Abstract
Free full text

Mouse NAIP1 detects the type III secretion system needle protein
Abstract
The NAIP/NLRC4 inflammasomes activate caspase-1 in response to bacterial type III secretion systems (T3SS). Inadvertent injection of the T3SS rod protein and flagellin into the cytosol are detected through murine NAIP2 and NAIP5/6, respectively. Here, we identify the agonist for the orphan murine NAIP1 receptor as the T3SS needle protein. NAIP1 is poorly expressed in resting mouse bone marrow-derived macrophages (BMMs), however, priming with poly(I:C) induces it, and confers needle protein sensitivity. Further, overexpression of NAIP1 in immortalized BMMs by retroviral transduction enabled needle detection. In contrast, peritoneal cavity macrophages basally express NAIP1 and respond to needle protein robustly independent of priming. Human macrophages are known to only express one NAIP gene, which detects the needle protein, but not rod or flagellin. Thus, murine NAIP1 is functionally analogous to human NAIP.
Introduction
The detection of virulence factors is a key step in the early innate immune response to infection. Type III or type IV secretion systems (T3SS and T4SS) are commonly used bacterial virulence apparati that inject effector proteins into the cytosol of target host cells, where they reprogram cellular functions to benefit the pathogen. Salmonella typhimurium is a flagellated bacterium that uses its SPI1 T3SS to induce macropinocytosis and invasion in epithelial cells. Structurally, the T3SS apparatus is a hollow multiprotein complex, composed of a basal body with an interior rod (PrgJ in SPI1) and a hollow needle (PrgI in SPI1) that protrudes from the bacterial surface. During assembly, the secretion apparatus exports rod subunits into the interior of the basal body, where they polymerize. Subsequently, needle subunits are exported, and polymerize to form a direct channel that links the bacteria to the host cell (1), enabling delivery of the effectors into the host cytosolic compartment.
There are many cytosolic surveillance pathways, including the Nod-like receptors (NLRs). Several NLRs are inflammasomes that act as a platform to activate caspase-1, which carries out two major functions: 1) processing of two inflammatory cytokines pro-IL-1β and pro-IL-18 into their mature secreted forms, and 2) induction of a pro-inflammatory lytic cell death termed pyroptosis. NLRC4 is an inflammasome that primarily responds to infection by bacteria that utilize T3SS and T4SS. NLRC4 was first shown to respond to flagellin in the host cell cytosol (2, 3), which is inadvertently injected by the T3SS (4). However, NLRC4 responded to naturally aflagellated S. flexneri (5), as well as S. typhimurium and Pseudomonas aeruginosa flagellin mutants (6, 7). This flagellin-independent response was later attributed to detection of the type III secretion system (T3SS) rod protein (8), which presumably is also accidentally translocated into the host cytosol (9, 10). Interestingly, both flagellin and rod were detected through the same NLRC4 inflammasome (8). The detection of these two different proteins though the same NLRC4 inflammasome was recently explained by the existence of distinct upstream NLRs in the NAIP family.
C57BL/6 mice have four functional NAIP transcripts (NAIP1, 2, 5, and 6), while only one NAIP gene has been described in humans (11). In mice, NAIP2 detects rod proteins while NAIP5 and NAIP6 detect flagellins, and NAIP1 remains an orphan receptor (12, 13). Although the single human NAIP is highly homologous to mouse NAIP5, it does not activate NLRC4 in the presence of flagellin. Instead, human NAIP detects the needle protein, while flagellin and rod are not detected (13).
Here, we identify the T3SS needle protein as the agonist for the orphan mouse NAIP1, which is functionally homologous to human NAIP. This detection event was previously not observed because murine bone marrow derived macrophages do not express NAIP1 in the resting state.
Materials and Methods
Tissue Culture
BMMs were prepared from the femurs of C57BL/6 mice by culturing with L-cell conditioned supernatants. Human U937 monocytes were obtained from ATCC cultured in RPMI-1640 containing 10% FBS and treated with 50 ng ml−1 PMA for 48 h to induce differentiation on plastic plates. Differentiated cells were lifted with PBS containing 1mM EDTA and sub-cultured for assays. Resident peritoneal cavity macrophages were harvested from naïve mice using ice cold PBS containing 1mM EDTA. Peritoneal lavages from 2–3 mice were pooled for each experiment.
Retroviral transduction
Cloning Salmonella PrgI and PrgJ into pMXsIG retroviral system and ensuing retroviral lethality screen has been described elsewhere (8). Expression of bacterial proteins was assessed on Day 2 post transduction by flow cytometry. For NAIP1 forced expression, Platinum-E cells (American Type Culture Collection) were transfected with MSCV2.2 retrovirus based NAIP1-IRES-GFP or empty vector (12). Retroviral supernatants were collected at 48 h and spinfections were performed on Myc-immortalized BMMs (iBMMs). GFP+ population was sorted using Beckman Coulter Mo-Flo XDP and sub-cultured for further experiments.
Protein Transfections
Cells were stimulated for 3–4 h with 50 ng/ml LPS (List Biological) unless indicated otherwise. PrgI-6XHis and PrgJ—6XHis were purified using Talon beads (Clontech). Purified proteins were transfected into macrophages using transfection reagent profect (P2) as described previously (8). Lactate dehydrogenase (LDH) activity in the supernatant was determined with the CytoTox 96 assay (Promega), and IL-1β secretion was determined by ELISA (R&D Systems).
RQ-PCR analysis
Total RNA was isolated from indicated tissues, BMMs or peritoneal cavity macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed (Invitrogen) and quantitative Taqman PCR. The primers and probes used are described elsewhere (2). Amounts of mRNA analyzed were normalized to GAPDH or Rps17.
Results and Discussion
We wanted to verify that human macrophages respond to cytosolic needle and not rod protein (13). Cytosolic delivery of purified PrgI needle protein induced robust pyroptosis and IL-1β release from THP-1 and U937 cells, two differentiated human macrophage-like cell lines (Fig. 1A–D). However, there was no significant response to the PrgJ rod protein (Fig. 1A–D).
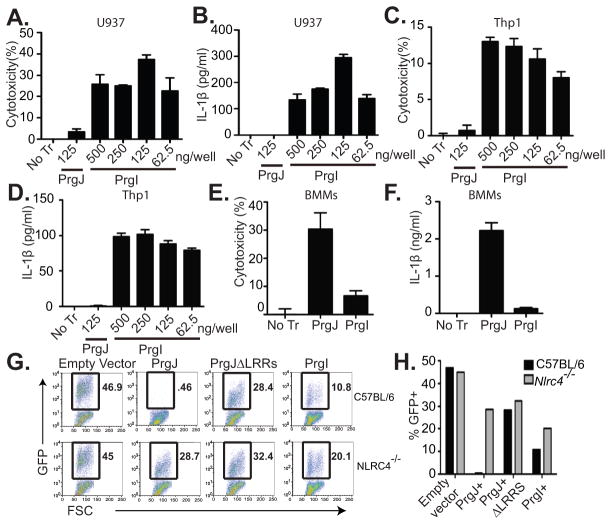
Human monocytic cell lines U937 (A, B) and Thp1 (C, D) were differentiated into macrophages with phorbol myristate acetate (PMA), primed with 50 ng/ml LPS, and then transfected with purified PrgI or PrgJ. 2 h post transfection, cytotoxicity (A, C) and IL-1β secretion (B, D) were determined by LDH release and ELISA, respectively. BMMs primed with LPS (50ng/ml) for 4 hours were transfected with 125ng/well PrgI or PrgJ for 1 h and (E) cytotoxicity or (F) IL-1β secretion were determined. (G, H) Retroviral lethality screen was performed by transducing C57BL/6 and Nlrc4−/− BMMs with retroviruses expressing GFP alone (empty vector), prgJ-GFP, prgJΔLRRs-GFP, or prgI-GFP. Survival of GFP+ cells was determined by flow cytometry 2 days later by flow cytometry. Part of data in G and H (GFP alone and prgJ-GFP) were previously published in (8); prgJΔLRRs-GFP and prgI-GFP transduction data from the same experiment is now presented here. (H) Quantification of G. Graphs show mean and standard deviation of triplicate wells and are representative of at least 3 independent experiments.
Interestingly, while we previously showed mouse bone marrow-derived macrophages (BMMs) cannot not detect PrgI (2, 8), higher doses of cytosolic PrgI protein resulted in a weak response (Fig. 1E, 1F). To further confirm this finding, we used a retroviral lethality screen (8). BMMs from C57BL/6 or Nlrc4−/− mice were retrovirally transduced with GFP only (empty vector), prgJ-GFP, or prgJΔLRRs-GFP, or prgI-IRES-GFP and the surviving cells were analyzed by flow cytometry for GFP expression 2 days following transduction. While GFP positive BMMs can be recovered from empty vector or, prgJΔLRRs-GFP transduction, all prgJ-GFP transduced cells undergo pyroptotic cell death (Fig. 1G, 1H). Some prgI-GFP BMMs could be recovered, but at somewhat reduced percentages (Fig. 1G, 1H). Together these findings suggest a weak needle detection by BMMs. This reduction was dependent on NLRC4, as Nlrc4−/− BMMs showed a higher recovery of prgI-GFP transduced cells (Fig. 1G, 1H), while empty vector was unaffected in the same experiment (47% in WT and 45% in Nlrc4−/−(8). These findings further establish that BMMs are weakly responsive to cytosolic PrgI in an NLRC4 dependent manner.
We hypothesized that PrgI was detected through the orphan NAIP1 receptor. But why is this response so inefficient? Activation of some inflammasomes occurs in a two-step process. For example, NLRP3 requires a priming step achieved by a TLR agonist and AIM2 inflammasome response is enhanced by type I interferon priming (15, 16). In contrast, NLRC4 inflammasome activation occurs without priming as basal expression of all required components are sufficient to generate robust responses to flagellin and rod in vitro and in vivo. However, BMMs primed with LPS for 4 hours show only a weak response to PrgI delivered into the cytosol (Fig. 1E, 1F). We hypothesized that needle detection by BMMs could be augmented by different priming conditions. Therefore, we tested TLR3 ligand, poly(I:C) which is known to prime a the non-canonicalcaspase-11 inflammasome pathway (17–19). Indeed, longer stimulation with LPS or poly(I:C) significantly enhanced Naip1 expression in BMMs (Fig. 2A). Consequently, BMMs primed with poly(I:C) showed increased pyroptosis in response to cytosolic PrgI (Fig. 2B). LPS enhances the pool of pro-IL-1β, type I interferon induced by poly(I:C) treatment inhibits transcription of pro-IL-1β mRNA (20), so IL-1β secretion cannot be examined under these conditions. Therefore, while NAIP2, NAIP5, and NAIP6 are basally expressed in BMMs, NAIP1 induction requires additional priming.
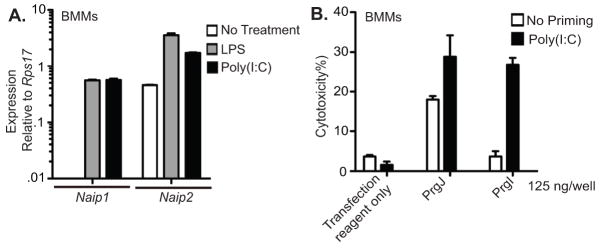
(A) BMMs were left untreated or primed with LPS or poly(I:C) 23 h and RQ-PCR was performed to assess Naip1 and Naip2 transcript levels relative to Rps17. (B) Cytotoxicity in response to cytosolic PrgI or PrgJ protein was determined at 2 h in unstimulated BMMs or BMMs primed with 6μg/ml Poly(I:C) for 2 days. Data representative of 2–3 independent experiments.
To show that Naip1 induction was sufficient amongst poly(I:C) primed genes, we transduced iBMMs with retroviruses expressing GFP alone or Naip1-IRES-GFP. Untransduced iBMMs closely resemble primary BMMs and respond robustly to PrgJ but not to PrgI (Fig. 3A, 3B). As expected, NAIP1 expression enabled detection of PrgI, resulting in significantly enhanced pyroptosis and IL-1β secretion (Fig. 3C, 3D).
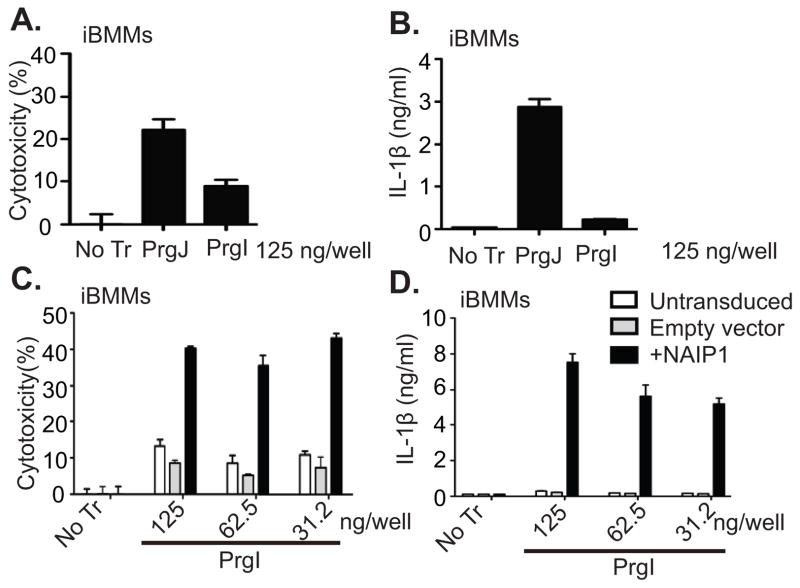
PrgI or PrgJ were transfected into the cytosol of iBMMs and (A) cytotoxicity and (B) IL-1β secretion were measured at 2 h. (C–D) iBMMs were transduced with retroviruses expressing either GFP alone (Empty vector) or Naip1-IRES-GFP and GFP+ cells were purified by FACS. (C) Cytotoxicity and (D) IL-1β secretion were determined 2 h after PrgI transfection. Data in (A–B) and (C–D) are representative of 2 and 3 independent experiments respectively.
Naip1 is poorly expressed in BMMs as determined by quantitative PCR, however, it can be detected in some macrophage rich tissues including the spleen, small intestine, and colon (Fig. 4A–B). Interestingly, even under steady state conditions, macrophages from the peritoneal cavity of mice express substantial amounts of Naip1 (Fig. 4B). These findings are further corroborated in gene expression profiling of immune cells by the ImmGen consortium (21). We therefore investigated whether peritoneal cavity (PerC) macrophages respond to PrgI without poly(I:C) priming. Indeed, PrgI induced robust IL-1β secretion from PerC macrophages compared to BMMs, and this was dependent upon NLRC4 (Fig. 4B). We also confirmed that the flagellar hook (FlgE), which is structurally analogous to the needle, is not detected by PerC macrophages (Fig. 4B). Therefore, primary macrophages that express NAIP1 in detectable amount are fully capable of sensing the needle protein without poly(I:C) priming. The physiological relevance of priming of the Naip1 pathway in vivo in other macrophage populations remains to be investigated.
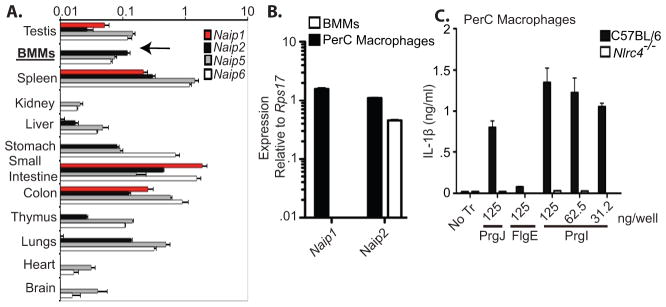
(A) Quantitative PCR was performed to assess the amounts of Naip1 (red bars), Naip2 (black bars), Naip5 (grey bars) and Naip6 (white bars) transcripts expressed in BMMs or the indicated organs and normalized to GAPDH. (B) Levels of Naip1 and Naip2 transcripts were determined in BMMs (white bars) or peritoneal cavity macrophages (PerC) by quantitative PCR.(C) PerC macrophages from C57BL/6 (black bars) or Nlrc4−/− mice (white bars) were primed with LPS (50ng/ml) before transfection with PrgI, PrgJ, or FlgE. IL-1β in the supernatants was measured at 2 h. Data representative of at least 2–3 independent observations.
Together, our studies show that detection of the T3SS needle in the cytosol requires NAIP1. These results underscore the importance of redundancy in the detection of T3SS. In mice, three distinct agonists, flagellin, rod, and needle, are targeted by four innate immune NAIP receptors, which converge upon a single NLRC4 inflammasome (Supplemental Fig. 1). Therefore the ability to detect a variety of components of this important virulence structure ensures a thorough surveillance on the part of innate immune phagocytes.
Our findings not only reveal an unappreciated role for NAIP1 in the detection of T3SS needle but also raise interesting question about mouse versus human pathogen recognition systems. Data published previously (13) and verified here indicate that humans seem to detect only the needle protein. Does the lack of rod and flagellin detection make humans more susceptible to Gram-negative pathogens that use T3SS? Future studies will examine the importance of flagellin, rod, and needle detection through NAIP5/6, NAIP2, and NAIP1, respectively.
References
Full text links
Read article at publisher's site: https://doi.org/10.4049/jimmunol.1301549
Read article for free, from open access legal sources, via Unpaywall:
https://www.jimmunol.org/content/jimmunol/191/8/3986.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/170409044
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4049/jimmunol.1301549
Article citations
NLRC4, inflammation and colorectal cancer (Review).
Int J Oncol, 65(4):99, 06 Sep 2024
Cited by: 0 articles | PMID: 39239759 | PMCID: PMC11387119
Review Free full text in Europe PMC
Flagellin-modulated inflammasome pathways characterize the human alveolar macrophage response to <i>Burkholderia pseudomallei</i>, a lung-tropic pathogen.
Infect Immun, 92(5):e0006024, 15 Apr 2024
Cited by: 0 articles | PMID: 38619302 | PMCID: PMC11075458
Strategies of bacterial detection by inflammasomes.
Cell Chem Biol, 31(5):835-850, 17 Apr 2024
Cited by: 0 articles | PMID: 38636521
Review
Programmed cell death and Salmonella pathogenesis: an interactive overview.
Front Microbiol, 14:1333500, 04 Jan 2024
Cited by: 0 articles | PMID: 38249488 | PMCID: PMC10797706
Review Free full text in Europe PMC
NAIP/NLRC4 inflammasome participates in macrophage responses to Trypanosoma cruzi by a mechanism that relies on cathepsin-dependent caspase-1 cleavage.
Front Immunol, 14:1282856, 06 Dec 2023
Cited by: 3 articles | PMID: 38124741 | PMCID: PMC10731265
Go to all (119) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome.
Proc Natl Acad Sci U S A, 114(50):13242-13247, 27 Nov 2017
Cited by: 98 articles | PMID: 29180436 | PMCID: PMC5740664
Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation.
Proc Natl Acad Sci U S A, 110(35):14408-14413, 12 Aug 2013
Cited by: 246 articles | PMID: 23940371 | PMCID: PMC3761597
The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus.
Nature, 477(7366):596-600, 14 Sep 2011
Cited by: 781 articles | PMID: 21918512
Emerging Concepts about NAIP/NLRC4 Inflammasomes.
Front Immunol, 5:309, 02 Jul 2014
Cited by: 38 articles | PMID: 25071770 | PMCID: PMC4078251
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: AI097518
Grant ID: R01 AI097518
Grant ID: R01 AI075039




