. 5).

In Panels A through D, the graphs at the left show the endogenous secretion of luteinizing hormone over a period of up to 12 hours. Patient 6 was studied on two occasions, 15 months apart (Panels A and B). Arrowheads indicate pulses of luteinizing hormone secretion, and boxes duration of sleep; the shading indicates the reference range for healthy men and women. Concentrations of estradiol (E2) and testosterone (T), measured from pooled samples obtained during the study, are indicated. In Panels A, B, and D, the graphs at the right show the response to exogenous pulsatile gonadotropin-releasing hormone (GnRH) over the course of up to 7 days. The dose of GnRH was 75 ng per kilogram of body weight, with the exception of the first dose of GnRH on day 1 for Patient 6 (Panel A), which was 165 ng per kilogram. (Note the difference in the y axis scales in Panels A and B.) In Panel C, the graph at the right shows the secretion of lutein-izing hormone in response to varying doses of GnRH (black circles and regression line). The data for the patient fall to the right of the 95% confidence interval (dashed red lines) for the mean amplitude of the response to a range of GnRH doses in 6 other men with idio-pathic hypogonadotropic hypogonadism (solid red line).
In Patient 8, in whom endocrine function was initially assessed before the onset of neurologic symptoms, there was an absence of endogenous pulsatile luteinizing hormone secretion (Fig. 5). After escalating doses of exogenous GnRH (from 25 ng per kilogram of body weight to 600 ng per kilogram every 2 hours), the patient’s testicular volume increased from 2 ml to 8 ml but did not increase further, despite these very high doses. Although his pituitary response to exogenous treatment with GnRH was impaired (Fig. 5), direct gonadal stimulation with human chorionic gonadotropin normalized the testosterone level, at 459 ng per deciliter (15.9 nmol per liter).
Pituitary responsiveness to GnRH was lost in patients late in the course of their disease. In Patients 1 and 2, who were bedridden when pituitary responsiveness was assessed, there was no detectable luteinizing hormone secretion and no measurable change in the gonadotropin level in response to the administration of pulsatile exogenous GnRH (Fig. 5).12
DISCUSSION
The underpinnings for the association of ataxia with hypogonadotropic hypogonadism have eluded investigators for more than a century. This report shows that ataxia with hypogonadotropic hypogonadism can be caused by mutations in RNF216 either singly or in combination with mutations in OTUD4. Both RNF216 and OTUD4 encode proteins that regulate ubiquitination, indicating that abnormalities in this fundamental cellular process can have pathologic effects on the cerebellum and hippocampus, the cerebral white matter, and the hypothalamic and pituitary components of the reproductive endocrine cascade.
The compound heterozygous termination mutations in RNF216 in Patient 4 firmly implicate RNF216 as a causative gene for this syndrome. Heterozygous RNF216 mutations were found in Patients 5 through 8 but did not cause disease in their parents. Oligogenicity offers a parsimonious explanation for these observed patterns. Oligogenic inheritance has been described in the Bardet–Biedl and Bartter syndromes, Hirsch-sprung’s disease, and isolated hypogonadotropic hypogonadism.19,22–27 In such an oligogenic model, RNF216 mutations can act with mutations at other genetic loci to cause disease.
Indeed, the phenotype in the index pedigree appears to have been caused by the interaction of hypomorphic mutations in RNF216 and OTUD4. Inhibition of either RNF216 or OTUD4 in zebrafish resulted in cerebellar hypoplasia, microphthalmia, and small optic tecta. The concordance of these phenotypes suggests that RNF216 and OTUD4 operate in the same pathway. This possibility is bolstered by the observation of epistatic interactions between RNF216 and OTUD4, with simultaneous knockdown of both genes resulting in more severe phenotypes. Taken together, these findings support a digenic model in which the OTUD4 mutation, in conjunction with the RNF216 mutation, played an essential role in causing disease in the index pedigree. By extension, in Patients 5 through 8, it is possible that mutations in other, currently unidentified loci may have acted in conjunction with the heterozygous mutations in RNF216 to cause disease. Oligogenicity is likely to be increasingly recognized as methods for detecting this genetic architecture, such as exome sequencing, are more widely adopted.
RNF216 encodes an E3 ubiquitin ligase that attaches ubiquitin to protein substrates, marking them for proteasome-mediated degradation. Known targets of RNF216 include upstream activators of nuclear factor κB signaling, which regulates diverse cellular processes.28–31 RNF216 is structurally similar to parkin, an E3 ubiquitin ligase that is mutated in a recessive form of Parkinson’s disease.32–34 The finding of neuronal intranuclear inclusions in Patient 2 may indicate that RNF216-associated neurodegeneration has similarities not only with Parkinson’s disease but also with other neurodegenerative disorders in which protein aggregates are found, such as Huntington’s disease and Alzheimer’s disease.35
OTUD4 encodes a deubiquitinating enzyme. Deubiquitinases allow target proteins and ubiquitin itself to be recycled and often function in partnership with specific E3 ligases. For example, the deubiquitinase ataxin-3 counteracts the ability of parkin to ubiquitinate itself.36 On the basis of this and other examples,37 OTUD4 and RNF216 may be similarly linked in a coregulatory partnership.
The progressive and debilitating dementia observed in the patients with RNF216 mutations (Patients 1 through 8) distinguishes them from the other patients with ataxia and hypogonadotropic hypogonadism. Furthermore, we observed changes in cerebral white matter in all the patients with RNF216-associated neurodegeneration, suggesting that such changes may constitute a consistent feature of this syndrome. None of these patients had oculomotor abnormalities such as the nystagmus and ophthalmoplegia seen in Patients 9, 10, and 12, who did not have RNF216 mutations.
The patients with RNF216-associated neuro-degeneration had dysfunction at multiple levels of the reproductive endocrine axis. In Patients 6 and 8, reproductive function was restored with extended GnRH treatment, which suggests that hypothalamic GnRH deficiency was the primary cause of their reproductive endocrine dysfunction. However, these two patients also appeared to have an element of pituitary dysfunction, given the diminishing responses to GnRH over time in Patient 6 and the observation of a right-shifted dose–response curve in Patient 8. In Patients 1 and 2, who were evaluated late in the course of their disease, the complete absence of response after 7 days of treatment with GnRH may represent progression of this pituitary dysfunction. The basis for the selective vulnerability of particular neuronal and pituitary cell types is currently unexplained.
In conclusion, we have identified loss-of-function mutations in RNF216 that cause a syndrome of ataxia, dementia, and hypogonadotropic hypogonadism. Genetic and in vivo evidence suggests that mutations affecting RNF216, an E3 ubiquitin ligase, and OTUD4, a deubiquitinase, can synergize to cause this syndrome, reinforcing the notion that the mutational load within biologic pathways can drive disease manifestation.20 Taken together, these data highlight a hitherto unknown role of the ubiquitination system in disorders of combined neurodegeneration and reproductive dysfunction. More broadly, our findings show the value of combining individual whole-exome sequencing with in vivo functional studies to identify disease-causing gene mutations and epistatic interactions.
Acknowledgments
Supported by grants from the Eunice K. Shriver National Institute for Child Health and Human Development (K24 HD067388, R01 HD043341, R01 HD042601, and U54 HD028138); from the National Institute of Diabetes and Digestive and Kidney Diseases (P50 DK096415); by Harvard Catalyst and the Harvard Clinical and Translational Science Center (funded by the National Center for Research Resources and the National Center for Advancing Translational Sciences [UL1TR025758 and M01RR01066] and Harvard University and its affiliated academic health care centers); by a grant from the National Human Genome Research Institute to the Broad Institute (U54 HG003967); and by grants from the National Heart, Lung, and Blood Institute for the Exome Sequencing Project (HL102923, HL102924, HL102925, HL102926, and HL103010). Dr. Chan is the recipient of a Charles A. King Trust postdoctoral fellowship and a Career Development Award from Boston Children’s Hospital.
We thank the patients and their families for their cooperation in these studies; Dr. Fawzi al-Hammouri and the physicians and staff of the Specialty Hospital, Amman, Jordan, for assisting with clinical care for Patients 1, 2, and 3; Dr. Momen al-Hadidi and the staff of the Forensic Medicine Department at Al-Basheer Government Hospital, Amman, Jordan, for performing the autopsy and obtaining the brain after the death of Patient 2; Drs. Daniel Metzger, Susan Ratzan, Meriel McEntagart, and Sandra Sirrs for patient referrals; Dr. J. Michael Andresen for performing the initial linkage analysis; Drs. Christopher Walsh and Timothy Yu at Boston Children’s Hospital for sharing data from the Middle Eastern control patients; Carlotta Fitch and Dr. Kathy Newell for assisting with the neuropathological analyses; Dr. Omar Abu Hijleh for providing the early endocrine history of Patients 1 and 2; the staff of the Massachusetts General Hospital (MGH) Clinical Research Center for assisting with clinical protocols; members of the MGH Reproductive Endocrine Unit; and Eric Lander and the Broad Institute for generating high-quality sequence data.
Footnotes
The views expressed in this article are those of the authors and do not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1056/nejmoa1215993
Read article for free, from open access legal sources, via Unpaywall:
https://www.nejm.org/doi/pdf/10.1056/NEJMoa1215993?articleTools=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Hypogonadotropic Hypogonadism as First Presentation of the Severe Neuroendocrine Disorder Caused by RNF216.
JCEM Case Rep, 2(11):luae195, 23 Oct 2024
Cited by: 0 articles | PMID: 39444518 | PMCID: PMC11497609
The role of ubiquitination in health and disease.
MedComm (2020), 5(10):e736, 25 Sep 2024
Cited by: 0 articles | PMID: 39329019 | PMCID: PMC11424685
Review Free full text in Europe PMC
An E3 ligase and autophagy adaptor regulates tau proteostasis through nested phase separation.
Nat Cell Biol, 26(8):1231-1232, 01 Aug 2024
Cited by: 0 articles | PMID: 39030428
The autophagy adaptor TRIAD3A promotes tau fibrillation by nested phase separation.
Nat Cell Biol, 26(8):1274-1286, 15 Jul 2024
Cited by: 0 articles | PMID: 39009640
Gordon Holmes Syndrome Model Mice Exhibit Alterations in Microglia, Age, and Sex-Specific Disruptions in Cognitive and Proprioceptive Function.
eNeuro, 11(1):ENEURO.0074-23.2023, 25 Jan 2024
Cited by: 0 articles | PMID: 38164552 | PMCID: PMC10849025
Go to all (124) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
dbGaP - The database of Genotypes and Phenotypes
- (1 citation) dbGaP - phs000475
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP.
Hum Mol Genet, 23(4):1013-1024, 09 Oct 2013
Cited by: 87 articles | PMID: 24113144 | PMCID: PMC3900109
RNF216 mutations as a novel cause of autosomal recessive Huntington-like disorder.
Neurology, 84(17):1760-1766, 03 Apr 2015
Cited by: 35 articles | PMID: 25841028
Whole-Exome Sequencing Identified a Novel Mutation in RNF216 in a Family with Gordon Holmes Syndrome.
J Mol Neurosci, 72(4):691-694, 28 Jan 2022
Cited by: 4 articles | PMID: 35088240
Clinical and genetic spectrum of RNF216-related disorder: a new case and literature review.
J Med Genet, 61(5):430-434, 19 Apr 2024
Cited by: 1 article | PMID: 38050071
Review
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1TR025758
NCRR NIH HHS (3)
Grant ID: M01 RR001066
Grant ID: M01RR01066
Grant ID: UL1 RR025758
NHGRI NIH HHS (1)
Grant ID: U54 HG003967
NHLBI NIH HHS (15)
Grant ID: UC2 HL102926
Grant ID: RC2 HL102924
Grant ID: UC2 HL102923
Grant ID: UC2 HL102924
Grant ID: HL102924
Grant ID: HL102925
Grant ID: RC2 HL102923
Grant ID: RC2 HL102926
Grant ID: HL102926
Grant ID: UC2 HL103010
Grant ID: HL102923
Grant ID: RC2 HL103010
Grant ID: UC2 HL102925
Grant ID: HL103010
Grant ID: RC2 HL102925
NICHD NIH HHS (4)
Grant ID: R01 HD042601
Grant ID: R01 HD043341
Grant ID: U54 HD028138
Grant ID: K24 HD067388
NIDDK NIH HHS (1)
Grant ID: P50 DK096415





